Abstract
Lysine methylation of the p65 subunit of nuclear factor κB (NF-κB) on K218 and K221 together or K37 alone strongly enhances gene expression in response to cytokines. We analyzed the effects of K-to-Q mutations in the REL homology domain of p65 on the response to IL-1β in 293 cells with low levels of p65. The K218/221Q mutation greatly reduced the expression of 39 of 82 genes, whereas the K37Q mutation reduced the expression of 23 different genes. Enhanced expression of the lysine demethylase FBXL11, which catalyzes the demethylation of K218 and K221 specifically, inhibited the expression of most of the genes that were inhibited by the DKQ mutation. CHIP-Seq analysis showed that the K218/221Q mutation greatly reduces the affinity of p65 for many promoters and that the K37Q mutation does not. Structural modeling showed that the newly introduced methyl groups of K218 and K221 interact directly with DNA to increase the affinity of p65 for specific κB sites. Thus, the K218/221Q and K37Q mutations have dramatically different effects because methylations of these residues affect different genes by distinct mechanisms.
Keywords: protein methylation, histone modifying enzyme
Members of the nuclear factor κB (NF-κB) family are central coordinators of innate and adaptive immune responses. Of the five family members in mammals [RelA (p65), RelB, c-Rel, NF-κB 1 (p50), and NF-κB2 (p52)], the p65/p50 heterodimer functions most often, in the “classic” signaling pathway (1). In unstimulated cells, NF-κB dimers are retained in an inactive state in the cytoplasm through binding to a member of the Inhibitor of κB (IκB) family. Activation of NF-κB is catalyzed by IκB kinase (IKK), followed by the phosphorylation and degradation of IκB, the phosphorylation of NF-κB itself, and the liberation of phosphorylated NF-κB dimers, which translocate to the nucleus and activate the transcription of target genes (2). The p65 subunit of NF-κB can be phosphorylated on more than a dozen different serine and threonine residues in response to activating stimuli, leading to differential gene regulation and dependent biological effects. Mutation of these phosphorylation sites modulates activity, affecting the ability of p65 to transactivate some promoters, but leaving others essentially unaffected (3). It seems likely that additional posttranslational modifications of p65 also occur in a signal-dependent manner, and that differential posttranslational modifications of p65 are likely to be responsible for the differences in p65 function that have been observed in response to different activating stimuli (4). The activity of NF-κB is regulated by many different stimuli in all cell types, with many different functional consequences (4). Regardless of the stimulus, all pathways leading to NF-κB activation involve posttranslational modifications of IKK, IκB, and NF-κB itself, including phosphorylation, ubiquitination, acetylation, sumoylation, and nitrosylation, and the specific modifications that occur depend on the nature of the inducing stimulus (5). Recent discoveries that several different lysine residues of p65 are methylated in response to activating signals have added a new level of complexity to the regulation of NF-κB function (6–10).
As reviewed recently (11), lysine methylation has recently emerged as a novel posttranslational modification of several different transcription factors, with profound effects on their functions. Reversible methylation is carried out by various histone lysine methyltransferases and demethylases, usually in a signal-dependent manner. The di-methylation of K140 of signal transducer and activator of transcription 3 (STAT3) follows the STAT3-dependent recruitment of the histone lysine methyltransferase SET9 to the suppressor of cytokine signaling 3 (SOCS3) promoter (12). Lysine methylation affects not only the ability of a transcription factor to bind to DNA but also specific protein–protein interactions. For example, for p53, methylations occur on K370, K372, and K382, with consequences for function that depend on the site and the degree of methylation. K370 is monomethylated by the H3K4 methylase SMYD2, repressing transcription (13), and is di-methylated at this site by an unknown methylase. Di-methylation of K370 provides a binding site for the coactivator 53BP1 and thus is strongly activating. K370me and K370me2 are both demethylated by the H3K4 demethylase LSD1 (14). K372 is methylated by the H3K4 methylase SET7/9, which enhances subsequent acetylation of this residue, in turn stabilizing p53 (15).
For p65, six different lysine residues (K37, K218/K221, K310, and K314/K315) can be methylated by different histone lysine methyltransferases in response to activating signals (6, 7, 9, 10). In response to IL-1β, p65 is reversibly methylated on K218 and K221, with profound effects on its function (7, 8). The activating monomethylation of K218 and di-methylation of K221 are both catalyzed by the H3K36 methyltransferase nuclear receptor-binding SET domain-containing protein (NSD1) and the methyl groups can be removed, leading to loss of NF-κB function, by the H3K36 demethylase FBXL11. Recently, the Hur group (16) reported that glioma-expressed antigen 2 (PHF20) interacts with p65 by recognizing methylation at K218 and K221. This methylation-dependent interaction of PHF20 with p65 leads to persistent NF-κB phosphorylation and function by limiting the recruitment of protein phosphatase PP2A to p65 (16). This important observation further confirms the biological importance of lysine methylation of p65.
