Abstract
Although the circadian clock is a self-sustaining oscillator having a periodicity of nearly 1 d, its period length is not necessarily 24 h. Therefore, daily adjustment of the clock (i.e., resetting) is an essential mechanism for the circadian clock to adapt to daily environmental changes. One of the major cues for this resetting mechanism is light. In the unicellular green alga Chlamydomonas reinhardtii, the circadian clock is reset by blue/green and red light. However, the underlying molecular mechanisms remain largely unknown. In this study, using clock protein-luciferase fusion reporters, we found that the level of RHYTHM OF CHLOROPLAST 15 (ROC15), a clock component in C. reinhardtii, decreased rapidly after light exposure in a circadian-phase–independent manner. Blue, green, and red light were able to induce this process, with red light being the most effective among them. Expression analyses and inhibitor experiments suggested that this process was regulated mainly by a proteasome-dependent protein degradation pathway. In addition, we found that the other clock gene, ROC114, encoding an F-box protein, was involved in this process. Furthermore, we demonstrated that a roc15 mutant showed defects in the phase-resetting of the circadian clock by light. Taken together, these data strongly suggest that the light-induced degradation of ROC15 protein is one of the triggers for resetting the circadian clock in C. reinhardtii. Our data provide not only a basis for understanding the molecular mechanisms of light-induced phase-resetting in C. reinhardtii, but also insights into the phase-resetting mechanisms of circadian clocks in plants.
Keywords: LUCnc, light pulse, phase shift
Circadian rhythms, observed ubiquitously in organisms from prokaryotic cyanobacteria to humans, are generated by the circadian clock, which is thought to rely on transcriptional/translational feedback loops and posttranslational biochemical oscillations of some genes and their protein products called clock genes/proteins (1–4). Clock genes/proteins have been identified in several organisms, including mammals, insects, fungi, land plants, cyanobacteria, and, recently, green algae (1, 2, 5–8). Except for general kinases and phosphatases (9), most of the components of circadian clocks are not conserved among evolutionarily divergent organisms. On the other hand, the components are conserved to some extent between organisms that are relatively close evolutionarily (i.e., mammals and insects, land plants and green algae) (6–8, 10).
The clock components in algae were first identified in the model green alga Chlamydomonas reinhardtii. There are the RNA-binding protein complex CHLAMY1 (11), a casein kinase (12), and RHYTHM OF CHLOROPLAST (ROC) proteins, including putative DNA-binding proteins (ROC15, ROC40, ROC66, and ROC75), an F-box protein (ROC114), and a leucine-rich repeat protein (ROC55) (13). The DNA-binding motifs of ROC proteins are homologous to those of Arabidopsis thaliana proteins associated with the circadian clock and photoperiodic flowering (13). However, because the evolutionary conservation of clock components between C. reinhardtii and A. thaliana seems to be weak compared with the case in animals (Mus musculus and Drosophila melanogaster), algal and plant clocks are an excellent model for studying evolutionary origin and divergence of the circadian system. In addition, because C. reinhardtii is a unicellular model organism that has been called “green yeast” because of its suitability for genetic and biochemical studies (14), it provides a powerful platform for studying the molecular mechanisms of the circadian system in a single-celled eukaryote.
Although circadian clocks can generate self-sustained oscillation without environmental cues, their period length is not necessarily 24 h. Therefore, daily adjustment of the clock (i.e., resetting) is an essential mechanism for the circadian clock to adapt to daily environmental changes. One of the major cues for resetting the circadian clock is light. In C. reinhardtii, it has been demonstrated that the action spectrum for the phase resetting of the circadian clock shows two peaks around green (520 nm) and red (660 nm) light in dark-adapted cells or around blue (450–480 nm) and red (650–670 nm) light in illuminated cells (15, 16). However, the underlying molecular mechanisms remain largely unknown.
In this study, we show that ROC15, a clock-related putative DNA-binding protein, is rapidly decreased by light in a circadian-phase–independent manner. The rapid decrease is induced by blue, green, and red light and is mainly the result of a proteasome-dependent protein degradation pathway in which the clock-related F-box protein ROC114 is probably involved. In addition, we demonstrate that roc15 mutant shows defects in the phase-resetting of circadian clock by light. Our data provide not only a basis for understanding the molecular mechanisms of the light-induced phase-resetting in C. reinhardtii, but also insights into phase-resetting mechanisms of circadian clocks in green plants.
Results
Bioluminescence of the ROC15-LUC Reporter Decreased Rapidly After Light Exposure.
