Abstract
In acute promyelocytic leukemia, granulocytic differentiation is arrested at the promyelocyte stage. The variant t(11;17) translocation produces two fusion proteins, promyelocytic leukemia zinc finger-retinoic acid receptor α (PLZF-RARα) and RARα-PLZF, both of which participate in leukemia development. Here we provide evidence that the activity of CCAAT/enhancer binding protein α (C/EBPα), a master regulator of granulocytic differentiation, is severely impaired in leukemic promyelocytes with the t(11;17) translocation compared with those associated with the t(15;17) translocation. We show that RARα-PLZF inhibits myeloid cell differentiation through interactions with C/EBPα tethered to DNA, using ChIP and DNA capture assays. Furthermore, RARα-PLZF recruits HDAC1 and causes histone H3 deacetylation at C/EBPα target loci, thereby decreasing the expression of C/EBPα target genes. In line with these results, HDAC inhibitors restore in part C/EBPα target gene expression. These findings provide molecular evidence for a mechanism through which RARα-PLZF acts as a modifier oncogene that subverts differentiation in the granulocytic lineage by associating with C/EBPα and inhibiting its activity.
Keywords: APL, granulocyte differentiation, transcription inhibition, histone modification, protein interaction
Acute promyelocytic leukemia (APL) epitomizes a rare disease that can now be successfully treated with targeted therapy (1). APL is invariably associated with chromosomal translocations involving the retinoic acid receptor α (RARα) locus and genes encoding proteins with self-aggregation motifs: promyelocytic leukemia (PML), PLZF/ZBTB16, nucleophosmin (NPM1), nuclear mitotic apparatus protein 1 (NuMA1), and signal transducer and activator of transcription 5B (STAT5b) (2, 3). The importance of the translocation partners is underscored by the fact that disruption of RARα and RARγ signaling is not sufficient to induce leukemia in mice (4, 5). PML-RARα, the most frequent fusion protein activated by the t(15;17) translocation (reviewed in refs. 1–3), caused differentiation arrest at the promyelocyte stage and aberrant self-renewal in myeloid progenitors (6). This disease responds to retinoic acid treatment due to proteasomal degradation of PML-RARα (reviewed in ref. 1). Among the variant translocations, the t(11;17) translocation is the most frequent, and the disease is resistant to all-trans retinoic acid (ATRA) treatment (7). In this translocation, RARA is fused to PLZF (8), encoding a protein with nine Krüppel-like zinc fingers in its C-terminal moiety and a POZ domain in the N-terminal region. The translocation produces two fusion proteins, PLZF-RARα and RARα-PLZF, harboring the POZ domain together with two or three Nt zinc fingers and seven or six Ct zinc fingers of PLZF, respectively (8, 9). It was initially thought that the disease is RA-resistant due to PLZF-RARα interaction with many proteins including PML and itself, as well as several corepressors, nuclear receptor corepressor 1 (NCOR1), NCOR2/SMRT, SIN3 transcription regulator homolog A (mSIN3A), histone deacetylase (HDAC) (reviewed in ref. 2), and the PRC1 polycomb group complex (10). Nonetheless, the reverse fusion protein, RARα-PLZF, was later shown to also contribute to RA resistance. RARα-PLZF modifies the disease induced by PLZF-RARα, i.e., chronic myeloid leukemia (CML) (11), into APL (12). Moreover, the presence of RARα-PLZF increases the proliferation and resistance to RA treatment in double transgenic mice (12). At the molecular level, RARα-PLZF binds to DNA via the PLZF binding site and derepresses PLZF target genes such as cyclin A, Hoxb2a and c-Myc (13–15), thereby causing increased proliferation. In addition, RA resistance has been associated with an up-regulation of cellular retinoic acid binding protein 1 (CRABPI) (16), although additional mechanisms could also contribute to this resistance. Furthermore, how RARα-PLZF modifies the tumor phenotype remains to be clarified.
Somatic mutations in genes encoding lineage-restricted transcription regulators were identified in cytogenetically normal acute myeloblastic leukemias (AML) (17, 18), supporting the view that disruption of lineage regulatory mechanisms is an important event in leukemogenesis. C/EBPα is a member of the basic region–leucine zipper (bZIP) family of transcription factors which is important for hematopoietic stem cells and for differentiation in the granulocytic lineage (19, 20). In human AML, C/EBPα function and activity are frequently disrupted via several mechanisms: dominant-negative point mutations (17), transcriptional inhibition by oncoproteins [acute myeloid leukemia 1-eight twenty one (AML1-ETO), myelodysplasia syndrome associated protein 1-ecotropic virus integration site 1 (AML1-MDS1-EVI1), and core binding factor, beta subunit-myosin heavy chain 11 (CBFB-MYH11)], or promoter hypermethylation (reviewed in refs. 18 and 19).
In the present study, we aim to define the cellular and molecular anomalies induced by RARα-PLZF, using the model cell line 32D, primary fetal liver cells, and primary leukemic cells from APL patients with t(11;17) or t(15;17) translocations. We show that RARα-PLZF recruits HDAC1 and severely impairs C/EBPα function, thereby contributing to differentiation arrest in APL.
Results
RARα-PLZF Impairs G-CSF–Induced Survival.
To define the role of RARα−PLZF in the myeloid lineage, we ectopically expressed the fusion gene in 32D myeloid cells. This cell line was chosen because it retains two essential properties of primary myeloid cells: growth factor requirements for survival and terminal granulocytic differentiation in response to G-CSF (Fig. S1A). In the presence of IL-3, the cells self-propagate and remain undifferentiated whereas G-CSF–induced typical granulocyte differentiation, Gr-1 surface expression, and Csf3r up-regulation (Fig. S1 B and C), reproducing the pattern of differentiation in primary myeloid cells, whereby Csf3r expression also increased as the cells progress from the immature myeloid stage (CD11b+Gr1−) to mature granulocytes (CD11b+Gr1+) (Fig. S1D).