The p65 subunit of NF-κB has two functional domains (5), the REL homology domain (RHD, amino acid residues 19–301) and the transactivation domain (TA, amino acid residues 428–551). Three of the six methylated lysine residues of p65 are in the RHD (K37, K218, and K221) and the other three (K310, K314, and K315) are in the region linking RHD and TA. Here, we compare the differential effects on gene regulation of the methylation of the three lysine residues in the RHD, namely, K218 and K221, which are modified together, or K37, which is modified alone. We expressed the K218/221Q double mutant (DKQ) or the K37Q single mutant of p65 at levels similar to that of the wild-type protein in 293IL1R cells, which have robust expression of the IL-1 receptor, and analyzed the differential induction of mRNAs in response to IL-1β, and also the ability of the mutant p65 proteins to bind to promoters. Individual NF-κB–responsive genes are affected quite differently by the methylation of K218/K221 or K37. Furthermore, the DKQ, K37Q, and WT proteins have quite different preferences for binding to specific promoters. We conclude that the methylations of different lysine residues of p65 have quite distinct functional consequences for the IL-1β–induced expression of individual genes. Because K218 and K221 are known to bind to DNA in κB sites (17), we analyzed the effects on binding of the methyl groups that are added to these two lysine residues in response to NF-κB activation, finding that they are likely to increase the affinity of NF-κB for DNA by making new hydrophobic contacts.
Results
Gene Induction as a Function of Time in 293IL1R Cells.
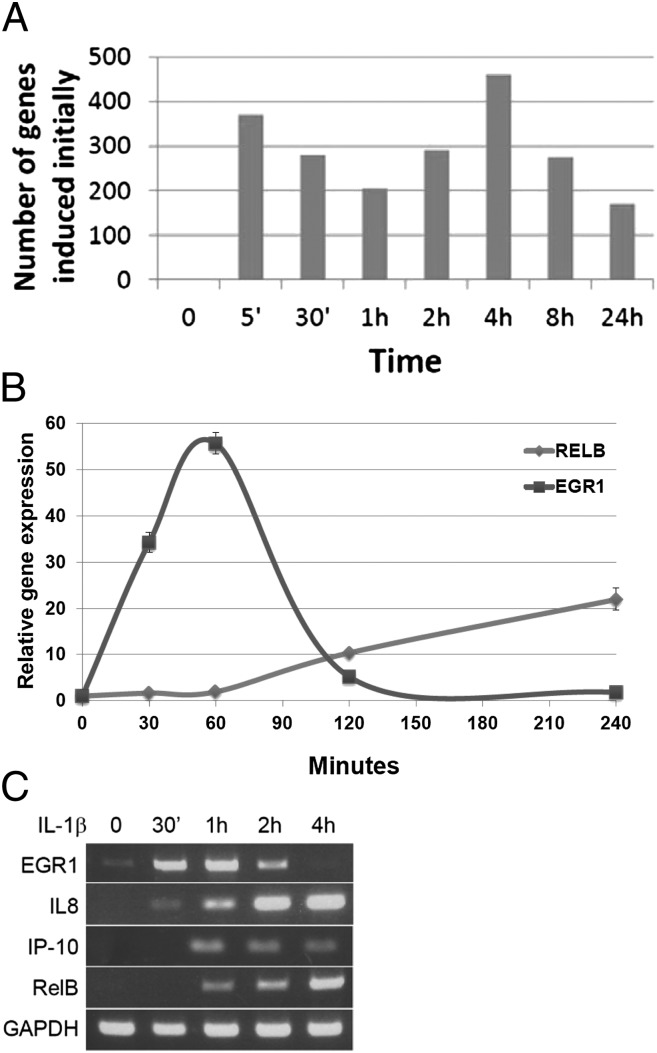
Because the primary response of individual NF-κB–induced genes can occur at different times (18), we analyzed whether the time of induction correlated with the effect of lysine methylation of the two different subregions of the RHD. We treated 293IL1R cells with IL-1β for times ranging from 5 min to 24 h and analyzed the expression of the induced mRNAs by using Illumina arrays (Fig. 1A). About 650 genes were induced by 30 min (350 genes at 5 min and an additional 300 genes at 30 min) were induced by twofold or more, and over 1,500 additional genes were induced between 1 and 24 h. PCR-based assays (Fig. 1 B and C) confirmed the parameters of induction for several of these genes. Adelman et al. (18) have classified NF-κB primary response genes as fast or slow. Upon stimulation with IL-1β, the expression of EGR1, a fast gene, was induced within 30 min and began to decrease after 2 h (Fig. 1 B and C), whereas the slow genes RELB (Fig. 1 B and C), IP10, and IL-8 (Fig. 1C) were induced after 1 h or more, and their expression was sustained for a much longer time. These data confirm the dynamic complexity of gene induction in response to IL-1β in 293IL1R cells.
