Abstract
Annual influenza vaccinations aim to protect against seasonal infections, and vaccine strain compositions are updated every year. This protection is based on antibodies that are produced by either newly activated or memory B cells recalled from previous encounters with influenza vaccination or infection. The extent to which the B-cell repertoire responds to vaccination and recalls antibodies has so far not been analyzed at a genetic level—which is to say, at the level of antibody sequences. Here, we developed a consensus read sequencing approach that incorporates unique barcode labels on each starting RNA molecule. These labels allow one to combine multiple sequencing reads covering the same RNA molecule to reduce the error rate to a desired level, and they also enable accurate quantification of RNA and isotype levels. We validated this approach and analyzed the differential response of the antibody repertoire to live-attenuated or trivalent-inactivated influenza vaccination. Additionally, we analyzed the antibody repertoire in response to repeated yearly vaccinations with trivalent-inactivated influenza vaccination. We found antibody sequences that were present in both years, providing a direct genetic measurement of B-cell recall.
Every year, influenza viruses cause the deaths of an average of 36,000 individuals in the United States alone (1). Although the immunological memory created through vaccination can confer decade-long protection against a particular viral strain, antigenic drift in the original strain and the occurrence of distinct viral strains can enable the virus to evade the immune system (2). As a result, influenza vaccination formulations have to be reevaluated, adjusted, and administered annually to best match the annual influenza strain. Vaccine-induced immunity against influenza is primarily antibody-based, and as such, it relies on the activation of naive B cells or the reactivation (recall) of memory B cells to produce high levels of antibody specific to the vaccine strain. Prior studies approached recall memory responses by measuring plasma antibody levels and specificity or sequencing antibody loci of isolated B cells, with one study concluding that the response to influenza vaccination is pauciclonal (i.e., composed of only a few distinct clones) (3, 4). However, this study and others were limited in the number of B cells that they were able to analyze and not able to show that the same clone recurs during recall. The strength of the recall response, the isotype distribution, and the clonal relationship to others have been unclear.
Recently, methods to sequence antibody repertoires of whole organisms and human blood samples were developed and applied to investigate several features of B-cell repertoires (5, 6). This approach has been used to investigate a variety of phenomena, including effects of influenza vaccination, residual disease in leukemia, effects of immune suppression, and differences between memory and naive B-cell compartments (5–11).
Analyzing vaccine recall response requires the detection of antibody sequences shared between separate blood samples taken over 12 mo apart. Because of the limited throughput and high error rate of next generation sequencing approaches, it is challenging to query a human blood sample exhaustively and accurately identify these shared sequences. To address these problems, we developed a highly accurate high-throughput approach that relies on the labeling of individual RNA molecules (12–14). We used these labels to generate multiple sequencing reads for each RNA molecule and compose a consensus read for each molecule. First, we validated this approach by sequencing the immunoglobulin heavy chain (IGH) repertoire of a blood sample. We found that this approach was highly accurate, quantitative, and reproducible. Second, we used the consensus read approach to estimate the size of the B-cell repertoire, determining a refined estimate for different B-cell populations. Third, we dissected immune responses to live-attenuated (LAIV) and trivalent-inactivated (TIV) influenza vaccines. LAIV and TIV are known to show distinct immune responses, and we could clearly distinguish the effects of the two vaccine types on the antibody repertoire. Finally, we analyzed the nature of the recall response of individuals to TIV administration in two consecutive years. We found hundreds of unique antibody lineages originating from distinct B-cell memory clones that were activated by vaccination in both consecutive years.
Results
Labeling of RNA Molecules with Random Nucleotide Unique Identifiers.
The sequencing approach that we used relied on labeling each RNA molecule during cDNA synthesis and preserving this nucleotide label throughout PCR amplification. Using these labels, we could identify group reads originating from the same RNA molecule. Therefore, both isotype- and V segment-specific primers were designed to include a stretch of 8 random nt followed by a partial paired-end adapter sequence on their 5′ end (Fig. S1). We used total RNA extracted from the B cell-containing peripheral blood mononuclear cells (PBMCs) as input; reverse transcription and subsequent primer extension resulted in a pool of double-stranded cDNA, in which initial IGH RNA molecules were labeled with a 16-nt unique identifier (UID). This pool was then amplified in a PCR where the primers completed the sequencer adapter sequences (Fig. S1). The resulting libraries were multiplexed and sequenced on the Illumina HiSeq2000 Sequencer. We used a paired-end sequencing protocol, sequencing 100 bp on the first read and 120 bp on the second read to cover the whole amplicon. The sequencing reads included the 16-nt UID, making it possible to retrieve the unique labels of the initial IGH RNA molecules. The high raw read number enabled us to sequence uniquely labeled IGH RNA molecules multiple times and build a highly accurate consensus before downstream analysis (Fig. S2A).
