Abstract
The Ets-Related Gene (ERG) belongs to the Ets family of transcription factors and is critically important for maintenance of the hematopoietic stem cell population. A chromosomal translocation observed in the majority of human prostate cancers leads to the aberrant overexpression of ERG. We have identified regions flanking the ERG Ets domain responsible for autoinhibition of DNA binding and solved crystal structures of uninhibited, autoinhibited, and DNA-bound ERG. NMR-based measurements of backbone dynamics show that uninhibited ERG undergoes substantial dynamics on the millisecond-to-microsecond timescale but autoinhibited and DNA-bound ERG do not. We propose a mechanism whereby the allosteric basis of ERG autoinhibition is mediated predominantly by the regulation of Ets-domain dynamics with only modest structural changes.
More than 50% of all prostate cancers harbor a chromosomal translocation that results in the transcription of ERG, or “Ets-related gene,” being controlled by the promoter of androgen-regulated TMPRSS2 (1). This translocation leads to aberrant overexpression of nearly the entire ERG protein, including the DNA-binding domain, in the prostate epithelium. In combination with deletion of the Phosphatase and Tensin Homolog (PTEN) or up-regulation of the oncogene Akt, ERG overexpression induces a progression to prostate cancer (2). This combination of aberrant ERG expression, which blocks differentiation, and PTEN deletion, which enhances proliferation, is consistent with a “two-hit” model proposed for leukemia in which a hit on differentiation, often a transcription factor (such as RUNX, CBFβ, or MLL) cooperates with a hit on proliferation (often kinase signaling pathways such as KIT, FLT3, or Ras) to induce disease (3). Interestingly, fusions of ERG have also been identified in a subset of acute myeloid leukemia patients (TLS/FUS-ERG) (4) and in Ewing’s sarcoma (EWS-ERG) (5).
The Ets family of transcription factors to which ERG belongs is essential for numerous functions in normal hematopoiesis and immune development (6). ERG in particular is critical for maintenance of the hematopoietic stem cell (HSC) population. In a mouse model, the HSC population cannot recover after being challenged by sublethal radiation in heterozygous ERG animals, whereas the loss of both functional copies of ERG is embryonic lethal (7). Overexpression of the ERG gene product, located on chromosome 21, has also been implicated in the myeloproliferative disorders frequently seen in Down Syndrome patients (8), has been shown to have a dedifferentiating effect (9), and confers greater invasiveness to cell lines (10, 11). Importantly, the aberrant expression or deletion/loss of function of a single copy of the ERG allele can produce profound phenotypes (8, 12, 13), indicating that a change in ERG activity of as little as twofold compared with wild type can have a tremendous impact.
All Ets family proteins contain a highly conserved DNA-binding “Ets” domain, which recognizes a GGA(A/T) motif. A degree of specificity is conferred by bases flanking this core sequence, but multiple mechanisms have been identified as playing a role in the regulation of Ets protein activity. Some family members possess a second structured domain known as the pointed (PNT) domain that can, in some Ets proteins, oligomerize and alter DNA-binding affinity (14). In addition, it has been shown that protein–protein interactions and cooperative DNA binding (15, 16), posttranslational modifications (6, 17), and autoinhibition (18), have a role in regulating Ets protein activity.
We show that ERG DNA-binding is allosterically regulated by autoinhibitory regions both N- and C-terminally adjacent to the Ets domain. By solving the crystal structures of uninhibited, autoinhibited, and DNA-bound ERG and combining these with NMR- and electron paramagnetic resonance (EPR)-based dynamics data, we demonstrate that ERG DNA-binding affinity is altered through only moderate structural changes in the Ets domain. We propose a mechanism whereby a combination of slight conformational alterations and the restriction of Ets domain dynamics propagates this allosteric effect.
Results
Mapping of the Regions Mediating ERG Autoinhibition.
We first cloned and expressed seven ERG constructs in addition to the full-length protein, each containing the Ets domain (Fig. 1). We examined the binding affinity of each of these constructs for target DNA by isothermal titration calorimetry (ITC) using DNA containing a sequence based on one previously identified to be a high-affinity target of the Ets protein Fli-1, a relative of ERG bearing 96% sequence identity across the Ets domain (16). This DNA sequence contains a single copy of the canonical Ets-binding site GGAA at the center of the oligo, flanked by bases showing selectivity for Fli1 binding. All of our ITC experiments yielded isotherms indicating strong binding of protein to DNA (Fig. S1), and thermodynamic analysis indicated that ERG bound to DNA with a stoichiometry of 1:1 through an endothermic, entropically driven interaction (Table S1).
Fig. 1.
ERG protein constructs and ITC DNA-binding measurements. The seven truncation constructs of ERG are shown with corresponding residues numbered. The Ets DNA-binding domain common to all constructs is illustrated in light gray, flanked by the N- and C-terminal autoinhibitory domains in black. The PNT domain is shown in dark gray.
