Abstract
Sirtuin 2 (SIRT2) is a sirtuin family deacetylase that directs acetylome signaling, protects genome integrity, and is a murine tumor suppressor. We show that SIRT2 directs replication stress responses by regulating the activity of cyclin-dependent kinase 9 (CDK9), a protein required for recovery from replication arrest. SIRT2 deficiency results in replication stress sensitivity, impairment in recovery from replication arrest, spontaneous accumulation of replication protein A to foci and chromatin, and a G2/M checkpoint deficit. SIRT2 interacts with and deacetylates CDK9 at lysine 48 in response to replication stress in a manner that is partially dependent on ataxia telangiectasia and Rad3 related (ATR) but not cyclin T or K, thereby stimulating CDK9 kinase activity and promoting recovery from replication arrest. Moreover, wild-type, but not acetylated CDK9, alleviates the replication stress response impairment of SIRT2 deficiency. Collectively, our results define a function for SIRT2 in regulating checkpoint pathways that respond to replication stress through deacetylation of CDK9, providing insight into how SIRT2 maintains genome integrity and a unique mechanism by which SIRT2 may function, at least in part, as a tumor suppressor protein.
Keywords: DNA damage, cell cycle checkpoint, genome maintenance
Members of the sirtuin NAD+-dependent deacetylase family regulate multiple biological processes, including genome maintenance, aging, tumorigenesis, differentiation, and metabolism (1–3). There are seven mammalian homologs of yeast silent information regulator 2 (Sir2). Whereas sirtuin 1 (SIRT1) and sirtuin 6 (SIRT6) have been linked directly to DNA repair or DNA damage response (DDR) signaling pathways (4–12), the role for other sirtuin family members, including SIRT2, is less clear. Mice deficient in Sirt2 develop breast, liver, and other cancers (13, 14), and SIRT2 expression is decreased in human breast and liver cancers (14), suggesting that SIRT2 is a tumor suppressor protein (TSP). In support of a role for SIRT2 in the DDR, Sirt2 deficiency in mouse embryo fibroblasts (MEFs) results in a mitotic checkpoint deficit, increased DNA damage, and genomic instability (13). In addition, SIRT2 deficiency in chicken DT40 cells induces hypersensitivity to cisplatin (15). SIRT2 also deacetylates histone H3 lysine 56, a marker for packaging of newly replicated and repaired DNA into chromatin (16), implying that SIRT2 may function in the maintenance of genome integrity during DNA replication.
The replication stress response (RSR) is a subset of the DDR signaling network that recognizes challenges to DNA replication and mobilizes diverse DNA repair and cell cycle checkpoint pathways. The RSR is critical for the prevention of cancer by acting as a barrier against genomic instability and tumorigenesis (17, 18). At the apex of the RSR signaling cascade is the ataxia-telangiectasia and Rad3-related protein (ATR) checkpoint kinase (19). ATR function is essential to stabilize stalled replication forks and promote recovery. Disruption of ATR causes early embryonic lethality in mice (20), and cells lacking ATR have defects associated with DNA replication (21), chromosomal instability, and expression of fragile sites of mammalian chromosomes (22). ATR senses stalled replication forks that result from uncoupling that exposes replication protein A (RPA)-bound single-stranded DNA (ssDNA) (23). Finally, RSR mobilizes several activities necessary to maintain genome integrity, including arresting the cell cycle, inhibiting the onset of mitosis, stabilizing the stalled replication fork, and facilitating replication recovery.
Although cyclin-dependent kinase 9 (CDK9) is a well-characterized component of the positive transcription elongation factor b complex with cyclin T or K (24, 25), it also has a direct role in promoting RSR activities, which is functionally conserved in yeast (26–29). Loss of CDK9 activity increases spontaneous levels of DNA damage signaling in replicating cells and a decreased ability to recover from a transient replication arrest. CDK9 localizes to chromatin to limit the amount of ssDNA after replication stress and interacts in a complex with ATR as well as other checkpoint signaling proteins. CDK9 also phosphorylates and activates ubiquitin-conjugating enzyme 2A (UBE2A) (30–32), which directs H2B and PCNA ubiquitination (33, 34). The role for CDK9 in the RSR is independent of cyclin T (29), and we and others have shown that cyclin K also functions to maintain genome integrity (29, 35); however, recent evidence suggests that cyclin K functions primarily with CDK12 (35–37). Similar to other RSR proteins, CDK9 activity is regulated by posttranslational modification, including acetylation (38). CDK9 is acetylated on lysine 44 by the p300 acetyltransferase (39), which promotes its activity, and lysine 48 by the general control non-depressible 5 (GCN5) and P300/CBP-associated factor (P/CAF) acetyltransferases (40), which inhibits its activity. Lysine 44 of CDK9 is also deacetylated by histone deacetylase 3 (HDAC3) and histone deacetylase 1 (HDAC1) (39); however, the deacetylase for lysine 48 of CDK9 is not known. It is also not known whether the acetylation status or kinase activity of CDK9 is regulated by DNA damage. In this study, we show that SIRT2, and by extension the cellular acetylome, has a role in promoting replication stress response activities by regulating CDK9 activity via changes in lysine acetylation.
Results
Silencing SIRT2 Causes Sensitivity to Replication Stress.