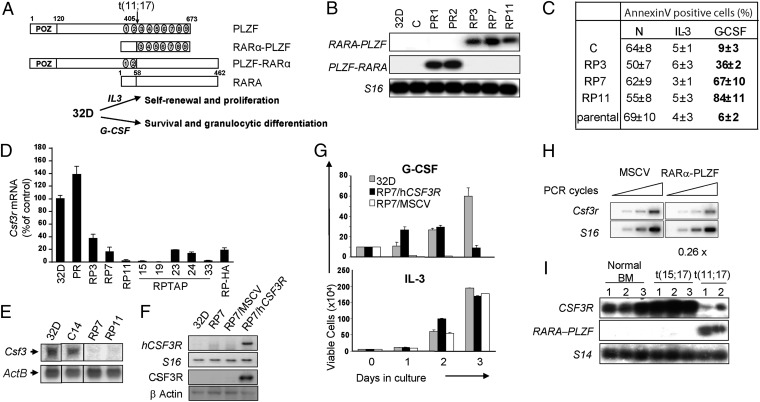
We generated several independent 32D clones expressing the RARα-PLZF (RP3, RP7, and RP11) or the PLZF-RARα (PR1 and PR2) fusion genes (Fig. 1B). All clones survived and proliferated in the presence of IL-3 (Fig. 1C) and were apoptotic after 24 h of growth factor withdrawal (Fig. 1C). Surprisingly, G-CSF failed to suppress apoptosis in RARα-PLZF–expressing cells which showed elevated apoptosis whereas parental 32D and PLZF-RARα –expressing cells (Fig. 1C and Fig. S2A) responded to G-CSF treatment.
Fig. 1.
RARα-PLZF impairs Csf3r expression and cell survival in response to G-CSF. (A) Schematic representation of PLZF, RARα, and the reciprocal fusion proteins of the t(11;17) translocation, RARα-PLZF, and PLZF-RARα. (B) Confirmation of ectopic PLZF-RARα or RARα-PLZF expression in 32D clones. S16 is shown as a control for variations in input RNA loading. (C) Flow cytometry analysis of 32D clones in the presence or absence of IL-3 or G-CSF for 24 h. Apoptotic cells were labeled with Annexin V, and dead cells were excluded by propidium iodide staining. Shown are the average ± SD of two to five independent experiments. (D) Csf3r mRNA levels in RARα-PLZF expressing clones (RP7, RP11, and RP-TAP) and in polyclonal PLZF-RARα expressing cells grown in IL-3 medium. Csf3r mRNA levels were determined by real-time PCR analysis, normalized to Hprt levels and calculated as percent of 32D parental cells. Data shown are the average ± SD of two independent experiments performed in quadruplates. (E) Northern blot analysis of Csf3r in 32D parental cells, control cells (C), and RARα-PLZF expressing clones 7 and 11. ActB is shown as a loading control. (F) Confirmation of Csf3r expression in RP7 transfected cells. RP7 cells were either transduced with the huCSF3R expression vector (RP7/huCSF3R) or the empty MSCV vector (RP7/MSCV). Expression levels of huCSF3R mRNA were measured by semiquantitative RT-PCR (Upper) and of CSF3R protein were measured by Western blotting (Lower). S16 is shown as a control for input RNA and β-actin, for input protein. (G) huCSF3R expression rescues G-CSF–induced cell survival in RP7 cells up to 48 h but fails to provide long-term cell survival by G-CSF. Cells were grown in the presence of G-CSF or IL-3, and viable cells were evaluated by trypan blue exclusion (average ± SD, n = 2 in triplicates). (H) Csfr3 expression in primary fetal liver cells expressing RARα-PLZF. Primary fetal liver cells from 12.5 dpc embryos were transduced with the empty vector or RARα-PLZF. Total RNA extracted from CD11b+GFP+ cells were analyzed by semiquantitative RT-PCR (n = 2). (I) CSF3R mRNA levels are decreased in leukemic cells from APL patients with t(11;17) translocation. Total bone marrow cells from three normal donors and three t(15;17) and two t(11;17) APL patient samples were analyzed by RT-PCR for the presence of CSF3R transcripts. S14 is shown as a control for input RNA.
The induction of STAT DNA binding activity is an early event in cytokine signaling (21). Compared with control cells, G-CSF–induced STAT activity, assessed by electromobility shift assay, was impaired in RP7 and RP11 cells and was reduced in RP3 cells, indicating that the defective survival/differentiation response to G-CSF is upstream of Janus kinase-STAT (JAK-STAT), possibly occurring at the receptor level (Fig. S2B).
RARα-PLZF Decreases Expression of CSF3R.
Csf3r mRNA expression levels in RP7, RP11, and in five clones expressing RARα-PLZF fused to the tag for tandem affinity purification (RP-TAP15, 19, 23, 24, 33) as well as RP-HA were less than 20% that of parental 32D cells by Northern blotting and/or quantitative RT-PCR analysis (Fig. 1 D and E). In contrast, PLZF-RARα expression did not affect Csf3r mRNA levels (Fig. 1D). Furthermore, RP3 cells that showed a partial response to G-CSF also exhibited a partial reduction of Csf3r mRNA (Fig. 1D). Therefore, Csf3r expression levels concurred with the biological assay and indicated that RARα-PLZF, but not PLZF-RARα, inhibits Csf3r gene expression in 32D myeloid cells (Fig. 1D).