Fig. 1.
Gene induction as a function of time in 293IL1R cells. (A) The number of genes newly induced by twofold or more at each time point is shown. The cells were treated with IL-1β and Illumina array analysis was carried out on the isolated RNAs. (B) Confirmation of array data. qPCR analysis. The EGR1 and RELB genes were induced with very different kinetics. (C) Confirmation of array data. 293IL1R cells treated with IL-1β for different times were analyzed by RT-PCR for expression of the EGR1, IL8, IP10, and RELB genes.
Differential Regulation of Gene Expression by DKQ or K37Q.
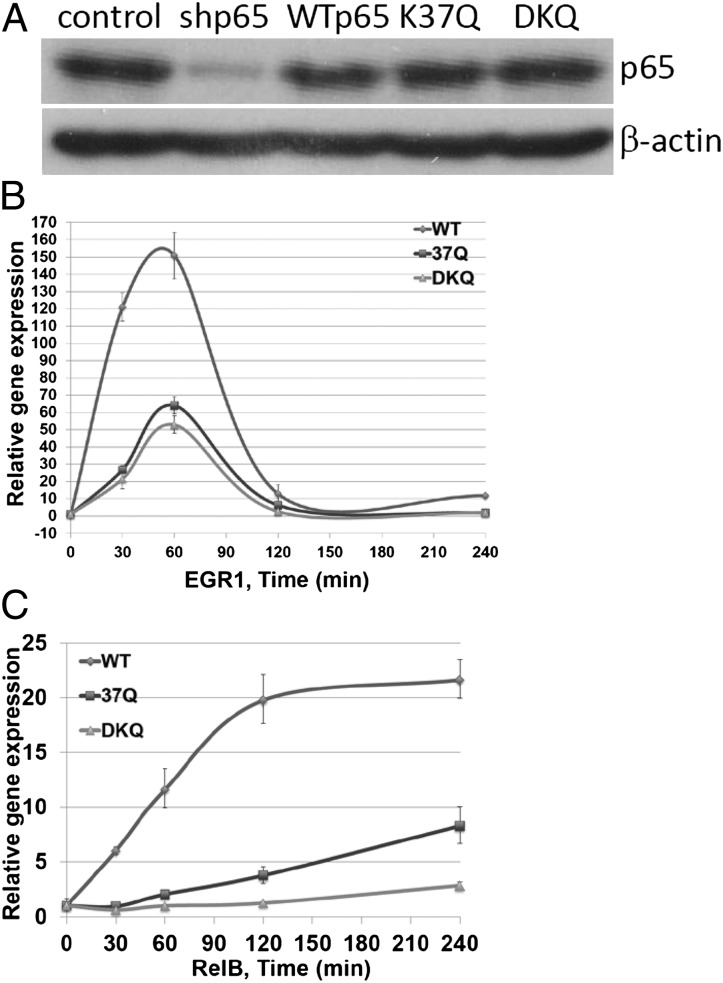
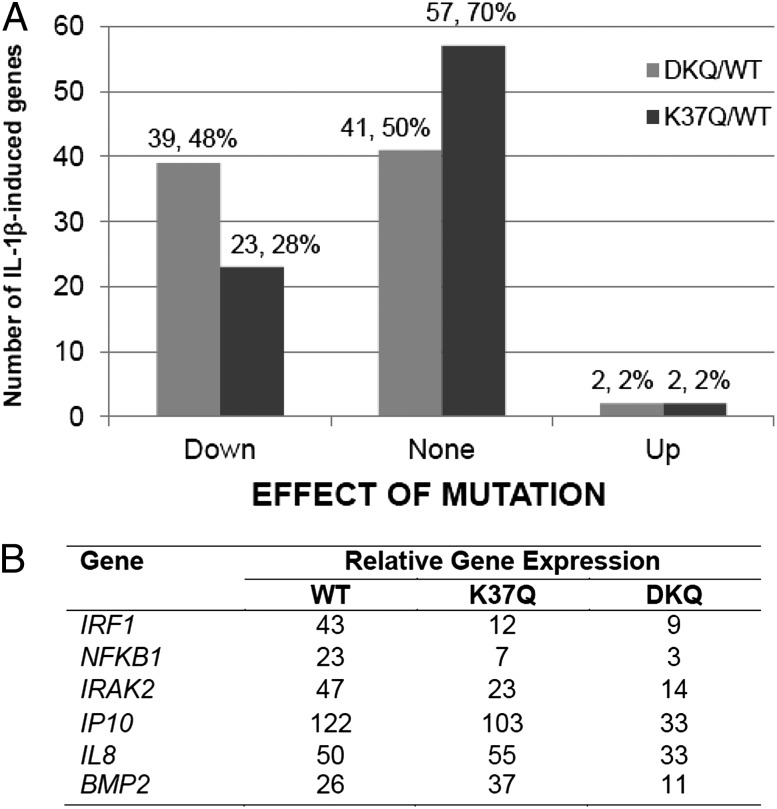
To examine the differential effects on gene regulation by methylation of K37 and K218/221, we knocked the expression of the p65 subunit of NF-κB down and expressed wild-type p65, DKQ, or the K37Q single mutant at levels similar to that of endogenous p65 in the parental cells (Fig. 2A). The induction of the EGR1 and RELB genes was tested in these three cell lines in response to IL-1β, revealing that the K37Q and DKQ mutations decreased the induction of both genes. However, the kinetics of gene induction in the mutant and wild-type cell lines were similar (Fig. 2 B and C). An Illumina array analysis was carried out with IL-1β–treated wild-type, K37Q, and DKQ cells (Fig. 3A). The DKQ mutation inhibited the expression of about half of the genes that were induced by fivefold or more in 4 h and did not affect the expression of the other half. In contrast, the K37Q mutation inhibited the expression of about a quarter of the induced genes, increased the expression of only two genes, and did not affect the expression of about three quarters of the induced genes. Among the 39 genes that were down-regulated by DKQ, 18 were also down-regulated by K37Q. Therefore, the methylations at each lysine residue affects a specific subset of genes, but there is also a subset of genes whose expression is affected by both mutations. DKQ and K37Q had quite different effects on some target genes (Fig. 3B). For example, the IRF1, NFKB1, IRAK2, and IP10 genes were down-regulated by both DKQ and K37Q, with DKQ having the greater effect. The IL8 gene was down-regulated by DKQ, but slightly up-regulated by K37Q, and the BMP2 gene was down-regulated DKQ, but substantially up-regulated by K37Q. We have classified all of the NF-κB target genes into nine groups, based on the effects of DKQ or K37Q (up-regulated, down-regulated, or insensitive). Short lists of typical genes in each subgroup are shown in SI Appendix, Tables S1–S9.
