Abstract
Sulfate-reducing bacteria (SRB) colonize the guts of ∼50% of humans. We used genome-wide transposon mutagenesis and insertion-site sequencing, RNA-Seq, plus mass spectrometry to characterize genetic and environmental factors that impact the niche of Desulfovibrio piger, the most common SRB in a surveyed cohort of healthy US adults. Gnotobiotic mice were colonized with an assemblage of sequenced human gut bacterial species with or without D. piger and fed diets with different levels and types of carbohydrates and sulfur sources. Diet was a major determinant of functions expressed by this artificial nine-member community and of the genes that impact D. piger fitness; the latter includes high- and low-affinity systems for using ammonia, a limiting resource for D. piger in mice consuming a polysaccharide-rich diet. Although genes involved in hydrogen consumption and sulfate reduction are necessary for its colonization, varying dietary-free sulfate levels did not significantly alter levels of D. piger, which can obtain sulfate from the host in part via cross-feeding mediated by Bacteroides-encoded sulfatases. Chondroitin sulfate, a common dietary supplement, increased D. piger and H2S levels without compromising gut barrier integrity. A chondroitin sulfate-supplemented diet together with D. piger impacted the assemblage’s substrate utilization preferences, allowing consumption of more reduced carbon sources and increasing the abundance of the H2-producing Actinobacterium, Collinsella aerofaciens. Our findings provide genetic and metabolic details of how this H2-consuming SRB shapes the responses of a microbiota to diet ingredients and a framework for examining how individuals lacking D. piger differ from those who harbor it.
Keywords: artificial human gut microbiota/microbiome, determinants of microbial fitness, hydrogenotrophs, microbial foodwebs, hydrogen sulfide
A major function of the human gut microbiota is to aid in the harvest of nutrients and energy from our varied diets. Dietary components that are not absorbed in the proximal intestine reach the distal gut, where they are metabolized through processes that involve trophic interactions among members of the microbial community. Proteins and carbohydrates are broken down by primary fermenters, yielding short-chain fatty acids (e.g., acetate, propionate, and butyrate) and gases (e.g., H2 and CO2). These fermentation products not only impact the host but also serve as sources of carbon and energy for other community members (1).
One challenge microbes face during fermentation is to maintain redox balance while maximizing their energy production. Many species have branched fermentation pathways that allow for disposal of reducing equivalents; producing H2 is an energetically efficient way of doing so, yielding higher levels of ATP. Because H2 buildup inhibits reoxidation of pyridine nucleotides and forces primary fermenters to accumulate reduced compounds (e.g., butyrate, ethanol) (2), hydrogenotrophic (H2 consuming) species are key to the energy-extracting capacity of primary fermenters in microbial food webs and contribute to more efficient and complete oxidation of substrates (3).
In the human gut, H2-consuming microbes include methanogens, acetogens, and sulfate-reducing bacteria (SRB). These organisms produce methane, acetate, and hydrogen sulfide (H2S), respectively. Among these, SRB are the most efficient hydrogenotrophs when using sulfate as the electron acceptor: under pure culture conditions, the H2 threshold (lowest concentration of H2 that can be used) of SRB is significantly lower than the average threshold of acetogens and methanogens (4). Although sulfate-reducing activity is not limited to a particular phylogenetic group, SRB in the human colon are predominantly members of the genus Desulfovibrio in the class δ-Proteobacteria (5). They can use H2 or organic compounds (e.g., lactate, formate) as electron donors for reduction of sulfate or other oxidized sulfur compounds to generate H2S (6). Sulfate and sulfite are used as preservatives and antioxidants in a variety of foods (bread, preserved meat, dried fruit, wine). Sulfate is also present in the dietary supplement chondroitin sulfate, and in food additives (carrageenan).
Various studies using different analytic approaches have identified SRB in the fecal microbiota of healthy adults (7, 8) and in the distal gut mucosa (9). Although sulfate/sulfite-reducing bacteria are positively associated with inflammation (10, 11), both pro- and anti-inflammatory signaling have been attributed to H2S (12–14).
Little is known about the metabolic activities and requirements of SRB in vivo. In this study, we used gnotobiotic mice harboring a defined artificial human gut microbiota composed of sequenced representatives of the four most prominent bacterial phyla present in healthy adults (Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria) (15, 16), to dissect the metabolic niche and contributions of a prominent SRB, Desulfovibrio piger, to microbial and host metabolism.
Results and Discussion
D. piger Is a Common SRB Present in the Fecal Microbiota.
Using PCR primers directed against aprA, which encodes the alpha-subunit of the adenosine-5′-phosphosulfate reductase present in all known SRB, we generated amplicons from fecal samples previously collected from 34 healthy individuals known to harbor SRB (8). Multiplex pyrosequencing of the PCR products [Titanium chemistry; 2,406 ± 1,696 reads per sample (mean ± SD); 361 ± 6 nt per read] revealed that D. piger was the most frequent SRB present [21/34 (60%)]. D. piger was the sole detectable SRB in 12 of these 21 subjects (57%) and coexisted with one or two other sulfate reducers, Desulfovibrio intestinalis and an unclassified SRB, in the other individuals (Fig. S1). The prominence of D. piger in this and other populations (5), coupled with the fact that we had previously isolated and sequenced a D. piger strain from human feces (D. piger GOR1) (17), led us to focus on characterizing its niche in gnotobiotic mice.
A Diet with Low Levels of Fermentable Carbohydrates Is Associated with Increased Utilization of Host-Derived Glycans and Increased Levels of D. piger.