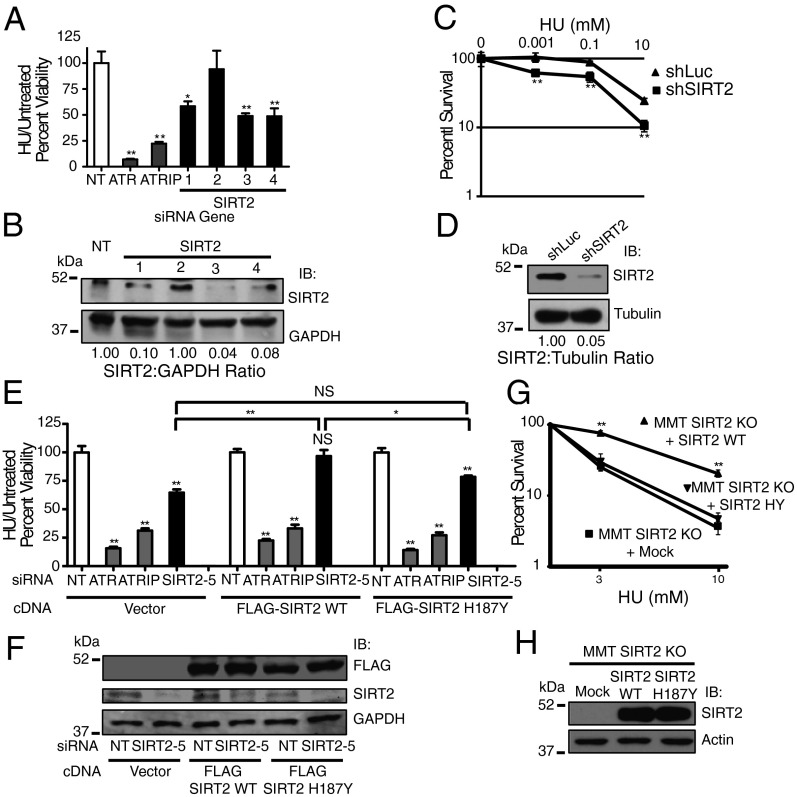
To investigate the role of SIRT2 in mediating recovery following replication fork arrest, we measured the ability of U2OS human osteosarcoma cells, silenced for SIRT2, to recover from a transient high dose hydroxyurea (HU) exposure, a drug that stalls DNA replication forks by inhibiting ribonucleotide reductase. U2OS cells were treated with or without HU for 24 h, allowed to recover for 24 h, and assayed for cell viability using water soluble tetrazolium salt (WST)-1 reagent. siRNA knockdown of ATR or ATR-interacting protein (ATRIP) caused HU hypersensitivity, compared with nontargeting (NT) controls as has been previously shown (30) (Fig. 1 A and E). Similarly, SIRT2 knockdown resulted in HU hypersensitivity (Fig. 1 A and E). Western blot analysis confirmed decreased levels of SIRT2 following siRNA knockdown (Fig. 1 B and F). The HU hypersensitivity of cells after SIRT2 knockdown was observed between 24 and 72 h following recovery from the HU challenge (Fig. S1A). SIRT2 silencing also caused hypersensitivity to gemcitabine, cisplatin, and mitomycin C (Fig. S1 B–D), implying that SIRT2 responds generally to replication stress.
Fig. 1.
SIRT2 deacetylase activity is required for the replication stress response. (A) U2OS cells were transfected with nontargeting (NT), ATR, ATRIP, or SIRT2 siRNA, split 1:4 48 h later and treated 24 h later with or without 3 mM HU for 24 h. Cell viability was analyzed 24 h after recovery from treatment. HU treated/untreated cell viability relative to NT siRNA was determined. (B) Western blot analysis demonstrating efficiency of SIRT2 knockdown with indicated siRNAs. (C) U2OS cells stably transfected with shRNA against SIRT2 or luciferase were seeded for colony formation, treated with indicated concentrations of HU for 24 h, and assayed for surviving colonies 8–12 d later. Percent survival of colonies from treated vs. untreated cells is indicated. (D) Western blot analysis demonstrating efficiency of SIRT2 knockdown with shSIRT2 relative to sh-luciferase. (E) U2OS cells stably expressing an empty vector, FLAG-SIRT2 wild-type (WT) or FLAG-SIRT2 H187Y were transfected with NT, ATR, ATRIP, or SIRT2 siRNA targeting the 3′ untranslated (UTR) region, split 1:4 48 h later, and treated 24 h later with or without 3 mM HU for 24 h before assaying for cell viability 24 h later. Mean and SEM from four replicas is shown. (F) Western blot analysis of stable cells lines demonstrating expression of FLAG-SIRT2 WT and H187Y and efficiency of SIRT2 silencing. (G) MMT Sirt2 KO (MMT S2KO) cells mock transfected or stably transfected with SIRT2 WT or H187Y were seeded for colony formation, treated with indicated concentrations of HU for 24 h, and assayed for surviving colonies 8–12 d later. Percent survival of colonies from treated vs. untreated cells is indicated. (H) Western blot analysis of stable MMT S2KO cell lines demonstrating expression of SIRT2 WT and H187Y. For A, C, E, and G, mean and SEM from at least three replicas is shown. *P < 0.05 and **P < 0.01. See also Fig. S1.
As SIRT2 can affect the production of NADH, which is measured by WST-1 reagent, we also determined HU sensitivity of SIRT2-depleted cells using a colony formation assay. U2OS cells stably silenced for SIRT2 using shRNA (Fig. 1D) demonstrated a significantly reduced percentage of surviving colonies following a 24-h pulse of HU in a dose-dependent manner compared with silencing with a sh-luciferase control (Fig. 1C), confirming the HU hypersensitivity of SIRT2-depleted cells observed using WST-1 reagent.
SIRT2 Deacetylase Activity Is Necessary for the Replication Stress Response.
To determine if the deacetylase activity of SIRT2 is essential for mediating sensitivity to replication stress, we generated U2OS cell lines stably expressing exogenous wild-type (WT) FLAG-SIRT2 or FLAG-SIRT2 H187Y and silenced endogenous SIRT2 accomplished by using siRNA targeting the 3′ untranslated region. Wild-type FLAG-SIRT2 but not FLAG-SIRT2 H187Y expression complemented the HU hypersensitivity of SIRT2-depleted cells (Fig. 1E), suggesting that the deacetylase activity of SIRT2 directs aspects of the RSR. Western blot analysis confirmed expression of the exogenously expressed fusion proteins and efficiency of SIRT2 silencing (Fig. 1F). As a second measure for determining the significance of SIRT2 deacetylase activity in the RSR, we generated mouse mammary tumor Sirt2 knockout (MMT S2KO) cell lines (14), stably expressing SIRT2 WT or SIRT2 H187Y (Fig. 1H) and assessed colony formation following a 24-h pulse of HU. SIRT2 WT but not SIRT2 H187Y significantly increased the percentage of surviving colonies following HU treatment in a dose-dependent manner (Fig. 1G), confirming that the deacetylase activity of SIRT2 is required for the RSR.