ITC results showed that full-length ERG binds its target DNA with a dissociation constant (KD) of ∼120 nM, whereas the Ets domain alone showed a lower KD of 37 nM (Table 1). This high-affinity construct consisting of residues 289–378 encompasses only the ERG Ets domain. We named this construct “ERGu,” because it was the minimal stable protein construct that showed uninhibited DNA binding activity. As we narrowed down our ERG constructs from the full-length protein, the KD peaked at 176 nM in a construct spanning residues 272–388, indicating nearly fivefold weaker binding compared with ERGu and 45% weaker than the full-length protein. We refer to this maximally inhibited construct as “ERGi.” These extra regions immediately flanking the Ets domain (the N-terminal inhibitory domain and C-terminal inhibitory domain, or NID and CID) both appear to be necessary for full autoinhibition of ERG. Altering the length of these sequences in any significant way, either by extending or truncating them, reduced the total effect of autoinhibition. NMR-based 15N-relaxation measurements (19) indicate a rotational correlation time (τc) of 7.6 ns for the 12-kDa ERGu construct, 9.4 ns for ERGi with a mass of 15 kDa, and 11.5 ns for the ERGi:DNA complex, which has a total combined mass of 23 kDa, all of which are consistent with monomeric species in solution.
Table 1.
ERG construct ITC results
| Construct | n | KD | Fold inhibition |
| 1–462 | 1.05 (±0.04) | 121 (±5) | 3.3 |
| 210–462 | 1.10 (±0.02) | 104 (±6) | 2.8 |
| 272–412 | 0.94 (±0.03) | 101 (±8) | 2.7 |
| 272–388 (ERGi) | 0.98 (±0.03) | 176 (±10) | 4.8 |
| 289–412 | 0.98 (±0.06) | 51 (±10) | 1.4 |
| 272–378 | 0.99 (±0.03) | 33 (±3) | 0.9 |
| 289–388 | 0.99 (±0.02) | 41 (±4) | 1.1 |
| 289–378 (ERGu) | 1.04 (±0.02) | 37 (±3) | 1.0 |
| ERGu Y354F | 1.00 (±0.03) | 106 (±6) | 2.9 |
| ERGi Y354F | 1.01 (±0.04) | 200 (±13) | 5.4 |
| ERGi S283A | 1.01 (±0.04) | 97 (±11) | 2.6 |
Stoichiometry, dissociation constant, and relative inhibition of ERG truncation constructs and point mutants. KD is given in nanomolar. Numbers in parentheses are ±SE.
EPR Spectroscopy Reveals Conformational Exchange in the N-Terminal Inhibitory Domain.
Although the ERGi protein is apparently structured and quite stable in solution, careful examination of the NMR spectra revealed that 12 nonproline residues corresponding to the NID could not be observed, although residues N- and C-terminal to these 12 were readily identified and unambiguously assigned. We suspected this line-broadening was due to conformational exchange and investigated this with EPR spectroscopy.
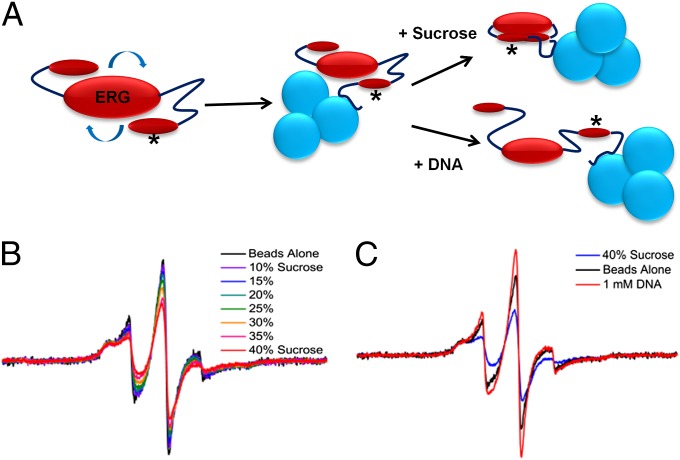
Because of the relatively small size of ERGi, EPR spectra of spin-labeled ERG were influenced by rapid rotational diffusion of the protein, producing narrow, motionally averaged lines and obscuring the effects of any local conformational equilibrium. To eliminate the effect of rotational diffusion, we devised a convenient method to slow the global correlation time. López et al. (20) have shown the utility of cross-linking a protein to a solid support in EPR. All of our ERG protein constructs were expressed with hexa-histidine tags for nickel resin purification, so we retained the histidine tag and bound the purified, (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate-labeled (MTSL) protein to nickel-nitrilotriacetic acid (Ni-NTA) beads to increase the correlation time. To observe the effects of various buffers on spin label motion, we loaded a slurry of protein-laden nickel beads directly into capillary tubes and proceeded with standard continuous wave EPR experiments (SI Materials and Methods).
This method proved to be highly effective. We tested spin labels at two different sites in the NID of ERGi: Tyr276 and Ala286. Both of these residues were mutated to cysteine for spin labeling, but the Y276C mutant was insoluble; therefore, we focused our experiments on the A286C mutant (denoted with an asterisk in Fig. 2A). When bound to beads, we saw broad hyperfine features corresponding to an incompletely averaged hyperfine interaction, as well as a narrow component corresponding to a more mobile motional mode for the spin-labeled side chain (Fig. 2B). These two-component spectra could arise either from two rotameric states of the spin-labeled side chain or from protein conformational exchange between ordered and disordered states; however, these spectra are sensitive to sucrose addition, indicating that they result from conformational equilibria and not multiple side-chain rotamers. The implication is that the peaks missing from our NMR spectra were due to conformational exchange (and were not a result of, for example, solvent exchange effects). The timescale of this motion is most likely on the order of hundreds of nanoseconds or slower, consistent with what we see by NMR (20, 21). Upon the addition of increasing concentrations of sucrose, the mobile population decreased, and addition of dsDNA to our EPR samples induced sharper lines (Fig. 2 B and C), indicating that although osmolyte favored the NID adopting a more ordered state, DNA shifted this equilibrium toward a more disordered state. This possible conformational exchange is also consistent with our preliminary NMR results, wherein we were able to make assignments of the missing NID residues when ERGi spectra were recorded in the presence of saturating DNA.