Adult germ-free male NMRI (Naval Medical Research Institute) mice were colonized with D. piger GOR1 and eight other sequenced human gut bacterial species. Together, their genomes contain 36,822 predicted ORFs that encode major metabolic functions present in the distal gut microbiome of healthy adults (15, 16) including the ability to break down proteins, plant, and host-derived polysaccharides (Bacteroides thetaiotaomicron, Bacteroides caccae, Bacteroides ovatus); consume oligosaccharides and simple sugars (Eubacterium rectale, Marvinbryantia formatexigens, Collinsella aerofaciens, Escherichia coli); and ferment amino acids (Clostridium symbiosum, E. coli). Dataset S1 lists the wide range of known or predicted proteases and carbohydrate active enzymes (glycoside hydrolases, polysaccharide lyases, carbohydrate esterases) encoded by this artificial microbiome and their distribution among community members.
Mice colonized with these nine species were fed one of two different diets ad libitum: one low in fat (4% wt/wt) and high in plant polysaccharides (LF/HPP), the other high in fat (20% wt/wt) and simple sugars (47% wt/wt sucrose) (HF/HS) (Dataset S2) (n = 5 mice/diet). COmmunity PROfiling by shotgun Sequencing (COPRO-Seq) of DNA isolated from fecal samples collected 7 and 14 d after introduction of this bacterial consortium revealed that the relative abundances of five of the nine members were significantly different between mice fed the different diets (P value < 0.05; two-tailed t test followed by Bonferroni correction). These five species included D. piger, which was present at higher levels when mice were consuming the HF/HS diet (Fig. S2A).
To identify microbial functions that changed with diet, we performed microbial RNA-Seq analysis of fecal samples collected 14 d after colonization. mRNA transcripts were functionally grouped based on enzyme commission numbers (ECs) assigned to their protein products (Fig. S2B, Dataset S3 A and B). Among the 1,191 ECs detected, we identified 96 whose representation in the fecal metatranscriptome was significantly different between the two diet groups (Dataset S3A). Many of these enzymes participate in various facets of carbohydrate metabolism. For example, the microbiota of mice fed the LF/HPP diet exhibited significantly higher expression of genes encoding ECs involved in the breakdown of plant-derived polysaccharides (Fig. S2B, Dataset S3A). In contrast, the microbiota of mice fed the HF/HS diet exhibited higher levels of expression of genes involved in the metabolism of sucrose, breakdown of host-derived mucus glycans, and removal of sulfate from sulfated glycans (Fig. S2B, Dataset S3A, SI Results and Discussion). The contributions of individual bacterial species to the pool of ECs differentially represented in the fecal metatranscriptome are shown in Dataset S3B and discussed in SI Results and Discussion.
Chemostat experiments have suggested that liberation of sulfate from sulfated mucins promotes SRB growth (18, 19). Consistent with these observations, we found that the increased sulfatase (EC3.1.6.14) gene expression in Bacteroides species observed in mice consuming the HF/HS diet was accompanied by significantly higher proportional levels of D. piger and significantly higher concentrations of H2S in cecal contents (Fig. S2 A–C, Dataset S3B). Targeted gas chromatography mass spectrometry (GC-MS) of cecal contents revealed significantly lower levels of bacterial fermentation products (acetate, propionate) on the HF/HS versus LF/HPP diet (Fig. S2D; P < 0.05; two-tailed t test).
These results suggest that D. piger benefits from diets that provide low levels of complex carbohydrates to the distal gut. We hypothesized that this benefit reflects the fact that consumption of the polysaccharide-poor HF/HS diet prompts increased utilization of host sulfated glycans by members of the defined artificial human gut community, thereby providing free sulfate to D. piger. We tested this hypothesis in several ways: first, by identifying the genetic determinants for D. piger fitness in vivo and in vitro, then by performing cross-feeding experiments in vitro and in vivo, and finally through a series of additional diet manipulations where sulfur content was varied.
Transposon Mutagenesis Identifies Key Determinants for D. piger Fitness in Vivo.
We used a genome-wide transposon mutagenesis method known as INsertion Sequencing (INSeq) (20) to define D. piger fitness determinants in various nutrient contexts. An isogenic library composed of ∼30,000 unique inter- and intragenic transposon insertion mutants of D. piger was constructed [Tn inserts identified in 2,181 of the organism’s 2,487 predicted ORFs; 10.6 ± 0.24 (mean ± SE) Tn insertions/ORF] (see SI Results and Discussion for details concerning library construction, and its initial in vitro characterization, plus Dataset S4, Dataset S5, and Fig. S3).