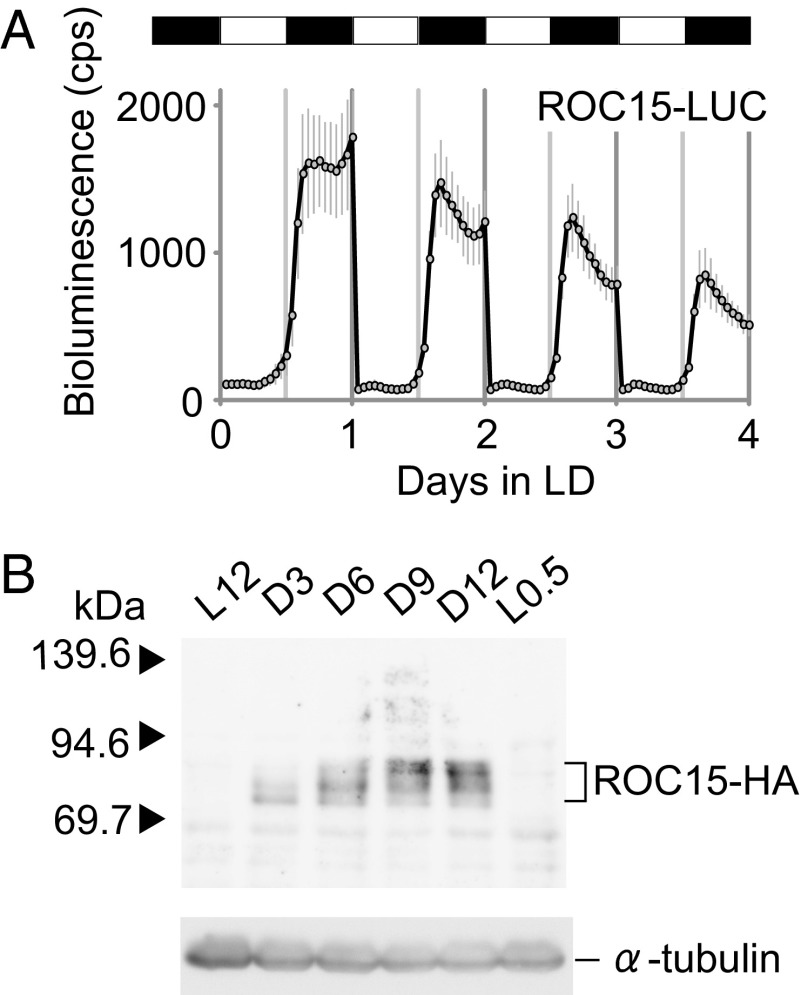
To investigate the regulatory mechanisms controlling clock protein abundances in C. reinhardtii, we generated clock protein-luciferase translational fusion reporter genes (SI Appendix, Fig. S1A) and introduced them into the nuclear genome of a reporterless WT strain (BR strain; Materials and Methods). Several transformants showed bioluminescence [ROC15-LUC, 26/480 (number of luminescent transformants/all transformants); ROC40-LUC, 19/576; ROC66-LUC, 14/480; and ROC75-LUC, 16/480]. The rates of luminescent transformants were reasonable because they were comparable to the complementation rates of their mutant phenotypes by the corresponding genomic DNA fragments (13). Under constant light conditions (LL), the transformants showed circadian bioluminescence rhythms with different phases (SI Appendix, Fig. S1B). Because the bioluminescence traces were comparable with those of their mRNA rhythms (13), their bioluminescence seemed to reflect their circadian expression features under LL conditions. To find unique features in clock protein expressions, we monitored bioluminescence rhythms in constant darkness (DD) and in a 12-h light/12-h dark (LD) cycle (SI Appendix, Fig. S1 C and D). Among these conditions and the investigated reporters, the most remarkable bioluminescence profile was that of ROC15-LUC in LD (Fig. 1A). Under this condition, ROC15-LUC activity was increased in the dark phase and immediately decreased at the beginning of the light phase (Fig. 1A). This suggested that ROC15 was directly responding to the light and dark signals, and implied that ROC15 may have some role in mediating light/dark information to the circadian clock in C. reinhardtii. Therefore, in this study, we focused on the regulation mechanisms and biological roles of ROC15.
Fig. 1.
Analysis of ROC15 expression in LD. (A) Bioluminescence trace of ROC15-LUC reporter under an LD condition. Asynchronous spot cultures of ROC15-LUC reporter strain was prepared in white 96-well plates as described previously (13). White and black bars represent light (2 μmol⋅m−2⋅s−1) and darkness, respectively. Each point represents the mean ± SD of bioluminescence counts from six independent cultures. (B) Western blot analysis of ROC15-HA protein. Asynchronous high-salt medium cultures (17) of the ROC15-HA strain were exposed to LD conditions, and samples were harvested at the time points indicated. Similar results were obtained in more than five independent experiments and in experiments with another transgenic line (A and B). White light was used in all experiments in this figure.
First, we confirmed whether the increase and rapid decrease in ROC15-LUC activity reflected changes in ROC15 protein abundance. To investigate ROC15 protein expression, we generated a transgenic strain expressing an HA epitope-tagged ROC15 protein (SI Appendix, Fig. S2 A and B). This ROC15-HA protein was able to fully complement roc15 mutant phenotypes (SI Appendix, Fig. S2 C and D), supporting that it was functional. Western blot analysis indicated that the temporal pattern of ROC15-HA protein expression were similar to that of ROC15-LUC bioluminescence in LD: very low expression at the end of the light phase (L12), increased in darkness (D3–D12), and decreased within 30 min after lights-on (L0.5) (Fig. 1B). This result strongly suggested that the bioluminescence of ROC15-LUC accurately reflected the dynamics of ROC15 protein abundance in LD. Moreover, we found that the apparent size of ROC15-HA increased during the night phase (Fig. 1B). The mobility changes were not observed in cells treated with the general kinase inhibitor 6-dimethylaminopurine [6-DMAP (18)] (SI Appendix, Fig. S3A), and were abolished in protein extracts treated with the lambda protein phosphatase (λPPase) (SI Appendix, Fig. S3B). These results indicated that ROC15 was gradually phosphorylated during the night phase.
In addition, we confirmed that the ROC15-LUC protein was also functional as a clock component because strains having ROC15-LUC in the roc15 mutant background showed a normal circadian rhythm (SI Appendix, Fig. S4).