Fig. 2.
EPR experimental setup and results. (A) The rotational correlation time of the fast-tumbling ERG molecule can be greatly reduced by binding the His-tagged protein to Ni-NTA beads before EPR spectroscopy. Asterisk denotes site of MTSL spin label on the ERG NID. (B) Increasing sucrose concentration causes the ERG spin label spectrum to shift from sharp and highly mobile (black trace, no sucrose) to broad and immobile [red trace, 40% (wt/vol) sucrose solution]. (C) Solution containing a high concentration of sucrose shows a shift in the ERG NID toward the ordered state (blue trace), whereas the addition of DNA instead induces the sharper linewidths of the more disordered state (red trace).
Structure of the ERG Ets Domain.
We originally sought to determine the NMR solution structure of ERG, but crystallization trials set up in parallel with our initial NMR studies yielded crystals before the completion of an NMR-based structure determination. After obtaining our initial crystals, we were able to use our NMR data to optimize the protein constructs by removing excessively flexible residues from ERG, which improved both the size and diffraction quality of our crystals.
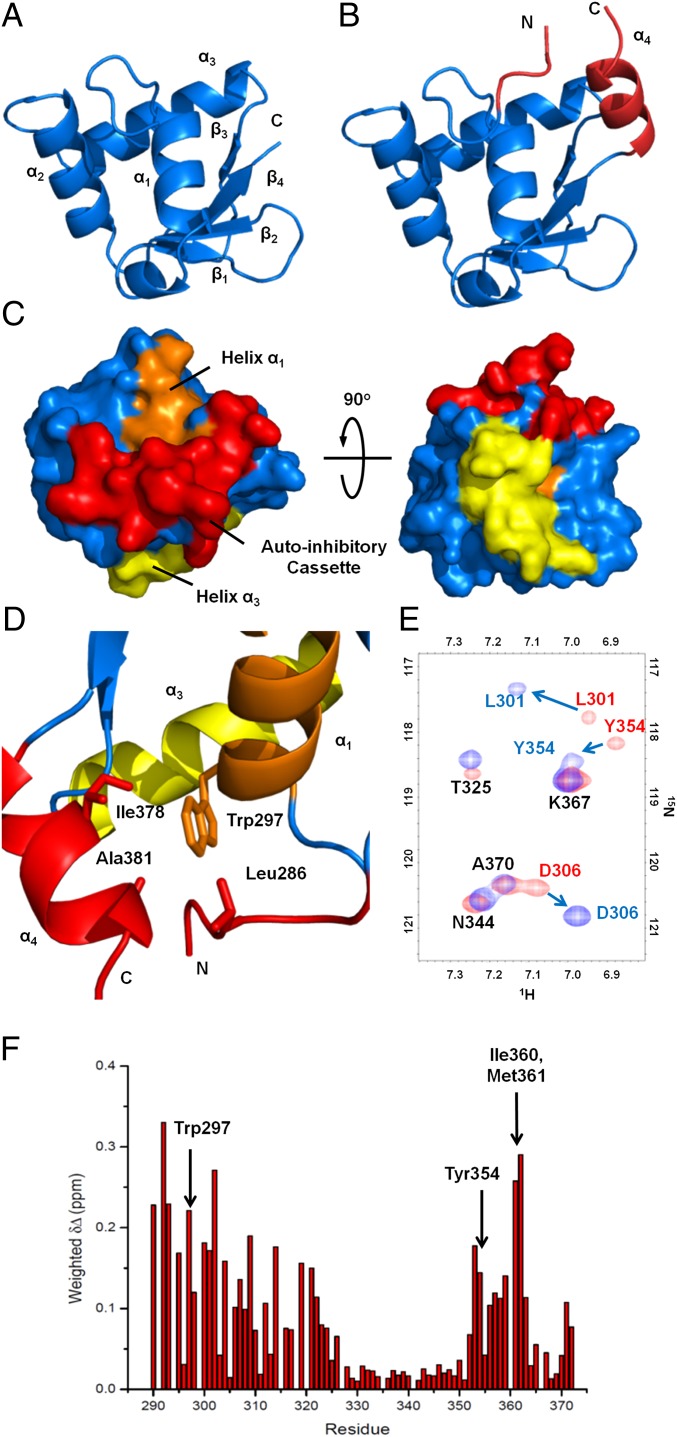
The minimal ERGu construct did not crystallize; however, we were able to grow large, high-quality crystals of ERG(289–388). Although ERG(289–388) contains the CID, it has a similar dissociation constant to the ERGu construct and is thus a good substitute for understanding the structure of ERGu. In addition, ERG(289–388) showed much improved solubility and stability in solution. This 1.7-Ǻ resolution structure is shown in Fig. 3A. For clarity, a vector-derived Ile-Arg-Gln peptide fragment and the CID are not displayed. The structure of the ERG Ets domain is quite similar to a number of other Ets domain proteins that have already been solved by NMR and X-ray crystallography methods, consisting of mixed α-helical/β-sheet content (18, 22–24) in what has been termed a winged helix-turn-helix motif. Residues Leu296–Ser305, Asp328–Lys338, and Tyr345–Lys358 form α-helices α1, α2, and α3, respectively, with a small helical turn between helix α1 and strand β1. The β-sheet consists of antiparallel strands β1–β4, spanning residues Ile313–Glu316, Glu321–Met324, Met361–Lys363, and Ala370–Phe373. Helix α3, which serves as the DNA recognition helix, is oriented across the back of the Ets domain with several residues key to the DNA interaction exposed, including Lys347, Arg350, Arg353, and Tyr354.