To assess whether Csf3r is indeed downstream of RARα-PLZF, we performed a rescue experiment by ectopically expressing CSF3R in RP cells. Because G-CSF is not species-specific, we delivered the human CSF3R gene to distinguish the transgene from the endogenous gene (Fig. 1F, Upper, RNA levels; Fig. 1F, Lower, protein levels). In clone RP7 expressing hCSF3R, cell survival in response to G-CSF was restored for a 24 h period whereas control RP7 cells (empty vector) as well as parental RP7 cells underwent apoptosis (Fig. 1G and Fig. S2D). However, the survival of RP7 rescued by hCSF3R could not be sustained for longer periods in G-CSF–containing medium (Fig. 1G, Upper), whereas parental 32D cells continued to survive and proliferate in response to G-CSF. As expected, all cells survived and proliferated in IL-3 control cultures, consistent with the specificity of RARα-PLZF for the CSF3R pathway (Fig. 1G, Bottom).
These results suggest that RARα-PLZF is upstream of Csf3r. Nonetheless, the inability of cells rescued with hCSF3R to survive beyond 2 d in G-CSF medium suggests additional defects due to the presence of RARα-PLZF. Cell death was not due to early differentiation, as assessed by morphological analysis (Fig. S2D).
Next, we assessed whether CSF3R expression is impaired in primary murine fetal liver cells expressing RARα-PLZF and in leukemic cells from APL patients. To rule out the possibility that differences in gene expression could be due to an imbalance in cell populations induced by the transgene in primary murine hematopoietic cells, we purified CD11b+ cells after retroviral gene transduction (Fig. S3). Ectopic expression of RARα-PLZF resulted in a twofold repression of Csf3r mRNA levels in CD11b+ fetal liver cells compared with control cells (empty vector) (Fig. 1H). Furthermore, high levels of CSF3R mRNA were detected by RT-PCR in three APL samples with the t(15;17) translocation, comparable to those expressed in primary bone marrow cells from three healthy donors (Fig. 1I). In contrast, CSF3R transcripts were low in two t(11;17) APL patient samples (Fig. 1I). Therefore, RARα-PLZF expression correlates with impaired CSF3R expression in leukemic promyelocytes.
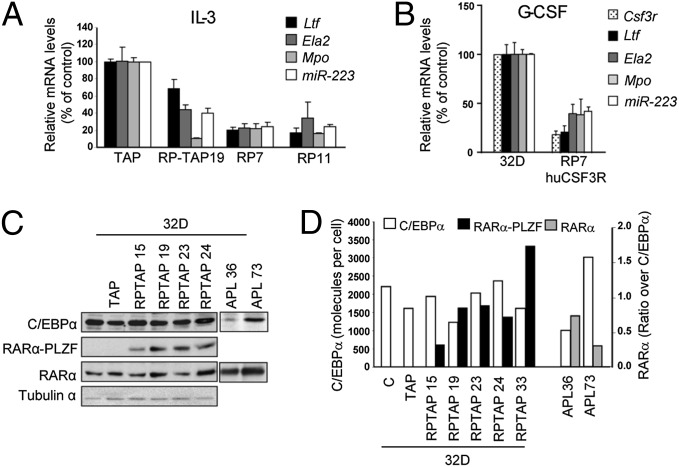
RARα-PLZF Decreases the Expression of C/EBPα Target Genes Without Affecting C/EBPα Levels.
C/EBPα is important for optimal Csf3r expression in vivo (22), although the induction of granulocytic differentiation by C/EBPα could be Csf3r-independent (23). Therefore, we analyzed the expression levels of additional C/EBPα target genes that are involved in myeloid differentiation, i.e., Ltf, Mpo, Ela2 as well as miR-223 (24–27), all produced at the promyelocyte stage of differentiation. Interestingly, these C/EBPα target genes were strongly reduced in RP7, RP11, and RP-TAP19 clones compared with parental cells or control cells expressing the empty TAP vector (Fig. 2A) and in RP7 ectopically expressing hGCSFR upon G-CSF stimulation (Fig. 2B). Decreased C/EBPα target genes were not due to decreased C/EBPα protein levels compared with control cells (Fig. 2C). In addition, we monitored RARα protein levels and found little clonal variation (Fig. 2C). Together, our results indicate that RARα−PLZF acts in parallel to or downstream of C/EBPα.
Fig. 2.
RARα-PLZF impairs the expression of CEBPα target genes without affecting CEBPα expression levels. (A) Decreased expression of C/EBPα target genes in RARα-PLZF expressing clones compared with control 32D cells. mRNA levels for the indicated C/EBPα target genes were determined by real-time PCR as in Fig 1D. (B) Decreased expression of C/EBPα target genes in RP7/huCSF3R expressing clones compared with control 32D cells, as determined by real-time PCR analysis. (C) Protein levels of RARα-PLZF and C/EBPα in 32D clones and of RARα and C/EBPα in t(15;17) APL patient samples. Total protein extracts from 32D clones stably transduced with a TAP-RARα-PLZF (RP-TAP) or from t(15;17) APL blasts were subjected to Western blot analysis (representative of two experiments). (D) Molecular levels of C/EBPα and RARα-PLZF in 32D clones and in APL patients. Western blot signals were compared with signals from tagged protein standards expressed in BOSC cells (HA-hC/EBPα, hJUN-HA, hJUN-TAP) with antibodies against HA, TAP, C/EBPα, and RARα. Shown are the number of C/EBPα molecules per cell calculated based on the number of cells corresponding to 30 μg of extracts per lane. The number of RARα-PLZF and RARα molecules per cell was calculated as above and expressed as molecular ratio of RARα-PLZF to C/EBPα levels in 32D clones and of RARα to C/EBPα in APL patients.