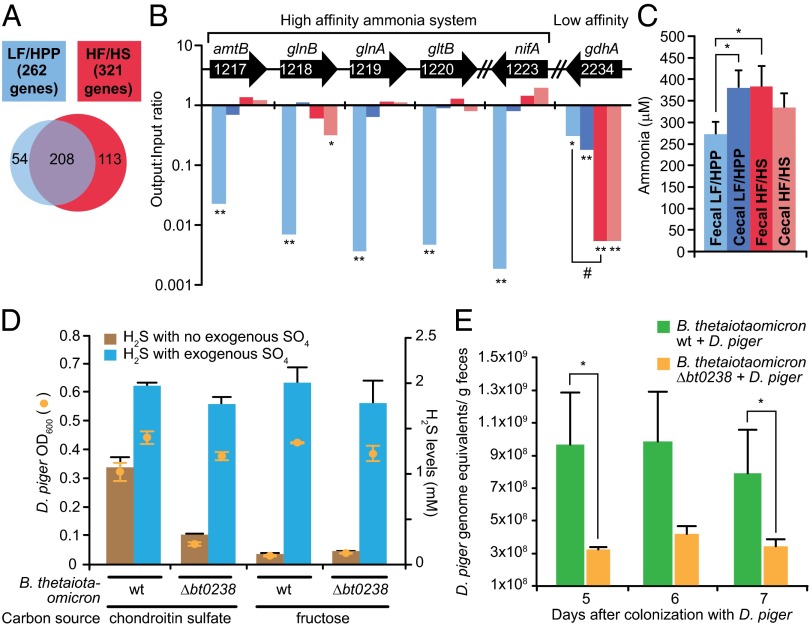
Mice colonized with the eight-member assemblage of gut species were fed the LF/HPP or HF/HS diet for 14 d before introduction of the D. piger mutant library via a single oral gavage, and remained on these diets for the duration of the experiment. COPRO-Seq analysis of fecal pellets obtained 7 d after gavage indicated that the relative abundance of the mutant library was not significantly different from levels achieved by wild-type D. piger in mice fed the same diets (Fig. S4). INSeq analysis of fecal pellets obtained from mice consuming LF/HPP and HF/HS diets revealed mutations in 262 and 321 D. piger genes, respectively, that produced a significant reduction in invasiveness/fitness (FDR padj < 0.05, output:input ratio < 0.3). Two hundred and eight of these fitness determinants were common to both diet selections, and their fitness effects were comparable in the cecal and fecal microbiota in both diet contexts (Fig. 1A, Dataset S6, and Dataset S7). These 208 genes include loci known or predicted to be involved in amino acid metabolism, ATP synthesis, membrane transport, and nucleotide metabolism (Dataset S6), leading us to conclude that they likely represent core fitness determinants for D. piger in the gut at least with these diets.
Fig. 1.
Genetic identification of determinants important for D. piger acquisition of nitrogen and sulfate in the gut. (A) Venn diagram of the number of D. piger fitness determinants identified by INSeq analysis of fecal microbiota obtained 7 d after introduction of the D. piger mutant library into mice harboring the eight-member consortium of human gut bacterial species and fed the LF/HPP or HF/HS diets (threshold criteria for significant fitness effect; output–input ratio of mutant strain <0.3; padj < 0.05) (n = 4 mice/diet). (B) Ammonia assimilation genes identified by INSeq analysis of fecal and cecal microbiota whose fitness effects exhibit diet specificity as well as specificity for cecal versus fecal microbiota. The representation of the indicated mutant locus in the output population is compared with representation in the input library: *Padj < 0.05; **Padj < 0.001 (negative binomial test from DESeq package; SI Methods). Significance of the difference observed in fecal samples obtained from mice on the LF/HPP versus HF/HS diets; #, Padj < 0.001. See C for color code. (C) Ammonia levels in fecal and cecal samples collected from mice colonized with the nine-member artificial community containing wild-type D. piger and fed the LF/HPP versus HF/HS diets. Mean values ± SEM are plotted. *P < 0.05 (Student t test). (D) In vitro test of sulfate cross-feeding. Left y-axis plots D. piger growth (final OD600) in filter-sterilized medium harvested from cultures of the B. thetaiotaomicron sulfatase maturation mutant (Δbt0238) or the isogenic wild-type (WT) strain after their growth in minimal medium with chondroitin sulfate or fructose. H2S levels during D. piger growth in B. thetaiotaomicron–conditioned medium are plotted on the right y-axis. Mean values ± SEM are shown (n = 3 replicates per condition; representative of two independent experiments). (E) Quantitative PCR analysis of D. piger levels in the feces of mice co-colonized with either wild-type or Δbt0238 B. thetaiotaomicron. Mean values ± SEM are plotted (n = 3–5 mice/time point/group). *P < 0.05 (Student t test).
The fitness effects of 167 other genes were diet-dependent (Fig. 1A, Dataset S8). A notable example involves ammo assimilation (Fig. 1B). Ammonia can serve as a source of nitrogen that is incorporated into glutamate and glutamine and then transferred to other nitrogen-containing compounds. Incorporation of ammonia can occur in an energy-dependent or -independent manner depending upon whether ammonia concentrations are low or high, respectively. We found that genes involved in assimilation when ammonia concentrations are low (high-affinity system) are important for fitness when mice were fed the LF/HPP but not the HF/HS diet; these genes included an ammonia transporter [DpigGOR1_1217 (amtB)], two nitrogen regulatory proteins [DpigGOR1_1218 (glnB), DpigGOR1_1223 (nifA)], glutamine synthase [DpigGOR1_1219 (glnA)], and glutamate synthase [DpigGOR1_1220 (gltB)] (Fig. 1B). In contrast, transposon disruption of the glutamate dehydrogenase gene [DpigGOR1_2234 (gdhA)], which is involved in ammonia assimilation when levels are high (low-affinity ammonia system), resulted in a strong fitness defect in mice fed the HF/HS diet, but had a significantly smaller effect in mice consuming the LF/HPP diet (Fig. 1B). Consistent with these findings, we detected significantly lower levels of ammonia in feces from mice fed the LF/HPP diet compared with mice consuming the HF/HS diet (Fig. 1C).