Fig. 3.
Uninhibited and autoinhibited ERG crystal structures and allosteric packing of the autoinhibitory cassette. (A) Structure of the uninhibited ERG Ets domain showing the canonical winged helix-turn-helix fold. For clarity, three N-terminal vector-derived residues and eight C-terminal residues have been omitted. (B) Structure of the autoinhibited ERGi construct, showing the Ets domain with appended autoinhibitory regions in red. (C) Surface representation of ERGi illustrating the packing of the autoinhibitory cassette (red) against helix α1 (orange). DNA-binding helix α3 (yellow) is not obscured by the autoinhibitory cassette. (D) Key residues in the autoinhibitory cassette–Leu286 in the NID, and Ala381 and Ile378 in the CID–pack around Trp297 of the ERG Ets domain. (E) 15N-1H HSQC spectra of 15N-labeled ERGi (blue) and ERGu (red). Residues in ERGi that see large changes from ERGu are labeled in colors corresponding to the respective spectra. (F) Net weighted chemical shift perturbations for residues in ERGu compared with ERGi.
ERG Autoinhibition Is Allosterically Modulated.
The structure of ERGi is remarkably similar to ERG(289–388). The overarching secondary structure of the core Ets domain is largely unchanged, with a total rmsd between the shared residues of 0.38 Ǻ. The ERGi structure reveals the CID of ERG to be a well-formed α helix (α4) appended directly to the C terminus of the Ets domain, containing residues Phe375–Gln383, whereas the NID adopts a largely random-coil conformation (Fig. 3B). Although the polypeptide construct extends beyond this structure, we could see no electron density for residues N-terminal to Ser283 or C-terminal to Pro384, most likely due to these residues being largely disordered.
Both the NID and CID pack primarily against the N-terminal end of helix α1 and each other, which serves to partially bury α1 in a hydrophobic bundle. Central to this bundle is Trp297 on α1, which is available to make van der Waals interactions with Leu286 of the NID, and Ile378 and Ala381 of the C-terminal autoinhibitory helix α4 (Fig. 3D). Interestingly, when the autoinhibitory cassette is folded in such a way, the DNA-binding surface of α3 is still exposed and available to interact with DNA (Fig. 3C), indicating that ERG autoinhibition is allosteric. This mechanism is similar to the autoinhibitory mechanism of Ets-1 (24), although the system is somewhat “stripped down” in ERG, which lacks N-terminal secondary structure elements.
To more closely examine the packing of the NID in solution, we compared the 15N-1H heteronuclear single-quantum coherence (HSQC) spectra of ERGi and ERGu. A number of peaks show substantial perturbations, some of which were quite dramatic (Fig. 3E). By calculating the average chemical shift differences, we identified regions of large peak movements at the N terminus of the Ets domain spanning residues Gly290–Thr325 as well as the very C terminus with residues Leu352–Phe373 (Fig. 3F). When we mapped these chemical shift differences onto the structure of the ERG Ets domain, we saw that they largely coincided with α1 and the adjacent 310 helix, the β-sheet including the near-side “wing” between strands β1 and β2, and the C-terminal end of α3 (Fig. S2). All of these regions fall on one face of the protein and overlay closely with the packing surface of the ERGi autoinhibitory cassette. Notably, Trp297 shows a net shift difference of >0.22 ppm, which we would expect given its close packing in ERGi compared with the solvent-exposed environment it experiences in ERGu. Those regions that are not in close proximity to the autoinhibitory cassette, including the majority of α3, the far wing between β3 and β4, and the entirety of α2 are left largely unperturbed, as are residues Leu296, Phe299, and Leu303, which lie on α1 but face away from the autoinhibitory packing site.
The ERG Autoinhibitory Cassette Rearranges upon DNA Binding.
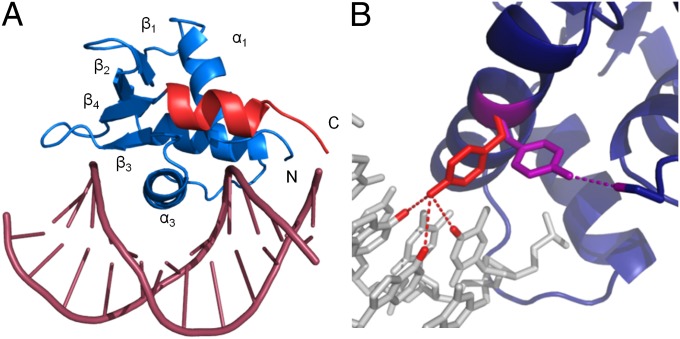
Using a combination of models from our structure of ERGi and a published structure of the c-fos promoter DNA (25), we solved the structure of ERGi in complex with a 12-bp DNA fragment containing the purine-rich Ets-binding site (Fig. 4A). As expected based on other Ets protein:DNA complexes (26, 27), helix α3 of ERG contacts the major groove of the DNA double helix, inducing a slight bend in the DNA strand. The core Ets domain of ERGi changes surprisingly little upon binding to DNA, with slight shifts occurring mainly in the wings of the β-sheet and the linker between α2 and α3.