We next verified the relative expression levels of RARα-PLZF to C/EBPα in our clones and compared this to the ratio observed in primary APL samples. To this end, we designed a semiquantitative Western blot analysis for C/EBPα and for RARα-PLZF (Fig. S4), using a single purified recombinant protein of known concentration. Briefly, we used purified JUN as a standard to determine the concentrations of HA- or TAP-tagged JUN in transfected cell extracts with an antibody against JUN (Fig. S4A). Then the concentrations of HA- or TAP-tagged proteins of interest (HA-C/EBPα, RP-TAP, or HA-PR), determined using JUN-HA or JUN-TAP as standards (Fig. S4 B, C, and E), served to estimate the concentrations of endogenous C/EBPα (Fig. S4C) and transfected RARα-PLZF (Fig. S4D) or PLZF (Fig. S4E) in our clones as well as of endogenous RARα (Fig. S4F) in patient samples.
C/EBPα protein levels were comparable in cells expressing RARα-PLZF and control cells, i.e., in the range of ∼1600–2200 molecules per cell (Fig. 2D). Next, we compared the ratio of RARα-PLZF to C/EBPα in our RP clones and found a molecular ratio of 0.3–0.9 (Fig. 2D). We compared these results in primary APL samples. Due to the scarcity of the t(11;17) translocation, we used two t(15;17) APL samples, with the assumption that expression levels driven from the endogenous RARα promoter in these two translocations, i.e., RARa and RARα-PLZF, would be comparable (Fig. 2D). The ratio of RARα to C/EBPα in two primary t(15;17) APL samples was 0.3 and 0.8, respectively (Fig. 2D). Thus, the levels of expression of our constructs compared with C/EBPα levels are reminiscent of the ratio between RARα and C/EBPα in t(15;17) APL patients, confirming the validity of our model.
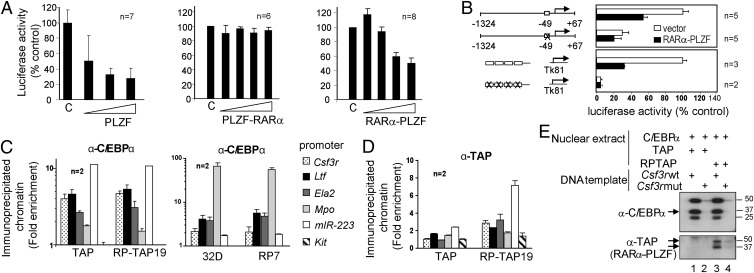
RARα-PLZF Occupies C/EBPα Target Loci.
Our results so far indicate that RARα-PLZF inhibits the expression of C/EBPα target genes without affecting C/EBPα expression levels. We therefore tested the possibility that RARα-PLZF directly inhibits C/EBPα transcriptional activity. We first assessed C/EBPα activity in myeloid cells using the CSF3R promoter reporter (22). The CSF3R promoter is active in 32D cells (Fig. S5A), and this activity was not affected by the empty MSCV vector (control) (Fig. 3A). However, CSF3R promoter activity was consistently repressed by RARα-PLZF (two- to threefold) and PLZF but not by PLZF-RARα (Fig. 3A). Moreover, RARα-PLZF did not inhibit the activity of the promoterless reporter vector pXPII (Fig. S5A), indicating that RARα-PLZF–dependent repression was specific to CSF3R promoter sequences.
Fig. 3.
RARα-PLZF is recruited to the promoters of C/EBPα target genes in vivo and in vitro via a consensus C/EBP binding site. (A) RARα-PLZF inhibits CSF3R promoter activity in a dose-dependent manner. The 32D cells were electroporated with the CSF3R promoter (−1324 + 67) luciferase construct and increasing amounts of PLZF, PLZF-RARα, or RARα-PLZF expression vectors. Data shown are the means ± SD of n experiments and represent percent of basal CSF3R promoter activity in 32D cells. (B) The C/EBP binding site is necessary and sufficient for transcriptional repression by RARα-PLZF. Wild-type CSF3R promoter construct or a mutant at the C/EBP consensus sequence as well as of a multimer of wild-type or mutated C/EBP binding site constructs were tested in 32D with RARα-PLZF or the empty vector. The basal activity of constructs containing wild-type C/EBPα binding sites were taken as 100%, respectively (average ± SD of n experiments). (C) RARα-PLZF does not affect promoter occupancy by C/EBPα in myeloid cells. Cross-linked chromatin extracts from control cells (32D and TAP) or RARa-PLZF expressing cells (RP7 and RP-TAP19) were subjected to chromatin immunoprecipitation (ChIP) with antibodies against C/EBPα or a control rabbit IgG. Myeloid promoters, amplified by qPCR, are shown as fold enrichment over control IgG and over Hprt promoter sequences (means ± SD of at least two independent experiments done in triplicates). The Kit promoter serves as an additional negative control. (D) RARα-PLZF occupies C/EBPα target promoters in 32D cells, as determined by ChIP with an antibody against the TAP tag or a control rabbit IgG. Data are presented as fold enrichment over control cells transduced with the empty vector (TAP). (E) C/EBPα and RARα-PLZF binding to the Csf3r promoter requires the integrity of the C/EBP binding site. Nuclear extracts (NE) from BOSC cells overexpressing C/EBPα and from 32D overexpressing TAP or RP-TAP were incubated with immobilized wild-type (lanes 1 and 3) or mutated Csf3r (lanes 2 and 4) promoter templates. Bound proteins were revealed by Western blotting.
