Abstract
Biofilms represent the predominant mode of microbial growth in the natural environment. Bacillus subtilis is a ubiquitous Gram-positive soil bacterium that functions as an effective plant growth-promoting agent. The biofilm matrix is composed of an exopolysaccharide and an amyloid fiber-forming protein, TasA, and assembles with the aid of a small secreted protein, BslA. Here we show that natively synthesized and secreted BslA forms surface layers around the biofilm. Biophysical analysis demonstrates that BslA can self-assemble at interfaces, forming an elastic film. Molecular function is revealed from analysis of the crystal structure of BslA, which consists of an Ig-type fold with the addition of an unusual, extremely hydrophobic “cap” region. A combination of in vivo biofilm formation and in vitro biophysical analysis demonstrates that the central hydrophobic residues of the cap are essential to allow a hydrophobic, nonwetting biofilm to form as they control the surface activity of the BslA protein. The hydrophobic cap exhibits physiochemical properties remarkably similar to the hydrophobic surface found in fungal hydrophobins; thus, BslA is a structurally defined bacterial hydrophobin. We suggest that biofilms formed by other species of bacteria may have evolved similar mechanisms to provide protection to the resident bacterial community.
Keywords: biofilm surface protein, in situ immunofluorescence, biofilm hydrophobicity
Biofilms are communities of microbial cells encased in a self-produced extracellular matrix (1–3). They are implicated in the majority of chronic infections (4) but conversely have critical roles in bioremediation (5) and biocontrol processes (6, 7). Biofilms are also thought to be one of the main repositories of bacteria in natural environments such as soil and water (8). It is well established that biofilm formation and disassembly are tightly regulated. The genetic pathways responsible, and the corresponding impact on biofilm structure, have been elucidated for many species of Gram-positive (9–11) and Gram-negative bacteria (12, 13). A defining feature common to biofilms from different species is the production of the extracellular matrix that is typically composed of proteins, exopolysaccharides, and nucleic acids (1, 14). Little is known about the 3D organization of components of the matrix, how they interact with the cells in the biofilm, and how they interact with each other (1). However, recent examination of the “microanatomy” of Escherichia coli rugose colonies has started to elucidate the organization and architecture of the matrix components in these biofilms (15, 16).
Many bacterial species reside in the rhizosphere in direct contact with plant roots. In this environment bacteria can be either pathogenic or symbiotic (17). The Gram-positive soil bacterium Bacillus subtilis is one such symbiont. It produces compounds that stimulate plant growth and defense mechanisms, as well as more traditional antibacterial compounds (as reviewed in refs. 7 and 18). Furthermore, it seems that the ability of B. subtilis to function as a biocontrol agent in the rhizosphere and reduce infection by fungal and bacterial pathogens is dependent on its biofilm formation capability (18, 19). In the laboratory, B. subtilis has the ability to form different types of biofilms: complex colonies on the surface of agar plates and floating biofilms (pellicles) at the air-to-liquid interface. The biofilm matrix produced by B. subtilis is needed for each biofilm type and has two main components: an exopolysaccharide (EPS) and an amyloid fiber-producing protein, TasA. The matrix assembles with the aid of a small protein, BslA (previously called YuaB) (20–23). The complex colony biofilms formed by B. subtilis have been shown to be highly hydrophobic (24), evidenced by the nonwetting nature that is observed upon the addition of a water droplet. This behavior extends to wetting by aqueous solutions of organic solvents, including 60% ethanol (24), suggestive of a protective role of the biofilm matrix against environmental threats. The hydrophobicity of the colony has been attributed to both the EPS (24) and BslA (22) components that are needed for biofilm formation. It has also been proposed that surface hydrophobicity may play a role in the protective nature of the B. subtilis biofilm formed on plant roots (24).
Here we show that native BslA forms an elastic film at the interfaces of the B. subtilis biofilms and that purified BslA can spontaneously self-assemble at interfaces in vitro. We reveal that the structure of BslA contains an unusual type of Ig-like fold and possesses a striking hydrophobic “cap” with physiochemical properties reminiscent of the hydrophobic surfaces found in fungal hydrophobins. A combination of in vivo genetic analysis and in vitro biophysical analyses demonstrates that the hydrophobic domain of BslA is responsible for the hydrophobicity of the colony biofilms by influencing the stability of the surface layer structures. Taken together, the data presented herein define BslA as a member of a unique class of bacterially produced hydrophobins.
Results
BslA Coats the Air–Cell and Agar–Cell Interfaces of a Complex Colony.
