Abstract
The ubiquitous inducible transcription factor NF-κB plays central roles in immune and inflammatory responses and in tumorigenesis. Complex posttranslational modifications of the p65 subunit (RelA) are a major aspect of the extremely flexible regulation of NF-κB activity. Although phosphorylation, acetylation, ubiquitination, and lysine methylation of NF-κB have been well described, arginine methylation has not yet been found. We now report that, in response to IL-1β, the p65 subunit of NF-κB is dimethylated on arginine 30 (R30) by protein-arginine methyltransferase 5 (PRMT5). Expression of the R30A and R30K mutants of p65 substantially decreased the ability of NF-κB to bind to κB elements and to drive gene expression. A model in which dimethyl R30 is placed into the crystal structure of p65 predicts new van der Waals contacts that stabilize intraprotein interactions and indirectly increase the affinity of p65 for DNA. PRMT5 was the only arginine methyltransferase that coprecipitated with p65, and its overexpression increased NF-κB activity, whereas PRMT5 knockdown had the opposite effect. Microarray analysis revealed that ∼85% of the NF-κB–inducible genes that are down-regulated by the R30A mutation are similarly down-regulated by knocking PRMT5 down. Many cytokine and chemokine genes are among these, and conditioned media from cells expressing the R30A mutant of p65 had much less NF-κB–inducing activity than media from cells expressing the wild-type protein. PRMT5 is overexpressed in many types of cancer, often to a striking degree, indicating that high levels of this enzyme may promote tumorigenesis, at least in part by facilitating NF-κB-induced gene expression.
Keywords: histone, mass spectrometry
The NF-κB family is comprised of two protein subfamilies: c-Rel, RelB, and RelA (p65), which include transactivation domains in their C termini, and p100 (p52) and p105 (p50), which include a number of ankyrin repeats in their C termini and have transrepressive functions. All family members include an N-terminal DNA-binding region, the Rel homology domain (1). In the absence of an activating stimulus, NF-κB is located in the cytoplasm in a complex with the inhibitory IκB protein. In the classic pathway of NF-κB activation, extracellular signals activate IκB kinase, which phosphorylates IκBα, leading to its ubiquitination and degradation by proteasomes. NF-κB, liberated from IκB, translocates to the nucleus to regulate its target genes (2). The NF-κB pathway is regulated by means of several different posttranslational modifications, including ubiquitination, phosphorylation, acetylation, sumoylation, and nitrosylation (3). The recent work of several laboratories has revealed the reversible methylation of nonhistone proteins by enzymes that were discovered on the basis of their activities toward histones. For NF-κB, we found that in response to an activating signal, such as treatment with IL-1β, the p65 subunit is reversibly methylated on two specific lysine residues by chromatin remodeling enzymes in ways that profoundly affect its function (4). The activating monomethylation of K218 and dimethylation of K221 are both catalyzed by the H3K36 methyltransferase, nuclear receptor-binding SET domain-containing protein 1 (NSD1), and these methyl groups are removed, leading to the inactivation of NF-κB, by the H3K36 demethylase, F-box leucine repeat rich protein 11 (FBXL11). Additional work of other laboratories has shown that lysines 37, 310, 314, and 315 of p65 can also be methylated, albeit by different histone methyltransferases, revealing that this posttranslational modification plays a key role in regulating NF-κB activity (4–9).
We have now found that the p65 subunit of NF-κB is methylated on R30 by protein arginine methyltransferase 5 (PRMT5). PRMTs catalyze the methylation of the three nitrogen atoms of the guanidinium side chains of arginine residues (10), using S-adenosylmethionine (AdoMet) as the common methyl donor (11). To date, 10 mammalian PRMTs have been identified, PRTM1, -2, -3, -4 (also called CARM1), -5, -6, -7, -8, -9 (also called FBXO11), and -10 (12). PRMTs are classified according to the types of methylarginines that are formed. Type I enzymes catalyze the formation of ω-NG-monomethylarginine and asymmetric ω-NG-dimethylarginine; type II enzymes catalyze the formation of ω-NG-monomethyl- and symmetrical ω-NG-dimethylarginine; type III enzymes catalyze the formation of ω-NG-monomethylarginine only; type IV enzymes uniquely catalyze the formation of δ-NG-monomethylarginine (11). Arginine methylation of proteins is well known to play important roles in RNA processing, transcriptional regulation, signal transduction, and DNA repair (13).