We located the essential contribution of the C/EBPα binding site at position −49 of the CSF3R promoter (22) (Fig. S5A) which is required for promoter activity in 32D cells (Fig. 3B) and for transcriptional repression by PLZF and RARα-PLZF in these cells (Fig. S5A). In addition, RARα-PLZF did not inhibit C/EBPα DNA binding by gel shift assay (Fig. S5B, lanes 1 and 6) nor C/EBPα occupancy of its target loci by ChIP using an anti-C/EBPα antibody (Fig. 3C). Finally, RARα-PLZF and C/EBPα cooccupied the promoters of all five C/EBPα target genes tested in Fig. 1E (Fig. 3 C and D), which were two- to sixfold higher in chromatin extracts immunoprecipitated with anti-TAP (RP-TAP 19, Fig. 3D) or with anti-RARα (RP7, Fig. S5C) compared with their respective controls (TAP or 32D). Of note, myeloid promoter sequences were less than twofold enriched in control TAP cells compared with control immunoglobulins and irrelevant promoter sequences (Kit) (Fig. 3D, Left), confirming the specificity of the ChIP, whereas all myeloid promoters were 5- to 25-fold enriched in anti-RARα–immunoprecipitated chromatin extracts in 32D cells, suggesting that RARα and C/EBPα may coregulate target genes. Because transcription repression by RARα-PLZF requires the integrity of the C/EBPα binding site (Fig. 3B), our data indicate that RARα-PLZF inhibits the transcriptional activity of C/EBPα tethered to its cognate binding sites on target promoters.
RARα-PLZF Interacts with C/EBPα and Recruits HDAC1.
The Csf3r promoter lacked the typical consensus [TACT/AGTAC] that recruits RARα-PLZF or PLZF to DNA (28). The above results led us to address the question whether C/EBPα actually recruits RARα-PLZF to target promoters. To this end, we designed a DNA capture assay (29) in which the −74 to +67 region of the Csf3r promoter was immobilized on magnetic beads and incubated with nuclear extracts from C/EBPα− and control TAP- or RP-TAP–expressing cells (Fig. 3E). Csf3r promoter sequences recruited both C/EBPα and RARα-PLZF, consistent with the ChIP assay, and both associations required the integrity of the C/EBP binding site (Fig. 3E, lanes 2 and 4). Together, these observations raise the possibility that RARα-PLZF is recruited to DNA by interaction with C/EBPα and inhibits its transcriptional activity, an issue that we addressed using in vitro and in vivo protein–protein binding assays.
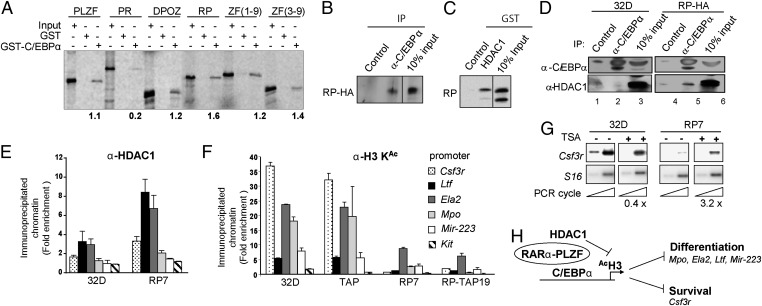
In GST–pull-down assays, in vitro translated RARα-PLZF and PLZF specifically interacted with GST-C/EBPα whereas PLZF-RARα did not (Fig. 4A), consistent with their respective capacities to inhibit or not CSF3R promoter activity in 32D cells (Fig. 3A). Furthermore, the seven C-terminal zinc fingers of PLZF were sufficient for this association (Fig. 4A). The interaction between RARα-PLZF and C/EBPα was further confirmed by coimmunoprecipitation using in vitro translated radiolabeled proteins (Fig. S6A). The antibody against C/EBPα brought down RARα-PLZF whereas a control preimmune rabbit serum did not. This interaction also occurred in vivo with endogenous C/EBPα, as observed by coimmunoprecipitation of HA-tagged RARα-PLZF with an anti-C/EBPα antibody using 32D nuclear extracts (Fig. 4B). The absence of coimmunoprecipitation in parental 32D cells confirmed the specificity of the procedure.
Fig. 4.
RARα-PLZF binds to C/EBPα and HDAC1 and inhibits C/EBPα target gene expression in myeloid cells via decreased histone H3 acetylation at their promoters. (A) RARα-PLZF binds to C/EBPα in vitro, as determined by pull-down assays with GST-C/EBPα and in vitro translated 35S-labeled PLZF, PLZF-RARα, RARα-PLZF, or the indicated PLZF deletion mutants. Data are typical of four (wild type) or two (truncated proteins) experiments. (B) Coimmunoprecipitation of endogenous C/EBPα with RARα-PLZF in myeloid cells. Cross-linked cellular extracts were immunoprecipitated with the indicated antibodies (n = 2) before Western blotting. (C) RARα-PLZF binds to HDAC1 in vitro. Pull-down assays were performed as in A with GST-HDAC1 and 35S-RARα-PLZF (n = 2). (D) HDAC1 coimmunoprecipitates with endogenous C/EBPα in myeloid cells expressing RARα-PLZF. Cross-linked cellular extracts were immunoprecipitated with anti-C/EBPα or a rabbit preimmune serum (control) before Western blotting. (E) RARα-PLZF–induced HDAC1 occupancy of C/EBPα promoters. Data are expressed as fold enrichment in HDAC1 chromatin association over control Ig observed by ChIP of 32D-RP7 or control 32D cells with anti-HDAC1 (average ± SD of at least two independent experiments). (F) Quantitative analysis of acetylated histone H3 bound to C/EBPα targets in parental 32D, TAP, RP7, and RP-TAP19 cell lines by ChIP. Chromatin immunoprecipitation assays were performed as in Fig. 3B with antibodies against acetylated histone H3 or a control rabbit IgG. (G) TSA restores Csf3r expression in RARα-PLZF expressing cells. Cells were exposed or not to TSA (10 ng/mL) for 6 h. Csf3r mRNA levels, assessed by RT-PCR (as in Fig. 1E), in untreated cells were set as 1 after normalization to S16 levels (n = 2). (H) A model of repression of C/EBPα target genes by RARα-PLZF. RARα-PLZF is recruited on C/EBP consensus sequences by its ability to physically interact with C/EBPα, resulting in HDAC1 recruitment, histone H3 hypoacetylation, and decreased expression of C/EBPα target genes.