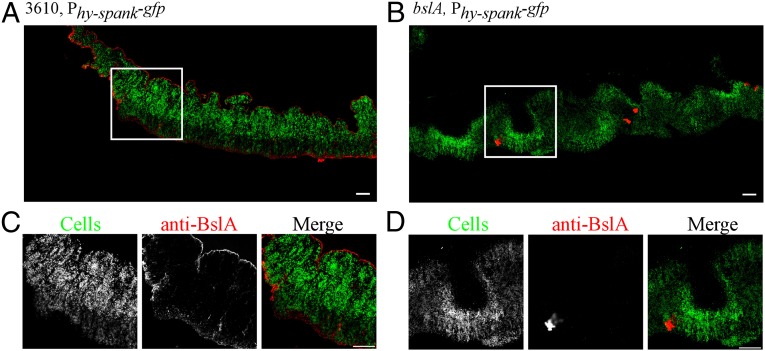
To gain functional insight regarding the role of BslA, the native protein was localized in situ within the colony biofilm using an immunofluorescence-based detection method (SI Materials and Methods). Strikingly, the majority of BslA formed a discrete layer at both the agar–cell and air–cell interfaces, surrounding the cells within the biofilm colony (Fig. 1). These findings are consistent with recent studies investigating the localization of purified fluorescently labeled BslA after heterologous addition to the colony biofilm (22). The BslA “coat” seems more compact at the top air–cell edge of the colony compared with the base of the colony (Fig. 1 A and C). On the top surface, only small protrusions of fluorescence from the BslA staining were found to penetrate deeper into the colony, whereas at the base of the colony BslA was slightly more diffuse, but still more prevalent at the surface (Fig. 1 A and C). The distinctive surface-associated BslA labeling was not visible upon analysis of the bslA mutant, indicating specificity of the labeling reaction (Fig. 1 B and D).
Fig. 1.
In situ analysis of BslA localization in the complex colony biofilm. Confocal scanning laser microscopy images of cross-sections through complex colonies formed by either (A and C) wild-type cells (3610, sacA::Phy-spank-gfp; NRS1473) or (B and D) the bslA mutant strain (3610, bslA::cat, sacA::Phy-spank-gfp; NRS3812). The smaller images show the region highlighted by the white box at higher magnification. Fluorescence from the GFP within the cells is shown in green in the large panels and in the merged images and the fluorescence associated with DyLight594, representing immuno-labeled BslA staining, is shown in red. (Scale bar, 50 µm.)
BslA Is Localized at the Liquid–Cell Interface in a Floating Biofilm.
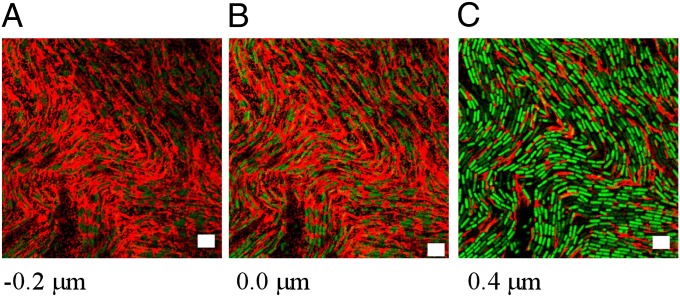
The in situ immunofluorescence technique used above was modified to analyze the localization of BslA in the pellicle biofilm. The mature pellicle was immobilized on a microscope coverslip using concanavalin A (25) and immunofluorescence analysis revealed a distinctive pattern of BslA-specific staining at the base of the pellicle structure (i.e., at the liquid–cell interface). BslA-specific staining was seen from 0.4 µm below the first layer of cells in the pellicle to a height of 4 µm into the pellicle biomass (Fig. 2 and quantification in Fig. S1A). BslA was found within the intercellular spaces and associated with the outer surface of the cells (Fig. 2B). Typically 25% of the surface area at the base of the pellicle was composed of BslA (Fig. S1B). Control experiments without the primary antibody showed no specific staining, indicating that the antibodies are not trapped in the biofilm matrix and/or accumulating at the base of the pellicle as a result of the experimental procedure (compare Fig. S1 B and C). Similarly, limited background fluorescence staining was observed in the bslA mutant pellicles (Fig. S1D) and a staining pattern similar to that in the wild type was observed in the complemented bslA mutant, indicating specificity of the labeling reaction (Fig. S1E). The abundance of the BslA-specific fluorescent staining at the base of the floating pellicle implies that BslA may form a protein raft carrying the biofilm mass and is suggestive of formation of higher-order structures by BslA in specific locations within the biofilm.
Fig. 2.
In situ analysis of BslA localization in the floating biofilm. Confocal scanning laser microscopy images of xy sections through a typical pellicle of wild-type strain NRS1473 (3610, sacA::Phy-spank-gfp) after immunofluorescence staining. (A) −0.2 μm, (B) 0 μm, and (C) 0.4 μm into the height of the pellicle. Fluorescence from the GFP within the cells is false-colored green and fluorescence associated with DyLight594, representing immuno-labeled BslA staining, is false-colored red in the merged image. (Scale bar, 5 μm.)