To investigate the time-of-day dependence on the increase and decrease of ROC15-LUC bioluminescence, we exposed ROC15-LUC–expressing algae to darkness at different times of day and to light at different times of night, respectively. In cells exposed to a 3- to 9-h premature offset of light, a dramatic increase in ROC15-LUC activity occurred around 36 h, corresponding to the dusk of the previous LD cycle (SI Appendix, Fig. S5A). In contrast, premature onset of light induced a rapid decrease in bioluminescence at all time points tested (SI Appendix, Fig. S5B). These results indicated that the observed increase and decrease in ROC15-LUC activity were time-of-day dependent and independent phenomena, respectively. To examine the circadian profile of the light-induced decrease in ROC15 expression, light pulses were given at various time points to cells free-running in DD. At all time points tested, ROC15-LUC bioluminescence was decreased after exposure to light pulses (SI Appendix, Fig. S5C), indicating that the timing of this phenomenon was not regulated by the circadian clock. In contrast, the recovery of bioluminescence after exposure to light pulses was observed only during the subjective night (SI Appendix, Fig. S5C), suggesting that the ROC15 reaccumulation was regulated by the circadian clock.
Wavelength Dependence of the Light-Induced Rapid Decrease in ROC15-LUC Bioluminescence.
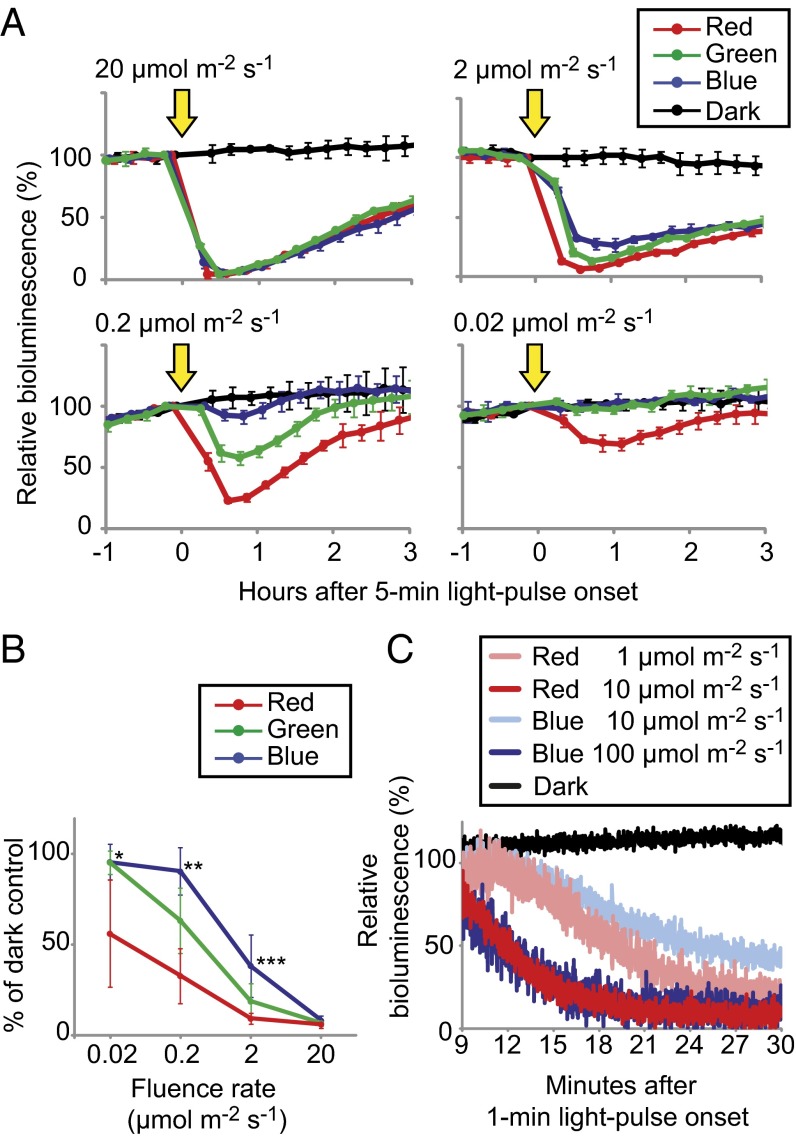
Next, we investigated the wavelength dependence of the light-induced rapid decrease in ROC15-LUC activity. Dark-adapted cells were exposed to light pulse with blue, green, and red light-emitting diodes (LEDs), and ROC15-LUC activity was monitored. At fluence rates of 20 and 2 μmol⋅m−2⋅s−1, ROC15-LUC bioluminescence was decreased immediately after exposure to light pulses for all colors tested (Fig. 2 A and B). At lower fluence rates, however, the magnitude of the effect was in the order of red > green > blue LEDs at 0.2 μmol⋅m−2⋅s−1, and only the red LED was still effective at 0.02 μmol⋅m−2⋅s−1 (Fig. 2 A and B). These results demonstrated that this phenomenon was induced by a wide range of wavelengths, but that its sensitivity varied depending on the wavelength. To exclude the possibility of any effects of the light emitted from the luciferase reaction on this process, we confirmed that the decrease in ROC15-HA protein followed the same wavelength-dependent pattern (SI Appendix, Fig. S6).
Fig. 2.
The effects of light pulses of different wavelengths on ROC15-LUC reporter activity. Asynchronous Tris-acetate-phosphate cultures (19) of the ROC15-LUC reporter strain were transferred to black 24-well plates and exposed to darkness for 9 h to induce bioluminescence of the reporter. (A) Representative data of responses to light pulses (arrows) by blue, green, and red LEDs. Fluence rates are indicated in the graph. The value just before the light pulse was normalized to 100. Each point represents the mean ± SD of the normalized bioluminescence traces from three independent cultures. (B) Fluence rate response of ROC15-LUC bioluminescence. Data shown are the mean ± SD of values relative to the dark controls (% of dark control) at the bottom of the bioluminescence traces after light pulse from two to five independent experiments of two transgenic lines. Asterisks indicate significant changes: *red vs. green and blue (Student t test; P < 0.01); **red vs. green and blue (P < 0.01) and green vs. blue (P < 0.05); ***red vs. blue (P < 0.05). (C) Detailed time courses of the bioluminescence decline after exposure to light pulses. Bioluminescence was monitored every second after a 1-min light-pulse with blue or red LEDs. The value just before the light pulse was normalized to 100 (not shown).