Fig. 4.
Crystal structure of ERGi bound to DNA and change in Tyr354 conformation. (A) ERGi bound to a 12-bp DNA fragment. The Ets domain is shown in blue and the C-terminal autoinhibitory helix in red. No electron density was seen for the N-terminal autoinhibitory region in this structure. (B) Tyr354 on DNA-binding helix a3 in the presence of DNA (red) can form hydrogen bonds with DNA bases Ade7, Ade8, and Thy17. When not bound to DNA (purple), this residue adopts an alternative rotameric state and is positioned to hydrogen bond to Ser283 of the NID, or perhaps other protein binding partners.
Moderate movement occurs in the CID, as α4 positions itself downward toward the DNA with an associated rmsd of more than 1.9 Ǻ for the nonhydrogen atoms in residues 375–383 (Fig. S3). Notably, there was no detectable electron density for any of the NID when bound to DNA.
Another intriguing difference between bound and unbound ERGi is the positioning of Tyr354. This residue lies on the solvent-exposed surface of α3 and makes a key direct interaction with DNA by hydrogen bonding with Ade7, Ade8, and Thy17 in the GGAA motif. In unbound ERGi, this residue adopts a different rotamer that shifts the side-chain phenol nearly 90° away from a DNA-facing conformation and into a position where it can hydrogen bond with Ser283 of the NID (Fig. 4B). The NMR data shows that Tyr354 displays a noticeable chemical shift perturbation (Fig. 3 E and F) in the presence or absence of the autoinhibitory domain, as would be expected from such a large conformational change in the aromatic side chain. To test the contribution of this interaction to DNA-binding affinity, we introduced a point mutation at Tyr354, and at the hypothesized hydrogen-bonding partner, Ser283. As expected, our Y354F mutant greatly weakened the binding of ERGu to DNA, increasing the KD by 186% from 37 nM to over 100 nM (Table 1). Binding is weaker in ERGi Y354F as well, although by a much smaller margin of 176 nM vs. 200 nM, a change of only 14%. Conversely, mutation of Ser283 to alanine in ERGi resulted in tighter DNA binding of 97 nM.
NMR Dynamics Measurements Probe Protein Backbone Motion.
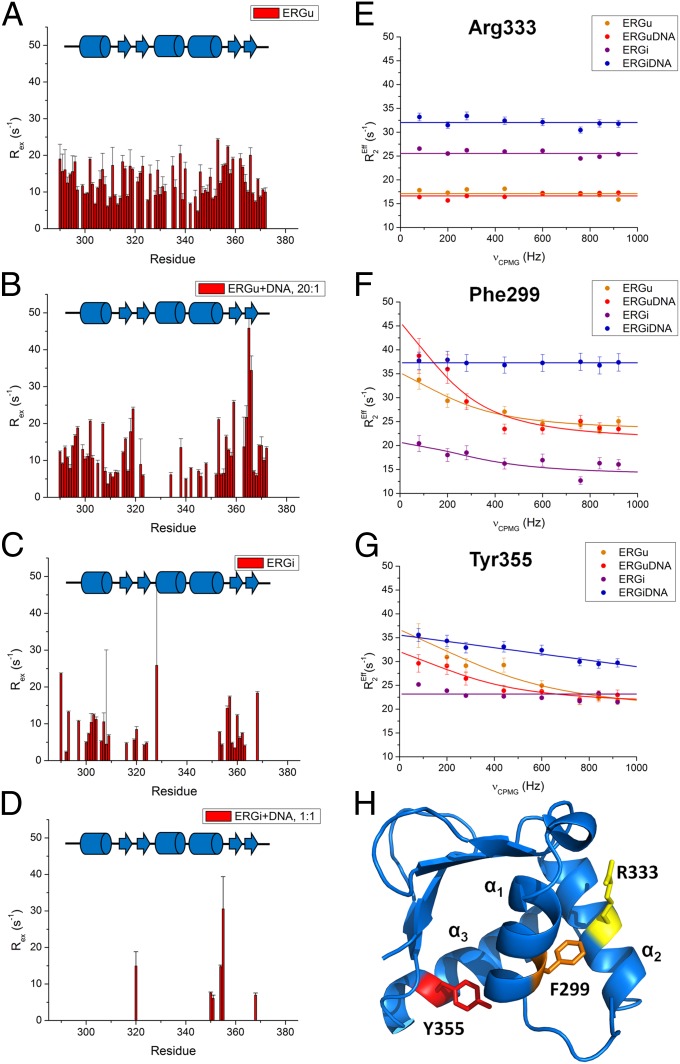
To explore the solution dynamics properties of ERG, we conducted a suite of NMR backbone dynamics experiments for the backbone amides of ERGu, ERGi, and the ERGi:DNA complex. 15N-1H-heteronuclear nuclear Overhauser effect (hetNOE) data shows the core Ets domain to be well-structured (Fig. S4 A and B). The 15N R1R2 product identifies residues undergoing chemical exchange with no influence from motional anisotropy (28) unlike R2/R1, allowing us to identify residues with altered dynamic behavior (Figs. S4 C–H and S5). Consistent with our hetNOE data, residues in the core regions of all three samples were largely ordered with those in the very N- and C-termini showing very low R1R2 values, indicative of nanosecond-to-subnanosecond motion. In ERGu, residue Trp297, crucial to the packing of the autoinhibitory cassette, and neighboring Gln298 both show significantly increased R1R2 values associated with microsecond-millisecond time scale motion. Similarly, much of the C terminus of ERGu including the C terminus of α3 and β3 and β4 also have heightened R1R2 values, which peak at residues Ile360 and Met361 (Fig. S5A). These residues all cluster on the face of the protein where the autoinhibitory cassette packs on the Ets domain (Fig. S6A). In contrast, unbound ERGi does not show elevated R1R2 values on the packing surface of the Ets domain, and only slightly elevated values at residues Ser308, Leu348, Tyr356, and His365 (Figs. S5 and S6B). The ERGi:DNA complex also shows overall reduced R1R2 values similar to unbound ERGi, with some exceptions (Figs. S5B and S6C). Intriguingly, the NID of ERGi bound to DNA shows R1R2 values suggestive of conformational exchange on the microsecond-millisecond timescale, rather than the fast, subnanosecond motions that might be expected were this region completely unstructured (Fig. S5B).