Although transposon disruption of genes involved in the high-affinity ammonia assimilation pathway resulted in lower D. piger abundance in the fecal microbiota of LF/HPP-fed mice, we observed no fitness defect in their cecal microbiota (Fig. 1B). In contrast, disruption of the gene encoding glutamate dehydrogenase from the low-affinity system had a significantly larger effect on fitness in the cecal compared with fecal microbiota of LF/HPP-fed mice (Fig. 1B). The differential effects of diet and location can be explained by the significantly lower ammonia levels in feces compared with cecal contents in mice consuming the LF/HPP diet (Fig. 1C).
Genes involved in H2 consumption and sulfate reduction are required for fitness in vivo in both diet contexts; they include (i) a predicted periplasmic [NiFeSe] hydrogenase complex (DpigGOR1_1496-DpigGOR1_1497) important in other Desulfovibrio species for growth with H2 (21); (ii) hydrogenase maturation genes (DpigGOR1_0739-DpigGOR1_0740); (iii) a predicted transport system for nickel, which functions as an important cofactor for the hydrogenase (DpigGOR1_1393-DpigGOR1_1398); (iv) two electron transport systems involved in sulfate reduction in other microbial species, a high molecular weight cytochrome complex, Hmc (DpigGOR1_0741-DpigGOR1_0744) and the QmoABC complex (DpigGOR1_0790-DpigGOR1_0792) (22, 23); plus (v) components of sulfite reductase (DpigGOR1_0170-DpigGOR1_0174). These results emphasize the importance of hydrogen metabolism and respiration of sulfate and/or other oxidized sulfur compounds for survival of D. piger in the distal gut and underscore the restricted metabolic options that D. piger has to efficiently generate energy in this environment.
Comparison of these in vivo INSeq results with results obtained from a series of in vitro experiments that used defined electron donors for sulfate reduction identified other fitness determinants that exhibit in vivo specificity; these are described in SI Results and Discussion, Dataset S9, and Dataset S10.
B. thetaiotaomicron Boosts D. piger Growth in Vitro and in Vivo Through Provision of Free Sulfate.
We next sought to identify the sources of sulfate required for optimal D. piger growth and survival in the gut. Potential in vivo sources of sulfate for D. piger include the diet, sulfated oligosaccharide side chains of glycosaminoglycans in host mucins, and sulfonic acid moieties in bile acids. Accessing these host sources of sulfate requires their liberation by sulfatases, an enzymatic activity encoded by members of the microbiota (24, 25). As our in silico analysis of the D. piger GOR1 genome did not identify any sulfatases, we hypothesized that it may benefit from other bacterial species that liberate sulfate from an array of sulfated host glycans. One member of the artificial community, B. thetaiotaomicron, expresses sulfatases that are required for its adaptive foraging on mucosal glycans when the diet lacks complex polysaccharides (25). B. thetaiotaomicron has 28 putative sulfatase genes and only one sulfatase maturation enzyme (BT0238) that is essential for all of its sulfatase activity (25).
Because it was unclear whether sulfate liberated by B. thetaiotaomicron from host mucosal glycans is available to D. piger, we initially used a simplified in vitro system to determine the potential for cross-feeding between these two bacterial species. A B. thetaiotaomicron strain (Δbt0238) that lacks a functional sulfatase maturation enzyme and hence any sulfatase activity, and the isogenic wild-type strain were inoculated into separate cultures containing minimal medium plus a sulfated or nonsulfated carbon substrate (chondroitin sulfate and fructose, respectively). Wild-type B. thetaiotaomicron grew in minimal medium containing chondroitin sulfate, whereas the Δbt0238 strain failed to grow. In contrast, both the wild-type and Δbt0238 strains grew in minimal medium containing fructose as the carbon source. Medium from each inoculated culture was harvested, filter-sterilized, supplemented with lactate, and used as a potential source of sulfate for D. piger. In this experimental design, lactate provided the sole carbon and electron source for D. piger; it does not support growth of D. piger without sulfate (electron acceptor).
We observed growth of D. piger in filter-sterilized conditioned medium obtained from wild-type B. thetaiotaomicron cultured in the presence of chondroitin sulfate. The lack of growth of D. piger in the fructose-containing conditioned medium was not due to inhibitory effects, as addition of exogenous sulfate restored its growth (Fig. 1D). Furthermore, the inability of D. piger to grow in chondroitin sulfate-containing medium that had been inoculated with B. thetaiotaomicron Δbt0238 showed that D. piger is unable to metabolize chondroitin sulfate. H2S measurements confirmed that the growth observed with conditioned chondroitin sulfate-containing medium correlated with sulfate reduction (Fig. 1D). Together, these in vitro results indicate that B. thetaiotaomicron can liberate sulfate from glycans, which then becomes available for D. piger.
To examine the role of sulfate cross-feeding between B. thetaiotaomicron and D. piger in vivo, we monocolonized adult germ-free mice with a single oral gavage of the wild-type or Δbt0238 strains of B. thetaiotaomicron. Mice were fed the HF/HS diet for 1 wk before a second gavage with wild-type D. piger. This diet was chosen because it results in increased expression of B. thetaiotaomicron sulfatase genes as well as genes involved in utilization of host glycans (Fig. S2B), thus permitting adaptive foraging of sulfated host glycans. qPCR analysis of feces collected 5, 6, and 7 d after introduction of D. piger revealed that its abundance in mice co-colonized with B. thetaiotaomicron Δbt0238 was significantly lower than in mice cocolonized with the isogenic wild-type B. thetaiotaomicron strain (Fig. 1E). These observations indicate that sulfate cross-feeding by bacteria with sulfatase activity supports higher levels of intestinal colonization by D. piger. However, because D. piger was still able to colonize mice containing the mutant B. thetaiotaomicron strain, there appear to be other available sources of oxidized sulfur, including the diet.