SIRT2 Is a Replication Stress Response Protein.
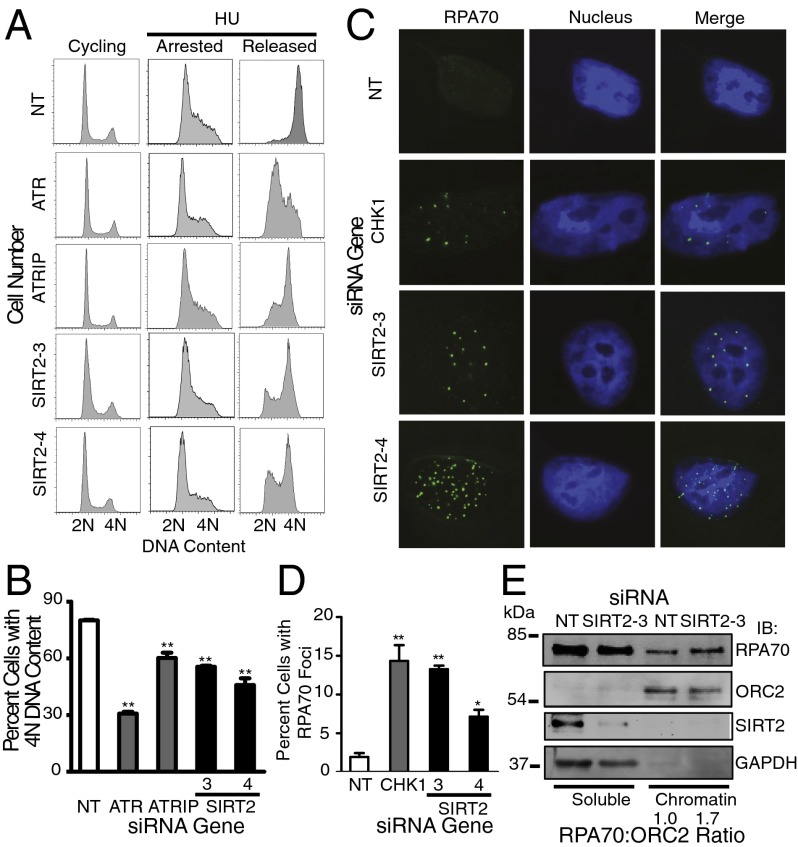
The ability of SIRT2-silenced cells to recover from transient replication fork arrest was assessed using cell cycle recovery by flow cytometry. Eight to 10 h after removing HU, U2OS, telomerase immortalized human retinal pigment epithelium (hTERT RPE-1), and human colorectal carcinoma cell line HCT-116 cells treated with NT siRNA progressed through S phase and accumulated 4N DNA content, whereas cells treated with ATR, ATRIP or SIRT2 siRNA exhibited delayed S-phase progression (Fig. 2 A and B and Fig. S2 A–D). Depletion of SIRT2 also caused a similar impairment in cell cycle recovery following a replication challenge of aphidicolin, a DNA polymerase inhibitor (Fig. S2 A and B).
Fig. 2.
SIRT2 is a replication stress response protein. (A and B) hTERT RPE-1 cells were transfected with NT, ATR, ATRIP, or SIRT2 siRNA, treated with 3 mM HU for 24 h (arrested), and released into nocodazole for 10 h (released). DNA content was analyzed by using flow cytometry. (B) The percentage (mean and SEM) of cells that completed DNA synthesis in three replicate experiments is shown. **P < 0.01. (C and D) U2OS cells were transfected with NT, CHK1, or SIRT2 siRNA and processed 72 h later for RPA70 staining by indirect immunofluorescence microscopy. (D) The percentage (mean and SEM) of RPA70 foci positive cells from four replicate experiments is shown. *P < 0.05 and **P < 0.01. (E) Soluble and chromatin fraction of U2OS cells transfected with NT or SIRT2 siRNA for 72 h. Chromatin-bound RPA70:ORC2 ratio of representative blot from three independent experiments is shown. See also Fig. S2.
The induction of spontaneous RPA foci by indirect immunofluorescence following SIRT2 knockdown was done as a second measure to determine the response to replication stress. In the absence of exogenous damage, RPA accumulates on single-stranded DNA formed by stalled replication forks. Similar to CHK1, at 72 h after SIRT2 knockdown, accumulation of RPA70 foci was significantly increased, compared with NT controls (Fig. 2 C and D). Moreover, the amount of chromatin-bound RPA70 is increased 1.7-fold in SIRT2-depleted cells (Fig. 2E), implying that SIRT2 limits the amount of single-stranded DNA available for RPA binding. Collectively, these results suggest a model in which SIRT2 functions as a replication stress response protein that maintains genome integrity by preventing the breakdown of stalled replication forks.
Proteins that function in the RSR often are involved in regulating cell cycle checkpoints. The G2/M checkpoint prevents cells from entering mitosis to allow time for removal of damaged DNA or replication blocks. As such, U2OS cells treated with 2 Gy of ionizing radiation (IR) displayed a strong G2/M checkpoint response after exposure, as determined by the low percentage of cells entering mitosis measured by phosphorylation of histone H3 Ser10 (Fig. S2 E and F). In contrast, following SIRT2, ATR, or ATRIP knockdown, the percentage of cells escaping the G2/M checkpoint and entering mitosis after IR was significantly increased (Fig. S2 E and F).
SIRT2 Interacts in a Complex with and Deacetylates CDK9 in Vitro and in Cells.
