Abstract
Leishmaniasis, a parasitic disease caused by protozoa of the genus Leishmania, affects millions of people worldwide. Aminoglycosides are mostly known as highly potent, broad-spectrum antibiotics that exert their antibacterial activity by selectively targeting the decoding A site of the bacterial ribosome, leading to aberrant protein synthesis. Recently, some aminoglycosides have been clinically approved and are currently used worldwide for the treatment of leishmaniasis; however the molecular details by which aminoglycosides induce their deleterious effect on Leishmaina is still rather obscure. Based on high conservation of the decoding site among all kingdoms, it is assumed that the putative binding site of these agents in Leishmania is the ribosomal A site. However, although recent X-ray crystal structures of the bacterial ribosome in complex with aminoglycosides shed light on the mechanism of aminoglycosides action as antibiotics, no such data are presently available regarding their binding site in Leishmania. We present crystal structures of two different aminoglycoside molecules bound to a model of the Leishmania ribosomal A site: Geneticin (G418), a potent aminoglycoside for the treatment of leishmaniasis at a 2.65-Å resolution, and Apramycin, shown to be a strong binder to the leishmanial ribosome lacking an antileishmanial activity at 1.4-Å resolution. The structural data, coupled with in vitro inhibition measurements on two strains of Leishmania, provide insight as to the source of the difference in inhibitory activity of different Aminoglycosides. The combined structural and physiological data sets the ground for rational design of new, and more specific, aminoglycoside derivatives as potential therapeutic agents against leishmaniasis.
Keywords: Leishmania growth inhibition, ribosomal decoding site, X-ray analysis
Aminoglycosides (AGs) are well known broad-spectrum antibacterial agents, which inhibit bacterial growth by interfering with translation processes during protein synthesis. AGs were shown to bind bacterial ribosomes at a rather conserved rRNA helical core located at the top of helix 44 that comprises part of the ribosomal A site in the small ribosomal subunit (1). The molecular mechanisms of AG action in bacterial cells have been extensively investigated over the last decade; and it is well documented that their binding to the A site of bacterial ribosomes affects translation accuracy and reduces translational rate (1–4).
In addition to the aforementioned studies on prokaryotes, the action of AGs is not limited to bacterial ribosomes. Indeed, AGs were long shown to interfere with several aspects of eukaryote translation. Such works highlighted the potential of some AGs for treating nonsense mediated human genetic disorders by encoding near-cognate tRNA molecules at premature termination codon positions (5–7). Other studies have shown that specific AGs can be used as alternative treatments for infections caused by human parasitic protozoa such as trypanosomiasis, giardiasis, amoebiasis and leishmaniasis (8–12). Nevertheless, despite the great potential use of AGs for additional therapeutic purposes, very little information is available regarding their molecular mechanisms of action in eukaryotes.
Leishmaniasis is a spectrum of diseases caused by parasitic protozoa belonging to the genus Leishmania. It affects millions of people worldwide, appearing mainly in tropical and subtropical areas (12). The parasite is transmitted by sandflies and, depending on the type of disease, can be fatal if untreated. The current state-of-the-art in treating leishmaniasis is based on chemotherapy using a limited array of drugs such as antimony containing agents, amphotericin B, and recently Miltefosine (12). However, due to the emergence of pronounced parasite drug resistance in some regions, relatively high costs, and/or the severe toxic effects, there has been an extensive search over the last few years for new therapeutic agents. Paromomycin, a clinically approved AG for the treatment of various bacterial and parasitic infections, is the major component of a topical ointment (Leishcutan) used to treat cutaneous leishmaniasis caused by several species of parasites, and attempts have been made to further improve existing formulations (13–15). Paromomycin is also effective against visceral leishmaniasis, the fatal form of this disease (16), and it is registered in India and Nepal. Clinical trials using Paromomycin in combination with other antileishmanial drugs are underway to prevent development of parasite resistance (17).
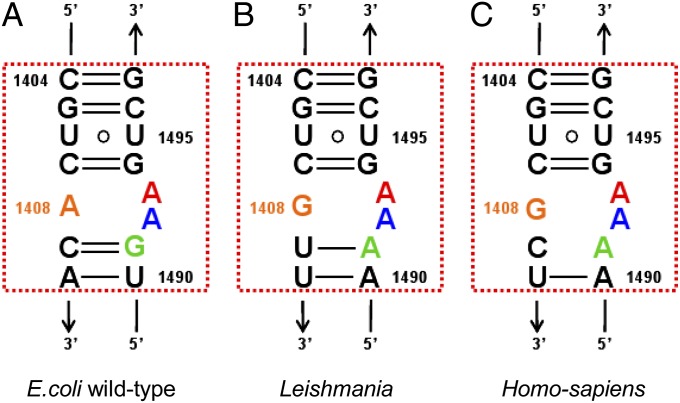
Despite the focus on AGs as antileishmanial agents their mechanism(s) of action in these parasites are still rather obscure. However, the rRNA A site is highly conserved among organisms belonging to all five kingdoms of life (Fig. 1), and together with recent evidence showing that AGs interfere with the leishmanial protein synthesis machinery (9, 18, 19), this implies that the putative binding site of AGs in Leishmania is similar to that present in bacteria; namely, the leishmanial A site. Recent works by Kondo and coworkers (10, 20) described the binding pattern of several natural and semisynthetic AGs to an A1408G mutant Mycobacterium A-site construct. These works demonstrate that the similar interaction patterns, as well as A-site conformational changes, occur upon AGs binding to wild-type and mutant bacterial strains. Due to higher sequential similarity between the mutant bacterial A sites and several eukaryotic parasites, the authors implied that a similar mode of interaction exists in protozoa.