To test how different dietary sulfur sources affect D. piger colonization levels, we generated 12 diets, all based on the HF/HS diet that contains 0.12% (wt/wt) sulfate. In these diets we deliberately modified the sulfate concentration over a 600-fold range (from 0.001 to 0.6% wt/wt), and introduced sulfur compounds with different redox states (e.g., sulfate versus thiosulfate versus sulfite). Because the gut has a large absorptive capacity for sulfate and likely related compounds (26), we also manipulated sulfate availability by constructing a diet with a glycan-bound source of sulfate (chondroitin sulfate) that is poorly absorbed in the small intestine (27) (see Dataset S2 for the composition of all diets). Six groups of gnotobiotic mice, each composed of two cohoused animals colonized with the nine-member community were fed one of the 13 diets (the unmodified HF/HS diet served as a reference control). A sequence of five different diets was administered to each set of mice. Each diet was given for 1 wk. All mice began with the baseline HF/HS diet and received ultrapurified water (sulfate concentration <0.0005 g/L) for the duration of the experiment. The order of presentation of the four subsequent diets and diet type were randomized among the six groups so that in the end each diet had been administered to two different sets of mice (n = 4 animals; Dataset S11).
We found that a 600-fold change in dietary sulfate levels did not affect the relative abundance of D. piger in the nine-member artificial human microbiota (Fig. S5). The lack of a reduction in D. piger levels with administration of the lowest sulfate diet (0.001% wt/wt) suggested that D. piger either predominately uses host-derived sulfate or that under these dietary conditions an alternative pathway for energy generation is used instead of sulfate reduction. To differentiate between these possibilities, mice were colonized with the eight-member species assemblage and fed the low-sulfate diet (0.001% wt/wt) or the control HF/HS diet before and for 7 d after gavage with the D. piger mutant library. INSeq analysis of fecal samples obtained 7 d after introduction of the mutant library revealed 291 genes as important fitness determinants for both the low and standard sulfate diets (out of a total of 384 unique fitness determinants; see Dataset S12 for a list of shared as well as diet-specific fitness factors). Importantly, we found that all of the sulfate reduction and hydrogenase genes are important for fitness in the low-sulfate diet context, just as they are with the standard HF/HS diet.
Together, these results indicate that although the ability to reduce sulfate is critical for D. piger colonization of the intestine, free sulfate in the diet is not a required determinant of D. piger colonization levels and that at least in our artificial community, D. piger can use sulfate from sources other than diet, such as the host, without a decrease in its proportional representation. Supplementation of the HF/HS diet with 3% (wt/wt) chondroitin sulfate significantly increased D. piger levels in the fecal microbiota relative to the HF/HS diet (Fig. S5; P < 0.05; one-way ANOVA and Dunnett’s post hoc test). Thus, chondroitin sulfate provided us with a means to test the effects of manipulating levels of D. piger on other members of the community and on host physiology.
Co-Colonization with D. piger Impacts Fecal Community Composition and Oxidative Metabolism in the Mouse Distal Gut.
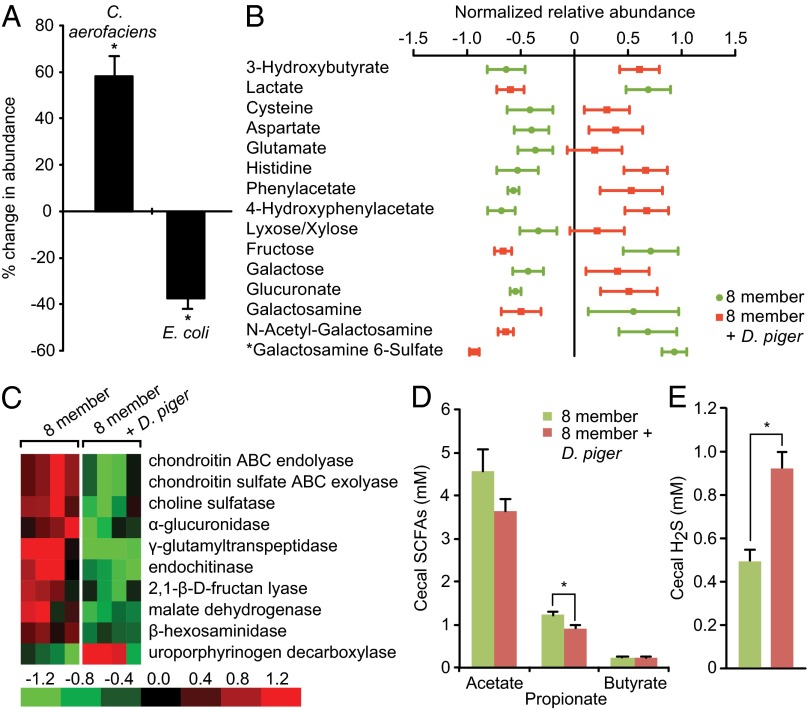
Adult (7–8-wk-old) germ-free mice were colonized with the eight-member artificial community alone or plus D. piger and fed the HF/HS diet supplemented with 3% (wt/wt) chondroitin sulfate for 2 wk. The presence of D. piger was associated with a significant increase in the representation of C. aerofaciens and a decrease in E. coli (Fig. 2A). Furthermore, Spearman correlation analysis of the relative abundance of D. piger and C. aerofaciens in the fecal microbiota of mice containing the nine-member community who were fed all of the diets described above (LF/HPP plus the 13 HF/HS-based diets) revealed a significant positive association between the levels of these two species [r = 0.376, P = 0.001 (r = 0.562, P = 0.003 if only the 2-wk diet exposures with LF/HPP, HF/HS, and HS/HS+3% chondroitin sulfate are considered)]. The main products of C. aerofaciens fermentation are lactate, H2, and formate, all of which serve as substrates for D. piger growth (6). GC-MS disclosed that lactate levels were lower in the cecal contents of mice harboring D. piger (Fig. 2B). Higher levels of D. piger may contribute to increased levels of C. aerofaciens by promoting more efficient fermentation through removal of H2 and formate.