Next, we compared the detailed time course of the ROC15-LUC bioluminescence decrease after red and blue light pulses. At higher fluence rates (red: 10 μmol⋅m−2⋅s−1, blue: 100 μmol⋅m−2⋅s−1), ROC15-LUC bioluminescence decreased rapidly within 20 min, with a similar time course after both red and blue light pulses (Fig. 2C). At lower fluence rates (red: 1 μmol⋅m−2⋅s−1; blue: 10 μmol⋅m−2⋅s−1), the decline tended to be delayed, but the time courses were comparable between blue and red lights (Fig. 2C). In sum, the time course changed depending on the light intensity, but there was no obvious wavelength dependence in the time course of ROC15-LUC bioluminescence decrease. This implied that a common mechanism was involved in the rapid decrease of ROC15 after blue and red light pulses.
The Rapid Decrease in ROC15-LUC Bioluminescence Occurred Mainly Because of Proteasome-Mediated Protein Degradation.
We next investigated the mechanisms underlying the rapid decrease in ROC15-LUC activity. Northern blot analysis indicated that endogenous ROC15 mRNA abundance was decreased by ∼50% after 20- and 60-min light exposures compared with the dark controls (SI Appendix, Fig. S7A). This decline in ROC15 mRNA abundance following exposure to light was consistent with a recent report (20). Western blot analysis indicated that ROC15-HA protein expression disappeared almost completely within 20 min under the same experimental conditions (SI Appendix, Fig. S7B). These results indicated that the rapid and complete disappearance of bioluminescence could not be explained only by the decrease in mRNA.
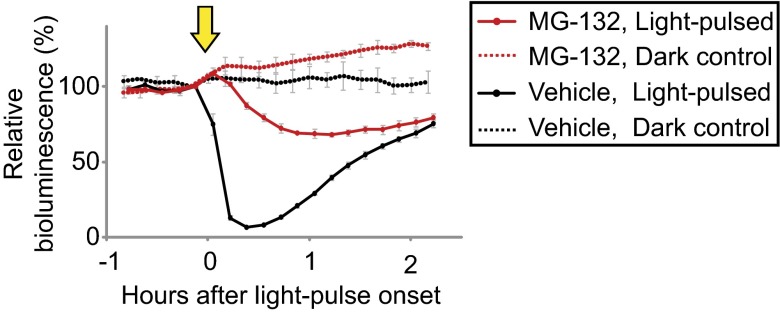
Protein accumulation is determined by the balance between the rate of synthesis and the rate of degradation. To evaluate the degradation of ROC15, the decay of ROC15-LUC bioluminescence was measured after treatment with the translational inhibitor cycloheximide (CHX). In DD, bioluminescence was sustained for more than 10 h after the addition of CHX (SI Appendix, Fig. S7C), indicating that this protein was very stable in darkness and that decreased synthesis alone could not be a cause of the decline in bioluminescence. In contrast, bioluminescence disappeared rapidly after the addition of CHX in LL (SI Appendix, Fig. S7D). From these data, we expected that light may induce ROC15 degradation; therefore, we tested whether inhibitors for protein degradation could affect the light-induced decrease in ROC15-LUC bioluminescence. As expected, this process was suppressed by pretreatment with the proteasome inhibitor MG-132 (Fig. 3) and others (SI Appendix, Fig. S7E), indicating that the rapid decrease in ROC15-LUC bioluminescence was due, at least in part, to protein degradation via the proteasome. The permeability of these drugs to C. reinhardtii cells seemed to be low because they had weaker effects on this process in cells that had not been subjected to permeabilization (SI Appendix, Fig. S7F). Thus, we could not exclude the possibility that the remaining activity of the ROC15 decline in the inhibitor treated cells (Fig. 3) was due to insufficient permeabilization of the cell.
Fig. 3.
The effect of the proteasome inhibitor MG-132. Media were changed to gametolysin-containing media (21, 22) to remove the cell wall for enhanced drug permeability, and then MG-132 was added to a final concentration of 100 μM. After a 30-h treatment in darkness, cells were exposed to a 0.5-min white light pulse (2 µmol⋅m−2⋅s−1, indicated by the arrow). The values just before the light pulses were normalized to 100. Each point represents the mean ± SD of the normalized bioluminescence traces from three independent cultures. Similar results were obtained in three independent experiments and in experiments with another transgenic line.
Interestingly, we found that this process was strongly inhibited by pretreatment with the general kinase inhibitor 6-DMAP (SI Appendix, Fig. S8A). Because this suggested an involvement of protein phosphorylation in the process, we examined the phosphorylation of ROC15 after light pulses. Although ROC15 at the mid-night phase is already phosphorylated (Fig. 1B and SI Appendix, Fig. S3), a much slower migration of ROC15-HA bands were observed after a light pulse (SI Appendix, Fig. S8B). The mobility changes were not observed in 6-DMAP–treated cells (SI Appendix, Fig. S8B) and were abolished in protein extracts treated with λPPase (SI Appendix, Fig. S8C). These results indicated that ROC15 was further phosphorylated by light.
In addition, pretreatment with CHX or 3-(3, 4-dichlorophenyl)-1,1-dimethylurea (an inhibitor of photosynthetic electron transport) did not significantly affect this process (SI Appendix, Fig. S9), suggesting that this process did not require newly synthesized proteins and was independent of photosynthesis, respectively.
The F-Box Protein Gene ROC114 Was Essential for the Light-Induced Decrease of ROC15.