Previously, NF-κB was reported to be regulated by other PRMTs. Hassa et al. (14) reported that PRMT1 coactivates NF-κB–dependent gene expression synergistically with PRMT4 and poly (ADP ribose) polymerase 1. Covic et al. (15) suggested that PRMT4 is a promoter-specific regulator of NF-κB–dependent gene expression in concert with the transcriptional coactivators p300/cyclic AMP-responsive element binding (CREB)-binding protein and the p160 family of steroid receptor coactivators. PRMT5, also called Janus kinase-binding protein 1, is found on human chromosome 14q11.2 (10). Human PRMT5, a type II enzyme with 637 amino acid residues, was the first found to synthesize symmetrical ω-NG-dimethylarginine (16). PRMT5 plays important roles in transcriptional modulation by methylating histones H2A, H3R8, and H4R3, negatively regulating the expression of genes such as suppressor of tumorigenicity 7 (ST7), and nonmetastatic 23 (NM23) (17). PRMT5 can methylate the tumor suppressor p53 on R333, R335, and R337 in the oligomerization domain to increase the response to damaged DNA (18).
We find that the signal-dependent dimethylation of R30 of the p65 subunit NF-κB by PRMT5 has a major effect on the ability of NF-κB to bind to κB elements and on gene expression. Overexpressing PRMT5 increases NF-κB activity, but knocking this enzyme down has the opposite effect. Therefore, the arginine methylation of p65 is an important posttranslational modification that profoundly regulates NF-κB activity.
Results
NF-κB Is Activated by Dimethylation of R30 of the p65 Subunit.
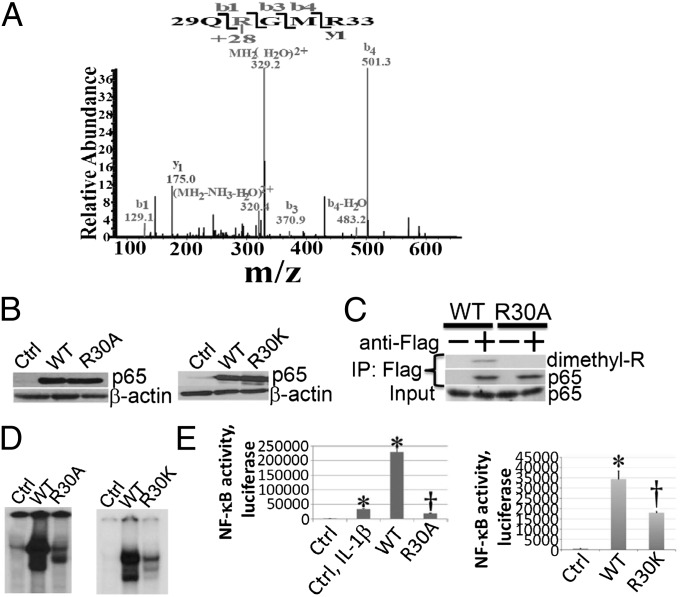
To explore the arginine methylation of NF-κB, we overexpressed Flag-tagged p65 in 293IL1R cells (4) and isolated p65 by using an anti–Flag-M2 antibody. Increased expression of p65 is well known to activate κB-dependent gene expression (4). Analysis by mass spectrometry revealed that p65 is dimethylated on R30, with an increase of 28 mass units (Fig. 1A). To test the functional significance of R30 dimethylation, we overexpressed the Flag-tagged R30A or R30K mutants, or wild-type p65 (WTp65) as a control, at similar levels in 293IL1R cells (Fig. 1B). To confirm that p65 is dimethylated on R30, we used anti-Flag to pull down either WT or R30A p65, followed by Western analysis of the level of total arginine methylation, using anti-dimethyl arginine (Fig. 1C). Overexpression of WTp65 induced the dimethylation of p65 and this modification was completely abolished when the R30A mutant protein was used. To assess the effect of the R30A mutation on NF-κB DNA binding activity, we carried out EMSAs. Overexpressing WTp65 strongly increased DNA binding activity in 293IL1R cells, but overexpressing the R30A or R30K mutants did not (Fig. 1D). An analysis of the activation of an NF-κB–dependent luciferase reporter in 293IL1R cells (Fig. 1E) showed strong activation in response to treatment with IL-1β or upon overexpression of WTp65, but much less activation when the R30A or R30K mutant proteins were overexpressed. Similar results were obtained in p65-null mouse embryo fibroblasts (MEF) (Fig. S1).
Fig. 1.
R30 of p65 is dimethylated upon NF-κB activation, and this modification is essential for NF-κB function. (A) An analysis of tryptic peptides by MS shows that p65 is dimethylated on R30 upon NF-κB activation. A pure single band of p65 was digested in the gel and samples were analyzed by LC-MS/MS. A mass shift of +28 was observed for peptides 29–33. Tandem MS analysis further revealed that R30 on the N-terminal side of the peptide is dimethylated. (B) Western analysis of p65. (Left) Both WTp65-Flag and R30A-Flag were overexpressed at similar levels in 293IL1R cells. (Right) Both WTp65-Flag and R30K-Flag were overexpressed at similar levels in 293IL1R cells. (C) A coimmunoprecipitation experiment confirmed that R30 of p65 is dimethylated upon NF-κB activation. WTp65-Flag and R30A-Flag proteins were expressed in 293IL1R cells. Anti-Flag was used to pull the proteins down, and arginine methylation was detected by using anti-dimethyl arginine. (D, Left) EMSA was performed with extracts of 293IL1R cells, and of these cells in which WTp65 and R30A were overexpressed. The ability of NF-κB to bind to DNA was greatly enhanced upon over-expression of the WTp65 protein, but the R30A mutation severely decreased this ability. (Right) An EMSA assay showed that 293IL1R cells overexpressing the R30K mutant protein had less NF-κB binding activity than 293IL1R cells overexpressing WTp65. (E) Luciferase assay of NF-κB activity. 293IL1R cells were treated with 10 ng/mL of IL-1β for 4 h as a positive control. Expression of WTp65 significantly activated NF-κB activity in 293IL1R cells, but the R30A (Left) or R30K (Right) mutations greatly decreased this activity. The data represent the means ± SD from three independent experiments. *P < 0.01 vs. Control (Ctrl) group; †P < 0.01 vs. WT group.