To better quantify the conformational exchange in ERG, we recorded Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments on ERGu, ERGi, the ERGi:DNA complex, and a substoichiometric solution of ERGu:DNA. As shown in Fig. 5A, the majority of the Ets domain of ERGu experiences dispersion indicative of conformational exchange (Table S2). When combined with DNA, even at a low ratio of 20:1 (protein:DNA), we begin to see a reduction in exchange particularly for the region corresponding to helix α2, whereas regions on the face of the protein adjacent to the autoinhibitory cassette see increased Rex values (Fig. 5B). In ERGi, which includes the autoinhibitory domain (Fig. 5C), these values are further reduced, suggesting that the presence of the AID restricts conformational exchange of the Ets domain. Finally, we observed a drastic reduction in dispersion when ERGi is completely bound to DNA (Fig. 5D).
Fig. 5.
Relaxation dispersion plots for the Ets domain of ERGu, ERGi, and the ERGi:DNA complex. (A–D) Exchange values (REx) and the corresponding residues in ERGu, ERGu:DNA (20:1), ERGi, and the ERGi:DNA complex, respectively, recorded at 18.8 T. (D–F) Representative relaxation dispersion profiles for residues in ERG. (E) Arg333 is located in a region of the protein that does not show exchange in any of our ERG constructs and is illustrated here as a control. (F) Phe299 packs near the core of the AID and shows dispersion in the ERGu and ERGi constructs, but not when ERGi is bound to DNA. (G) Tyr355 lies on DNA-binding helix α3 and shows dispersion in unbound ERGu, but limited or no dispersion in DNA-bound or free ERGi, respectively. (H) Cartoon representation of our ERGu structure. Key residues described in E–G are highlighted in yellow (Arg333), orange (Phe299), and red (Tyr355).
On a local scale, we observed different behavior for different residues in the protein and changes in exchange at residues in key positions in the Ets domain between the four different samples. For Arg333 in helix α2, we see no relaxation dispersion for any of the ERG samples we tested (Fig. 5E). Because of spectral overlap in one or more of the samples examined, we chose to display data for Phe299, which is very near Trp297 in helix α1 at the AID-docking surface, as described above, and Tyr-355 in the DNA-binding helix α3 next to Tyr354, which could both be characterized for all four samples. Both of these residues show dramatically increased R2,Eff values at low νCPMG frequencies (vs. higher νCMPG values) in ERGu and the ERGu:DNA (20:1) sample compared with both ERGi and the ERGi:DNA complex (Fig. 5 F and G).
Discussion
Using a variety of biophysical methods, we have demonstrated that the DNA-binding activity of the Ets family transcription factor ERG is allosterically autoinhibited. By coupling structural and dynamic studies, we were able to explore the nature of this mechanism and show that ERG autoinhibition is a consequence of a combination of subtle conformational alterations and changes in protein dynamics. Although ERG autoinhibition has been biochemically characterized in a recent study (29), our current report presents structures of ERG and characterization of the molecular mechanism of autoinhibition. The changes in ERG activity we identified here, ∼fivefold weaker binding in ERGi and threefold weaker binding in full-length ERG, are highly relevant in a biological context, because a change in ERG expression of as little as 50% is sufficient to alter angiogenesis (13), myeloproliferation (8), and maintenance of the hematopoietic stem cell population (7, 12).
Given the allosteric nature of this system and a lack of large structural alterations in the core Ets domain, we propose that the basis of ERG autoinhibition results from a combination of factors. The first component is Tyr354, which we identified in our crystal structures as adopting two rotameric states. In one state, this highly conserved tyrosine side chain is positioned to hydrogen bond with specific bases in the target DNA by orienting itself orthogonal to helix α3. When not bound to DNA, the Tyr354 hydroxyl can hydrogen bond to Ser283 in the NID, which pulls this side chain into an orientation less amenable to binding with this particular DNA sequence. Our calorimetry data show that the S283A mutation partially relieves autoinhibition, as we would expect if Ser283 was no longer available to interact with Tyr354. This concept is supported by our NMR data, because we see a large chemical shift change for Tyr354 in the uninhibited ERGu (which lacks Ser283) vs. ERGi.
It is also possible that the flexibility of the NID would allow it to be displaced entirely, possibly as a result of DNA binding as we see in the structure of our protein:DNA complex, or perhaps by interactions with other binding partners. Indeed, a similar mechanism has been identified in the cooperative binding of the transcription factors Ets-1 and Pax5 to DNA. In a previously published structure, the equivalent residue to Tyr354 in Ets-1 was found to hydrogen bond to Gln22 of Pax5, inducing a rotameric change almost identical to that which we have identified in ERG. By shifting this tyrosine to a different rotamer, which is less favorable to interact with the high-affinity Ets binding site, however, Pax5 allows this residue to form a different network of van der Waals interactions that improve the binding of Ets-1 to a suboptimal DNA sequence. Mutation of Gln22 to alanine in Pax5 disrupts this interaction (6), just as we see with our S283A mutation.