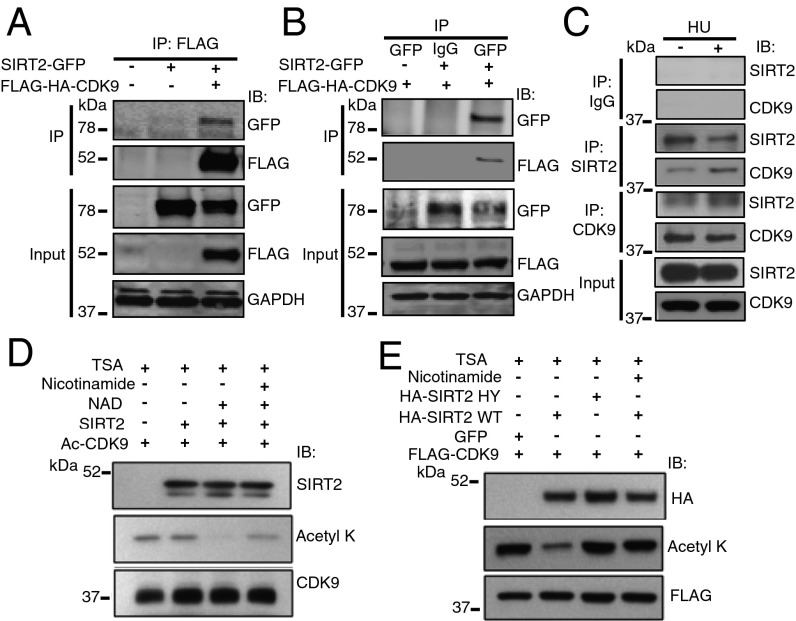
FLAG-SIRT2 was purified from HeLa cells, and proteomic analysis by mass spectrometry identified multiple interacting proteins, including CDK9, which we previously characterized as a RSR protein (29). SIRT2-GFP and FLAG-HA-CDK9 were expressed in HEK293T cells and reciprocal coimmunoprecipitation (co-IP) analyses suggested that the proteins interact in a complex (Fig. 3 A and B). The endogenous interaction of SIRT2 and CDK9 was also validated by co-IP in HeLa cells and increased in response to HU despite similar protein levels, suggesting that the interaction of SIRT2 and CDK9 is regulated by replication stress (Fig. 3C).
Fig. 3.
SIRT2 Interacts in a complex with and deacetylates CDK9. (A and B) 293T cells were transfected with Flag-HA-CDK9 and/or SIRT2-GFP, harvested and immunoprecipitated (IPed) with either anti-Flag M2 beads (A) or anti-GFP antibody. (B) Bound proteins were washed, separated by SDS/PAGE, and immunoblotted with antibodies against GFP, Flag, and GAPDH. (C) Endogenous SIRT2 or CDK9 was immunopreciptated from HeLa cell lysates treated with or without HU. Immunocomplexes were washed, separated by SDS/PAGE, and immunoblotted with antibodies against SIRT2 and CDK9. (D) Acetylated CDK9 was isolated from 293T cells transfected with FLAG-CDK9, p300/CBP, and pCAF and incubated in an in vitro deacetylation assay with FLAG-SIRT2 isolated from 293T cells in the presence of TSA with or without NAD and nicotinamide. The reaction mixtures were separated by SDS/PAGE and immunoblotted with antibodies against acetyllysine, SIRT2, and CDK9. (E) HeLa cells were transfected with FLAG-CDK9 and HA-SIRT2 WT or HY in the presence of TSA with or without nicotinamide, IPed with an anti-FLAG antibody, separated by SDS/PAGE, and immunoblotted with antibodies against HA, acetyllysine, and FLAG. See also Fig. S3.
To determine if SIRT2 deacetylates CDK9, we purified acetylated FLAG-CDK9 from HEK293T cells and performed an in vitro deacetylation assay with purified wild-type FLAG-SIRT2 or FLAG-SIRT6 in the presence and absence of NAD+. SIRT2 but not SIRT6 deacetylated FLAG-CDK9 in an NAD+-dependent manner (Fig. 3D, lane 3 and Fig. S3), and deacetylation was inhibited by nicotinamide (Fig. 3D, lane 4). To determine if SIRT2 deacetylates CDK9 in cells, we transfected HeLa cells with FLAG-CDK9 along with wild-type HA-SIRT2 or HA-SIRT2 HY, a deacetylase mutant (14). Transfection with wild-type HA-SIRT2, but not HA-SIRT2 HY, deacetylated FLAG-CDK9 in a similar manner as our cell-free biochemical experimental system (Fig. 3E), suggesting that CDK9 is a legitimate SIRT2 downstream deacetylation target.
SIRT2 Deacetylates CDK9 at Lysine 48 in Response to Replication Stress.