Fig. 1.
Secondary structures of the bacterial (A), leishmanial (B), and human (C) cytoplasmic A sites. Orange, A/G1408; green, G/A1491; blue, A1492; red, A1493. The rRNA residues are numbered according to the numbering used in E. coli 16S rRNA.
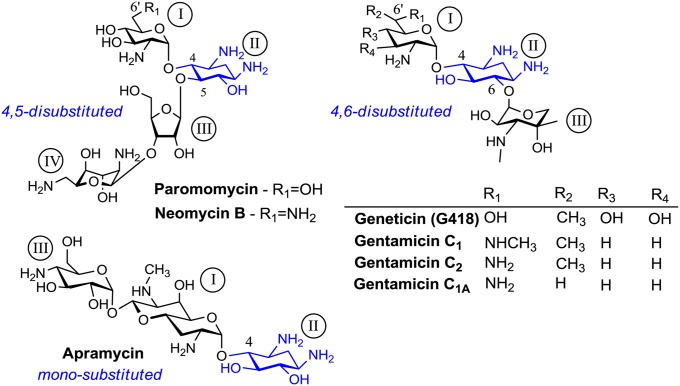
In the present study, we show the crystal structures of two AGs, Geneticin (G418) and Apramycin (Fig. 2), bound to rRNA duplexes mimicking their putative leishmanial ribosome binding site at 2.65-Å and 1.40-Å resolutions, respectively. The crystallographic studies imply for a great similarity in the decoding mechanisms of leishmanial and bacterial ribosomes; along with a rather distinct mechanism of action upon binding of AGs that differ at their substitution pattern around the conserved aminocyclitol–2-deoxystreptamine (2-DOS) ring (ring II). Our Leishmania susceptibility assays indicate that the induced conformational change upon AGs binding is highly important for their antiparasitic activity. These studies set ground for the understanding the decoding mechanism, as well as other mechanisms of AG action in eukaryotes at the molecular level.
Fig. 2.
Chemical structures of aminoglycosides used in this study. The common 2-deoxystreptamine ring (ring II) along with the substitution patterns are highlighted in blue.
Results
Leishmania A Site and AGs: An Overview.
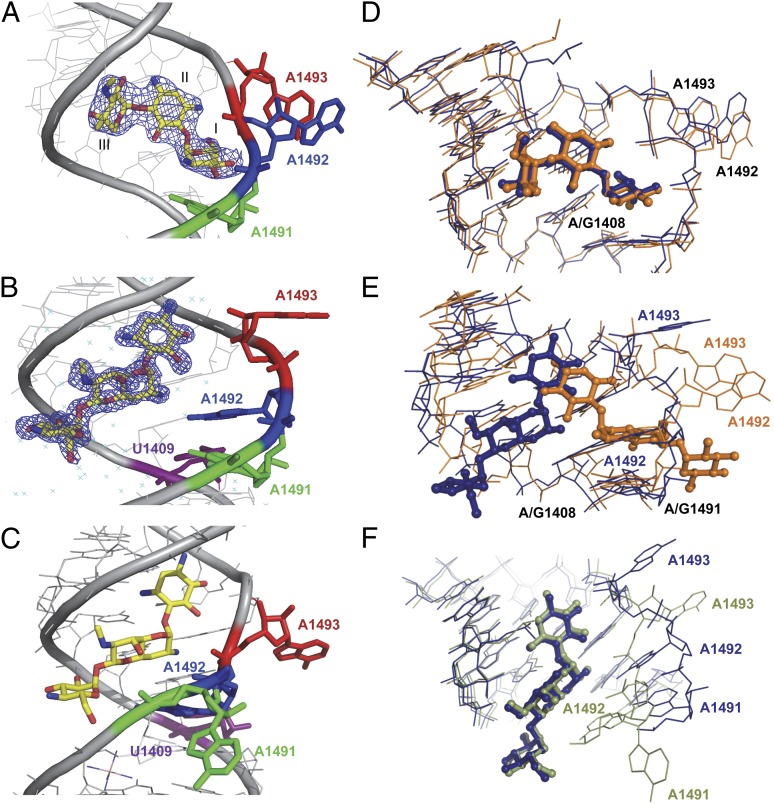
To explore the interactions of AGs with their putative leishmanial binding site, we determined the 3D structures of two AG representatives, G418 and Apramycin (Fig. 2), bound to rRNA duplexes that mimic their putative parasite binding site at 2.65-Å and 1.4-Å resolutions, respectively (G418–Leish, Apra–Leish; Fig. 3 and Figs. S1 and S2). The rRNA constructs contain two A sites corresponding to the sequences of leishmanial A site separated by four G–C pairs (Fig. S1). Similar constructs representing bacterial and human A sites have been previously used to explore AGs binding pattern within the A site by crystallographic means (21–23).
Fig. 3.