Fig. 2.
Impact of D. piger on the artificial human gut microbiota and host. (A) Bacterial species from the eight-member artificial community that showed significant changes in abundance in the fecal microbiota when D. piger was present versus absent. Mice (n = 19–20/treatment group; three independent experiments) were fed the HF/HS diet supplemented with 3% chondroitin sulfate; *P < 0.05 (Mann–Whitney test). (B) GC-MS and UPLC-MS (*) analysis of cecal contents from the mice described in A. Metabolites that were significantly changed when D. piger was present in mice consuming the HF/HS diet supplemented with chondroitin sulfate are listed. Normalized MS peak areas were mean centered and unit variance scaled. Scores ± SEM are plotted (P < 0.05, Student t test). (C) Microbial RNA-Seq analysis of the fecal metatranscriptome in response to colonization with D. piger. The heat map shows selected ECs encoded by mRNA that were differentially represented between the two conditions [fold-change <–2 or >2; P < 0.01, posterior probability of differential expression (PPDE) > 0.95]. Each column represents a different mouse in the indicated treatment group sampled 14 d after colonization. The maximal relative expression across a row is red; the minimum is green. (D and E) Targeted GC-MS analysis of cecal short chain fatty acid and H2S levels [n = 19–20 mice; mean values ± SEM are plotted; *P < 0.05 (Student t test)].
Microbial RNA-Seq analysis of the fecal metatranscriptome revealed that genes encoding malate dehydrogenase (EC1.1.1.37; Fig. 2C, Dataset S13) exhibited lower levels of expression in the presence of D. piger. This change was largely attributable to changes in expression in B. caccae, B. ovatus, and B. thetaiotaomicron. Malate dehydrogenase is involved in the NADH-consuming step that converts oxaloacetate into malate, which in turn is used for the production of succinate or propionate in Bacteroides spp. Consistent with this finding, levels of propionate, a major end-product of fermentation generated by Bacteroides spp., were lower in the fecal microbiota of mice cocolonized with D. piger (Fig. 2D).
Untargeted GC-MS and Ultra High-Performance Liquid Chromatography (UPLC)-MS analyses of cecal contents harvested from mice colonized for 2 wk with the eight-member versus nine-member communities indicated that D. piger impacted microbial metabolism of amino acids and carbohydrates. Levels of phenylacetate and 4-hydroxyphenylacetate, two microbial metabolites derived from phenylalanine and tyrosine, respectively, were increased with D. piger colonization. Cecal levels of fructose, N-acetyl galactosamine (one of the alternating sugars of chondroitin sulfate), galactosamine, and galactosamine-6-sulfate were lower with D. piger, whereas glucuronate (the other alternating sugar of chondroitin sulfate) was present at higher levels (Fig. 2B). Glucuronate is more oxidized than N-acetyl galactosamine, and its fermentation results in lower biomass yields per mole of carbohydrate metabolized compared with more reduced carbon sources (28). Although there were no differences in microbial biomass between the groups of mice (defined by fecal DNA content), microbial RNA-Seq identified several enzymes involved in the degradation of chondroitin sulfate that were expressed at lower levels in the presence of D. piger: (i) chondroitin sulfate lyase (EC4.2.2.20; EC4.2.2.21), which degrades chondroitin sulfate to sulfated disaccharides; (ii) a glucuronidase (EC3.2.1.139), which cleaves the disaccharides to monosaccharides; and (iii) N-acetyl-β-hexosaminidase (EC3.2.1.52), which is involved in the degradation of compounds containing terminal N-acetyl hexosamine residues such as chondroitin sulfate (Fig. 2C, Dataset S13). Altogether, these results suggest that in the presence of D. piger, community members require less chondroitin sulfate and prioritize the use of its more reduced carbohydrate moiety (N-acetyl-galactosamine). Utilization of more reduced carbon sources in the presence of D. piger may be facilitated via interspecies formate/hydrogen transfer.
We next assessed the effects of D. piger on host physiology. At high concentrations (mM range), H2S impairs oxygen consumption by inhibiting cytochrome c oxidase, the terminal oxidase of the mitochondrial respiratory chain (29). Mice containing D. piger and consuming the HF/HS diet supplemented with chondroitin sulfate had significantly increased cecal levels of H2S (Fig. 2E) compared with mice consuming the same diet but with the eight-member consortium. Besides short-chain fatty acids, amino acids and ketone bodies (e.g., 3-hydroxybutyrate generated via ketogenesis) serve as respiratory fuels for the gut epithelium (30–32). GC-MS of cecal contents disclosed that levels of glutamate, cysteine, aspartate, histidine, and 3-hydroxybutyrate were significantly increased in the presence of D. piger (Fig. 2B). RNA-Seq of mouse gene expression in the proximal colon provided evidence of decreased host consumption of amino acids in HF/HS diet-fed mice colonized with the nine-member compared with the eight-member consortium that lacked this SRB. Oxidation of amino acids results in the production of intracellular ammonia that is subsequently detoxified via the urea cycle. Levels of mRNA encoding carbamoyl-phosphate synthase 1, the enzyme that catalyzes the first committed step of the urea cycle, were 3.8-fold lower (P < 0.005; Dataset S14) in mice harboring D. piger. Moreover, because there were no significant differences in expression of microbial genes involved in the metabolism of these compounds between the two groups of mice, as judged by microbial RNA-Seq, we surmised that the increased cecal levels of amino acids, particularly glutamate, or 3-hydroxybutyrate were not a consequence of reduced microbial consumption or increased production of these metabolites brought about by the presence of D. piger but rather a reflection of reduced host metabolism.