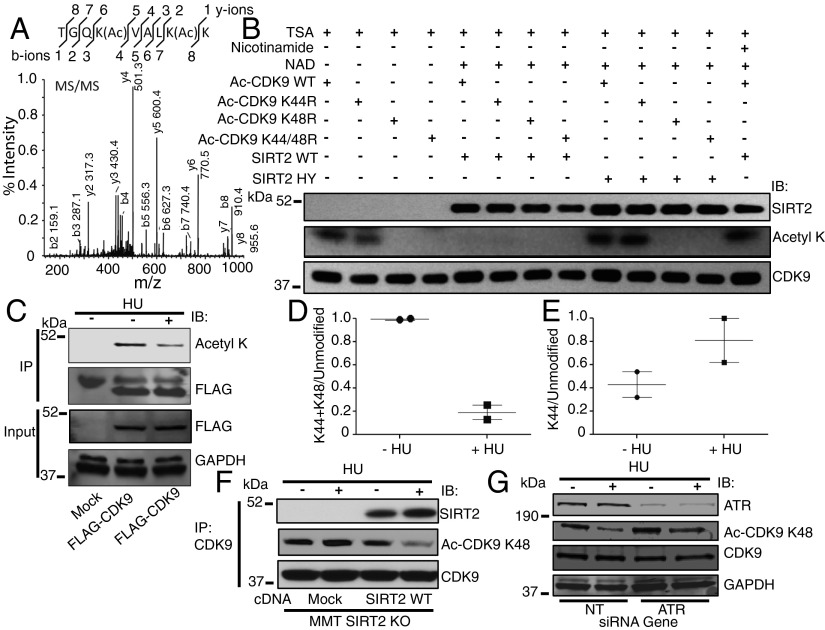
To determine the specific SIRT2 downstream acetyllysine target, mass spectrometry analysis was performed from purified FLAG-CDK9 expressed in 293T cells, which identified amino acids 44 and 48 as potential reversible acetyllysines (Fig. 4A and Fig. S4A), consistent with findings using alternative approaches (39, 40). Mutation of lysine residue 44 to arginine (K44R) caused no significant difference in acetylation of FLAG-CDK9 as determined by Western blot analysis with an antiacetyllysine antibody (Fig. 4B). In contrast, mutation of lysine residue 48 to arginine (K48R) or both lysine residues 44 and 48 to arginine (K44/48R) reduced FLAG-CDK9 acetylation to undetectable levels (Fig. 4B), suggesting that CDK9 K48 is a legitimate SIRT2 deacetylation target lysine. Recombinant wild-type SIRT2, but not SIRT2-HY, also deacetylated purified acetylated wild-type FLAG-CDK9 and FLAG-CDK9 K44R (Fig. 4B). The results of these experiments imply that SIRT2 deacetylates CDK9 at lysine residue 48.
Fig. 4.
SIRT2 deacetylates CDK9 at lysine residue 48 in response to replication stress. (A) MS/MS peptide spectra of IPed FLAG-CDK9 expressed in 293T cells shows site-specific CDK9 diacetylation at lysine residues 44 and 48. (B) Acetylated CDK9 WT or deacetylated mutants CDK9 K44R, CDK9 K48R, or CDK9 K44/48R was isolated from cells and incubated in an in vitro deacetylation assay with HA-SIRT2 WT or HY in the presence of TSA with or without NAD and nicotinamide. The reaction mixtures were separated by SDS/PAGE and immunoblotted with antibodies against acetyllysine, SIRT2, and CDK9. (C) The 293T cells were transfected with vector or FLAG-CDK9 together with acetyltransferases, treated with or without HU, harvested, IPed with an antibody against FLAG, separated by SDS/PAGE, and immunoblotted with antibodies against acetyllysine, FLAG, and GAPDH. (D and E) Quantitation of acetylated peptide intensities across control and HU-treated samples from C using LC-MS/MS. The propensity of a peptide to ionize, a prerequisite for detection, is dependent on its physicochemical properties such as peptide length, in which representative tryptic peptides have a mass between 600 and 6,000 Da (46). In this study, the expected fully tryptic monoacetyl K48 CDK9 peptide would be only 5 amino acids (K.VALK(Ac)K.V), with an expected mass of 557.39, making this short peptide challenging to detect by LC-MS/MS. However, we were able to observe the double acetylated K44 and K48 peptide and infer that the reduction of this signal after HU treatment was likely driven by deacetylation at K48 specifically because no significant decrease was observed on K44 aceytlation alone (see also Fig. S4 for K44 MS/MS spectra). (F) MMT S2KO cells mock transfected or stably transfected with SIRT2 WT were treated with or without HU, harvested, IPed with an anti-CDK9 antibody, separated by SDS/PAGE, and immunoblotted with antibodies against SIRT2, Ac-CDK9 K48, and CDK9. (G) The 293T cells were transfected with NT or ATR siRNA and treated with or without HU, harvested, separated by SDS/PAGE, and immunoblotted with antibodies against ATR, Ac-CDK9 K48, CDK9, and GAPDH. See also Fig. S4.
FLAG-CDK9 from 293T cells, treated with or without HU, was immunoprecipitated (IPed) and Western blotting with an antiacetyllysine antibody also exhibited a significant decrease in acetylation of CDK9 from HU-treated cells, compared with nontreated cells (Fig. 4C). Subsequent analysis of purified FLAG-CDK9 from these cells by mass spectrometry identified that acetylation of CDK9 K48 is decreased at least fivefold after treatment with HU (Fig. 4 D and E). To determine if endogenous CDK9 K48 is deacetylated by SIRT2 in response to replication stress, we generated and validated an antiacetyl CDK9 K48 antibody (Fig. S4B). Whereas endogenous CDK9 IPed from MMT S2KO cells was acetylated at K48 before and after HU treatment, K48 was deacetylated in response to HU treatment when these cells were complemented with SIRT2 WT, suggesting that endogenous CDK9 is present in the acetylated form at K48 in cells and is subsequently deacetylated by SIRT2 in response to replication stress (Fig. 4F).
CDK9 K48 deacetylation in response to HU treatment was also observed in HeLa cells and alleviated by nicotinamide treatment (Fig. S4C). As sirtuins are known to regulate a network of proteins, we also examined if SIRT2 deacetylates α-tubulin, a well-characterized substrate (41), in response to replication stress. Similar to CDK9 K48, α-tubulin K40 was also deacetylated in response to HU treatment in MMT S2KO cells when complemented with SIRT2 WT (Fig. S4D), providing further support for SIRT2 as a RSR protein and demonstrating that SIRT2 may regulate other proteins in response to replication stress.
CDK9 K48 Deacetylation in Response to Replication Stress Is Partially Dependent on ATR but Not Cyclin T or K.