IGH molecules with the same nucleotide sequence were grouped into IGH sequences. The abundance of an IGH sequence is defined by the number of unique UIDs that share the same consensus sequence (Fig. S1). Biologically, the abundance of an IGH sequence is determined by the number B cells that express this sequence convolved with the distribution of expression levels across all these cells and represents a proxy for overall antibody abundance.
Characterization of the Consensus Read Approach.
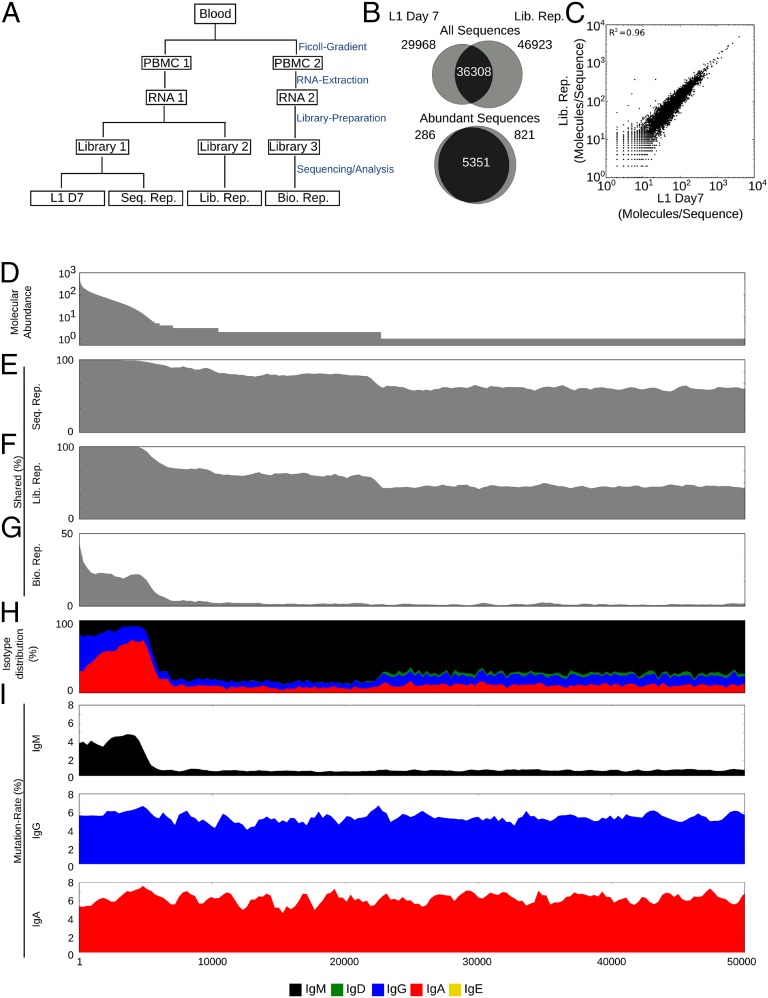
To validate the sequencing protocol, a blood sample was collected from a young adult individual (L1) taken 7 d after vaccination with the LAIV vaccine. A sequencing library (L1 D7) (Fig. 1A) was prepared and then sequenced. We determined the reproducibility of this IGH sequence abundance information by preparing a second sequencing library (Lib. Rep.) (Fig. 1A) from a separate aliquot of the same RNA extraction and sequenced it on an independent run. We leveraged the exceptional accuracy of our approach to find IGH sequences shared between the sequenced libraries (L1 D7 and Lib. Rep.) and compare their abundances (Fig. 1 B and C); 36,308 of 66,307 IGH sequences were shared between L1 D7 and Lib. Rep. and showed very strong similarity in their abundance (R2 = 0.96) (Fig. 1C), illustrating the robustness of the approach. The overlap between the libraries was almost complete when we focused on abundant sequences, defined to be those sequences with more than 5 IGH molecules per IGH sequence (abundance ≥5); 5,351 of 5,637 abundant sequences in L1 D7 were shared by Lib. Rep. (Fig. 1B).
Fig. 1.
Consensus read approach validation. (A) Experimental setup. Blood is drawn from an individual. Two separate PBMC aliquots are prepared, and RNA is extracted. Several sequencing libraries are prepared and sequenced. (B) Venn diagrams illustrating the IGH sequence overlap between L1 D7 and Lib. Rep. for all IGH sequences or only abundant IGH sequences represented by more than 5 IGH molecules. (C) The abundances of IGH sequences shared between L1 D7 and Lib. Rep. are shown as a scatterplot. (D–I) IGH sequences in L1 D7 are ordered by abundance. (D) Abundance (IGH molecules/IGH sequence) of sequence. In bins of 300 sequences from left to right, percents of shared sequences between L1 D7 and indicated samples [(E) Seq. Rep., (F) Lib. Rep., and (G) Bio. Rep.] are shown. (H) Isotype distribution of sequences. (I) Mutation rate (%) of L1 D7 sequences separated by isotypes (IgM, IgG, and IgA).