Together, our metabolic profiling and microbial and mouse RNA-Seq analyses suggest that high levels of H2S generated by D. piger in the presence of dietary chondroitin sulfate result in lower host metabolic activity in the colon and possibly reduced uptake of nutrients from luminal contents. These results are consistent with a previous study that showed that daily colonic infusions of mM levels of H2S significantly diminished the ex vivo oxidative capacity of colonocytes (33). We did not observe significant differences in total body weight or epididymal fat pad weights between mice colonized with and without D. piger and consuming the HF/HS diet supplemented with chondroitin sulfate (n = 19–20 mice/treatment group; three independent experiments; assayed 14 d after introduction of D. piger).
The reported effects of H2S on gut mucosal barrier function and immune activation in preclinical animal models vary from promotion of inflammation to prevention of colitis (12–14). In vitro experiments have also demonstrated that H2S reduces proliferation of colonic epithelial cells (34). Moreover, a severe decrease in oxidative metabolism in the colonic mucosa of rats is associated with inflammation (35). In our study, in vivo bromodeoxyuridine (BrdU) labeling followed by flow cytometry of isolated colonocytes (SI Methods) did not reveal differences in epithelial turnover rates between mice colonized with the eight-member versus the D. piger–containing nine-member artificial community. RNA-Seq analysis of the proximal colons of these two groups of mice revealed that D. piger colonization is associated with significantly lower levels of mRNAs encoding several Ig subclasses and the tight junction protein claudin-4, plus higher levels of matrix metalloproteinase-7 (P < 0.005; Dataset S14). Histological analysis did not show evidence of an ongoing inflammatory process in the distal colons of either group of mice. Thus, in these short-term experiments involving the HF/HS diet, increasing D. piger and H2S levels with chondroitin sulfate did not have detectable effects on these measures of gut barrier integrity.
Prospectus.
Given that a significant fraction of humans lack detectable SRB in their gut microbiota (8) and the substantial representation of D. piger in our sampled healthy adult population that do harbor SRB, these findings raise the question of what differences exist in the microbiota of individuals with and without this organism. For example, what other organisms occupy its niche in different diet contexts, or is its niche unoccupied? What are the functional consequences to the microbiota and host in different diet contexts? Sulfur is a precious resource and its limitation, notably in malnourished children and experimental animal models, is associated with diminished growth (36, 37). Differences in the utilization of carbon sources observed in the artificial gut community we constructed in gnotobiotic mice, coupled with the capacity of D. piger to invade the community, evoke the question of what effects administration of this organism would have on human biology. These questions emanate from and illustrate the benefits of studying gnotobiotic mouse models of the type described here.
Methods
Gnotobiotic Husbandry.
All experiments involving mice were performed using protocols approved by the Washington University Animal Studies Committee. Mice belonging to the NMRI inbred strain were maintained in plastic flexible film gnotobiotic isolators under a strict 12 h light cycle (lights on at 0600) and fed diets ad libitum. Diets (Dataset S2) were sterilized by irradiation.
Other Procedures.
Methods related to (i) multiplex pyrosequencing of amplicons generated from the D. piger aprA gene, (ii) COPRO-Seq, (iii) qPCR measurements of D. piger colonization, (iv) microbial and mouse RNA-Seq, (v) transposon mutagenesis of D. piger GOR1, (vi) in vitro and in vivo INSeq analysis, (vii) the in vitro B. thetaiotaomicron–D. piger sulfate cross-feeding model, (viii) GC-MS and UPLC-MS analysis, and (ix) BrdU staining/flow cytometry are described in SI Methods. See Dataset S15 for media used for growing bacterial strains.
Supplementary Material
Acknowledgments
We thank David O’Donnell, Maria Karlsson, Sabrina Wagoner, and Jessica Hoisington-López for technical support; Mike Morrison and Mark Roth for generously sharing their protocols for measuring hydrogen sulfide; Barbara Mickelson (Harlan Teklad) for guidance in the preparation of diets; and members of the Gordon lab for their invaluable suggestions during the course of these studies. This work was supported by grants from the National Institutes of Health (DK30292, DK70977, and DK078669) and the Crohn’s and Colitis Foundation of America.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE48809). The INSeq data are available at http://gordonlab.wustl.edu/inseqreads/dpiger.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312524110/-/DCSupplemental.