PLZF recruits histone deacetylase complexes via its C-terminal zinc fingers to repress transcription (30, 31). Here we show that RARα-PLZF interacts with HDAC1 in vitro (Fig. 4C). Moreover, HDAC1 coimmunoprecipitated with C/EBPα in RARα-PLZF transfectants but not in parental 32D cells (Fig. 4D, compare lanes 2 and 5) and was twofold enriched in immunoprecipitated chromatin from 32D RP7 cells compared with 32D control cells (Fig. 4E). This was reproducibly and specifically associated with a reduction in histone H3 acetylation at all of the C/EBPα target promoters (Fig. 4F). The absence of histone H3 acetylation at the Kit promoter in these cells correlated with the absence of Kit expression and confirmed the specificity of the ChIP procedure. In addition, inhibition of histone deacetylase activity with trichostatin A (TSA) induced a three- to fourfold increase in Csf3r mRNA levels in clone RP7, while not affecting parental 32D cells (Fig. 4G), concurring with the view that transcription repression by RARα-PLZF involves a histone deacetylase activity. Moreover, TSA treatment reversed the repression of the CSF3R proximal promoter in 32D cells by RARα-PLZF in transcription assays, without affecting the activity of the promoter alone (Fig. S6B). Whereas TSA was fully efficient in transient assays with luciferase reporters (Fig. S6B), the expression of the endogenous Csf3r gene was only partially rescued by TSA (Fig. 4G). This difference is currently not known but could possibly be due to more complex regulatory mechanisms in situ. We therefore conclude that transcription inhibition by RARα-PLZF is sensitive to the HDAC inhibitor TSA.
Together, our data support a model where RARα-PLZF is recruited on C/EBP consensus sequences by its ability to physically interact with C/EBPα, resulting in HDAC1 recruitment, histone H3 hypoacetylation, and decreased expression of C/EBPα target genes (Fig. 4H).
Discussion
In the present study, we provide evidence for a unique role of RARα-PLZF in APL, via direct interaction with and inhibition of C/EBPα activity.
C/EBPα is a master regulator of granulocytic differentiation. Decreased C/EBPα transcription, translation, or loss of function mutations is commonly observed in AML (mostly M1, M2, and M4), in familial AML with variable morphology (M1, M2-Eo, and M4-Eo), as well as in CML (reviewed by ref. 18). We now define a distinct mechanism of inhibition of C/EBPα transcriptional activity via direct protein–protein interaction with RARα-PLZF and recruitment of HDAC1.
RARα-PLZF, which is always present in t(11;17) APL, derepresses PLZF target genes involved in cell proliferation. Whereas this may explain deregulated cell proliferation, RARα−PLZF also causes differentiation arrest and modifies the phenotype of the disease induced by PLZF-RARα in transgenic mice (12). Thus, our observations unravel a previously undescribed mechanism through which RARα-PLZF can cause differentiation arrest and favor proliferation, by inhibiting C/EBPα activity.
PLZF has been shown to control myelopoiesis via binding to C/EBPA promoter, resulting in a two-fold decrease in C/EBPA mRNA levels (32). In contrast, RARα-PLZF does not affect C/EBPα levels as shown here by RT-PCR and quantitative Western blotting, possibly due to fusion with the RARα moiety as discussed above. Rather, we show that both PLZF and RARα-PLZF bind C/EBPα, and therefore, both can repress myeloid gene expression via direct protein–protein interactions and recruitment of corepressors, without binding DNA. Because the pool of stem cells and progenitors remains balanced despite the production of large numbers of differentiated myeloid cells during steady state and in stress response, we propose that the regulation of gene expression programs by protein–protein interactions provides an additional level of complexity that modifies the outcome of protein–DNA interactions and that networks of interacting transcriptional regulators can keep the hematopoietic system stable despite peripheral fluctuations.
The t(11;17) APL is mostly of poor prognosis, and the disease is relatively RA-resistant, possibly due to RA-resistant corepressor recruitment (33) and/or CRABPI derepression (16). Additionally, the inhibition of C/EBPε by PLZF-RARα (34) and the inhibition of C/EBPα by RARα-PLZF reported here, both required for terminal granulocytic differentiation, unequivocally contribute to this RA resistance and support the use of HDAC inhibitors for the treatment of t(11;17) APL. Furthermore, the presence of RARα-PLZF could facilitate leukemogenesis by eliminating the need for additional hits such as the deletion of the remaining Plzf allele that could dysregulate the control of cell proliferation and myeloid differentiation. Thus, in PLZF-RARα transgenic mice, loss of wild-type Plzf was sufficient to modify a CML-like disease into APL (12), in lieu of the coexpression of RARα-PLZF. The extensive network of protein–protein interaction entertained by both fusion proteins and the functional contribution of these fusion proteins to cell transformation could explain the fact that the incidence of APL does not increase with age, consistent with one or a limited number of rate-limiting mutation(s) (1, 35).
Materials and Methods
Cell Line and Cell Culture Condition.