Crystal structure visualization of G418 (A) or Apramycin (yellow; B) leishmanial A-site rRNA complexes. Electron density 2Fo-Fc maps are contoured at 1.0σ around the AG. (C) A representation of Apramycin bound to the human A-site construct (PDB ID code 2G5K). A1491 is highlighted in green, A1492 in blue, A1493 in red and U1409 in purple. (D) Superimposition of G418 bound to the bacterial A site (orange) and the leishmanial A site (blue). G418 is highlighted in ball-and stick representation. Superimposition was performed using the PyMol software align algorithm using all identical atoms (rmsd, 0.9 Å). PDB ID codes are 1MWL and 4K32 for the bacterial and Leishmania structures, respectively. (E) Superimposition of Apramycin (ball and stick) bound to the bacterial (orange) and leishmanial (blue) A sites (rmsd, 2.2 Å). PDB ID codes are 1YRJ and 4K31 for the bacterial and Leishmania structures, respectively. (F) Superimposition of Apramycin (ball and stick) bound to the human cytoplasmic (green) and leishmanial (blue) ribosomal A sites (rmsd, 1.4 Å). PDB ID codes are 2G5K and 4K31 for the human and Leishmania structures, respectively. In all figures A/G1491, A1492, A1493, and A/G1408 are highlighted in the relevant colors.
Based on its primary sequence, the leishmanial A site is quite distinct from the conserved bacterial one. As can be seen in Fig. 1, 5 out of 15 nucleotides located in positions 1408–1410 and 1490–1491 are different. However, our results indicate that despite the differences in the sequences between the two A sites, the overall geometric features dictating their 3D structures are rather conserved. In both A sites, two canonical base pairs are obtained between four nucleotides located in positions 1409:1491 and 1410:1490; therefore, the resulting helical core is composed of three noncanonically paired nucleotides of which two are highly conserved (A1492, A1493) and one, located at position 1408, that is varied (A in bacteria vs. G in Leishmania). This geometric similarity does not come as a surprise, because both sites are believed to be an important molecular switch dictating the tRNA recognition at the ribosomal A site. However, because AGs mostly interact with the nucleotide bases, and there are significant differences in sequence, we hypothesized that the obtained interaction patterns and conformational changes induced by different AGs might differ in Leishmania from the ones present in bacteria.
Overall, in both the G418–Leish and Apra–Leish structures, a single AG molecule was found to specifically interact with the deep/major groove of each A site (Fig. S1). In the Apra–Leish structure, two additional Apramycin molecules were found to interact with the G–C pairs region connecting the two putative binding sites present in the rRNA model (Figs. S1B and S3). Similar nonspecific interactions with the nonnatural constructed G–C pair region were previously reported in the studies of both natural and semisynthetic AG derivatives in complex with human and bacterial A-site constructs (24–26). In addition, in various crystal structures of human and bacterial models crystallized in the presence of Cobalt Hexammine (Co-Hex), two Co-Hex molecules were shown to occupy the very same positions as those obtained for the extra two Apramycin bound molecules in our leishmanial crystal structure (21, 27, 28). However, despite the fact that both ligands (G418 and Apramycin) were shown to bind the same region of their primary binding site, the two ligands demonstrated a significantly different interaction pattern with the rRNA construct, and thus each AG induces a different conformational change to the A site. The conformational change induced by G418 is similar to that already observed for the bacterial A site, where two conserved adenine residues, A1492 and A1493 (using the nucleotide numbering of Escherichia coli), are fully bulged out from the helical core (Fig. 3 A and D). In contrast, Apramycin induces a rather different conformation, in which the A1492 base moiety is sequestered within the helical core, forming a pair with G1408, whereas A1493 is bulged out (Fig. 3B and Fig. S1).
The Binding Mode of G418 to the Leishmanial A Site: “ON” State.
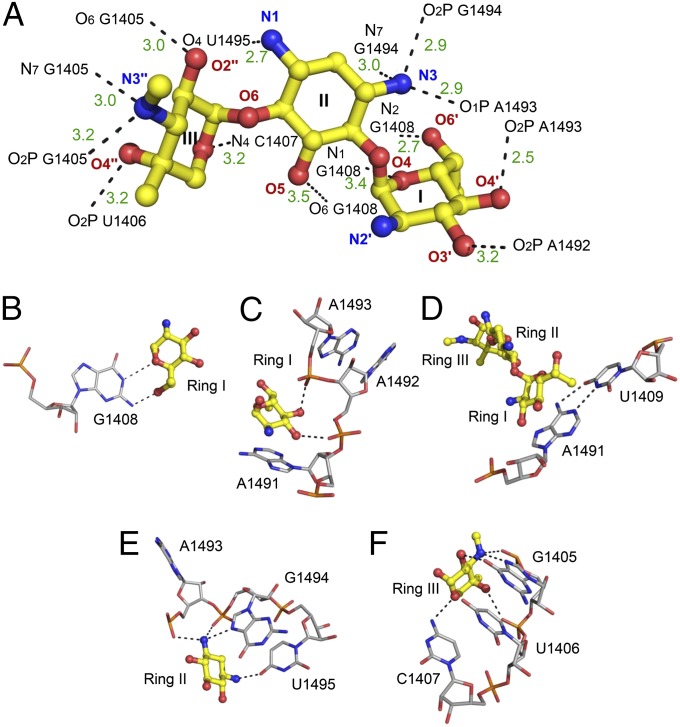
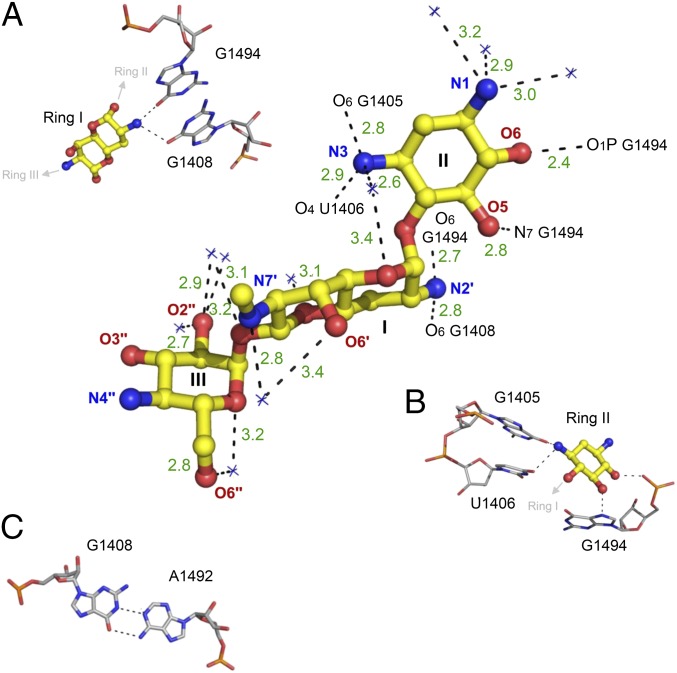