The lower rate of sharing among sequences that were not abundant was caused by either subsampling from the total RNA pool during library construction or insufficient raw read coverage during the sequencing reaction and data analysis quality filtering. To identify the main cause, we resequenced the initial sequencing library (Seq. Rep.). We then visualized the rate at which sequences were shared between these samples as a function of their abundances (Fig. 1 D–H). The rates at which sequences were shared between L1 day 7 and either Lib. Rep. or Seq. Rep. were very similar and clearly dependent on sequence abundance, ranging from ∼50% for sequences with abundance of one to almost 100% for sequences with an abundance more than five (Fig. 1 E and F). The fact that the construction of a separate library (Lib. Seq.) had only a small effect on the rate of shared sequences implicated raw read coverage and quality filtering, but not library construction, as the main cause for the reduced sharing rates among low-abundance sequences. IGH sequences represented by fewer IGH molecules have a higher chance that none of their molecules meet the raw read coverage or quality thresholds and are consequently dropped completely.
The resequencing of the initial library (Seq. Rep.) also allowed us to measure the sequencing accuracy of our approach. The unique labeling of molecules enabled us to use discrepancies between IGH molecules with barcodes that appeared in both L1 D7 and Seq. Rep. to determine the sequencing error of consensus reads to be ∼1/50,000 errors per 1 bp or Q47 (Fig. S2B). We used discrepancies between raw reads in UID groups in L1 D7 to determine the raw Illumina sequencing error to be ∼1/500 errors per 1 bp or Q27. This 100-fold higher raw error rate would, in the absence of barcodes, dramatically inflate the apparent number of unique IGH sequences (Fig. S2C), thereby illustrating the problem of determining the complexity of the antibody repertoire using a large number of error-prone raw sequencing reads.
These experiments showed that the consensus read approach that we developed is highly accurate and quantitative and detects practically all abundant sequences in a PBMC sample.
Identifying the Activated B-Cell Compartment.
To determine how representative a PBMC sample is of the complete peripheral blood and whether the abundance of an IGH sequence might help us identify sequences expressed by memory B cells and plasmablasts, we generated an additional sequencing library from a PBMC sample purified from blood taken at the same visit as L1 D7 (Bio. Rep.) (Fig. 1A).
Recapturing an IGH sequence in two separate PBMC aliquots requires at least two cells of the same B-cell clone expressing this IGH sequence to exist and partition into the separate aliquots. Because activated B cells have undergone clonal expansion, we expected the sequences shared between L1 D7 and Bio. Rep. to be enriched for sequences expressed by memory B cells and plasmablasts, which are known to make up ∼30% and 2% of peripheral B cells, respectively (15).
The Bio. Rep. sample shared 1,401 of 5,637 (25%) of its abundant sequences (>5 IGH molecules/IGH sequence) but only 974 of 66,307 (1.5%) low-abundance sequences (<5 IGH molecules/IGH sequence) with L1 D7.
We again visualized the rate at which sequences were shared between L1 D7 and Bio. Rep. as a function of their abundances. This visualization showed that the sequences shared between different PBMC samples were heavily biased to more abundant sequences (Fig. 1G). This bias to more abundant sequences was much more pronounced than in the Lib. Rep. sample, which was constructed from the same PBMC RNA as L1 D7.
Abundant sequences also showed a distinct isotype distribution. Although low-abundance sequences were mostly IgM, abundant sequences were mostly IgG and IgA (Fig. 1H). Furthermore, although abundance did not correlate with the mutation rate of class-switched IgG and IgA sequences, the mutation rate of IgM increased in abundant sequences to levels only slightly lower than IgG and IgA mutation rates (Fig. 1I).
The enrichment for IgG and IgA as well as mutated IgM in abundant sequences together with the heavily biased recapture by the Bio. Rep. sample indicated strongly that the abundant IGH sequences are heavily enriched for sequences expressed by activated B cells. Together, this data shows that we could use isotype and abundance information to identify activated B-cell sequences.
Estimating Repertoire Size.
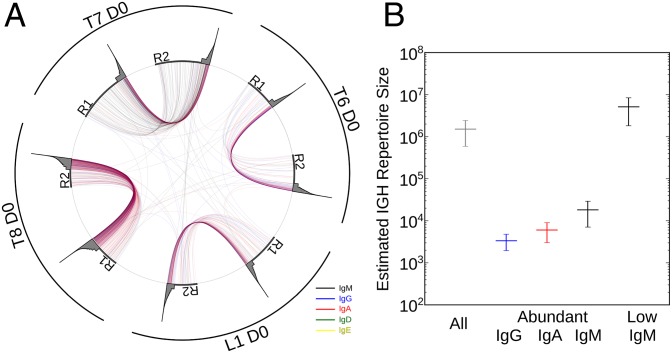
Next, we used this isotype and abundance information to refine capture–recapture analysis and estimate the number of distinct activated B-cell clones present in the peripheral blood at any given time. We first combined highly similar but distinct IGH sequences into IGH lineages that are highly likely to originate from the same original B-cell clone (Materials and Methods and Fig. S1B). We then performed capture–recapture analysis (Materials and Methods) on replicate pairings of four individuals at day 0 (before vaccination). Previous studies have estimated the overall size of the B-cell repertoire without taking abundance or isotype into account in the range of 2–9 million sequences (5, 10).