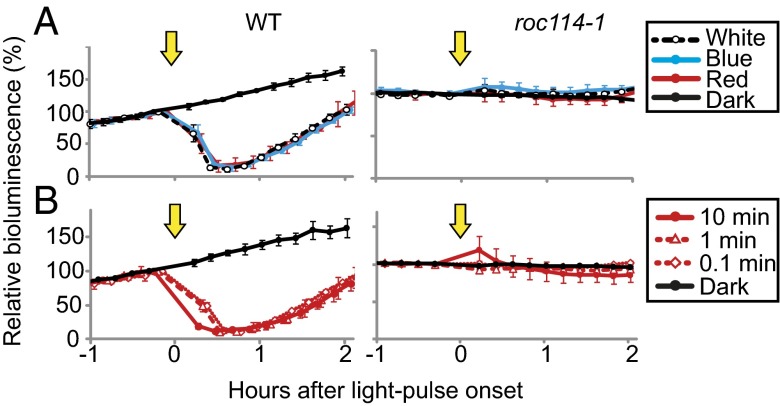
The data suggested that proteasome-dependent protein degradation and phosphorylation were involved in the rapid decrease in ROC15 expression. Thus, to find the molecular components involved in this process, we examined the light-induced decrease in ROC15-LUC bioluminescence in kinase and F-box protein mutants that have been isolated as rhythm mutants (13). F-box proteins are subunits of the SKP1-CUL1-F-box E3-ubiquitin ligase, mediating protein degradation via the ubiquitin-proteasome pathway (23). The ROC15-LUC reporter strain was crossed with kinase mutants [roc69, roc78, and roc94 (Mut-9)] and F-box protein mutants [roc114-1 and roc114-2 (previously called roc114 and roc108, respectively)]. In kinase mutant backgrounds, the light-induced decline in ROC15-LUC bioluminescence was observed, similar to that observed in WT siblings (SI Appendix, Fig. S10). On the other hand, the rapid decrease in bioluminescence was hardly detectable in the roc114-1 and roc114-2 backgrounds (Fig. 4A and SI Appendix, Fig. S10). Light pulses with both blue and red LEDs had little effect (Fig. 4A), even at the higher fluence of red light (Fig. 4B). These results indicated that the F-box protein ROC114 was essential for the blue and red light-induced decrease in ROC15.
Fig. 4.
The light response of ROC15-LUC reporters in the roc114-1 background. A ROC15-LUC reporter strain having the roc114-1 background was obtained by genetic crosses (SI Appendix, Fig. S10). WT siblings were used as a control. Cell cultures were prepared in black 24-well plates as described in Fig. 2. Cells were exposed to white (20 μmol⋅m−2⋅s−1, 3 min), blue (20 μmol⋅m−2⋅s−1, 3 min), or red (20 μmol⋅m−2⋅s−1, 0.5 min) light pulses (A) or to red light pulses (200 μmol⋅m−2⋅s−1) of different durations (B). Arrows indicate the times when the light pulses were given. The values just before the light pulse were normalized to 100. Each point represents the mean ± SD of the normalized bioluminescence traces from three independent cultures. Similar results were obtained in two independent experiments (A and B).
To address the relationship between ROC15 and ROC114, we examined the subcellular localization of these proteins by immunocytochemistry. ROC15-HA signals were observed at the nucleus just inside the signals of nuclear pore complex that we used as a marker for the nuclear envelope (SI Appendix, Fig. S11). No obvious changes in the localization of ROC15-HA were observed in roc114 mutant background (SI Appendix, Fig. S11). To investigate the localization of ROC114, we generated strains expressing a tandem affinity purification (TAP)-tagged ROC114 (ROC114-TAP; SI Appendix, Fig. S12 A and B) (24). The ROC114-TAP was functional because it restored the circadian rhythmicity and the light-induced degradation of ROC15-HA (SI Appendix, Fig. S12 C and D). Immunostaining of cells coexpressing ROC15-HA and ROC114-TAP detected specific signals of these proteins (SI Appendix, Fig. S13A). ROC15-HA and ROC114-TAP signals were colocalized at the nucleus in the dark (SI Appendix, Fig. S13B; 0 min). After a light pulse was given, ROC15-HA signals disappeared rapidly, whereas ROC114-TAP signals persisted at least until 20 min (SI Appendix, Fig. S13B). No obvious changes in their localization were detectable after the light pulse (SI Appendix, Fig. S13B). These results suggested that ROC15 and ROC114 were localized to the same region of the nucleus and that ROC15 was degraded in the region after light exposure.
The slower mobility bands of ROC15-HA were observed throughout the night phase in roc114 background even in the early night (SI Appendix, Fig. S14A). In addition, the mobility shift of ROC15-HA was observed after light pulses in roc114 (SI Appendix, Fig. S14B). These results suggested that phosphorylation of ROC15, at least after light pulses, was not affected by roc114 mutation. It was not clear whether the ROC15 in roc114 was phosphorylated in the dark or not because we could not exclude the possibility that the slower mobility bands observed in the dark in roc114 reflected the phosphorylation in the previous light phase (SI Appendix, Fig. S14A).
Abnormal Light Responses of the Circadian Clock in a roc15 Mutant.