The 32D cell line was a gift of P. J. Quesenberry (Brown Medical School, Brown University, Providence, RI). The cells were cultured at a concentration of 5 × 104 cells/mL in IMDM [Gibco (Invitrogen Life Technologies)] supplemented with 10% (vol/vol) FBS (FBS, Gibco) and WEHI-3–conditioned medium as a source of IL-3 in a 5% CO2–95% (vol/vol) air humified atmosphere at 37 °C. Cells were washed twice with PBS before G-CSF stimulation (5 ng/mL, Amgen, Inc.). APL blood samples were collected with informed consent and with approval of the project by the Research Ethics Board of Hôpital Maisonneuve-Rosemont, University of Montreal and Shanghai Jiao-Tong University School of Medicine.
Plasmids and antibodies are described in SI Materials and Methods.
Retroviral Mediated Gene Transfer.
Stable 32D or Ter119− fetal liver cells from 12.5 dpc C57 embryos expressing HA-tagged RARα-PLZF, GFP, or TAP were obtained by retroviral gene transfer or through Lipofectin-mediated DNA transfer as reported in ref. 36. Cells were selected in G-418 (1 mg/mL, Gibco) or puromycin (5 μg/mL) containing media or sorted on the basis of GFP expression 1 week postinfection. Cells were cloned immediately after gene transfer by limiting dilution.
Flow cytometry analyses were performed as described previously (37). Dead cells were excluded on the basis of staining with propidium iodide.
Northern Blotting and Electrophoretic Mobility Shift Assays.
Northern blotting was performed as previously described (36), using the Csf3r and ActB cDNA as probes. Electrophoretic mobility shift assays (EMSA) were performed as described in ref. 38 using nuclear extracts (10 μg) from 32D cells with the C/EBP site of the CSF3R (−57 to −37) promoter (22), as detailed in SI Materials and Methods.
GST–Pull-Down Assay, Coimmunoprecipitations, and Western Blot Analysis.
GST–protein purification and pull-down assay protocol were performed as described previously (38).
Coimmunoprecipitations were performed using either in vitro translated 35S radiolabeled protein, nuclear extracts, or cellular extracts from formaldehyde fixed cells as previously described (38). Individually synthesized proteins were preincubated for 30 min at 37 °C to allow for protein–protein interactions to occur. Protein complexes were incubated with 2 μg of antibody overnight at 4 °C in 1% Nonidet P-40, 50 mM NaCl, 50 mM Tris pH 8.0, 1mM MgCl2, 10 μM ZnCl2, 4% glycerol. Immune complexes were then recovered with Pansorbin cells (Calbiochem) (30 min at 4 °C), eluted, and resolved by SDS/PAGE. The 35S-labeled proteins were revealed using a PhosphoImager screen. Unlabeled proteins were analyzed by Western blotting using ECL plus (Amersham-Pharmacia Biotechnology).
Whole cell extracts from 2 × 106 cells in radio-immunoprecipitation assay (RIPA) lysis buffer (10mM Tris pH 8.0, 140mM NaCl, 1mM EDTA, 1% Triton ×100, 0.1% SDS, 0.1% Deoxycholate) were resolved by electrophoresis and analyzed by Western blotting. β-Actin was shown as a loading control.
Semiquantitative Western Blotting.
Semiquantitative Western blotting was performed using cellular extract from 32D clones lysed in hot Laemmli buffer as follows. Purified recombinant JUN (Promega, Medison) was quantified by Bradford assay using BSA. Total extract from 293T (human embryonic kidney cell line transformed with large T antigen) cells expressing either hJUN-HA or hJUN-TAP was quantified against standard curve of recombinant JUN by Western blotting using an antibody against JUN. Anti-HA or -TAP antibodies were used to create standard curves for HA- or TAP-tagged proteins, which subsequently served to quantify either RP-TAP, HA-PR, or HA-hC/EBPα in cellular extracts. Then, using antibodies against RARα or C/EBPα on the quantified HA-PR and HA-hC/EBPα expressed in BOSC cells, the endogenous levels of RARα in APL patients and C/EBPα in both APL patients and 32D clones were quantified.
Chromatin Immunoprecipitation Assay.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (29, 38) using 500 μg of protein per sample and anti-acetyl histone H3 (Upstate Biotechnologies) or normal rabbit serum as control. Primer sequences are shown in SI Materials and Methods and Table S1.
DNA Capture Assay.
DNA capture assay was performed as described previously (29) using PCR amplified Csf3r promoter fragments covering the C/EBPα binding site, followed by elution and Western blotting.
Supplementary Material
Acknowledgments
This work was supported by grants from the Leukemia Lymphoma Society of Canada (LLSC) and the Cancer Research Society, Inc. (T.H.); by studentship awards from the Canadian Institutes for Health Research (to N.G. and M.T.), the Fonds de Recherche du Québec-Santé (FRQS) (to M.T.), and the Cole Foundation (to M.T.); by postdoctoral fellowship awards from the LLSC (to J.L.) and the Swiss National Foundation (to M.H.); as well as by Visiting Scientist awards from the FRQS (to Z.C. and S.-J.C.). The Institute for Research in Immunology and Cancer is supported by the Canada Foundation for Innovation, the FRQS, and the Networks of Centres of Excellence through the Centre of Excellence for Commercialization and Research program. The Banque de Cellules Leucémiques du Québec is supported in part by the Cancer network of the FRQS.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310067110/-/DCSupplemental.