Fig. 2.
The DKQ and K37Q mutations do not affect the dynamics of gene induction. (A) Western analysis, showing knockdown of p65 expression in 293IL1R cells and reexpression of the wild-type, K37Q, and DKQ proteins at similar levels. (B) qPCR analysis. Upon IL-1β treatment, the DKQ and K37Q cells showed less EGR1 induction but similar kinetics, compared with wild-type cells. (C) qPCR analysis. Upon IL-1β treatment, the DKQ and K37Q cells showed less RELB induction but similar kinetics, compared with wild-type cells.
Fig. 3.
Differential gene regulation by DKQ and K37Q. (A) Cells expressing the WT, K37Q or DKQ proteins were treated with IL-1β for 4 h and an Illumina array analysis was carried out. The effect of the DKQ (left-hand columns) and K37Q (right-hand columns) mutations was assessed for the 82 NF-κB target genes that were induced by fivefold or more in wild-type cells. (B) qPCR analysis of the relative expression of several genes that were strongly affected by the K37Q and DKQ mutations.
CHIP-Seq Analysis Reveals Differential Effects on Promoter Binding by DKQ or K37Q.
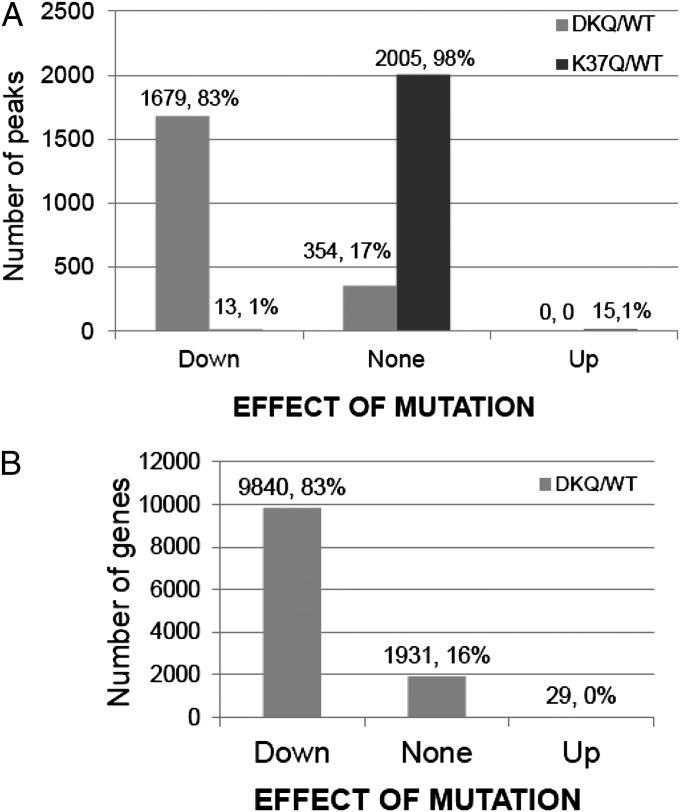
To assess the effects of the K-to-Q mutations on the affinity of p65 for κB sites, we carried out a CHIP-Seq analysis on IL-1β−treated wild-type, DKQ, and K37Q cells. We used the Partek Genomic Studio to detect enriched regions/peaks in our sequencing data (Fig. 4A). The DKQ mutation decreased promoter binding by twofold or more for 83% of the 2,033 regions scored and did not change promoter binding for 17%. In striking contrast, the K37Q mutation had virtually no effect on the ability of NF-κB to bind to DNA. We conclude that the methylation of K218 and K221, which lie in the DNA binding domain of p65, enhances gene expression mainly by facilitating binding to promoters, whereas the K37 methylation enhances gene expression by one or more different mechanisms. In the analysis of Fig. 4A, the peaks detected are not limited to promoters, making it difficult to compare the CHIP-Seq data with Illumina array data. To do this comparison, we generated total numbers of sequence read counts within the boundaries of each National Center for Biotechnology Information (NCBI) RefSeq gene. This second analysis shows that the DKQ mutation decreased promoter binding for 83% of 11,800 sequence read counts by twofold or more (Fig. 4B), identical to the percentage affected in the Peak Detection analysis (Fig. 4A).