Consistent with a transcription-independent function for CDK9 in the RSR (29), CDK9 complexed with IPed cyclin T1 (CCNT1) in 293T cells was not deacetylated in response to HU treatment (Fig. S4E), and no significant difference in deacetylation of CDK9 K48 after HU treatment was observed following CCNT1 or cyclin T2 (CCNT2) silencing in 293T cells compared with a NT control (Fig. S4 G and I). Similar to a previous report (37), we detected an interaction of CDK9 with cyclin K1 (CCNK1) only when overexpressed (Fig. S4F), and FLAG-HA-CDK9 complexed with IPed CCNK1 in 293T cells also was not deacetylated in response to HU treatment (Fig. S4F) nor was there a significant difference in deacetylation of CDK9 K48 following CCNK1 silencing in 293T cells (Fig. S4H). Thus, these results imply that CDK9 K48 deacetylation in response to replication stress is restricted to a cylin T- and K-independent fraction and is also not dependent on cyclin T or K. In contrast, ATR silencing in 293T cells alleviated the HU-regulated deacetylation of CDK9 K48 (Fig. 4G), suggesting that CDK9 K48 deacetylation in response to replication stress is at least partially dependent on ATR.
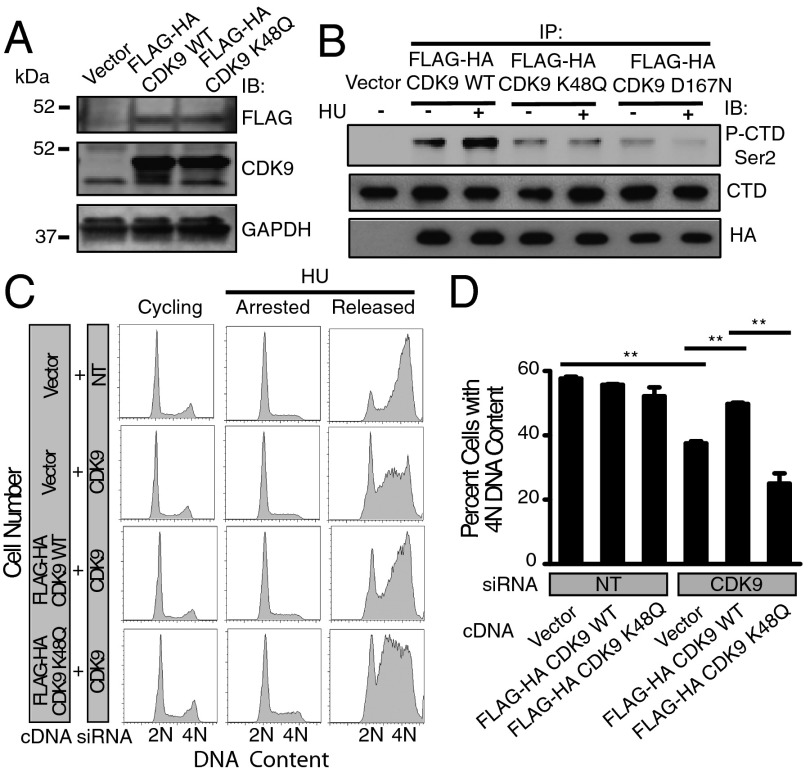
Lysine 48 Acetylation Impairs CDK9 Kinase Activity and Recovery from Replication Stress.
To examine the functional significance of the acetylation status of lysine 48, U2OS cell lines stably expressing wild-type FLAG-HA CDK9 or FLAG-HA CDK9 K48Q were generated (Fig. 5A). Lysine 48 mutated to glutamine (K48Q) has been shown to mimic an acetylated lysine state. Whereas no significant difference in stability of wild-type FLAG-HA CDK9 compared with FLAG-HA CDK9 K48Q was observed following treatment of cells with cyclohexamide (Fig. S5 A and B), the kinase activity of FLAG-HA CDK9 WT but not FLAG-HA CDK9 K48Q was significantly increased following treatment with HU using both a fragment of RNA polymerase II carboxyl terminal domain (CTD) and UBE2A as substrates (Fig. 5B and Fig. S5C). The basal kinase activity of FLAG-HA CDK9 K48Q was comparable to that of FLAG-HA CDK9 D167N, a kinase dead mutant (Fig. 5B). To determine the role of SIRT2 deacetylation in promoting CDK9 kinase activity, we performed the in vitro kinase assay using CDK9 IPed from MMT S2KO cells complemented with SIRT2 WT or H187Y and using a fragment of CTD as a substrate and found that endogenous CDK9 kinase activity was significantly increased following HU treatment in the MMT S2KO cells complemented with SIRT2 WT but not H187Y (Fig. S5D). These findings suggest that acetylation of lysine 48 decreases CDK9 kinase activity and deacetylation of lysine 48 by SIRT2 increases CDK9 kinase activity in response to replication stress.
Fig. 5.
Lysine 48 acetylation impairs CDK9 kinase activity and recovery from replication stress. (A) Western blot analysis of U2OS stable cell lines demonstrating expression of FLAG-HA CDK9 WT and K48Q. (B) FLAG-HA-CDK9 was purified from U2OS cells stably expressing an empty vector, FLAG-HA CDK9 WT, K48Q, or kinase dead D167N, treated with or without HU, and incubated in an in vitro kinase reaction with a recombinant fragment of RNA polymerase II CTD purified from bacterial cells as substrate. The reaction mixtures were separated by SDS/PAGE and immunoblotted with antiphospho-CTD Ser2, antipan-CTD, and anti-HA antibodies. (C and D) U2OS cells stably expressing an empty vector, FLAG-HA-CDK9 WT or K48Q were transfected with NT or CDK9 siRNA targeting the 3′ untranslated region, treated with 3 mM HU for 20 h (arrested), and released into nocodazole for 10 h (released). DNA content was analyzed by using flow cytometry. (D) The percentage (mean and SEM) of cells that completed DNA synthesis in three replicate experiments is shown. **P < 0.01. See also Fig. S5.
We have previously shown that the kinase activity of CDK9 is required for recovery from replication stress (29). Consistent with these findings, cells expressing wild-type FLAG-HA CDK9, but not FLAG-HA CDK9 K48Q, as well as silenced for endogenous CDK9 using siRNA targeting the 3′ untranslated region, partially complemented the HU recovery deficit (Fig. 5 C and D). These experiments demonstrate that CDK9 acetylation at lysine residue 48 impairs recovery from replication stress and suggest that CDK9 lysine 48 deacetylation by SIRT2 is critical for the replication stress response.