In the case of ERG, the role of Pax5 Gln22 is fulfilled intramolecularly by the NID of ERG, but it is possible that cooperative binding with other proteins that interact with Tyr354 could also be a major factor in determining ERG DNA targets. Several DNA-binding proteins, including the androgen receptor and the transcription factor RUNX1 (also known as AML1), have recently been shown to interact with ERG (6, 30). Even PU.1, which is one of only three Ets family proteins not having this conserved tyrosine (substituted with an asparagine that could also form hydrogen bonds), has been shown to interact with Bcl6 (31). Thus, regulation mediated through interaction partners could be a feature of Ets transcription factor activity in general.
The second major contribution to ERG autoinhibition is rooted in protein dynamics. As discussed above, the C-terminal portions of uninhibited ERGu experience extensive millisecond-microsecond time scale backbone dynamics, particularly on the face of the protein where the autoinhibitory cassette docks. In the presence of the NID and CID, the conformational exchange processes vanish, indicating that in the autoinhibited state, this dynamic regime is largely quenched. Because we see so few structural changes between autoinhibited and uninhibited ERG, we would expect that this dynamic behavior of the Ets domain is critical to ERG’s DNA-binding activity. Indeed, such a relationship has been suggested to be important to most, if not all, protein–DNA interactions (32).
Dynamics may also explain why our S283A ERGi mutation shows only partial release of autoinhibition. Although the nonnative alanine allows Tyr354 to return to a conformation more favorable to DNA binding, the autoinhibitory cassette is present and can quench the necessary conformational sampling required for DNA binding. By mutating Tyr354 to phenylalanine in ERGu, which lacks the autoinhibitory cassette altogether, we see weaker DNA binding than wild-type ERGu due to the loss of the hydrogen bonding network in which this residue participated. However, the dynamics of the Ets domain remain unchanged, thus binding is not as weak as wild-type ERGi. Along these same lines, our Y354F mutation in ERGi shows the weakest binding of all because it can exploit neither the hydrogen bond interactions bestowed by Tyr354 nor the increased dynamics allowed by removal of the autoinhibitory regions.
The frequent occurrence of aberrant ERG expression in a variety of cancers has stimulated a great deal of study of this transcription factor, with much of the interest focusing on the potential of ERG as a diagnostic (33). Until now, however, the molecular mechanisms of ERG function were poorly understood. With our exploration of the structure and dynamics of ERG function, it may be possible to not only better understand the role ERG plays in the regulation of hematopoiesis, but also the role of its misregulation in prostate cancer and leukemia.
Materials and Methods
Protein Expression and Purification.
Human ERG protein constructs were expressed in the pET-based pHis vector (34) for purification from Escherichia coli lysates by affinity chromatography. Isotopically labeled samples to be used for NMR studies and selenomethionine-labeled samples for crystallization were expressed as described in SI Materials and Methods. During the purification process, the protein was thoroughly washed with 1 M KCl to remove extraneous DNA fragments, and the affinity tag was removed by proteolysis before ion-exchange chromatography and storage at −80 °C until needed.
Oligonucleotides.
Single-stranded DNA oligonucleotides containing the 16-base sequence AGGACCGGAAGTAACT and its reverse complement (for calorimetry and NMR studies) or the 12-base sequence GACCGGAAGTGG and reverse complement (used in crystallographic studies) were annealed in-house and purified by ion-exchange chromatography.
ITC.
Calorimetry experiments were conducted at 290 K by using dsDNA at a concentration of ∼200 μM injected into 15 μM ERG. Experiments were performed in triplicate and corrected for dilution enthalpy. Raw data were processed in Origin and fit with a single-site binding model.
EPR Spectroscopy.
Protein samples to be used in EPR experiments were prepared as described in SI Materials and Methods. Protein-laden beads were loaded directly into capillary tubes, and spectra were recorded on a Bruker EMX X-band spectrometer by using an ER 4123D dielectric resonator. Spectra were collected with a 150-G scan width and a 1-G modulation amplitude. Spectra were baseline corrected and normalized by using the Bruker WinEPR software.
X-Ray Crystallography.
Purified ERG protein samples were set up in crystal screens by using the sitting-drop method. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. All data were indexed and processed by using HKL2000 (35); multiple rounds of model building and refinement were performed in Coot (36) and RefMac5 (37) as provided in the CCP4 program suite (38). Complete structural statistics are listed in Table S3.
NMR Spectroscopy.
Protein samples were exchanged into buffer containing 200 mM MgSO4, 20 mM KPi, 5 mM DTT, 0.01% sodium azide, and 5% (vol/vol) D2O at a final pH of 6.0 for ERG alone, or the same buffer lacking MgSO4 for samples containing the ERGi:DNA complex at pH 6.9. Protein backbone dynamics data were gathered from 15N {1H} NOE, 15N T1, and 15N T2 experiments by using the set of relaxation delays listed in SI Materials and Methods at 14.1 T. T1 and T2 relaxation rates were calculated by using SPARKY. Weighted chemical shift changes in parts per million were calculated by using the equation (39): δΔ = (((0.2 × ΔN)2 + ΔH2)/2)1/2. R1R2 and δΔ graphs were generated by using OriginLab software. Relaxation dispersion experiments were run at 18.8 T by using a CPMG delay of 50 ms as described in SI Materials and Methods. Errors were calculated based on an estimate of signal/noise by using absolute data heights at 10,000 points in each spectrum.