PRMT5 Binds to p65 and Dimethylates R30.
A mass spectrometric analysis revealed that PRMT5 was the only arginine methyltransferase that coimmunoprecipitated with p65, suggesting that PRMT5 is very likely to be the enzyme that dimethylates p65 on R30. For confirmation, we treated 293IL1R cells with IL-1β for 1 h, used anti-p65 to pull down endogenous p65, and analyzed Western transfers of the coprecipitated proteins with anti-PRMT5. The result clearly shows that PRMT5 binds to p65 in a signal-dependent manner (Fig. 2A) and the reciprocal coimmunoprecipitation experiment confirmed this result (Fig. 2B). PRMT1 and PRMT4 were previously reported to play a role in activating some NF-κB–inducible genes. Even though PRMT5 is the only PRMT that was identified in our mass spectrometry experiment, to further confirm that PRMT1 and -4 do not bind to p65 in response to IL-1β treatment, we performed a similar coimmunoprecipitation experiment, using anti-p65, and probed with antibodies to PRMT1 and PRMT4 (Fig. S2). The data clearly show that PRMT1 and PRMT4 do not bind to p65 in response to IL-1β treatment in 293IL1R cells.
Fig. 2.
PRMT5 binds to p65 and is responsible for NF-κB activation and dimethylation of R30. (A and B) Endogenous p65 coimmunoprecipitates with PRMT5. 293IL1R cells were treated with 10 ng/mL of IL-1β for 1 h or were untreated. PRMT5 binds to p65 only when NF-κB is activated. Reverse immunoprecipitation, using anti-PRMT5, gave a similar result. (C, Upper) Western assay showing either overexpression or shRNA-mediated knockdown of PRMT5 in 293IL1R cells. (Lower) Luciferase assay of NF-κB in 293IL1R cells treated with 10 ng/mL of IL-1β for different times, showing that reduced PRMT5 expression decreased and overexpression increased NF-κB activation. The data represent the means ± SD for three experiments. *P < 0.01 vs. Ctrl group. (D, Upper) Western assays, showing either overexpression (Upper Left) or shRNA-mediated knock down of PRMT5 (Upper Right) in colon cancer HT29 cells. (Lower) Luciferase assay of NF-κB in HT29 cells, showing that overexpression of PRMT5 further increased NF-κB activation, whereas reduced PRMT5 expression decreased NF-κB activation. The data represent the means ± SD for three experiments. *P < 0.01 vs. Ctrl group. (E) EMSA, showing that in response to IL-1β treatment, NF-κB DNA binding ability was significantly decreased in 293IL1R cells in which PRMT5 has been knocked down, compared with control cells (Left), whereas increased PRMT5 expression greatly enhanced NF-κB DNA binding ability (Right). (F) Coimmunoprecipitation and Western assays, showing that R30 of p65 is dimethylated by PRMT5 upon IL-1β treatment. Overexpression of PRMT5 increased the dimethylation of R30, but shRNA-mediated knock down had the opposite effect. (G) Luciferase assay of NF-κB activity, showing that the R30A mutation decreased NF-κB activation. Overexpression of PRMT5 activated NF-κB in control cells and in 293IL1R cells overexpressing WTp65, but not in these cells overexpressing the R30A mutant protein. The data represent the means ± SD from three experiments. *P < 0.01 vs. Ctrl group; †P < 0.01 vs. WTp65 group; #P < 0.01 vs. Ctrl, PRMT5 group; $P < 0.01 vs. WTp65, PRMT5 group.