Upon binding to the deep major groove of the leishmanial A site, G418 forms 14 electrostatic contacts with the rRNA bases and phosphate oxygen atoms (Fig. 4). Ring I forms a pseudo base pair with G1408, by forming two hydrogen bonds involving the ring oxygen and the 6′-OH group (Fig. 4B). The planar nature of these interactions is dominated by stacking interactions of ring I and A1491 (Fig. 4C). On the opposite side of ring I, two OH groups located at the 3′ and 4′ positions make additional contacts with the oxygen atoms belonging to the phosphate backbone of the two conserved adenine residues, A1492 and A1493 (Fig. 4C). These interactions prevent the two adenine residues from flipping back inside the helical core. This interaction is therefore deemed to be highly important for the induction of the flipped out conformation of A1492 and A1493. The chiral exocyclic methyl group at 6′ position, (R)-6′-Me, points in the middle of the A1491–U1409 pair (Fig. 4D); thus adding some hydrophobic stabilization as the one observed upon G418 ring I interaction with the G1491–C1409 pair in bacteria (23). Ring II interacts with three sequential residues A1493, G1494, and U1495 (Fig. 4E). Due to the high conservation of these residues, similar interactions were previously highlighted in the exploration of wild-type and mutant bacterial A sites in complex with AGs containing a 2-DOS ring (20, 23). Ring III forms five hydrogen bonds with G1405, U1406, and C1407; these interaction are typical to 4,6-disubstituted AGs containing a garoseamine ring (ring III) at position 6 (Fig. 4F). Similar interactions were previously reported for the interaction of 4,6- disubstituted garoseamine derivatives such as Gentamicin C1A and G418 with bacterial A sites (23, 24).
Fig. 4.
Description of the contacts between G418 and the leishmanial A site. (A) The 3D structure of bound G418. Ring numbers (I–III) and atom names are specified. rRNA atoms are numbered according to the E. coli numbering. Hydrogen bonds and salt bridges are presented as black dashed lines. Bond lengths are presented in dark green in ångström (Å). (B–F) The atomic details of the contacts involving the rings with conserved and nonconserved rRNA residues.
Superposition of the G418–Leish structure with the crystal structure of G418 bound to the bacterial A site (G418–Bact) indicated a rather similar binding mode with rmsd values of 0.9 Å (Fig. 3D). The overall conformation of the binding site upon G418 binding was nearly identical to the one present in the bacterial A site, where the two conserved adenines 1492 and 1493 are flipped out from the helical core prone to interact with both the mRNA and the tRNA codon:anticodon complex. This conformational state was earlier referred to as an ON state (27, 28) and is known to mimic the conformational change resulting from the complexation of a cognate aminoacylated-tRNA molecule and the mRNA at the bacterial ribosomal A site; thus, signaling for the continuation of translation process. The induced conformational change upon AGs binding to bacterial ribosomes is believed to play a major role in their ability to induce translational miscoding (24). The structural similarity between the two structures (G418–Leish and G418–Bact) together with documented evidence for AGs such as Paromomycin enhancing miscoding events in leishmanial ribosomes (18) might suggest a similar mechanism of action in Leishmania.
The Binding Mode of Apramycin to the Leishmania A-Site: “OFF” State.
In contrast to G418, Apramycin demonstrated a looser interaction network, maintaining only 6 direct contacts with the leishmanial A site (Fig. 5). Four of these contacts are mediated by ring II (2-DOS) that interacts with the RNA bases and phosphate backbones of the three conserved residues G1405, U1406, and G1494 (Fig. 5B). The two additional remaining bonds are between the 2'-NH2 group located at the bicyclic ring I to the O6 atoms of G1494 and G1408 (Fig. 5A). As can be seen from the superposition with the bacterial-A site–Apramycin complex (Apra–Bact) (Fig. 3E and Fig. S4) the binding pattern of Apramycin to the leishmanial A site is rather distinct from the one observed in bacteria (rmsd, 2.2 Å). As a matter of fact, when only the two ligand molecules are compared their atoms barely overlap; with the molecules oriented in a ∼180° horizontal flip one to another. As a result Apramycin induces a distinctive conformational change upon binding to the leishmanial A site; where only A1493 is flipped out from the helical core. A1492, which is directed toward the helical core forms pseudo canonical interactions with G1408 (Fig. 5C). A conformation where one of the two conserved adenines (A1492 and A1493) or both are directed toward the helical core was earlier referred as an OFF-state conformation (27, 28). This conformation usually occurs when an inappropriate or no tRNA molecule is present at the ribosomal A site; therefore it might signal for translational arrest.
Fig. 5.