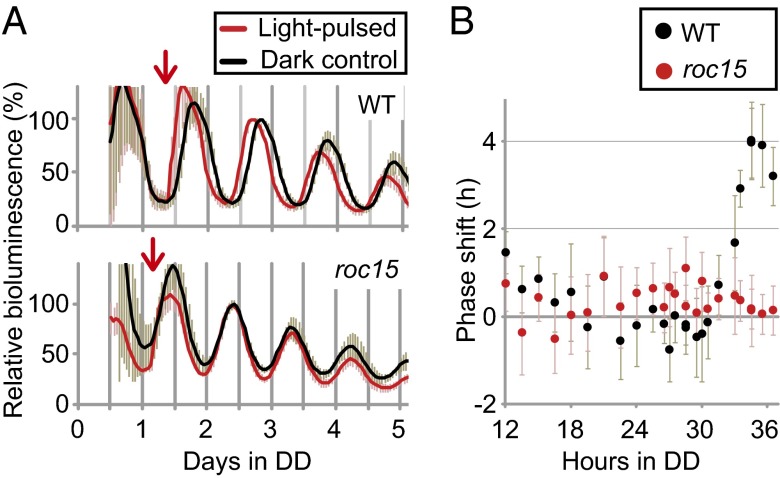
The rapid response of ROC15 to light encouraged us to speculate on the roles of ROC15 in the light response mechanisms of the circadian clock in C. reinhardtii. The roc15 mutant was originally isolated as a mutant showing short-period and advanced phase-angle phenotypes in the circadian rhythm of bioluminescence derived from the tufA-lucCP reporter in the chloroplast genome (13, 25). The abnormal phase angle of the circadian rhythm to the preceding environmental light-dark phase is thought to be an indication of defects in the light response mechanism of the circadian clock. To test this possibility, we examined the light-induced phase shifts of the circadian clock in the roc15 mutant. Light pulses were given to WT algae (CBR strain; Materials and Methods) and roc15 mutant algae at various time points in DD, and the degree of phase shift in the bioluminescence rhythm of the tufA-lucCP reporter was measured. Large phase advances were observed in WT algae but not in roc15 mutant algae, when light pulses were given just after the trough phase of the bioluminescence rhythm (corresponding to the late subjective night; Fig. 5A). Such large phase shifts were not detected throughout the circadian cycle in the roc15 mutant (Fig. 5B). These results indicated that ROC15 was involved in the phase-shifting of the circadian clock by acute light pulses.
Fig. 5.
Light-induced phase shift of circadian clock in roc15 mutant. Spot cultures of WT (CBR) and roc15 strains were prepared as described previously (13). Each plate was subjected to a 5-min white light pulse (85 μmol⋅m−2⋅s−1) at various time points between 12 and 36.5 h under DD conditions. (A) Representative bioluminescence traces. The light pulses were given at the times indicated by arrows. For phase comparisons, the value of the second peak after the light pulse was normalized to 100. Each trace represents the mean ± SD of the normalized traces from 8–10 independent cultures. (B) Phase-response curves to light pulses. Phases of each trace after exposure to light pulses were calculated by the cosinor method of the Rhythm-Analyzing Program (26). The amount of phase shift was calculated by subtracting the phase of the light-pulsed sample from that of the dark control. Positive and negative values indicate phase advances and delays, respectively. Each point represents the mean ± SD of 10 independent cultures.
Next, we investigated phase responses of the circadian clock to various durations of light exposure. Asynchronous cells were kept in darkness for 12 h, followed by exposure to light for various durations and were then released into DD (SI Appendix, Fig. S15A). In the WT strain, the phases of peaks and troughs in the tufA-lucCP reporter rhythm were affected by extended light periods (SI Appendix, Fig. S15A); phases were delayed or advanced compared with those observed after the normal LD cycle (see the plots at the top of each panel of SI Appendix, Fig. S15A). In contrast, in the roc15 mutant, the peaks and troughs occurred at nearly the same time, regardless of the extended light period (SI Appendix, Fig. S15A). Similarly, we investigated the effects of extended darkness on the subsequent circadian rhythm under LL conditions. Asynchronous cells were subjected to various durations of darkness and were then released into LL (SI Appendix, Fig. S15B). In this case, the circadian clock of WT strain appeared to be almost fully reset by the light onsets; phases were delayed depending upon the duration of darkness such that the timing of the peaks and troughs occurred in parallel with light onset (SI Appendix, Fig. S15B). On the other hand, this “full reset” was not observed in the roc15 mutant, although the phases were affected by extended darkness (SI Appendix, Fig. S15B). These results indicated an abnormality in the phase response function of the circadian clock in the roc15 mutant. Taken together, our results indicated that ROC15 was an important molecular component involved in the phase-resetting mechanisms of the circadian clock in C. reinhardtii.
The bioluminescence rhythm was hardly detectable when WT cultures were exposed to darkness for 24 h (SI Appendix, Fig. S15C). These cells were expected to be transferred into LL when the phase of their circadian clock was in the beginning of the subjective night (i.e., 180° out of phase between the circadian clock and the lighting condition). We speculated that desynchronization of the individual cellular rhythms was induced by this condition. Exposure to darkness for 48 h also induced a similar effect, although almost half of the cultures tested showed weak rhythms. Similarly, the arrhythmic phenotype was observed after exposure to darkness for 18 and 42 h in roc15. The difference of the effective length of dark exposure was thought to be due to its short period and advanced phase-angle phenotypes.
Discussion
In this study, by using a luciferase reporter assay, we observed a rapid decrease in the expression of the clock-related protein ROC15 after light exposure in a circadian-phase–independent manner. This process was induced by a relatively wide range of wavelengths and was mainly mediated through a proteasome-dependent protein degradation pathway in which the F-box protein ROC114 is probably involved. In addition, we demonstrated that the roc15 mutant exhibited defects in the light-induced phase resetting of the circadian clock. Our data provided a basis for understanding the molecular mechanisms of the light-induced phase resetting of the circadian clock in C. reinhardtii.
The action spectrum of the phase resetting of the circadian clock in dark-adapted C. reinhardtii cells has been shown to exhibit a major peak around 660 nm and a second peak around 520 nm (15). In this study, we found that the wavelength dependence of the ROC15 decline was well correlated with that of the phase-resetting mechanism. The red LED (660 nm) was the most effective; the green LED (525 nm) was the second most effective (Fig. 2B). In addition, the effective doses for green and red light pulses in ROC15 decline (Fig. 2B) were remarkably similar in order of magnitude to those of the phase-resetting mechanism reported in a previous study (15). These results strongly support the idea that ROC15 degradation is a key event required for phase resetting in C. reinhardtii.