When we neglect isotype differences and abundance effects, the peripheral IGH lineage repertoire seems to be 1,514,640 (±922,827), which is very close to what previous studies have estimated. However, as with IGH sequences (Fig. 1G), the rates at which IGH lineages were shared between the replicates were much higher for abundant lineages (at least 5 IGH molecules/IGH lineage), which was shown when we visualized the lineages shared between samples normalized by subsampling to 50,000 IGH molecules (Fig. 2A). These uneven sharing (recapture) rates are likely to skew repertoire size estimation. To take these uneven recapture rates into account, we separated lineages according to isotype and abundance (low <5 IGH molecules/IGH lineage ≤abundant) and repeated the analysis. This approach resulted in an estimation of, on average (±SD), only 18,169 (±11,130) IgM, 5,998 (±2,972) IgA, and 3,359 (±1,412) IgG abundant lineages in contrast to 5,102,725 (±3,279,203) low-abundance IgM lineages in the peripheral blood (Fig. 2B).
Fig. 2.
Repertoire size estimation. (A) Visualization of lineages shared between replicate sample pairs (R1 and R2) from individuals (L1, T6, T7, and T8). Data were subsampled to 50,000 IGH molecules to normalize for variability in sampling depth. Lineages of each time point are plotted on the circumference of the circle, with the gray area representing abundance of the respective lineages (logarithmic). Lineages present in two time points are connected with lines colored according to their isotype. (B) Capture–recapture estimation of different IGH lineage populations. Average (±SD) of four individuals is shown.
The estimate of ∼1,500,000 lineages when neglecting isotype and abundance is, therefore, the skewed result of a mixture of two distinct populations: thousands of activated B cells and millions of naive B cells. These data also suggest that only several thousand activated B-cell clones are circulating in the peripheral blood at any given time, which is far lower than previously thought.
Shared Sequences Between Samples and Individuals.
Interestingly, lineages were shared between individuals at a very low level (Fig. 2A). To test whether this very low level of lineage sharing was an effect of the process of clustering similar sequences into lineages, we additionally compared samples taken at different time points of several LAIV and TIV recipients on the amino acid level. Although different samples of the same individual shared ∼1,100 sequences, samples of different individuals shared ∼25 (of ∼100,000) sequences (Fig. S3 A and B). These sequences shared by different individuals were almost entirely of IgM isotype with low-abundance and -mutation levels (Fig. S3 C–E) and short complementarity determining region 3 (CDR3) (Fig. S3 F and G), suggesting that interindividual overlap is likely caused by the chance overlap of naive sequences; similar behavior was recently shown for T cells (16).
Identifying Vaccine-Induced Lineages.
Next, we wanted to investigate whether the analysis of the abundant lineages (at least 5 IGH molecules/IGH lineage) representing the activated B-cell compartment might allow us to identify lineages activated by seasonal influenza vaccines. We analyzed the IGH repertoire of 14 LAIV and 14 TIV recipients at three time points. Blood was collected at day 0 (right before vaccination) and days 7 and 28 after vaccination. Day 7 is known to show the most pronounced vaccine response (3); 84 resulting samples were prepared and analyzed as described above. To directly compare vaccine effects on the number of abundant lineages between samples, we limited the analysis to 50,000 randomly selected IGH molecules.
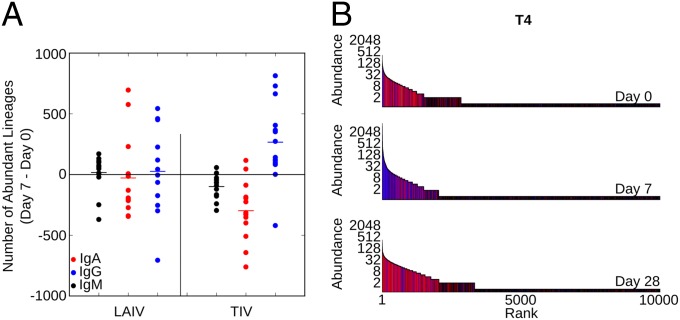
TIV is known to trigger a strong hemagglutinin inhibition (HAI) antibody response, IgG plasmablast release, and isotype class switching, whereas the effects of LAIV are less pronounced (8, 17, 18). We showed that TIV vaccination indeed strongly increases numbers of abundant IgG lineages (Fig. 3A). LAIV vaccination caused a much weaker response and on average, did not increase the numbers abundant lineages of IgG, IgA, and IgM isotypes (Fig. 3A).