We generated stable pools of 293IL1R cells in which PRMT5 was either overexpressed or knocked down by using shRNA (Fig. 2C, Upper) and tested the effects of varying the level of PRMT5 expression on the activation of NF-κB in response to IL-1β at different times. Cells in which PRMT5 was overexpressed showed enhanced IL-1β–induced NF-κB activation, but cells in which the expression of PRMT5 was knocked down showed the opposite effect (Fig. 2C, Lower). To determine the effect of PRMT5 in cells with constitutively active NF-κB, the enzyme was either overexpressed or knocked down in human colon cancer HT29 cells (Fig. 2D, Upper), which have high constitutive NF-κB activity, as found in many cancer cells (17). Overexpressing PRMT5 enhanced the constitutive activity of NF-κB, but knocking this enzyme down greatly reduced this activity (Fig. 2D, Lower). Therefore, PRMT5 is a strong NF-κB activator in these cells. To test the effect of PRMT5 on the ability of NF-κB to bind to DNA, 293IL1R cells and the corresponding cells in which PRMT5 was stably knocked down were treated with IL-1β for different times, followed by EMSAs. Reducing PRMT5 levels greatly reduced the ability of NF-κB to bind to DNA (Fig. 2E, Left). On the other hand, increased PRMT5 expression greatly enhanced this ability (Fig. 2E, Right).
To confirm the effect of PRMT5 on the arginine methylation of NF-κB, coimmunoprecipitation experiments were carried out using the above three 293IL1R-derived cell lines, followed by analysis of the proteins coimmunoprecipitated with p65, by the Western method, using anti-dimethylarginine. The level of arginine dimethylation of p65 was enhanced by overexpressing PRMT5, but substantially reduced in cells in which PRMT5 had been knocked down (Fig. 2F), revealing that PRMT5 is responsible for the arginine dimethylation of p65. As shown in Fig. 2G, the activation of NF-κB by overexpressing WTp65 was enhanced by coordinately overexpressing PRMT5. However, overexpressing the R30A mutant of p65 did not activate NF-κB, and coordinately overexpressing PRMT5 had no effect. These results reveal that PRMT5 activates NF-κB by dimethylating R30 of p65, and that there is very little methylation of additional arginine residues of p65 in this system.
Dimethylation of p65 on R30 Is Essential for NF-κB–Regulated Gene Expression.
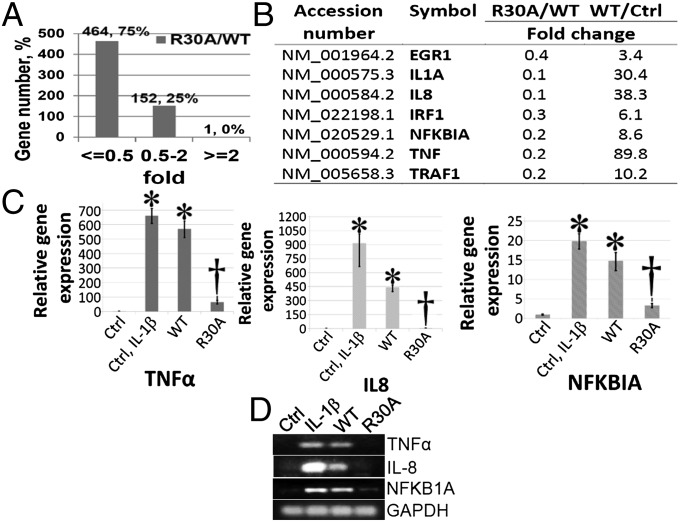
The 293IL1R cells, or these cells overexpressing WTp65 or the R30A mutant, were used to test the effect of this mutation on the expression of NF-κB target genes by microarray analysis. Compared with WTp65, 464 NF-κB target genes (∼75%) were induced less well by a factor of 2 or more by the R30A mutant protein, but 152 NF-κB target genes (∼25%) were not significantly affected. Only one gene was significantly up-regulated by the mutation (Fig. 3A). A short list of typical NF-κB–inducible genes whose induction is inhibited by the R30A mutation is shown in Fig. 3B. A full list of genes that are affected by the R30A mutation is given in Table S1. We performed quantitative PCR (qPCR) and RT-PCR analyses to confirm the expression of several candidate genes. The mRNA levels of TNF-α, IL-8, and NFKBIA were strongly induced in both IL-1β–treated cells and in cells overexpressing WTp65, but the induction was substantially less in cells with the R30A mutant protein (Fig. 3 C and D).
Fig. 3.
Regulation of NF-κB–dependent gene expression by R30 methylation. (A) Illumina array data from 293IL1R cells overexpressing WTp65 or the R30A mutant protein. 293IL1R cells were used as the control. Among the 617 NF-κB–dependent genes induced by twofold or more in cells overexpressing WTp65 (WTp65 vs. 293IL1R), 464 (75%) were induced less well, by twofold or more, in cells with the R30A mutant protein (R30A/WT ≤ 0.5); the remaining 25% were not affected significantly. (B) A short list of typical NF-κB–inducible genes, showing that some were up-regulated by WTp65 by twofold or more (WT/Ctrl ≥ 2), but were up-regulated less well, by twofold or more, by the R30A mutant protein (R30A/WT ≤ 0.5). (C) Confirmation of Illumina array data by qPCR analysis. The expression of TNF-α, IL-8, and NFKBIA was tested, confirming that these genes were strongly induced upon IL-1β treatment or by the overexpression of WTp65 but not by the R30A mutant protein. The data represent the means ± SD from three independent experiments. *P < 0.01 vs. Ctrl group; †P < 0.01 vs. WT group. (D) An RT-PCR experiment further confirmed that the TNF-α, IL-8, and NFKBIA genes were induced in response to IL-1β in 293IL1R cells or by the overexpression of WTp65, but not by the overexpression of the R30A mutant protein.