In D. melanogaster, light-induced degradation of TIMELESS protein, an essential clock component, is known to be one of the mechanisms required for resetting the circadian clock (27–30). Our findings indicated that a similar mechanism (light-induced rapid degradation of clock protein) was used in evolutionary divergent systems. On the other hand, in higher plants, it is not known whether such a mechanism is used in the phase resetting of the circadian clock, which includes some components similar to those in C. reinhardtii. ROC15 is a putative transcription factor that contains a GARP DNA-binding motif, similar to the A. thaliana clock protein PHYTOCLOCK1 (PCL1) [also called LUX ARRHYTHMO (LUX)] (6, 7, 13). Because PCL1 (LUX) is expressed in the evening (31, 32), its mRNA expression profiles do not correspond to those of ROC15, which peaks during the late subjective night (6, 13). In addition, because PCL1 (LUX) protein expression increases gradually during the day and declines gradually during the night (33), there seems to be no obvious acute light response at the protein level. These facts suggest that PCL1 (LUX) is not the counterpart of ROC15. Therefore, we speculated that other members of the higher plant GARP protein family may be light responsive. According to a public database of microarray data (DIURNAL) (34), the At3g10760 gene, a member of the PCL1 (LUX) family (31), is expressed in relatively similar phases as ROC15. Thus, it is possible that the At3g10760 gene product responds to light. Further studies are required to fully investigate this possibility.
ROC15-HA was gradually phosphorylated during the night phase (Fig. 1B and SI Appendix, Fig. S3) and was further phosphorylated by light (SI Appendix, Fig. S15 B and C). The latter may be necessary for the light-induced degradation of ROC15 because pretreatment with kinase inhibitor at the late night almost completely blocked the effect of light pulse (SI Appendix, Fig. S8A). On the other hand, the former seems to not affect the light-induced ROC15 degradation because the rapid decrease of ROC15-LUC bioluminescence was observed throughout the night phase, even in the early night (SI Appendix, Fig. S5B) when ROC15-HA was not sufficiently phosphorylated (Fig. 1B and SI Appendix, Fig. S3A). The phosphorylation of ROC15 in the dark may be for the purpose of its circadian oscillation. We speculate that these two phosphorylation events occur at different sites in the ROC15 protein. Further study is needed to clarify this point.
At present, the pathway or pathways that mediate light signals to ROC15 are largely unknown. The very similar time course of ROC15 decrease after blue and red light (Fig. 2C) implies that common mechanisms are involved in this process, even if the blue and red light signals are mediated through independent pathways. Because roc114 mutation blocked the effects of both blue and red light (Fig. 4 A and B), the degradation pathway for ROC15 is thought to be common to blue and red light. The photoreceptors mediating light-induced ROC15 degradation are also still not clear. In A. thaliana, phytochromes and cryptochromes are known to be involved in the light signaling to the circadian clock (35–37). Although the typical absorption peak of phytochromes corresponds well with the major peak of the action spectrum of the phase resetting of the circadian clock in C. reinhardtii (∼660 nm) (15), there is no obvious homolog for these phytochromes in the C. reinhardtii genome sequence (38). In addition, the phase resetting of the circadian clock of C. reinhardtii by red light is not diminished by subsequent exposure to far-red light (15). Therefore, red light-absorbing photoreceptor(s) other than typical phytochromes should exist in C. reinhardtii. Recently, it was demonstrated that aCRY, a cryptochrome homolog in C. reinhardtii, absorbs a wide range of wavelengths, including red regions (19). Although it is an attractive candidate of photoreceptor for light-induced ROC15 degradation, we could not exclude the possibility of the existence of other unknown red-light photoreceptors in C. reinhardtii. Another cryptochrome homolog, Chlamydomonas photolyase homolog 1 (CPH1), shows a rapid decrease in protein accumulation level, similar to ROC15, after exposure to blue or red light because of proteasome-dependent protein degradation (39). The light-induced degradation of ROC15 and CPH1 may occur through the same pathway.
The bioluminescence pattern of ROC15-LUC under DD conditions gave an unexpected result. On the first day, bimodal peaks occurring in the subjective night and subjective day were observed (SI Appendix, Fig. S1C, Top). The peak in the subjective night tended to decrease gradually, and the peak in the subjective day became the major peak (SI Appendix, Fig. S1C), although the reaccumulation rate of ROC15 was low in the subjective day (SI Appendix, Fig. S5C). However, because ROC15 protein was very stable in darkness (SI Appendix, Fig. S7C), the luciferase reporter would not be able to reflect the circadian profile of gene expression and it might be possible to be affected by factors other than the amount of luciferase protein (e.g., intracellular ATP or luciferin concentrations). Further experiments are needed to confirm the ROC15 expression pattern in DD (e.g., Western blotting of ROC15-HA). However, because the ROC15-LUC reporter proved to be a good reflection of the light-induced degradation of ROC15 protein, it will be a useful tool for further analysis of the mechanisms mediating light-induced ROC15 degradation, including a forward genetic screening of the components of this process. The other reporters developed in this study will also be useful tools for deepening our understanding of the molecular mechanisms of the circadian clock in C. reinhardtii.
Materials and Methods
Strains and Media.
Strains and growth conditions are described in SI Appendix.
Generation of Luciferase Reporter Strains.