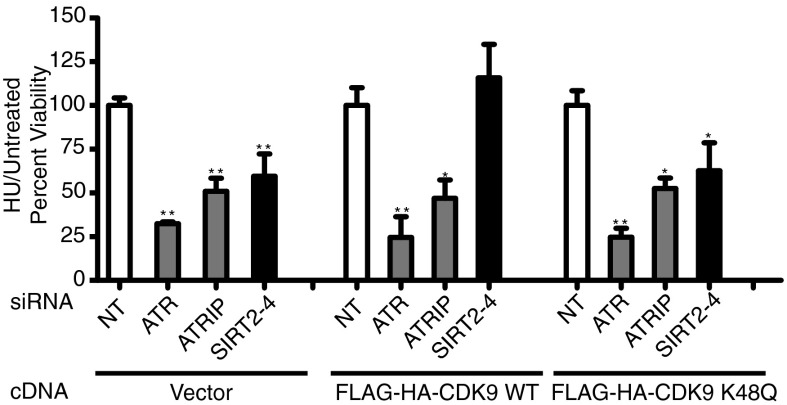
CDK9 Complements the Replication Stress Response Impairment of SIRT2 Deficiency.
To confirm that the role of SIRT2 in the replication stress response is mediated through CDK9 lysine 48 acetylation status, SIRT2 was silenced in U2OS cells stably expressing wild-type FLAG-HA-CDK9 or FLAG-HA-CDK9 K48Q. FLAG-HA CDK9 but not FLAG-HA CDK9 K48Q complemented the HU hypersensitivity of cells following SIRT2 knockdown (Fig. 6). These results indicate that CDK9 lysine 48 is a major SIRT2 downstream target in promoting the replication stress response.
Fig. 6.
CDK9 complements the replication stress response impairment of SIRT2 deficiency. U2OS cells stably expressing an empty vector, FLAG-HA-CDK9 WT or K48Q were transfected with NT, ATR, ATRIP, or SIRT2 siRNA, split 1:4 48 h later, and treated 24 h later with or without 3 mM HU for 24 h before assaying for cell viability using WST-1 reagent. Mean and SEM from four replicas is shown. *P < 0.05 and **P < 0.01.
Discussion
Our results reveal a unique function for SIRT2 providing unique scientific evidence connecting SIRT2, cell cycle checkpoint pathways, and the RSR, through deacetylation of CDK9, a protein required for cells to recover from replication arrest. We found that depletion of SIRT2 results in sensitivity to replication stress, impairment in recovery from replication arrest, spontaneous accumulation of RPA to foci and chromatin, and a G2/M checkpoint deficit. These findings establish SIRT2 as a replication checkpoint protein required to maintain genome integrity and help explain how SIRT2 deficiency leads to genetic instability and an in vivo tumor permissive phenotype. In this regard, we showed that SIRT2 interacts with and deacetylates CDK9 in vitro and in cells, thus identifying CDK9 as a unique interacting partner and substrate for SIRT2. We also showed that CDK9 is deacetylated by SIRT2 at lysine residue 48, in a manner that is partially dependent on ATR but not cyclin T or K, which increases its kinase activity, in response to replication stress, further validating CDK9 as a RSR protein, identifying lysine 48 as a specific SIRT2 target, and providing a mechanistic model for how SIRT2 maintains genomic integrity in response to replication stress. Finally, we showed that wild-type, but not acetylated CDK9, alleviates the RSR impairment of SIRT2 deficiency, firmly placing CDK9 as a downstream deacetylase target through which SIRT2 functions in the RSR.
Our findings support a model in which replication stress triggers SIRT2-dependent deacetylation of CDK9 at lysine 48, thereby stimulating CDK9 kinase activity and promoting recovery from replication arrest (Fig. S6). SIRT2 thus acts as a positive regulator of CDK9 function by removing its inhibitory lysine acetylation. This leads to several downstream effects necessary to maintain genome integrity including preventing the breakdown of stalled replication forks that leads to RPA accumulating on single-stranded DNA, arresting the cell cycle to allow additional time for repair, and facilitating recovery from replication arrest. Loss of SIRT2 leads to genomic instability and ultimately cancer progression.
A role for sirtuins in maintaining genome integrity by regulating the functions of nonhistone DNA damage response proteins through deacetylation has previously been demonstrated for SIRT1 and SIRT6. SIRT1 substrates include KU70 (5), NBS1 (42), WRN (7), XPA (4), and TIP60 (10, 11), whereas SIRT6 substrates include CtIP (6), PARP1 (8), and DNA-PK (9). Our results extend these ideas and now demonstrate that a third member of the human sirtuin family, SIRT2, also functions in maintaining genome integrity through deacetylation of CDK9.
Consistent with previous reports (39, 40), we found using proteomic analysis by mass spectrometry that CDK9 is acetylated in its catalytic core at lysine residues 44 and 48 with the major site of acetylation at lysine 48. We further provided evidence for SIRT2 as the deacetylase that targets lysine 48 and showed that the deacetylation of lysine 48 is regulated by replication stress. Lysine 48 deacetylation increases CDK9 kinase activity and promotes recovery from replication arrest. Thus, SIRT2 regulates CDK9 activity in part by removing its inhibitory acetylation to promote its kinase activity, which is required for its functions in the RSR (29). SIRT2 may be required for the CDK9-dependent phosphorylation and activation of UBE2A (31), which directs H2B and PCNA ubiquitination (33, 34). In addition, our finding that CDK9 K48 deacetylation by SIRT2 in response to replication stress is partially dependent on ATR but not cyclin T or K, which regulates DDR gene expression (35), is consistent with a transcription-independent function for CDK9 in the RSR that may be downstream of ATR.