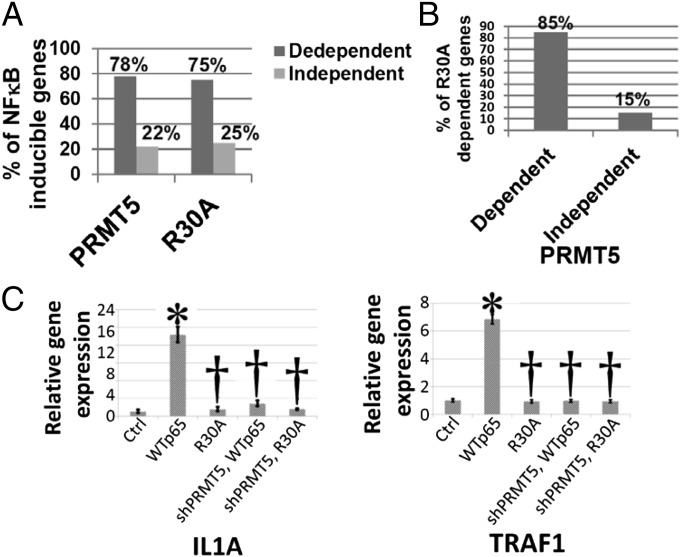
We also performed microarray analyses with 293IL1R cells and with the same cells in which PRMT5 was knocked down by shRNA (Fig. 4A). The induction of about 78% of the NF-κB target genes was down-regulated in the knockdown cells. In a cross comparison, we found that about 85% of the genes whose induced expression was reduced by the R30A mutation were also down-regulated by knocking the expression of PRMT5 down (Fig. 4B), suggesting that regulation of NF-κB–inducible genes through R30 is mainly regulated by PRMT5-dependent dimethylation. About 15% of PRMT5-independent genes were affected by the R30A mutation, perhaps indicating the existence of another interaction involving R30. A qPCR experiment was carried out to confirm data from the microarray analysis of PRMT5-regulated genes. Interleukin 1-α (IL1A) and TNF receptor-associated factor 1 (TRAF1) were induced by overexpressing WTp65 and the induction of both genes was inhibited by both the R30A mutation and knocking PRMT5 down, confirming that they are regulated by PRMT5 via dimethylation of R30 of p65.
Fig. 4.
Comparison of regulation of NF-κB dependent genes by PRMT5 and R30A. (A) Illumina array data, showing that 78% of genes were down-regulated by shPRMT5 by twofold or more, but the remaining 22% were not significantly affected. These percentages are similar to the percentages of R30A-dependent genes. (B) Illumina array data, suggesting that among the R30A dependent genes, about 85% were also PRMT5-dependent, meaning that they are also down-regulated by shPRMT5 by twofold or more, whereas the remaining 15% were not significantly affected. (C) A qPCR experiment was done to confirm that IL1A and TRAF1 were induced by the overexpression of WTp65, compared with control cells. However, this induction was greatly decreased after either R30A mutation or knockdown of PRMT5 by shRNA. Data represent the means ± SD for three independent experiments. *P < 0.01 vs. Ctrl group; †P < 0.01 vs. WT group.
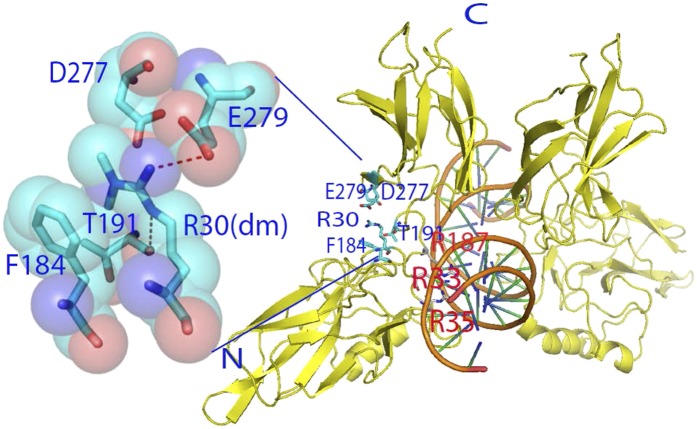
As shown in Fig. 3B, many of the genes whose expression was inhibited by the R30A mutation encode cytokines, chemokines, and growth factors. To test the effects of this mutation on the secretion of these factors, conditioned media from 293IL1R cells overexpressing either WTp65 or the R30A mutant were used to treat NF-κB reporter cells (19). Media from cells with the R30A mutant protein had significantly less NF-κB–inducing activity than media from WTp65 cells (Fig. S3A, Left). Similar results were obtained with the R30K mutant protein (Fig. S3B, Left) in 293IL1R cells and also in p65-null MEFs in which the expression of WT or the R30A or R30K mutant of p65 had been restored (Fig. S3 A and B, Right), further supporting the conclusion that R30 is a crucial site whose methylation regulates important biological function of NF-κB. We generated a model, based on the crystal structure of p65, to look at the effect of dimethylation of R30 on DNA binding (Fig. 5). Dimethylation of R30 is likely to stabilize domain interactions by creating new contacts between one of the methyl groups of modified R30 and the π electrons of the benzene ring of F184 at one end, and between the second methyl group and the aliphatic portion of D277 at the other end. This model indicates that methylation at R30 will have a major impact on DNA binding and thus on gene expression.