Fig. 4.
CHIP-Seq analysis. (A) Analysis by the peaks detection method. The binding to κB elements of most of the 2,033 peaks detected in the WT sample was decreased by twofold or more by the DKQ mutation (left-hand columns), whereas the K37Q mutation (right-hand columns) had virtually no effect on binding. Neither mutation caused a substantial increase in binding. (B) Analysis by the RefGene Search method. In agreement with the data of A, the promoter binding to 83% of κB elements were decreased by twofold or more by the DKQ mutation compared with wild-type.
When we compared the effects of mutations on promoter binding (CHIP-Seq data) with the effects on gene expression (Illumina array data) for specific genes, we found that 59 genes (72%) whose expression is either reduced or unaffected by the DKQ mutation (the same 82 genes analyzed in Fig. 3A) also have their ability to bind to promoters reduced by this mutation. For the remaining 23 genes, the DKQ mutation had no effect on promoter binding. Therefore, the methylations of K218 and K221 are essential for the binding of NF-κB to the promoters of most inducible genes.
We analyzed the patterns of gene expression for the 59 genes whose abilities to bind to promoters were decreased by twofold or more by the DKQ mutation. The expression of 29 of these genes was also reduced by twofold or more, but for the remaining 30 genes the expression was not significantly affected. These results suggest that, for the DKQ sites, promoter binding is a dominant factor in determining gene expression for many, but not all, NF-κB–inducible genes. Examples in which both promoter binding and gene expression are decreased by the DKQ mutation are shown in Table 1. Interestingly, both fast and slow genes are found in this group, revealing that there is no obvious relationship between lysine methylation and the kinetics of gene induction and, again, that promoter affinity is not the sole factor that determines why some genes are transcribed faster than others.
Table 1.
Examples of genes for which both promoter binding and gene expression were decreased by twofold or more by the DKQ mutation
| Gene | Number of binding events |
Relative gene expression |
Kinetic type | |||
| WT | K37Q | DKQ | K37Q/WT | DKQ/WT | ||
| MAP3K8 | 435 | 432 | 117 | 0.9 | 0.5 | Slow |
| MYB | 447 | 518 | 159 | 0.9 | 0.3 | Slow |
| NFKB1 | 609 | 847 | 289 | 1.0 | 0.5 | Slow |
| PTGES | 651 | 549 | 140 | 1.2 | 0.1 | Slow |
| TNFSF10B | 734 | 684 | 228 | 0.9 | 0.4 | Fast |
| IRF1 | 306 | 245 | 69 | 1.2 | 0.2 | Fast |
For this group of genes, promoter binding plays the dominant role in regulating gene expression. Both fast and slow genes were found in this group.
Overexpression of FBXL11 Down-Regulates the Expression of Most DKQ-Regulated NF-κB Target Genes.
The FBXL11 gene encodes a negative regulator of NF-κB that demethylates K218/K221. FBXL11 is itself an NF-κB target gene, contributing to negative feedback of NF-κB−induced gene expression (7, 8). To compare the effects of demethylating K218/K221 with the effects of mutating these residues, 293IL1R cells expressing a normal or high level of FBXL11 were treated with IL-1β for 4 h and the induced mRNAs were compared with those affected by the DKQ mutation. Of the 226 genes induced by twofold or more in normal cells after 4 h, 86 (38%) were down-regulated by twofold or more by increased expression of FBXL11. Furthermore, 65% of the IL-1β–induced NF-κB target genes that were down-regulated by DKQ were also down-regulated by increased expression of FBXL11. These results confirm that the methylation of K218 and K221 of p65 plays a major role in gene regulation.
Models of Gene Regulation by K218/221 Methylation and Prediction of Structure.