References
- 1.Fischbach MA, Sonnenburg JL. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10(4):336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolin MJ, Miller TL. Interactions of microbial populations in cellulose fermentation. Fed Proc. 1983;42(1):109–113. [PubMed] [Google Scholar]
- 3.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7(8):568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 4.Cord-Ruwisch R, Seitz HJ, Conrad R. The capacity of hydrogenotrophic anaerobic-bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron-acceptor. Arch Microbiol. 1988;149(4):350–357. [Google Scholar]
- 5.Scanlan PD, Shanahan F, Marchesi JR. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol. 2009;69(2):213–221. doi: 10.1111/j.1574-6941.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 6.Loubinoux J, et al. Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int J Syst Evol Microbiol. 2002;52(Pt 4):1305–1308. doi: 10.1099/00207713-52-4-1305. [DOI] [PubMed] [Google Scholar]
- 7.Stewart JA, Chadwick VS, Murray A. Carriage, quantification, and predominance of methanogens and sulfate-reducing bacteria in faecal samples. Lett Appl Microbiol. 2006;43(1):58–63. doi: 10.1111/j.1472-765X.2006.01906.x. [DOI] [PubMed] [Google Scholar]
- 8.Hansen EE, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6(1):57–70. doi: 10.1038/ismej.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loubinoux J, Bronowicki JP, Pereira IA, Mougenel JL, Faou AE. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol. 2002;40(2):107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 11.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitcher MC, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut. 2000;46(1):64–72. doi: 10.1136/gut.46.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine J, Ellis CJ, Furne JK, Springfield J, Levitt MD. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol. 1998;93(1):83–87. doi: 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- 14.Wallace JL, Vong L, McKnight W, Dicay M, Martin GR. Endogenous and exogenous hydrogen sulfide promotes resolution of colitis in rats. Gastroenterology. 2009;137(2):569–578, e1. doi: 10.1053/j.gastro.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333(6038):101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis CL, Cummings JH, Neale G, Gibson GR. In vitro effects of mucin fermentation on the growth of human colonic sulphate-reducing bacteria. Anaerobe. 1996;2(2):117–122. [Google Scholar]
- 19.Gibson GR, Cummings JH, Macfarlane GT. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl Environ Microbiol. 1988;54(11):2750–2755. doi: 10.1128/aem.54.11.2750-2755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6(3):279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caffrey SM, et al. Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J Bacteriol. 2007;189(17):6159–6167. doi: 10.1128/JB.00747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolla A, Pohorelic BK, Voordouw JK, Voordouw G. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch Microbiol. 2000;174(3):143–151. doi: 10.1007/s002030000183. [DOI] [PubMed] [Google Scholar]
- 23.Zane GM, Yen HC, Wall JD. Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 2010;76(16):5500–5509. doi: 10.1128/AEM.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salyers AA, O’Brien M. Cellular location of enzymes involved in chondroitin sulfate breakdown by Bacteroides thetaiotaomicron. J Bacteriol. 1980;143(2):772–780. doi: 10.1128/jb.143.2.772-780.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem. 2011;286(29):25973–25982. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curno R, Magee EA, Edmond LM, Cummings JH. Studies of a urinary biomarker of dietary inorganic sulphur in subjects on diets containing 1-38 mmol sulphur/day and of the half-life of ingested 34SO4(2-) Eur J Clin Nutr. 2008;62(9):1106–1115. doi: 10.1038/sj.ejcn.1602822. [DOI] [PubMed] [Google Scholar]
- 27.Barthe L, et al. In vitro intestinal degradation and absorption of chondroitin sulfate, a glycosaminoglycan drug. Arzneimittelforschung. 2004;54(5):286–292. doi: 10.1055/s-0031-1296972. [DOI] [PubMed] [Google Scholar]
- 28.Kotarski SF, Salyers AA. Effect of long generation times on growth of Bacteroides thetaiotaomicron in carbohydrate-induced continuous culture. J Bacteriol. 1981;146(3):853–860. doi: 10.1128/jb.146.3.853-860.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholls P, Kim JK. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can J Biochem. 1982;60(6):613–623. doi: 10.1139/o82-076. [DOI] [PubMed] [Google Scholar]
- 30.Ardawi MS, Newsholme EA. Fuel utilization in colonocytes of the rat. Biochem J. 1985;231(3):713–719. doi: 10.1042/bj2310713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura RE, Ilich JZ. The oxidation of 3-hydroxybutyrate in developing rat jejunum. J Pediatr Gastroenterol Nutr. 1991;13(4):347–353. doi: 10.1097/00005176-199111000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Firmansyah A, Penn D, Lebenthal E. Isolated colonocyte metabolism of glucose, glutamine, n-butyrate, and beta-hydroxybutyrate in malnutrition. Gastroenterology. 1989;97(3):622–629. doi: 10.1016/0016-5085(89)90633-1. [DOI] [PubMed] [Google Scholar]
- 33.Moore JW, Millard S, Babidge W, Rowland R, Roediger WE. Hydrogen sulphide produces diminished fatty acid oxidation in the rat colon in vivo: implications for ulcerative colitis. Aust N Z J Surg. 1997;67(5):245–249. doi: 10.1111/j.1445-2197.1997.tb01956.x. [DOI] [PubMed] [Google Scholar]
- 34.Wu YC, et al. Hydrogen sulfide lowers proliferation and induces protective autophagy in colon epithelial cells. PLoS ONE. 2012;7(5):e37572. doi: 10.1371/journal.pone.0037572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roediger WE, Nance S. Metabolic induction of experimental ulcerative colitis by inhibition of fatty acid oxidation. Br J Exp Pathol. 1986;67(6):773–782. [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MI, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori M, Manabe S, Uenishi K, Sakamoto S. Nutritional improvements of soy protein isolate by different levels of methionine supplementation in pregnant rats. Tokushima J Exp Med. 1993;40(1-2):35–42. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.