References
- 1.de Thé H, Chen Z. Acute promyelocytic leukaemia: Novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10(11):775–783. doi: 10.1038/nrc2943. [DOI] [PubMed] [Google Scholar]
- 2.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93(10):3167–3215. [PubMed] [Google Scholar]
- 3.Scaglioni PP, Pandolfi PP. The theory of APL revisited. Curr Top Microbiol Immunol. 2007;313:85–100. doi: 10.1007/978-3-540-34594-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Kastner P, et al. Positive and negative regulation of granulopoiesis by endogenous RARalpha. Blood. 2001;97(5):1314–1320. doi: 10.1182/blood.v97.5.1314. [DOI] [PubMed] [Google Scholar]
- 5.Labrecque J, et al. Impaired granulocytic differentiation in vitro in hematopoietic cells lacking retinoic acid receptors alpha1 and gamma. Blood. 1998;92(2):607–615. [PubMed] [Google Scholar]
- 6.Du C, Redner RL, Cooke MP, Lavau C. Overexpression of wild-type retinoic acid receptor alpha (RARalpha) recapitulates retinoic acid-sensitive transformation of primary myeloid progenitors by acute promyelocytic leukemia RARalpha-fusion genes. Blood. 1999;94(2):793–802. [PubMed] [Google Scholar]
- 7.Licht JD, et al. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17) Blood. 1995;85(4):1083–1094. [PubMed] [Google Scholar]
- 8.Chen Z, et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12(3):1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jovanovic JV, et al. Development of real-time quantitative polymerase chain reaction assays to track treatment response in retinoid resistant acute promyelocytic leukemia. Front Oncol. 2011;1:35. doi: 10.3389/fonc.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boukarabila H, et al. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23(10):1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng GX, et al. Distinct leukemia phenotypes in transgenic mice and different corepressor interactions generated by promyelocytic leukemia variant fusion genes PLZF-RARalpha and NPM-RARalpha. Proc Natl Acad Sci USA. 1999;96(11):6318–6323. doi: 10.1073/pnas.96.11.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He LZ, et al. Two critical hits for promyelocytic leukemia. Mol Cell. 2000;6(5):1131–1141. doi: 10.1016/s1097-2765(00)00111-8. [DOI] [PubMed] [Google Scholar]
- 13.Ivins S, et al. Regulation of Hoxb2 by APL-associated PLZF protein. Oncogene. 2003;22(24):3685–3697. doi: 10.1038/sj.onc.1206328. [DOI] [PubMed] [Google Scholar]
- 14.McConnell MJ, et al. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol Cell Biol. 2003;23(24):9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeyati PL, et al. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18(4):925–934. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- 16.Guidez F, et al. RARalpha-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc Natl Acad Sci USA. 2007;104(47):18,694–18,699. doi: 10.1073/pnas.0704433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pabst T, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27(3):263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 18.Löwenberg B. Diagnosis and prognosis in acute myeloid leukemia—the art of distinction. N Engl J Med. 2008;358(18):1960–1962. doi: 10.1056/NEJMe0802379. [DOI] [PubMed] [Google Scholar]
- 19.Paz-Priel I, Friedman A. C/EBPα dysregulation in AML and ALL. Crit Rev Oncog. 2011;16(1-2):93–102. doi: 10.1615/critrevoncog.v16.i1-2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21(6):853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Rajotte D, et al. Contribution of both STAT and SRF/TCF to c-fos promoter activation by granulocyte-macrophage colony-stimulating factor. Blood. 1996;88(8):2906–2916. [PubMed] [Google Scholar]
- 22.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88(4):1234–1247. [PubMed] [Google Scholar]
- 23.Wang QF, Friedman AD. CCAAT/enhancer-binding proteins are required for granulopoiesis independent of their induction of the granulocyte colony-stimulating factor receptor. Blood. 2002;99(8):2776–2785. doi: 10.1182/blood.v99.8.2776. [DOI] [PubMed] [Google Scholar]
- 24.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Ford AM, et al. Regulation of the myeloperoxidase enhancer binding proteins Pu1, C-EBP alpha, -beta, and -delta during granulocyte-lineage specification. Proc Natl Acad Sci USA. 1996;93(20):10,838–10,843. doi: 10.1073/pnas.93.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khanna-Gupta A, Zibello T, Sun H, Gaines P, Berliner N. Chromatin immunoprecipitation (ChIP) studies indicate a role for CCAAT enhancer binding proteins alpha and epsilon (C/EBP alpha and C/EBP epsilon ) and CDP/cut in myeloid maturation-induced lactoferrin gene expression. Blood. 2003;101(9):3460–3468. doi: 10.1182/blood-2002-09-2767. [DOI] [PubMed] [Google Scholar]
- 27.Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol. 1996;16(9):4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JY, et al. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272(36):22,447–22,455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 29.Grondin B, et al. c-Jun homodimers can function as a context-specific coactivator. Mol Cell Biol. 2007;27(8):2919–2933. doi: 10.1128/MCB.00936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guidez F, et al. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood. 1998;91(8):2634–2642. [PubMed] [Google Scholar]
- 31.Melnick A, et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol Cell Biol. 2002;22(6):1804–1818. doi: 10.1128/MCB.22.6.1804-1818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doulatov S, et al. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 2009;23(17):2076–2087. doi: 10.1101/gad.1788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He LZ, et al. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18(2):126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 34.Spicuglia S, et al. Characterisation of genome-wide PLZF/RARA target genes. PLoS ONE. 2011;6(9):e24176. doi: 10.1371/journal.pone.0024176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vickers M, Jackson G, Taylor P. The incidence of acute promyelocytic leukemia appears constant over most of a human lifespan, implying only one rate limiting mutation. Leukemia. 2000;14(4):722–726. doi: 10.1038/sj.leu.2401722. [DOI] [PubMed] [Google Scholar]
- 36.Hoang T, et al. Opposing effects of the basic helix-loop-helix transcription factor SCL on erythroid and monocytic differentiation. Blood. 1996;87(1):102–111. [PubMed] [Google Scholar]
- 37.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1(2):138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 38.Lécuyer E, et al. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100(7):2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.