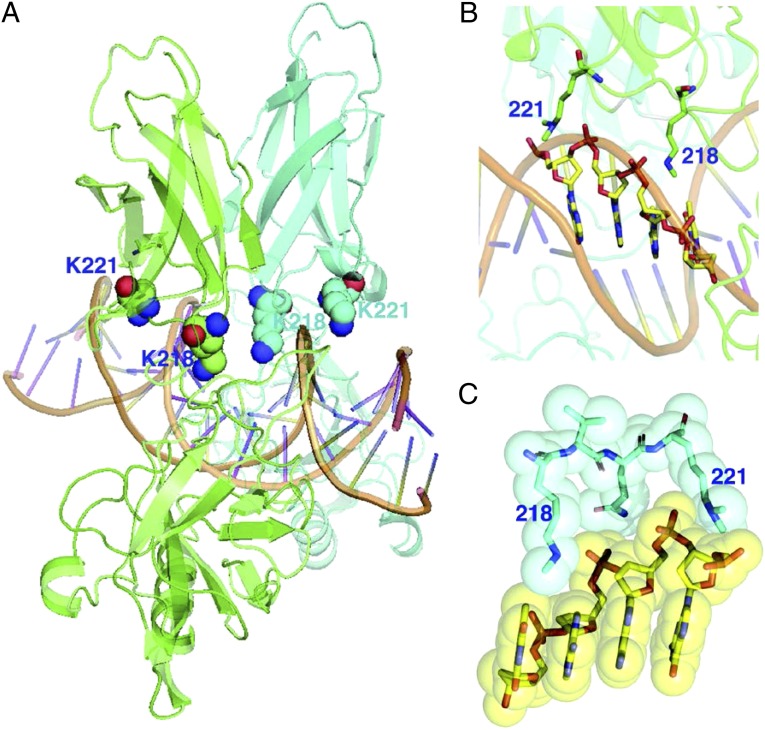
How might the methylation of K218/K221 affect the interaction of NF-κB with a κB element? The structural model shown in Fig. 5 reveals the importance of lysine methylation in facilitating the binding of p65 to DNA. Shown in Fig. 5A is a p65/p50 heterodimer (the two subunits are in different colors) bound to the κB site of the IL-8 gene (GGAAATTCC). K218 faces the major groove directly, whereas K221 lies near the backbone. Interactions of these two lysine residues from one monomer are shown close-up in Fig. 5B. Fig. 5C shows a space-filling surface view of the contacts. It is clear that both modified lysine residues interact directly with DNA through hydrophobic contacts. This predicted structure strongly supports the dramatic contributions made by methylation of K218/K221 to the function of NF-κB.
Fig. 5.
A structural model shows the interaction between K218me1 and K221me2 of p65 and the DNA of a κB element. (A) The p65/p50 heterodimer, with the two subunits in different colors, is shown bound to the κB site of the IL-8 gene (GGAAATTCC). K218 faces the major groove directly, whereas K221 lies near the backbone. (B) A close-up view of the interactions of the two lysine residues from one monomer. (C) A space-filling surface view of the contacts. Both methylated lysine residues are close to the DNA and make direct hydrophobic contacts.
We chose to study K-to-Q mutations to facilitate comparison with the data of Ea and Baltimore (6). However, no amino acid substitution is without potential problems. We consider here the extent to which glutamine residues might mimic monomethylated lysine residues. In the complex between p65 and DNA, lysine residues 218 and 221 are fully extended, without much room to change conformation, and the ε-amino groups make contacts with phosphates in the DNA backbone. Methylation further extends the length of the lysine side chain, allowing these residues to form new van der Waals contacts with the DNA, especially with sugar residues. In addition to eliminating the positive charges, glutamine substitutions of these lysine residues reduces the distance to DNA, weakening binding. Furthermore, the carbonyl oxygen of glutamine may make unfavorable contacts to further reduce binding energy.
Discussion
The activity of NF-κB is regulated in many different ways in response to many different stimuli. A major aspect of this regulation is posttranslational modification of the p65 subunit. Among the modifications that have been studied (including phosphorylation, ubiquitination, and acetylation), the contribution of lysine methylation to the function of NF-κB is least-well understood at present. In this study, we compared the roles of the methylated lysine residues K218 and K221 with that of K37, finding that the methylation of these two different regions of the RHD of p65 regulate different groups of genes, with the DKQ mutation affecting 48% and the K37Q mutation affecting 28% of NF-κB targets. Detailed comparison of the effects of DKQ and K37Q on individual genes reveals major differences. A global analysis by CHIP-Seq of the ability of NF-κB to bind to DNA provided a much more complete picture of how the methylations of lysine residues in two different subregions of the REL domain affect the regulation of gene expression. The DKQ mutation significantly decreases the ability of NF-κB to bind to most promoters, whereas the K37Q mutation has virtually no effect on binding. These data fit very well with structural information (17) showing that K218 and K221 make direct contact with DNA in κB sites, whereas K37 does not. Therefore, the mechanism of gene regulation by K218/221 and K37 methylation are fundamentally different. In our previous study, we showed that overexpression of the K218/221A mutant greatly decreased the ability of both the p65/p65 homo-dimer and the p65/p50 hetero-dimer to bind to DNA, compared with wild-type p65 in EMSAs (8). However, the ratio of p65/p50 to p65/p65 for the K218/221A mutant was similar to the ratio in cells with wild-type p65, suggesting that the K218/221A mutation did not affect the ability of mutant p65 to bind to p50.