It is noteworthy that CDK9 lysine 48, which is critical for orienting ATP and binding magnesium in the catalytic core (43, 44), is highly conserved among all members of the CDK family as well as subdomain II of most eukaryotic protein kinases (45), suggesting that SIRT2 deacetylation of CDK9 at lysine 48 may be a conserved mechanism of regulation that is generalized to other CDKs and protein kinases. Moreover, our finding that SIRT2 also deacetylates α-tubulin at K40 in response to replication stress suggests that SIRT2, like SIRT1 and SIRT6, may also regulate a network of proteins involved in genome maintenance.
In summary, our results define a function for SIRT2 in regulating checkpoint pathways that respond to replication stress by activating CDK9 through deacetylation, providing insight into how SIRT2 maintains genome integrity and loss of SIRT2 results in an in vitro tumor permissive phenotype.
Materials and Methods
Cell Culture.
U2OS, 293, HeLa cells were grown in DMEM supplemented with 7.5% (vol/vol) FBS. Stable U2OS cells were grown in 1ug/ml puromycin (Fisher).
Transfection, Immunofluorescence, Western Blot, IP, Deacetylation and Kinase Assay, Cell Survival and Colony Formation Assays, Cell Cycle Recovery Assay, LC-MS/MS.
Details of these methodologies can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the D.S.Y. laboratory for helpful discussion; Paul Doetsch for critical review of the manuscript; and Mauro Giacca for CDK9 K44R, K48R, and K44/48R plasmids. D.S.Y. is supported by National Institutes of Health/National Cancer Institute (NCI) K08CA143902, Department of Defense (DOD)/Breast Cancer Research Program BC085283, GCC11072, and Emory/Winship Cancer Institute Shared Resource Advisor Council pilot funding. D.S.Y. is a Georgia Cancer Coalition Distinguished Cancer Clinician and Scientist. D.G. is supported by NCI 1R01CA152601-01, 1R01CA152799-01A1, 1R01CA168292-01A1, 1R01CA16383801A1, and by BC093803 from the DOD and a Hirshberg Foundation for Pancreatic Cancer Research Seed Grant Award. H.-S.K. is supported by the National Research Foundation of Korea (NRF) Grant 2012M3A9C5048708, funded by the Ministry of Science, Information and Communication Technology and Future Planning; and by the Basic Science Research Program through the NRF, funded by the Ministry of Education, Science, and Technology (Grant NRF-2012R1A1A2005588).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301463110/-/DCSupplemental.
References
- 1.Guarente L. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N Engl J Med. 2011;364(23):2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- 2.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460(7255):587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders LR, Verdin E. Sirtuins: Critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26(37):5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 4.Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell. 2010;39(2):247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Jeong J, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 6.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Li K, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283(12):7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 8.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCord RA, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany, NY Online) 2009;1(1):109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;285(15):11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagata K, Kitabayashi I. Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem Biophys Res Commun. 2009;390(4):1355–1360. doi: 10.1016/j.bbrc.2009.10.156. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Z, Seto E. A functional link between SIRT1 deacetylase and NBS1 in DNA damage response. Cell Cycle. 2007;6(23):2869–2871. doi: 10.4161/cc.6.23.5026. [DOI] [PubMed] [Google Scholar]
- 13.Serrano L, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27(6):639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HS, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita N, et al. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells. 2005;10(4):321–332. doi: 10.1111/j.1365-2443.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- 16.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 18.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 19.Cimprich KA, Cortez D. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9(8):616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14(4):397–402. [PMC free article] [PubMed] [Google Scholar]
- 21.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294(5547):1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 22.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111(6):779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 23.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19(9):1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20(3):334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7(23):3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- 26.Clausing E, et al. The transcription elongation factor Bur1-Bur2 interacts with replication protein A and maintains genome stability during replication stress. J Biol Chem. 2010;285(53):41665–41674. doi: 10.1074/jbc.M110.193292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Herrmann CH. Differential localization and expression of the Cdk9 42k and 55k isoforms. J Cell Physiol. 2005;203(1):251–260. doi: 10.1002/jcp.20224. [DOI] [PubMed] [Google Scholar]
- 28.Yu DS, Cortez D. A role for CDK9-cyclin K in maintaining genome integrity. Cell Cycle. 2011;10(1):28–32. doi: 10.4161/cc.10.1.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu DS, et al. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO Rep. 2010;11(11):876–882. doi: 10.1038/embor.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morawska ME. A role for CDK9 in UV damage response. Cell Cycle. 2012;11(12):2229–2230. doi: 10.4161/cc.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shchebet A, Karpiuk O, Kremmer E, Eick D, Johnsen SA. Phosphorylation by cyclin-dependent kinase-9 controls ubiquitin-conjugating enzyme-2A function. Cell Cycle. 2012;11(11):2122–2127. doi: 10.4161/cc.20548. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Yu DS. One stone, two birds: CDK9-directed activation of UBE2A regulates monoubiquitination of both H2B and PCNA. Cell Cycle. 2012;11(13):2418. doi: 10.4161/cc.21068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 34.Pirngruber J, et al. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10(8):894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25(20):2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Q, et al. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J Biol Chem. 2012;287(30):25344–25352. doi: 10.1074/jbc.M111.321760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng SW, et al. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol Cell Biol. 2012;32(22):4691–4704. doi: 10.1128/MCB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho S, Schroeder S, Ott M. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle. 2010;9(9):1697–1705. doi: 10.4161/cc.9.9.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu J, Yoon HG, Qin J, Wong J. Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol Cell Biol. 2007;27(13):4641–4651. doi: 10.1128/MCB.00857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabò A, Lusic M, Cereseto A, Giacca M. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol Cell Biol. 2008;28(7):2201–2212. doi: 10.1128/MCB.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 42.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27(1):149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baumli S, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27(13):1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Bondt HL, et al. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363(6430):595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 45.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–596. [PubMed] [Google Scholar]
- 46. Eyers CE, et al. (2011) CONSeQuence: Prediction of reference peptides for absolute quantitative proteomics using consensus machine learning approaches. Mol Cell Proteomics 10(11):M110 003384. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.