Description of the contacts between Apramycin and the leishmanial A site. Ring numbers (I–III) and atom names are specified. rRNA atoms are numbered according to the E. coli numbering. Hydrogen bonds and salt bridges are presented as black dashed lines. Bond lengths are presented in dark green in angström (Å). (A and B) The atomic details of the contacts involving ring I (A) and the bicyclic ring II (B) with conserved and nonconserved rRNA residues. (C) Pseudo Watson–Crick interactions of A1492 and G1408.
Previously, two structures of Apramycin bound to different human cytoplasmic A-site constructs have been reported (21, 29). Interestingly, the binding pattern observed upon Apramycin binding to the leishmanial A site was found to be rather similar to the one observed in previous studies exploring the binding of Apramycin to human cytoplasmic double A-site constructs (superposition rmsd, 1.4 Å; ref. 21; Fig. 3 C and F). The human A site departs from the leishmanial one by a single nucleotide located in position 1409 (C1409 human vs. U1409 Leishmania; Fig. 1). However, although these two A sites share greater sequential similarity compared with the bacterial one, the geometry of the two A sites is more distinct due to the abrogation of the 1409:1491 Watson–Crick pair in the human A site; giving its helical core a more complex conformational arsenal. Nevertheless, despite the noticeable differences in the orientations of A1491–A1493, Apramycin binding and interaction with the leishmanial A site is nearly identical to the one observed in the human A site (Fig. 3F).
In Vitro Inhibition of Leishmania donovani and Leishmania major Promastigote Growth by AGs.
The results described above provide insight into the modes of interactions between AGs and RNA constructs derived from the leishmanial ribosomes. However, to date a comprehensive study comparing susceptibility to different AG derivatives was not carried out. To evaluate the effect of AGs on Leishmania growth, five representative derivatives were chosen. Paromomycin and Neomycin B were chosen as representatives of the 4,5-disubstituted 2-DOS group; G418 and Gentamicin as 4,6-disubstituted and Apramycin as monosubstituted. The representatives of each group differ by the nature of the substituent group at the 6′-position of ring I (Fig. 2), which is known to be an important component of selectivity toward eukaryotic species (30). Drug susceptibility was tested using two species, L. major and L. donovani, which induce cutaneous and visceral leishmaniasis in humans, respectively. The concentrations giving 50% of growth inhibition (LC50) are listed in Table 1. All AGs tested inhibited Leishmania growth in a dose-dependent manner. The LC50 values obtained for the 6′-OH AGs (Paromomycin and G418) were in the μM range, and showed good agreement with previous published values for these compounds (18, 35, 36). The obtained LC50 values for the 6′-NH2 derivatives (Neomycin and Gentamicin) were higher than those obtained for the 6′-OH derivatives, and are also in good agreement with previously reported work demonstrating the lower potency of Neomycin B, compared with Paromomycin, for treatment of leishmaniasis (18). These results are also correlated well with the fact that 6′-NH2 derivatives are less effective against AG resistant bacterial strains containing an A1408G substitution (30). Interestingly, Apramycin was the least effective compound and did not appear to inhibit Leishmania growth even at 2,000 μM. Moreover, it should be noted that all AGs tested were more effective against L. major growth than L. donovani; these differences might be attributed to the differences in the promastigote surface glycocalyx that affects AG permeability (36).
Table 1.
In vitro inhibition of Leishmania promastigotes by AGs along with their MIC values in E. coli
Discussion
Recent studies, demonstrating the potential of some AGs to influence translational processes of eukaryotes, highlighted these compounds as potential therapeutic candidates for the treatment of a wide variety of parasite related infections (8–12) and some genetic disorders caused by nonsense mutations (5–7). However, despite these promising clinical results, the mechanistic and structural information regarding AGs action in eukaryotes is currently very limited. In the present study, we deciphered the crystal structures of two AGs bound to their putative binding site in leishmanial ribosomes. Leishmania is a eukaryotic parasite causing mild to severe symptoms in humans, as well as dogs. It affects as many as 12 million people worldwide, with 1.5–2 million new cases each year (37). The leishmanial ribosome shares great similarity with human ribosomes, with only a single nucleotide differentiating between the AGs putative binding sites (U1409 in Leishmania vs. C1409 in human; Fig. 1); therefore, the results presented here not only shed light on the mechanistic parameters of AGs action against these protozoan parasites, but also serve as a potential model for the exploration of their binding mode to human ribosomes.
Our structural results demonstrate distinct modes of binding for the two AGs tested. G418 binding was similar to that present in bacteria and induced an ON-state conformation where the two conserved adenine residues, A1492 and A1493, are flipped out from the binding site (Figs. 3A and 4). In contrast, Apramycin demonstrated a rather unique binding pattern, inducing a different conformational change upon binding to the A site where only one adenine residue, A1493, is flipped out (Figs. 3B and 5). The leishmanial growth inhibition results showed an activity gap of at least 2 orders of magnitude between these compounds (Table 1), suggesting that the differences in induced conformational changes for the two compounds play an importance in their activity.
G418 Inhibits Leishmania Growth by Translatory Miscoding.