Fig. 5.
Crystal structure of p65 showing that methylated R30 can mediate van der Waals contacts with both F184 and D277 to enhance its binding to DNA. (Right) Ribbon presentation of the RelA:κB DNA complex. A protein–protein interface away from the protein-DNA interface is highlighted to reveal the amino acid residues that are involved. Residues in red make DNA contacts. (Left) Surface presentation of the residues at the protein–protein interface. Dotted lines show that the atoms are in H-bonding distance. Methylated R30 can mediate van der Waals contacts with both F184 and D277.
To summarize, in Fig. 6 we propose a model in which PRMT5 associates with NF-κB after it has been liberated from IκB, followed by the dimethylation of R30 of the p65 subunit, leading to increased affinity of NF-κB for the κB elements of many genes and enhanced induced expression of these genes, including many genes that encode cytokines, chemokines, and growth factors.
Fig. 6.
A model. In addition to previously known regulatory pathways, NF-κB is regulated by PRMT5-mediated methylation of p65 on R30, which affects the expression of many NF-κB–induced genes.
We also explored the expression levels of PRMT5 in various cancers (Fig. 7A), finding that this enzyme is substantially overexpressed in cancers of the colon, ovary, kidney, lung, bladder, liver, pancreas, breast, prostate, cervix, and skin, with especially high expression in colon cancer. Oncomine data (Fig. 7B) also show that the majority of the colon cancer patients evaluated express elevated levels of PRMT5. The data are strongly suggestive of the possibility that PRMT5 is a tumor promoter.
Fig. 7.
PRMT5 is substantially overexpressed in cancers. (A) GeneNote data showing that PRMT5 is overexpressed in cancers of the thymus, bone marrow, blood, brain, kidney, lung, colon, bladder, liver, pancreas, prostate, skin, breast, salivary gland, ovary, and cervix, with the most striking overexpression in colon cancer. (B) Comparison of PRMT5 expression in 19 colon/colorectal vs. normal datasets in Oncomine. The intensity of the red signal indicates the level of overexpression.
Discussion
Although the activity of NF-κB is regulated by several different posttranslational modifications, lysine methylation of the p65 subunit has been discovered only recently: We and other laboratories reported that NF-κB is methylated on lysines 37, 218, 221, 310, 314, and 315. We found that the combined activities of the histone methyl transferase NSD1 and the demethylase FBXL11 regulate NF-κB through the reversible methylation of K218 and K221 of p65. Remarkably, expression of the FBXL11 gene is induced in response to NF-κB activation, forming a unique negative feedback loop similar in effect to the one that involves the well-known negative regulator IκB (5). Recently, the Hur group (20) reported that glioma-expressed antigen 2 (PHF20) interacts with p65 by recognizing methylation at K218 and K221. This methylation-dependent interaction of PHF20 with p65 leads to persistent NF-κB phosphorylation and function by limiting the recruitment of protein phosphatase PP2A to p65 (20). Yang et al. (6) reported that K314 and K315 of p65 are monomethylated by SET7/9 in response to NF-κB activation, an inhibitory modification that stimulates proteosome-mediated degradation of promoter-associated p65. Ea and Baltimore (7) reported that SET7/9 methylates p65 at lysine 37, and that this methylation is restricted to the nucleus and regulates the promoter binding of p65. The methylation of p65 at K37 affects the stability of DNA-p65 complexes, which in turn regulates the recruitment of p65 to the promoter and the induction of a subset of NF-κB-regulated genes. Levy et al. (8) reported that SETD6 monomethylates p65 on K310, leading to induction of a repressed state at NF-κB target genes through the binding of G9a-like protein.
In this study, we report that p65 is dimethylated on R30 in a signal-dependent manner, leading to activation of NF-κB that is responsible for the expression of nearly 75% of the inducible genes. These genes encode cytokines, chemokines, kinases, and cell-adhesion molecules, including TNF-α (21), IL-8 (22), mitogen-activated protein kinase kinase kinase 8 (MAP3K8) (23), and integrin β2 (ITGB2) (24), whose roles are widely accepted to be essential in inflammation and tumorigenesis. Therefore, p65 is dimethylated on R30 by PRMT5, which profoundly affects its biological activity.