Decreased promoter binding in response to the DKQ mutation correlated with either decreased gene expression or no significant effect on gene expression. These two different effects can be explained by two different models of gene regulation: (i) promoter binding alone has the dominant role in regulating gene expression or (ii) other factors help to determine the level of expression. We confirmed our previous conclusion that the enzyme FBXL11 is a potent negative regulator of NF-κB–dependent gene expression through its ability to catalyze the demethylation of K218 and K221. However, for many genes, the DKQ mutation decreases but does not eliminate DNA binding or expression and, at least for some genes, DKQ does not affect either DNA binding or gene expression. An important question for future research is to determine the structures of NF-κB bound to the κB elements of genes for which the DKQ mutation either does or does not inhibit function. Whether promoter binding plays a major role in regulating the expression of a specific NF-κB–induced gene does not seem to be related to whether the expression of that gene is fast and slow, indicating that promoter binding is not the major factor determining how quickly a particular gene responds to the activating signal. As Adelman et al. (18) have suggested, the status of RNA polymerase II at each promoter is an important factor. In the context of LPS-induced gene expression in macrophages, RNA Polymerase II initiates transcription in resting macrophages, but stalls near the promoter of fast response genes, such as TNF-α, until LPS triggers rapid and transient release of the negative elongation factor complex and productive elongation. In contrast, no negative elongation factor or polymerase is detectible near the promoter of the slow genes, such as IP10, before induction, and LPS-dependent polymerase recruitment is rate-limiting for transcription.
Taken together, our observations reveal an interrelated set of factors that contribute to the complex regulation of gene expression in response to the activation of NF-κB. First, the methylation of K218 and K221 and K37 profoundly affect the function of NF-κB, altering transactivation potency and affinity for DNA, and thus affecting the strength and duration of induced gene expression. Second, the effects of methylations of K37 and K218/221 are gene-specific, leading to quite different effects on individual genes, helping to give plasticity to the dependent biological responses. As discussed by Stark et al. (11), evidence is emerging that methylation—and perhaps other posttranslational modifications as well—of transcription factors occur following their binding to promoters. The ability to methylate NF-κB differentially at each specific promoter is likely to be guided by the promoter-specific presence and arrangement of regulatory proteins, with different arrangements optimally suited to fine-tune each response. The strength of interaction of NF-κB with specific κB sequences is certain to be an important factor in determining how each gene is expressed. Thus, a single modified species of activated NF-κB can affect specific subclasses of promoters and gene expression in very different ways.
Materials and Methods
Antibodies, Plasmids, and Cells.
The following antibodies were from commercial sources: anti–NF-κB p65 (Santa Cruz Biotechnology) and anti–NF-κB p65-Chip grade (Abcam). p65 shRNAs were from Open Biosystems. The 293IL1R cells and 293IL1R cells overexpressing FBXL11 were described by Lu et al. (8).
Transfections.
Constructs were transfected into cell lines by using the Lipofectamine and PLUS Reagents (Invitrogen). To establish stable pools, cells were cotransfected with a plasmid encoding a puromycin resistance gene, and selected in 1 μg/mL of puromycin 48 h later. Transfections were carried out essentially as previously described (8).
Western Analyses.
Cells were cultured to about 95% confluence and samples were collected and assayed by the Western method, as described by Lu et al. (7).
Reverse PCR and Quantitative Real-Time PCR.
Human cDNA samples were made by reverse PCR from total RNA of 293IL1R cells by using the SuperScript III First-Strand Synthesis System (Invitrogen). For different experiments, samples were either amplified according to standard PCR procedure or analyzed by using FastStart Universal SYBR Green Master (ROX) (Roche Diagnostics) quantitative PCR (qPCR) reactions. Primers were designed by Primer Express 3.0 software.
Site Mutagenesis.
Using pcDNA3-p65 plasmid as a template, we generated the DKQ and 37Q mutants by using the QuikChange Site-Directed Mutagenesis Kit from Stratagene following the protocol provided by the manufacturer. Primers used were: DKQ-sense: 5′'-ttcctactgtgtgaccaggtgcagcaagaggacattgagg-3′; DKQ-antisense: 5′-cctcaatgtcctcttgctgcacctggtcacacagtaggaa-3′; 37Q-sense: 5′-cgcttccgctaccagtgcgaggggc-3′; and 37Q-antisense: 5′-gcccctcgcactggtagcggaagcg-3′.