The structural data for the G418–Leish structure reveals considerable similarity to the binding pattern of disubstituted AGs in bacterial A sites (Fig. 3D). The induced conformational change upon G418 binding is nearly identical in both systems, possessing an ON-state conformation; therefore, mimicking the conformational effect upon cognate tRNA binding to the A site. These finding suggest that G418 has a similar mechanism of action for the two systems, altering the ribosomes ability to distinguish between cognate and near/noncognate tRNAs, thus resulting in multiple miscoding events and eventually leading to cell death. Although no direct evidence is currently available showing that G418 induces miscoding by leishmanial ribosomes, G418 has long been marked as a potent miscoding agent in eukaryotes (38). In addition, a recent work has demonstrated the ability of a structurally related derivative, Paromomycin, to induce miscoding in Leishmania (18). Our modeling data with Paromomycin indeed indicate that the two compounds share a rather great similarity in their binding mechanisms and both might cause the induction of an ON-state conformation upon binding (Fig. S5).
Our in vitro viability assays indicated that both Paromomycin and G418 are capable of inhibiting leishmanial growth in the low micromolar range (Table 1); these values are similar to the minimum inhibitory concentration values reported in bacterial systems for the two compounds (Table 1); thus adding an additional support to the suggested similarity in their mechanism of action in bacteria and Leishmania. However, the obtained results indicate that G418 is ∼10 times more potent than Paromomycin against the two Leishmania strains tested. Although no such potency gap for the two derivatives has been documented in wild-type bacterial strains, similar tendency has been reported in A1408G mutant bacteria (23), sharing greater binding site similarity with Leishmania. These results also correlate well with the higher miscoding (38) and premature termination codon suppressing potency reported for G418 over Paromomycin in eukaryotes (39).
Apramycin Induces an OFF-State Conformation.
In contrast to G418, Apramycin seems to bind the leishmanial A site in a rather distinct binding mode, where A1492 is directed toward the helical core and A1493 is partially flipping out. This conformational state has previously referred as OFF state and was suggested to describe a nondecoding ribosomal state, which usually occurs in the absence of a cognate tRNA molecule at the A site. These structural insights are in good agreement with a recent solution NMR structure indicating the lack of A1492 and A1493 destacking upon Apramycin’s binding to an rRNA bacterial A-site model (40). The results also correlate with some recent evidences for Apramycin’s ability to inhibit translocation in bacterial ribosomes, but not to induce bacterial miscoding (34, 40) or premature termination codon suppression in eukaryotes (26).
However, unlike in bacterial systems, where Apramycin is conserved as a potent antibacterial agent, our LC50 results in Leishmania indicated a rather low activity of the compound (LC50 > 2,000 μM; Table 1). These results correlate well with previous studies demonstrating low potency of Apramycin to A1408G or A1491G mutant bacteria (34); and could be explained either by the distinct A-site conformation observed in X-ray experiments designed to explore Apramycin’s binding pattern in bacteria (34, 41) (Fig. 3E), or by the possibility of alternative binding sites at the bacterial ribosome. The conformational alternatives suggested for Apramycin in bacterial systems cannot be obtained in the leishmanial A site due to the alternations in the rRNA sequence preventing several H bonds from forming, therefore limiting the binding affinity of such conformations.
Interestingly, the conformation obtained upon Apramycin’s binding to the leishmanial A site shares great similarity with Apramycin’s interactions with human ribosomes (Fig. 3 C and F), thus indicating a rather conserved interaction pattern in eukaryotes. Such similarity could be attributed to the presence of the highly conserved guanine residue in position 1408, maintaining a preserved interaction with the OH moiety located in the bicyclic ring I (Fig. 5A). The differences obtained between the conformational changes induced upon Apramycin binding to the two eukaryotic A sites probably result from the disruption of the 1409:1491 Watson–Crick pair in the human A site enhancing the mobility of A1491, therefore resulting in its further direct stabilization outside the helical core by a flipped out A1493 of a neighboring molecule within the crystal packing (Figs. 3 C and F and 5C) (21, 26). The recent indications of the rather low translation inhibition of human ribosomes by Apramycin (42), together with the obtained structural similarity in Apramycin’s binding to eukaryotic A sites, further support our bioactivity results indicating a low susceptibility of eukaryotic parasites to Apramycin.
In summary, the presented data show the mechanistic parameters governing the interactions of AGs in leishmanial ribosomes, and supplies the molecular explanation for their potent activity against Leishmania. The data also reveal that in contrast to bacterial systems, the substitution pattern of AGs around the 2-DOS ring affects their binding mode and might induce an entirely different conformational change upon binding. Our data also indicate that the induction of an ON-state conformation is highly important for AGs potency as anti-Leishmania agents; thus implying the significance of miscoding events in the killing mechanism. Due to the high similarity to human ribosomes, our Leishmania structural data also supply indirect evidence regarding AGs action in humans. The structures will be highly beneficial in the rational development of new derivatives as potential therapeutic agents to treat leishmaniasis and genetic disorders.
Materials and Methods
Crystallization.
RNA duplexes containing two leishmanial A-site internal loops were used as models in the crystallographic studies (Fig. S1). The duplexes were composed of two identical ssRNA corresponding to the following sequence: 5′-UUG CGU CGU UCC GGA AAA GUC GC-3′. Before crystallization, RNA solution containing 2 mM RNA in 100 mM sodium cacodylate (pH 7.0) and 25 mM NaCl was denaturated at 90 °C for 2 min followed by a slow cool-down (∼2 h) to 37 °C. Equal volumes of RNA and a solution containing 4 mM AG were mixed and incubated at 37 °C for 10 min. Crystallization experiments were performed at 20 °C using the hanging-drop vapor diffusion method. Droplets were prepared by mixing 1 μL of RNA/AG solutions and 1 μL of crystallization solutions containing 50 mM sodium cacodylate pH 7.0, 1 mM spermine tetrahydrochloride, 1–20% (vol/vol) 2-methyl-2,4-pentanediol (MPD) and 100–250 mM KCl or NaCl, and were equilibrated against 500 μL of reservoir solution containing 40% (vol/vol) MPD. Crystals emerged after 4–6 d.