Fig. 3.
Influenza vaccination response. (A) Change in the number of abundant lineages (containing at least 5 IGH molecules) in response to vaccination by LAIV or TIV is shown for 14 individuals each. Data were subsampled to 50,000 molecules to normalize for variability in sampling depth. Data are shown for each isotype separately. P value is determined by a Mann–Whitney u test. (B) IGH repertoire at days 0, 7, and 28 for individual T4. Each sample was subsampled to 50,000 IGH molecules to normalize for variability in sampling depth. Sequences were ordered by abundance, and each sequence is shown as a vertical bar [the height depicts its abundance (logarithmic)]. The color of each bar indicates the isotype of the sequence.
To visualize these effects in more detail, we ordered the IGH lineages of one TIV recipient (T4) by rank and then plotted abundance (height) and isotype (color). Distinct patterns of vaccine response become clear in the day 7 sample, where several highly abundant IgG lineages and hundreds of moderately abundant IgG lineages dominate the TIV response (Fig. 3B and Fig. S4, inlets). A closer look at the most abundant lineages reveals that they can consist of thousands of IGH molecules grouped into hundreds IGH sequences often from more than one isotype (Fig. S4).
Overall, focusing on abundant lineages revealed isotype distributions distinct to TIV, with the highly abundant IgG lineages observed at day 7 likely corresponding to memory-derived IgG plasmablasts known to be released into the periphery at that time (3, 18).
Recall Response to TIV Vaccination.
We went on to analyze the recall response to repeated TIV vaccination. Because of its more pronounced effect on individuals (Fig. 3A), we chose to investigate the recall response to TIV. The individuals T1–T5 were vaccinated again with TIV in the year after their initial vaccination. The formulation of TIV was changed for the influenza A(H3N2) and influenza B strains, with only the A(H1N1)pdm09 strain remaining unchanged. Blood samples were again collected at days 0, 7, and 28 (Y2 D0, Y2 D7, and Y2 D28) relative to the second year vaccination.
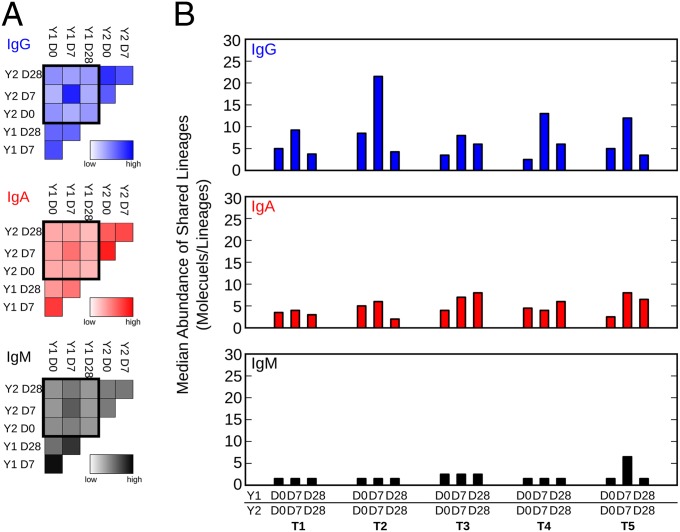
In all individuals, any pair of samples shared hundreds of lineages. Overall, the highest number of lineages was shared between samples taken in the same year, probably because of lineages that persist in the periphery during the duration of the blood draws. Fewer lineages were shared between samples taken in different years (Fig. 4A). When normalized to the overall number of lineages, the Y1 D7/Y2 D7 pair of samples showed the highest rate of shared lineages of IgG, IgA, and IgM isotypes (Fig. 4A). Additionally, compared with Y1 D0/Y2 D0 and Y1 D28/Y2 D28 pairs, the shared IgG lineages but not the shared IgA and IgM lineages were more abundant in all individuals (Fig. 4B). This finding highlighted two interesting findings. First, in the Y1 D0/Y2 D0 prevaccine sample, pairs were collected 12 mo apart but still shared IGH lineages, suggesting that the B-cell clones expressing these lineages either are reactivated by recurrent infection unrelated to influenza or persisted unchanged in the periphery for up to 12 mo in the absence of antigen, which would be consistent with the recent finding that germinal center reactions can persist for over 8 mo in mice (19). Second, highly abundant IgG lineages are preferably shared in the Y1 D7/Y2 D7 postvaccine sample pairs, likely representing vaccine-induced recalled lineages.
Fig. 4.
TIV recall response. (A) Number of lineages shared between indicated time points normalized to the overall amount of lineages. Average of five individuals is shown as heatmaps for IgG, IgA, and IgM. (B) Median abundance of IgG, IgA, and IgM lineages shared between the indicated time points.