PRMT5, a type II arginine methyltransferase, plays important roles in modulating transcription by methylating histones, such as H2A, H3R8, and H4R3 (16). For example, PRMT5 methylates histone H2AR3 (25), leading to its interaction with methylosome protein 50 (Mep50), which in turn represses differentiation-specific genes in embryonic stem cells. In addition to its role in histone modification, PRMT5 can also directly methylate nonhistone proteins to regulate their activities. Jansson et al. (18) reported that PRMT5 methylates p53 on R333, R335, and R337 in the oligomerization domain, leading to increased response to DNA damage. PRMT5 can also methylate transcription elongation factor SPT5 (26) to decrease its interaction with RNA polymerase II, suggesting that PRMT5 might be involved in regulating transcriptional elongation. The Epstein-Barr virus 1 protein (EBNA1) is methylated at multiple arginine residues, from R325 to R376, by PRMT5. Inhibition of arginine methylation of EBNA1 altered its localization, resulting in the formation of EBNA1 rings around nucleoli (27). Another example is the homeobox transcription factor HOXA9, which is methylated on R140 by PRMT5, a modification that is essential for the expression of endothelial cell leukocyte adhesion molecules during the endothelial cell inflammatory response (28). PRMT5 has been recently shown to promote cell growth and transformation by other mechanisms. For example, Wang et al. (29) reported that PRMT5 inhibited transcription of the retinoblastoma protein family of tumor suppressors in leukemia and lymphoma to promote cancer progression. Aggarwal et al. (30) suggested that the nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth by activating PRMT5. Ullman and colleagues reported that PRMT5 accelerates tumor growth by arginine methylation of tumor suppressor programmed cell death 4 (31). Gu et al. (32) suggested that PRMT5 is essential for the growth of lung cancer cells. Wei et al. (33) reported that PRMT5 is a potential oncoprotein that up-regulates G1 cyclins and cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. In the present study, we provide unique insight regarding how high levels of PRMT5 may promote tumorigenesis, at least in part by facilitating NF-κB–induced gene expression.
Using X-ray crystallography, several structural models of p65 dimers (p65 homodimers and p50:p65 heterodimers) bound to different κB sites have been generated over the last several years (34, 35). These structures provide a wealth of information concerning how p65 interacts with κB response elements in the DNA, revealing, in addition to specific protein-DNA contacts among several invariant amino acids and conserved DNA bases, protein–protein interactions between the N- and C-terminal domains and the linker region connecting the two domains. The most conserved of these interactions, seen in all structures, is between the base of loop L1 of the N-terminal domain and loop L5 of the C-terminal dimerization domain. A single residue from the domain-connecting loop L3 also participates in this multiside-chain assembly, away from the protein–DNA interface. The key residues that mediate the interaction are D277 and E279 from loop L5 and R30 and T191 from loops L1 and L3, respectively (Fig. 5). F184, emerging from the end of the last secondary structure of the N-terminal domain, lies in close proximity to R30 and T191. R30 is sandwiched between F184/T191 on one side and D277/E279 on the other side. It is important to note that the phosphorylation of Ser276, located in the same L5 loop, is known to significantly affect gene regulation, in part, by directly affecting protein-DNA contacts. This evidence strongly supports the important role of dimethyl R30 in increasing the ability of p65 to bind to DNA and thus to affect gene expression.
Histone-modifying enzymes have been shown to reversibly methylate a variety of transcription factors, with important functional consequences. Most of the examples involve mono- or dimethylation of lysine residues, as summarized by Stark et al. (36). In one report concerning the reversible lysine methylation of K140 of STAT3 and three reports concerning methylations of several different lysine residues of NF-κB (36), it was found that the reactions take place on chromatin-bound factors. In each of these cases and in the study of arginine methylation of NF-κB reported here, it is important to note that the steady-state fraction of methylated transcription factors is quite low, of the order of a few percent of the total protein, which seems counter-intuitive given the important functional consequences of these reactions. A hypothesis that explains this apparent paradox is that only promoter-bound transcription factors are methylated, and that this modification does not take place on every promoter. The clearest evidence supporting this idea is found in the study of Yang et al. (37) of the reversible methylation of K140 of STAT3. Similar experiments for the several different methylation sites of NF-κB will be needed to extend this model further. The small fraction of a given transcription factor that is methylated in steady state also helps to explain why this important class of posttranslational modifications has only recently been identified. Careful analysis of other cases will surely yield additional examples.