Crystal Handling, Data Collection, Structure Determination, and Refinement.
Crystals were soaked in 40% MPD and flash frozen in liquid nitrogen. X-ray data were collected at ID-14–4 in c. Data processing and refinement were performed as described in the SI materials and methods and the crystallographic data are summarized in Table S1. The atomic coordinates and structure factors of G418 and Apramycin A-site complexes have been deposited in the Protein Data Bank (PDB) with ID codes 4K32 and 4K31, respectively.
Leishmania Cell Culture and Promastigote Viability Assay (LC50).
Two strains of Leishmania were used in the present work: L. donovani (MHOM/SD/1962/1S-Cl2d) and L. major (MHOM/IL/2003/LRC-L1025). Promastigotes were grown in complete Schneider’s Drosophila medium containing 20% FCS and antibiotics at 26 °C. Compounds were screened for leishmanicidal activity using an alamarBlue (AbD Serotec) viability assay essentially as described (43); modification and materials are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of beamline 14-4, European Synchrotron Radiation Facility, for provision of synchrotron radiation facilities and assistance. This work was supported by National Institutes of Health/National Institute of General Medical Sciences Grant 1 R01 GM094792-01 A1 and German-Israeli Foundation for Scientific Research and Development Research Grant G-1048-95.5/2009 (to T.B.); and Bill and Melinda Gates Foundation Global Health Program Grant OPPGH5336 (to C.L.J.). C.L.J. holds the Michael and Penny Feiwel Chair in Dermatology. M.S. acknowledges the Schulich Fellowship for PhD students. This paper forms part of the PhD thesis of M.S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4K31 and 4K32).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307365110/-/DCSupplemental.
References
- 1.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407(6802):340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 2.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292(5518):897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 3.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111(5):721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 4.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7(2):104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 5.Bidou L, Allamand V, Rousset JP, Namy O. Sense from nonsense: Therapies for premature stop codon diseases. Trends Mol Med. 2012;18(11):679–688. doi: 10.1016/j.molmed.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Hainrichson M, Nudelman I, Baasov T. Designer aminoglycosides: The race to develop improved antibiotics and compounds for the treatment of human genetic diseases. Org Biomol Chem. 2008;6(2):227–239. doi: 10.1039/b712690p. [DOI] [PubMed] [Google Scholar]
- 7.Keeling KM, Wang D, Conard SE, Bedwell DM. Suppression of premature termination codons as a therapeutic approach. Crit Rev Biochem Mol Biol. 2012;47(5):444–463. doi: 10.3109/10409238.2012.694846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edlind TD. Susceptibility of Giardia lamblia to aminoglycoside protein synthesis inhibitors: Correlation with rRNA structure. Antimicrob Agents Chemother. 1989;33(4):484–488. doi: 10.1128/aac.33.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobbie SN, et al. Genetic reconstruction of protozoan rRNA decoding sites provides a rationale for paromomycin activity against Leishmania and Trypanosoma. PLoS Negl Trop Dis. 2011;5(5):e1161. doi: 10.1371/journal.pntd.0001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo J, Koganei M, Maianti JP, Ly VL, Hanessian S. Crystal structures of a bioactive 6′-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites. ChemMedChem. 2013;8(5):733–739. doi: 10.1002/cmdc.201200579. [DOI] [PubMed] [Google Scholar]
- 11.Loebenberg D, Counels M, Waitz JA. Antibiotic G418, a new micrommomospora-produced aminglycoside with activity apainst protozoa and helminths: Antiparasitic activity. Antimicrob Agents Chemother. 1975;7(6):811–815. doi: 10.1128/aac.7.6.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh N, Kumar M, Singh RK. Leishmaniasis: Current status of available drugs and new potential drug targets. Asian Pac J Trop Med. 2012;5(6):485–497. doi: 10.1016/S1995-7645(12)60084-4. [DOI] [PubMed] [Google Scholar]
- 13.Ben Salah A, et al. WR279,396, a third generation aminoglycoside ointment for the treatment of Leishmania major cutaneous leishmaniasis: A phase 2, randomized, double blind, placebo controlled study. PLoS Negl Trop Dis. 2009;3(5):e432. doi: 10.1371/journal.pntd.0000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben Salah A, et al. Topical paromomycin with or without gentamicin for cutaneous leishmaniasis. N Engl J Med. 2013;368(6):524–532. doi: 10.1056/NEJMoa1202657. [DOI] [PubMed] [Google Scholar]
- 15.El-On J, Bazarsky E, Sneir R. Leishmania major: In vitro and in vivo anti-leishmanial activity of paromomycin ointment (Leshcutan) combined with the immunomodulator Imiquimod. Exp Parasitol. 2007;116(2):156–162. doi: 10.1016/j.exppara.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Davidson RN, den Boer M, Ritmeijer K. Paromomycin. Trans R Soc Trop Med Hyg. 2009;103(7):653–660. doi: 10.1016/j.trstmh.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Sinha PK, et al. Phase 4 pharmacovigilance trial of paromomycin injection for the treatment of visceral leishmaniasis in India. J Trop Med. 2011;2011:645203. doi: 10.1155/2011/645203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández MM, Malchiodi EL, Algranati ID. Differential effects of paromomycin on ribosomes of Leishmania mexicana and mammalian cells. Antimicrob Agents Chemother. 2011;55(1):86–93. doi: 10.1128/AAC.00506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maarouf M, Lawrence F, Croft SL, Robert-Gero M. Ribosomes of Leishmania are a target for the aminoglycosides. Parasitol Res. 1995;81(5):421–425. doi: 10.1007/BF00931504. [DOI] [PubMed] [Google Scholar]