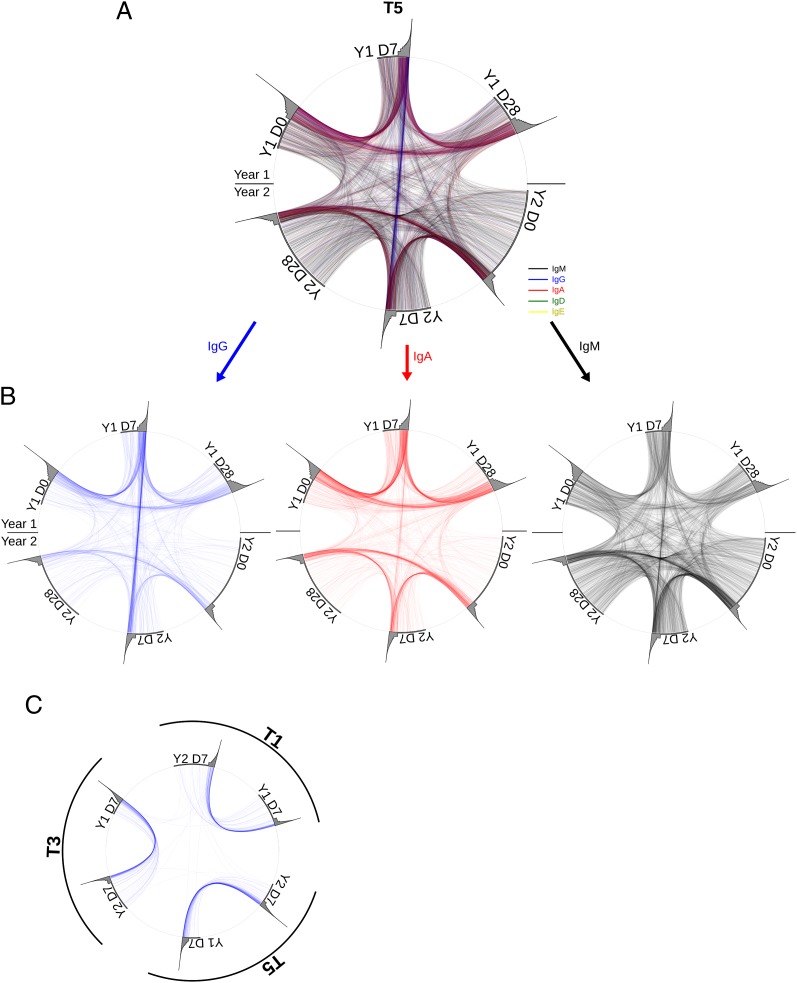
We visualized the shared lineages between time points in the individual with the strongest putative recall response (T5) (Fig. 5 A and B). These connection plots clearly show a surplus in shared highly abundant IgG lineages in the Y1 D7/Y2 D7 pair. This trend is also visible in the other individuals, albeit to a lesser extent (Fig. S5). If these shared IgG lineages had arisen independently in the 2 y sampled, one would expect IgG lineages to also be shared between individuals, assuming that different individuals have the same chance of generating the same antibody. The fact that IgG lineages show practically no overlap between individuals (Fig. 5C) suggests that our data provide direct genetic evidence for memory B-cell recall.
Fig. 5.
Lineage recall. (A and B) Visualization of lineages shared between time points. Data were subsampled to 50,000 IGH molecules for each time point to normalize for variability in sampling depth. Lineages of each time point are plotted on circumference of the circle, with the gray area representing the abundance of the respective lineages (logarithmic). Lineages present in two time points are connected with lines colored according to their isotype. All shared lineages are shown in A, and shared IgG, IgA, and IgM lineages are shown separately in B. (C) Shared IgG lineages between Y1 D7 and Y2 D7 time points of three individuals are shown as in A.
Discussion
Here, we present a method to investigate the IGH antibody repertoire in blood samples that is highly accurate, quantitative, and reproducible. This consensus read method allows us to determine the abundance, sequence, and isotype of IGH mRNA. Using this approach, we have shown that we can identify all abundant sequences in a blood sample. These abundant sequences are enriched for IgG, IgA, and mutated IgM, and they likely correspond to the activated B-cell compartment. The identification of compartments (naive vs. memory/plasmablasts) and isotype distributions of IGH sequences directly from PBMCs obviates the distortion and additional work associated with cell-sorting procedures. We used this compartment information to refine capture–recapture analysis to estimate the activated peripheral B-cell compartment to the surprisingly low number of ∼25,000 B-cell clones.
In a recent study, we outlined the age-dependent differential effects of TIV and LAIV vaccines on the IGH repertoire, showing that overall TIV causes a stronger class switch response than LAIV (8). Focusing on the analysis of the abundant lineages in the present study enabled us to create an even more detailed fingerprint of the B-cell response to influenza vaccine TIV, in which tens of highly abundant B-cell clones are accompanied by hundreds of moderately abundant B-cell clones.