Supplementary Material
Acknowledgments
We thank Dr. Zygmunt Derewenda for the use of his crystallography facilities and for his time and advice in our initial crystallography experiments, Dr. David Cooper for assistance in crystallization strategies and data refinement, and Dr. Michael Wiener for the use of his crystallography equipment and assistance from his postdoctoral fellows. This work was supported by Department of Defense Grant PC060195 and a University of Virginia Cancer Center grant. Use of the Advanced Photon Source was supported by the Department of Energy, Office of Basic Energy Sciences, under Contract W-31-109-Eng-30.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4IRG, 4IRG, and 4IRI) and the NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 19136, 19137, and 19138).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301726110/-/DCSupplemental.
References
- 1.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Squire JA. TMPRSS2-ERG and PTEN loss in prostate cancer. Nat Genet. 2009;41(5):509–510. doi: 10.1038/ng0509-509. [DOI] [PubMed] [Google Scholar]
- 3.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 4.Hsu T, Trojanowska M, Watson DK. Ets proteins in biological control and cancer. J Cell Biochem. 2004;91(5):896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Codrington R, et al. The Ews-ERG fusion protein can initiate neoplasia from lineage-committed haematopoietic cells. PLoS Biol. 2005;3(8):e242. doi: 10.1371/journal.pbio.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng AP, et al. Erg is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. Blood. 2011;118(9):2454–2461. doi: 10.1182/blood-2011-03-344739. [DOI] [PubMed] [Google Scholar]
- 8.Ng AP, et al. Trisomy of Erg is required for myeloproliferation in a mouse model of Down syndrome. Blood. 2010;115(19):3966–3969. doi: 10.1182/blood-2009-09-242107. [DOI] [PubMed] [Google Scholar]
- 9.Tsuzuki S, Taguchi O, Seto M. Promotion and maintenance of leukemia by ERG. Blood. 2011;117(14):3858–3868. doi: 10.1182/blood-2010-11-320515. [DOI] [PubMed] [Google Scholar]
- 10.Zong Y, et al. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci USA. 2009;106(30):12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klezovitch O, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105(6):2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loughran SJ, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9(7):810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds LE, et al. Tumour angiogenesis is reduced in the Tc1 mouse model of Down’s syndrome. Nature. 2010;465(7299):813–817. doi: 10.1038/nature09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green SM, Coyne HJ, 3rd, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J Biol Chem. 2010;285(24):18496–18504. doi: 10.1074/jbc.M109.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21(15):1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szymczyna BR, Arrowsmith CH. DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein-DNA recognition. J Biol Chem. 2000;275(37):28363–28370. doi: 10.1074/jbc.M004294200. [DOI] [PubMed] [Google Scholar]
- 17.Lee GM, et al. The affinity of Ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J Mol Biol. 2008;382(4):1014–1030. doi: 10.1016/j.jmb.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee GM, et al. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem. 2005;280(8):7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 19.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 20.López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18(8):1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy J, Hubbell WL. High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc Natl Acad Sci USA. 2011;108(4):1331–1336. doi: 10.1073/pnas.1017877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coyne HJ, 3rd, et al. Autoinhibition of ETV6 (TEL) DNA binding: Appended helices sterically block the ETS domain. J Mol Biol. 2012;421(1):67–84. doi: 10.1016/j.jmb.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang H, et al. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat Struct Biol. 1994;1(12):871–875. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- 24.Jonsen MD, Petersen JM, Xu QP, Graves BJ. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16(5):2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo Y, Vaessen B, Johnston K, Marmorstein R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: Insights into DNA sequence discrimination by Ets proteins. Mol Cell. 1998;2(2):201–212. doi: 10.1016/s1097-2765(00)80130-6. [DOI] [PubMed] [Google Scholar]
- 26.Pio F, et al. New insights on DNA recognition by ets proteins from the crystal structure of the PU.1 ETS domain-DNA complex. J Biol Chem. 1996;271(38):23329–23337. doi: 10.1074/jbc.271.38.23329. [DOI] [PubMed] [Google Scholar]
- 27.Babayeva ND, Baranovskaya OI, Tahirov TH. Structural basis of Ets1 cooperative binding to widely separated sites on promoter DNA. PLoS ONE. 2012;7(3):e33698. doi: 10.1371/journal.pone.0033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc. 2002;124(9):1852–1853. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- 29.Gangwar SP, Dey S, Saxena AK. Structural modeling and DNA binding autoinhibition analysis of Ergp55, a critical transcription factor in prostate cancer. PLoS ONE. 2012;7(6):e39850. doi: 10.1371/journal.pone.0039850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens JHA, et al. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood. 2012;120(19):4038–4048. doi: 10.1182/blood-2012-05-429050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei F, Zaprazna K, Wang J, Atchison ML. PU.1 can recruit BCL6 to DNA to repress gene expression in germinal center B cells. Mol Cell Biol. 2009;29(17):4612–4622. doi: 10.1128/MCB.00234-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzeng S-R, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488(7410):236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 33.Tomlins SA, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3(94):94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15(1):34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grzesiek S, et al. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3(4):340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.