Most methylations of transcription factors are likely to affect function in a manner similar to the same modifications of histones, by providing docking sites for accessory proteins (36). An excellent example is an analysis of the consequences of methylating p53, where different histone-modifying enzymes methylate different lysine (36) or arginine (18) residues to activate or repress activity, and sometimes mono- or dimethylation of the same lysine residue has opposing effects (36). For example, the dimethylation of K370 of p53 provides a docking site for the important accessory protein 53BP1, therefore strongly activating transcription, whereas monomethylation of the same residue leads to transcriptional repression (36). In our work with the p65 subunit of NF-κB, it is striking that the monomethylation of K218 and dimethylation of K221 (4) and, reported here, the dimethylation of R30, are important to facilitate tight binding to DNA. Coordinating methylation of all three sites on a single p65 molecule would provide the maximum increase in affinity, and modulation of affinity at specific κB sites might be achieved by limiting methylation to only one or two of these three lysine and arginine residues. Detailed analysis of the events at specific promoters will provide important information regarding the fine-tuning of NF-κB–dependent gene expression in the future.
Given the important role of NF-κB in cancer and knowing how PRMT5 regulates NF-κB by dimethylating R30 of the p65 subunit, together with evidence that PRMT5 is overexpressed in many types of cancer, our findings suggest that PRMT5 is an oncogene and raise the possibility of focusing on PRMT5 as a drug target and biomarker.
Materials and Methods
The 293IL1R cell line was described previously (4). The colon cancer HT29 cell line was purchased from the American Tissue Culture Collection. Luciferase assays were done by using the κB-luciferase construct p5XIP10 κB (4). EMSAs were done by using the oligomer for an NF-κB binding site (Santa Cruz Biotechnology). qPCR experiments were done by using FastStart Universal SYBR Green Master ROX (Roche). Detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Laiqing Hua (Indiana University School of Medicine) for his excellent technical support; Dr. Alexander Hoffmann (University of California at San Diego) for generously providing mouse embryonic fibroblast p65−/− cells; and the Cleveland Clinic Genomics Core for helping with microarray experiments;. This project was supported in part by Indiana University Grant 23-862-07TL, Indiana Biomedical Research Grant 2286226, Indiana Cancer Center Grant 4487513-LU, and a Case Western Reserve University Center for Bioinformatics and Proteomics pilot grant (all to T.L.). Work from the laboratories of G.R.S. and G.G. was supported by National Institutes of Health Grants P01CA062220 and GM08490, respectively.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311784110/-/DCSupplemental.
References
- 1.Gilmore TD. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene. 2006;25(51):6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 2.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 3.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25(51):6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 4.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu T, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci USA. 2009;106(38):16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XD, et al. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28(8):1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106(45):18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D, et al. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat Immunol. 2011;12(1):29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LF, et al. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25(18):7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause CD, et al. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113(1):50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn P, Xu W. Protein arginine methyltransferases: Nuclear receptor coregulators and beyond. Prog Mol Biol Transl Sci. 2009;87:299–342. doi: 10.1016/S1877-1173(09)87009-9. [DOI] [PubMed] [Google Scholar]
- 12.Yu MC. The role of protein arginine methylation in mRNP dynamics. Mol Biol Int. 2011;2011:163827. doi: 10.4061/2011/163827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18(3):263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Hassa PO, Covic M, Bedford MT, Hottiger MO. Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1. J Mol Biol. 2008;377(3):668–678. doi: 10.1016/j.jmb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 15.Covic M, et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 2005;24(1):85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Branscombe TL, et al. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276(35):32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 17.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansson M, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10(12):1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 19.Lu T, et al. Secreted transforming growth factor beta2 activates NF-kappaB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci USA. 2004;101(18):7112–7117. doi: 10.1073/pnas.0402048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang T, et al. PHF20 regulates NF-kB signaling by disrupting recruitment of PP2A to p65. Nature Communication. 2013;4:2062. doi: 10.1038/ncomms3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214(2):149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 22.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14(21):6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 23.Serebrennikova OB, et al. Tpl2 ablation promotes intestinal inflammation and tumorigenesis in Apcmin mice by inhibiting IL-10 secretion and regulatory T-cell generation. Proc Natl Acad Sci USA. 2012;109(18):E1082–E1091. doi: 10.1073/pnas.1115098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberyszyn TM, et al. Beta2 integrin/ICAM-1 adhesion molecule interactions in cutaneous inflammation and tumor promotion. Carcinogenesis. 1998;19(3):445–455. doi: 10.1093/carcin/19.3.445. [DOI] [PubMed] [Google Scholar]
- 25.Tee WW, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24(24):2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwak YT, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11(4):1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 27.Shire K, et al. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J Virol. 2006;80(11):5261–5272. doi: 10.1128/JVI.02682-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandyopadhyay S, et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32(7):1202–1213. doi: 10.1128/MCB.05977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28(20):6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aggarwal P, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18(4):329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powers MA, Fay MM, Factor RE, Welm AL, Ullman KS. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011;71(16):5579–5587. doi: 10.1158/0008-5472.CAN-11-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Z, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J. 2012;446(2):235–241. doi: 10.1042/BJ20120768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei TY, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012;103(9):1640–1650. doi: 10.1111/j.1349-7006.2012.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391(6665):410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 35.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell. 1998;95(6):759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 36.Stark GR, Wang Y, Lu T. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 2011;21(3):375–380. doi: 10.1038/cr.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107(50):21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.