Although TIV is a mix of inactivated viral proteins that is injected, LAIV is an attenuated live virus that is administered by nasal spray. Therefore, the absence of a measurable peripheral B-cell response is likely explained by a local response in the upper respiratory tract. Comparing these vaccine responses with WT influenza infections might help explain the differential protective mechanisms and effects of TIV and LAIV (4, 17, 20). The high IgG levels in response to TIV vaccination correlate well with the release of IgG plasmablasts around day 7 after vaccination. Because of their fast appearance and high mutation levels, these plasmablasts are thought to be recalled from B-cell memory (3).
Immunological memory is an area of great interest for the development of effective vaccinations that provide lifelong protection. For decades, B-cell memory has been analyzed using methods based on antibody affinities (21, 22). Although these methods can detect the recall of B cells with specific affinities, they do not provide insight into the composition of these recalled cells. Indeed, distinct B-cell clones expressing antibodies with similar affinities cannot be distinguished by these methods. A direct measurement and quantification of B-cell recall require the repeated identification of B-cell clones in separate immune responses to an antigen. The sequencing depth and accuracy of our approach enabled us to provide this direct genetic measurement of memory B-cell recall. We detected IGH lineages, enriched for abundant IgG lineages, that were shared by IGH repertoires after but not before two annual influenza vaccination, clearly showing that the B-cell clones expressing these sequences were recalled from B-cell memory as a response to two substantially different vaccine compositions. Sequences found at high levels in response to distinct vaccines present prime targets for the identifications of cross-specific antibodies.
Materials and Methods
Study Volunteers, PBMC Isolation, and RNA Extraction.
All study protocols were approved by the Institutional Review Boards at Stanford University. Informed consent was obtained from participants. Blood was taken before (day 0), 1 wk after (day 7), and 1 mo after (day 28) vaccination in the first year (2011) and after vaccination (day 7) in the second year (2012). Volunteers were 28 young adults (18–30 y) in generally good health and vaccinated with one dose of either seasonal LAIV (FluMist intranasal vaccine; MedImmune) or TIV (Fluzone; Sanofi-Pasteur); 60 mL peripheral blood were taken from individuals and heparinized. PBMCs were extracted from 10 mL blood using a Ficoll-Gradient and frozen in 10% (vol/vol) DMSO/40% (vol/vol) FBS according to the Protocols of the Stanford Human Immune Monitoring Center (HIMC). After thawing, total RNA was extracted from 5 million PBMCs using the Qiagen AllPrep Kit.
Library Preparation.
Five hundred nanograms total RNA were used as input for library preparation. Reverse transcription was performed according to the manufacturer’s instructions using SuperScript III Enzyme (Life) and primers for all five isotypes containing 8 random nt and partial Illumina adapters containing Illumina barcodes (Table S1). Second-strand synthesis was done using Phusion Polymerase (NEB) and primers containing 8 random nt and partial Illumina adapter sequences covering all V segments with a maximum of one mismatch (Table S1) (98 °C for 2 min, 52 °C for 2 min, and 72 °C for 10 min). Double-stranded cDNA was purified two times using Ampure XP beads at a ratio of 1:1. Double-stranded cDNA was amplified with Platinum Hifi Polymerase (Life) with two primers completing Illumina adapter sequences (95 °C for 2 min, 27 cycles of 95°C for 30 s, 65 °C for 30s, and 68 °C for 2 min, and 68 °C for 7 min). Final sequencing libraries were generated by purifying the PCR product using Ampure XP beads at a ratio of 1:1.
Sequencing and Data Analysis.
Libraries were sequenced on the Illumina HiSeq2000 using a custom 100 × 120-bp protocol. The sequencing runs yielded 3 and 15 million raw reads per sample. Raw reads were split into UID groups with unique 16-bp identifiers, and a consensus read was built for each UID group with at least two raw reads.
For the generation of consensus reads, raw bases with an Illumina Quality Score under 20 were discarded. Each base in the consensus read was determined by a majority call of raw bases, which was weighted by quality scores. UID groups containing any majority base making up <66% of all bases at that position were discarded. This requirement avoided draws and discarded UID groups with only two raw reads if they were not completely identical. Final quality filtering discarded low-quality consensus reads. Alignments to variable (V), diversity (D), and joining (J) segments as well as mutation analysis were then performed by the pipeline described in ref. 23 using IMGT references for V, D, J, and C segments. Furthermore, bases templated by V-segment primers as well as most of the constant region were truncated. This process generated ∼50,000–500,000 high-quality consensus reads per sample. Each consensus read represents one initial IGH molecule.
Identical IGH molecules were grouped into IGH sequences, which were then clustered into IGH lineages as described below (Fig. S1B). Abundance was then defined as the number of IGH molecules that an IGH sequence or IGH lineage contained. IGH sequences or lineages were defined as abundant if they contained at least 5 IGH molecules. For the determination of shared IGH sequences (Fig. S3), sequences shared by more than two people were removed from the analysis, because they might represent possible cross-contamination.