- 20.Kondo J. A structural basis for the antibiotic resistance conferred by an A1408G mutation in 16S rRNA and for the antiprotozoal activity of aminoglycosides. Angew Chem Int Ed Engl. 2012;51(2):465–468. doi: 10.1002/anie.201106084. [DOI] [PubMed] [Google Scholar]
- 21.Kondo J, François B, Urzhumtsev A, Westhof E. Crystal structure of the Homo sapiens cytoplasmic ribosomal decoding site complexed with apramycin. Angew Chem Int Ed Engl. 2006;45(20):3310–3314. doi: 10.1002/anie.200600354. [DOI] [PubMed] [Google Scholar]
- 22.Vicens Q, Westhof E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001;9(8):647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 23.Vicens Q, Westhof E. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J Mol Biol. 2003;326(4):1175–1188. doi: 10.1016/s0022-2836(02)01435-3. [DOI] [PubMed] [Google Scholar]
- 24.François B, et al. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: Role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005;33(17):5677–5690. doi: 10.1093/nar/gki862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo J, et al. Crystal structure of the bacterial ribosomal decoding site complexed with a synthetic doubly functionalized paromomycin derivative: A new specific binding mode to an a-minor motif enhances in vitro antibacterial activity. ChemMedChem. 2007;2(11):1631–1638. doi: 10.1002/cmdc.200700113. [DOI] [PubMed] [Google Scholar]
- 26.Kondo J, et al. Differential selectivity of natural and synthetic aminoglycosides towards the eukaryotic and prokaryotic decoding A sites. ChemBioChem. 2007;8(14):1700–1709. doi: 10.1002/cbic.200700271. [DOI] [PubMed] [Google Scholar]
- 27.Kondo J, Westhof E. The bacterial and mitochondrial ribosomal A-site molecular switches possess different conformational substates. Nucleic Acids Res. 2008;36(8):2654–2666. doi: 10.1093/nar/gkn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo J, Urzhumtsev A, Westhof E. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006;34(2):676–685. doi: 10.1093/nar/gkj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermann T, Tereshko V, Skripkin E, Patel DJ. Apramycin recognition by the human ribosomal decoding site. Blood Cells Mol Dis. 2007;38(3):193–198. doi: 10.1016/j.bcmd.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Pfister P, et al. Mutagenesis of 16S rRNA C1409-G1491 base-pair differentiates between 6’OH and 6’NH3+ aminoglycosides. J Mol Biol. 2005;346(2):467–475. doi: 10.1016/j.jmb.2004.11.073. [DOI] [PubMed] [Google Scholar]
- 31.Nudelman I, et al. Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations. J Med Chem. 2009;52(9):2836–2845. doi: 10.1021/jm801640k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokrovskaya V, Belakhov V, Hainrichson M, Yaron S, Baasov T. Design, synthesis, and evaluation of novel fluoroquinolone-aminoglycoside hybrid antibiotics. J Med Chem. 2009;52(8):2243–2254. doi: 10.1021/jm900028n. [DOI] [PubMed] [Google Scholar]
- 33.Nudelman I, et al. Repairing faulty genes by aminoglycosides: Development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg Med Chem. 2010;18(11):3735–3746. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 34.Matt T, et al. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci USA. 2012;109(27):10984–10989. doi: 10.1073/pnas.1204073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zufferey R, Bibis SS, Zhu T, Dhalladoo S. Characterization of a compensatory mutant of Leishmania major that lacks ether lipids but exhibits normal growth, and G418 and hygromycin resistance. Exp Parasitol. 2012;130(3):200–204. doi: 10.1016/j.exppara.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Jhingran A, Chawla B, Saxena S, Barrett MP, Madhubala R. Paromomycin: Uptake and resistance in Leishmania donovani. Mol Biochem Parasitol. 2009;164(2):111–117. doi: 10.1016/j.molbiopara.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvar J, et al. WHO Leishmaniasis Control Team Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7(5):e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burke JF, Mogg AE. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 1985;13(17):6265–6272. doi: 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kandasamy J, et al. Increased selectivity toward cytoplasmic versus mitochondrial ribosome confers improved efficiency of synthetic aminoglycosides in fixing damaged genes: A strategy for treatment of genetic diseases caused by nonsense mutations. J Med Chem. 2012;55(23):10630–10643. doi: 10.1021/jm3012992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai A, et al. The impact of aminoglycosides on the dynamics of translation elongation. Cell Rep. 2013;3(2):497–508. doi: 10.1016/j.celrep.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Q, et al. Molecular recognition by glycoside pseudo base pairs and triples in an apramycin-RNA complex. Angew Chem Int Ed Engl. 2005;44(18):2694–2700. doi: 10.1002/anie.200500028. [DOI] [PubMed] [Google Scholar]
- 42.Francis SP, et al. A novel role of cytosolic protein synthesis inhibition in aminoglycoside ototoxicity. J Neurosci. 2013;33(7):3079–3093. doi: 10.1523/JNEUROSCI.3430-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimony O, Jaffe CL. Rapid fluorescent assay for screening drugs on Leishmania amastigotes. J Microbiol Methods. 2008;75(2):196–200. doi: 10.1016/j.mimet.2008.05.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.