Chromatin Immunoprecipitation.
Cells were cultured to 90% confluence in 10-cm plates, and then 20% (vol/vol) formaldehyde was added directly to the media to a final concentration of 1%, followed by incubation at room temperature for 10 min. The cross-linking reaction was then stopped by adding glycine to a final concentration of 0.125 M. Media was removed from plates and cells were washed with an equal volume of cold 1 × PBS (PBS, Thermo Scientific). The buffer was then removed by aspiration and 5–8 mL of cold Farnham lysis buffer [5 mM Pipes, pH 8.0, 85 mM KCl, 0.5% (vol/vol) Nonidet P-40, plus Roche Protease Inhibitor Mixture] was then added to the dish. Cells were scraped off the dish and transferred into 50-mL conical tubes on ice and pelleted at 4 °C. The fresh pellet (108 cells) was then resuspended on ice in 5 mL of Farnham Lysis Buffer, and the crude nuclear preparation was collected by centrifugation at 4 °C. The pellets were then resuspended into 5 mL of RIPA buffer [1× PBS, 1% (vol/vol) Nonidet P-40, 0.5% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) SDS, plus Roche Protease Inhibitor Mixture] and 1 mL of aliquots were transferred to 1.5 mL Eppendorf tubes (2 × 107 cells/mL equivalent). Each 1.0 mL CHIP sample was sonicated on ice at Power output 5 W, four times for 30 s each, with at least 30 s of cooling on ice between each sonication, to obtain an average length of the sheared chromatin of about 250 bp or less, with a range of about 100–600 bp. The sonicated material was centrifuged at 14,000 × g in a microcentrifuge (Fisher Scientific, model microCL 21) for 15 min at 4 °C and the supernatant was collected and immunoprecipitated by using specific antibodies and the EZ-ChIP kit from Millipore.
Illumina Microarrays.
RNA (250 ng) was reverse-transcribed into cRNA and labeled with biotin-UTP using the Illumina TotalPrep RNA Amplification Kit (Ambion/Applied Biosystems). The amount of cRNA was determined by using a nanodrop spectrophotometer and the cRNA quality (size distribution) was further analyzed in a 1% (wt/vol) agarose gel. cRNA was hybridized to Illumina Human Ref-v3 v1 Expression BeadChips, and scanned in a BeadArray Reader using standard protocols (provided by Illumina). Illumina’s BeadStudio software was used for data analysis.
Data Analysis.
Partek Genomics Suite (Partek) was used to identify peaks in our CHIP-Seq data. Partek fits a zero truncated negative binomial model to the sequencing data and identifies enriched regions as peaks (Partek ChiPseq white paper: www.partek.com/Tutorials/microarray/User_Guides/ChIPSeqPeakDetection.pdf). R/Bioconductor packages Rsamtools and GenomicRanges were used to download NCBI RefSeq gene annotations and to count the total number of sequence reads in each annotated region.
Supplementary Material
Acknowledgments
We thank the Cleveland Clinic Genomics Core for performing the Illumina Array analyses; Ms. Banu Gopalan from the Cleveland Clinic Bioinformatics core for helping with statistical analyses; and the Ohio State University Biomedical Informatics Core for performing the CHIP-Seq analysis. This research was supported by National Cancer Institute Grant P01 CA062220 (to G.R.S.) and GM08490 (to G.G.), Ohio Cancer Research Associates Grant 036433730102 (to T.L.), and Indiana University School of Medicine Grant 23-862-07TL (to T.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311770110/-/DCSupplemental.
References
- 1.Karin M, Yamamoto Y, Wang QM. The IKK NF-κ B system: A treasure trove for drug development. Nat Rev Drug Discov. 2004;3(1):17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Anrather J, Racchumi G, Iadecola C. cis-acting, element-specific transcriptional activity of differentially phosphorylated nuclear factor-κ B. J Biol Chem. 2005;280(1):244–252. doi: 10.1074/jbc.M409344200. [DOI] [PubMed] [Google Scholar]
- 4.Perkins ND, Gilmore TD. Good cop, bad cop: The different faces of NF-kappaB. Cell Death Differ. 2006;13(5):759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 5.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor κ B pathway. Oncogene. 2006;25(51):6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 6.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106(45):18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu T, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci USA. 2009;106(38):16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XD, et al. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28(8):1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D, et al. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat Immunol. 2011;12(1):29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark GR, Wang Y, Lu T. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 2011;21(3):375–380. doi: 10.1038/cr.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107(50):21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov GS, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27(19):6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, et al. PHF20 regulates NF-κB signaling by disrupting recruitment of PP2A to p65. Nature Communication. 2013;4:2062. doi: 10.1038/ncomms3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391(6665):410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 18.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci USA. 2009;106(43):18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.