Lineage Clustering.
IGH sequences were clustered into IGH lineages according to similarity in their junctional region. Lineages were created according to the following steps.
A lineage is formed and populated with one IGH sequence (seed). Then, all IGH sequences in the lineages (initially only the seed) are compared with all other IGH sequences of the same length using the same V and J segments. If their junctional regions (untemplated nucleotides and D segments) are at least 90% identical, the IGH sequence is added to the lineage. This process is repeated until the lineage does not grow.
Subsampling.
For some figures, data were subsampled randomly to 50,000 IGH molecules/sample. These 50,000 IGH molecules were then grouped into IGH sequences and clustered into IGH lineages as for the nonsubsampled data.
Capture–Recapture Analysis.
Capture–recapture analysis was done using the following Chapman–Estimator formula: R = (((S1 + 1)(S2 + 1))/(C + 1)) − 1, where R = estimate of repertoire, S1 = total number of IGH sequences in sample 1, S2 = total number of IGH sequences in sample 2, and C = number of IGH sequences shared between samples.
Additional analysis was done using custom Python scripts in a Unix environment. The SeqPrep tool (https://github.com/jstjohn/SeqPrep) was used in the generation of consensus reads. Sequences were clustered into lineages if their junctional regions differed by less than one in 10 bases. Custom scripts are available on request. Figures were generated using the matplotlib Python library (24).
Supplementary Material
Acknowledgments
We thank Norma Neff, Ben Passarelli, and Gary Mantalas (Stanford Stem Cell Genome Center) for assistance with sequencing. We also thank Sally Mackey for coordinating clinical recruitment, Sue Swope and Michele Ugur for collecting samples, Xiaosong He for providing access to samples, and Holden Maecker, Jackie Bierre, and Ben Varasteh (Stanford Human Immune Monitoring Core) for sample banking. This research was supported by National Institutes of Health Grant U19 AI057229 (to S.R.Q.). The clinical project was supported by National Institutes of Health/National Center for Research Resources Clinical and Translational Science Award UL1 RR025744.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312146110/-/DCSupplemental.
References
- 1.Thompson WW, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 2.Couch RB, Kasel JA. Immunity to influenza in man. Annu Rev Microbiol. 1983;37:529–549. doi: 10.1146/annurev.mi.37.100183.002525. [DOI] [PubMed] [Google Scholar]
- 3.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453(7195):667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki S, et al. Influence of prior influenza vaccination on antibody and B-cell responses. PLoS One. 2008;3(8):e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324(5928):807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krause JC, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187(7):3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5(171):171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glanville J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci USA. 2009;106(48):20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnaout R, et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6(8):e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briney BS, Willis JR, McKinney BA, Crowe JE., Jr High-throughput antibody sequencing reveals genetic evidence of global regulation of the naïve and memory repertoires that extends across individuals. Genes Immun. 2012;13(6):469–473. doi: 10.1038/gene.2012.20. [DOI] [PubMed] [Google Scholar]
- 12.Fu GK, Hu J, Wang PH, Fodor SP. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc Natl Acad Sci USA. 2011;108(22):9026–9031. doi: 10.1073/pnas.1017621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108(23):9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiroguchi K, Jia TZ, Sims PA, Xie XS. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci USA. 2012;109(4):1347–1352. doi: 10.1073/pnas.1118018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Andres M, et al. Human peripheral blood B-cell compartments: A crossroad in B-cell traffic. Cytometry B Clin Cytom. 2010;78(Suppl 1):S47–S60. doi: 10.1002/cyto.b.20547. [DOI] [PubMed] [Google Scholar]
- 16.Murugan A, Mora T, Walczak AM, Callan CG., Jr Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA. 2012;109(40):16161–16166. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treanor JJ, et al. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine. 1999;18(9-10):899–906. doi: 10.1016/s0264-410x(99)00334-5. [DOI] [PubMed] [Google Scholar]
- 18.Nakaya HI, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dogan I, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10(12):1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 20.DiazGranados CA, Denis M, Plotkin S. Seasonal influenza vaccine efficacy and its determinants in children and non-elderly adults: A systematic review with meta-analyses of controlled trials. Vaccine. 2012;31(1):49–57. doi: 10.1016/j.vaccine.2012.10.084. [DOI] [PubMed] [Google Scholar]
- 21.Davenport FM, Hennessy AV. A serologic recapitulation of past experiences with influenza A; antibody response to monovalent vaccine. J Exp Med. 1956;104(1):85–97. doi: 10.1084/jem.104.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li GM, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci USA. 2012;109(23):9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang N, et al. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci USA. 2011;108(13):5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter JD. Matplotlib: A 2D graphics environment. Computing In Science & Engineering. 2007;9(3):90–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.