A codon-adapted firefly luciferase for the C. reinhardtii nuclear genome (LUCnc; GenBank database, European Molecular Biology Laboratory database, and DNA Data Base in Japan accession no. AB762768) was generated by artificial gene synthesis (Hokkaido System Science). Reporter plasmids were constructed by standard DNA manipulations, and transformation was carried out as described previously (13). Details are given in SI Appendix.
Bioluminescence Monitoring.
Bioluminescence was measured using the apparatus described by Okamoto et al. (40) and a newly developed, highly sensitive monitoring apparatus (model CL24-W; Churitsu Electric Corp.). Details are given in SI Appendix.
Light Sources.
We used white fluorescence tubes (Panasonic) and LEDs (CCS, Panasonic). Details are given in SI Appendix.
Further Information.
Details of methods for Northern blot, Western blot, phosphatase treatment, and immunocytochemistry are described in SI Appendix. Details for inhibitors used in this study are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank A. Kinoshita (Nagoya University) for technical assistance, G. Ito and H. Hashimoto (Hokkaido System Science) for synthesis of LUCnc, and Editage for providing editorial assistance. This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and the Daiko Foundation (to M.I.) and from MEXT, the Nakajima Foundation, and the Novartis Foundation (to T.M.). The Graduate School of Science of Nagoya University was supported by a Global Center of Excellence grant from MEXT. Development of the highly sensitive bioluminescence-monitoring apparatus CL24-W was supported by the Japan Science and Technology Agency.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database, European Molecular Biology Laboratory database, and DNA Data Base in Japan (accession no. AB762768).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220004110/-/DCSupplemental.
References
- 1.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 3.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulze T, et al. How the green alga Chlamydomonas reinhardtii keeps time. Protoplasma. 2010;244(1-4):3–14. doi: 10.1007/s00709-010-0113-0. [DOI] [PubMed] [Google Scholar]
- 6.Matsuo T, Ishiura M. New insights into the circadian clock in Chlamydomonas. Int Rev Cell Mol Biol. 2010;280:281–314. doi: 10.1016/S1937-6448(10)80006-1. [DOI] [PubMed] [Google Scholar]
- 7.Matsuo T, Ishiura M. Chlamydomonas reinhardtii as a new model system for studying the molecular basis of the circadian clock. FEBS Lett. 2011;585(10):1495–1502. doi: 10.1016/j.febslet.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 8.Corellou F, et al. Clocks in the green lineage: Comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21(11):3436–3449. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizoguchi T, Putterill J, Ohkoshi Y. Kinase and phosphatase: The cog and spring of the circadian clock. Int Rev Cytol. 2006;250:47–72. doi: 10.1016/S0074-7696(06)50002-6. [DOI] [PubMed] [Google Scholar]
- 10.Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol. 2003;54(1):111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- 11.Iliev D, et al. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142(2):797–806. doi: 10.1104/pp.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006;18(8):1908–1930. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuo T, et al. A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 2008;22(7):918–930. doi: 10.1101/gad.1650408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodenough UW. Green yeast. Cell. 1992;70(4):533–538. doi: 10.1016/0092-8674(92)90424-b. [DOI] [PubMed] [Google Scholar]
- 15.Kondo T, Johnson CH, Hastings JW. Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas: I. Cells in darkness. Plant Physiol. 1991;95(1):197–205. doi: 10.1104/pp.95.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson CH, Kondo T, Hastings JW. Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas: II. Illuminated cells. Plant Physiol. 1991;97(3):1122–1129. doi: 10.1104/pp.97.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1960;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comolli J, Taylor W, Hastings JW. An inhibitor of protein phosphorylation stops the circadian oscillator and blocks light-induced phase shifting in Gonyaulax polyedra. J Biol Rhythms. 1994;9(1):13–26. doi: 10.1177/074873049400900102. [DOI] [PubMed] [Google Scholar]
- 19.Gorman DS, Levine RP. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54(6):1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beel B, et al. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012;24(7):2992–3008. doi: 10.1105/tpc.112.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda Y, Saito T, Yamaguchi T, Kawase H. Cell wall lytic enzyme released by mating gametes of Chlamydomonas reinhardtii is a metalloprotease and digests the sodium perchlorate-insoluble component of cell wall. J Biol Chem. 1985;260(10):6373–6377. [PubMed] [Google Scholar]
- 22.Kubo T, Saito T, Fukuzawa H, Matsuda Y. Two tandemly-located matrix metalloprotease genes with different expression patterns in the chlamydomonas sexual cell cycle. Curr Genet. 2001;40(2):136–143. doi: 10.1007/s002940100239. [DOI] [PubMed] [Google Scholar]
- 23.Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9(6):631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 25.Matsuo T, Onai K, Okamoto K, Minagawa J, Ishiura M. Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period. Mol Cell Biol. 2006;26(3):863–870. doi: 10.1128/MCB.26.3.863-870.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto K, Onai K, Ishiura M. RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal Biochem. 2005;340(2):193–200. doi: 10.1016/j.ab.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84(5):677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 28.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380(6570):129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 29.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271(5256):1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271(5256):1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 31.Hazen SP, et al. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102(29):10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells. 2005;10(10):963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 33.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 35.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282(5393):1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 36.Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12(12):2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanovsky MJ, Mazzella MA, Whitelam GC, Casal JJ. Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J Biol Rhythms. 2001;16(6):523–530. doi: 10.1177/074873001129002213. [DOI] [PubMed] [Google Scholar]
- 38.Mittag M, Kiaulehn S, Johnson CH. The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol. 2005;137(2):399–409. doi: 10.1104/pp.104.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reisdorph NA, Small GD. The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 2004;134(4):1546–1554. doi: 10.1104/pp.103.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto K, Onai K, Furusawa T, Ishiura M. A portable integrated automatic apparatus for the real-time monitoring of bioluminescence in plants. Plant Cell Environ. 2005;28:1305–1315. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.