BslA Localization Is Not a Consequence of Site-Specific Transcription.
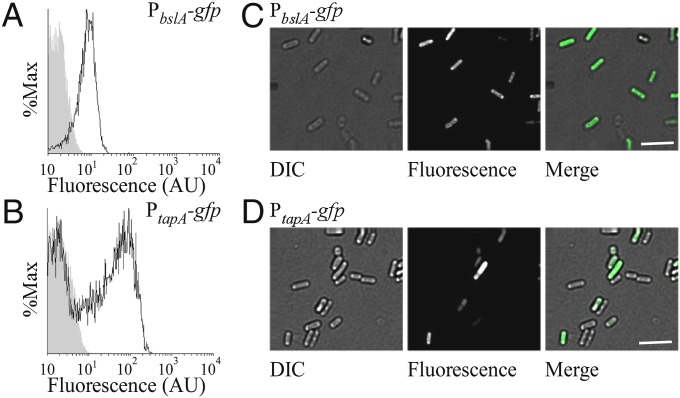
One mechanism by which BslA could occupy a spatially defined position in the biofilm is through localized transcription in the cells located at the biofilm surfaces. To test whether this was the case, the PbslA-gfp transcriptional reporter fusion was introduced into NCIB3610 and the fluorescence generated was analyzed by both single-cell microscopy and flow cytometry of cells isolated from an entire colony biofilm. These analyses showed homogenous expression from the bslA promoter in the population (Fig. 3 A and C), which is consistent with previous results (26, 27) and is in contrast to a PtapA-gfp reporter fusion that was used as a positive control for heterogeneous gene expression in the biofilm (Fig. 3 B and D) (27). Thus, we demonstrate that the localization of the mature BslA protein is not the result of localized transcription, but how BslA self-segregates in the biofilm remains unknown and most likely depends on the innate properties of the mature protein that are discussed below.
Fig. 3.
Analysis of bslA expression by cells forming complex colonies. (A and B) Flow cytometry analysis of expression of (A) bslA (NRS2289; 3610, sacA::PbslA-gfp) and (B) tapA-sipW-tasA (NRS2394; 3610, sacA::PtapA-gfp) in cells isolated from complex colonies after 18 h of growth at 37 °C. The gray-shaded zone represents the fluorescence observed for wild-type NCIB3610 containing no gfp. (C and D) Single-cell microscopy of cells isolated from complex colonies carrying either the (C) PbslA-gfp (NRS2289) or (D) PtapA-gfp (NRS2394) transcriptional reporters. (Scale bars, 10 µm.)
BslA Spontaneously Self-Assembles into a Protein Film in Vitro.
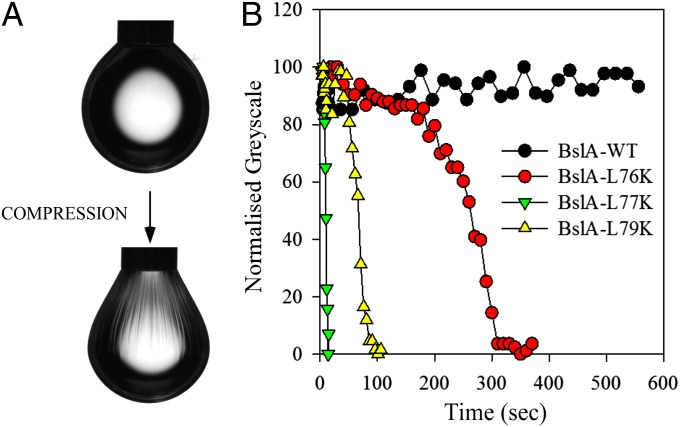
BslA forms a distinct layer at biofilm surfaces (Figs. 1 and 2) (22) and has been shown to floc in vitro (22). It was therefore investigated whether BslA could self-assemble into a protein layer in vitro or whether this higher level of protein organization was dependent on a protein or carbohydrate partner within the biofilm matrix. Recombinant BslA42–181 protein, purified from E. coli, has been shown to be biologically active when added to the B. subtilis biofilm (22). To assess self-assembly into a protein sheet at the interface recombinant BslA42–181 protein was subjected to analysis by the pendant droplet method (28, 29) (Fig. S2). For this a 40-μL droplet of BslA42–181 at 0.2 mg/mL in aqueous solution was expelled into an oil bath of glyceryl trioctanoate (Fig. 4A). Under these conditions a surface-active protein partitions to the oil/water interface. Upon retraction of 5 μL from the initial 40-μL droplet of BslA42–181 it was observed that BslA42–181 formed an elastic skin at the protein–oil interface (Fig. 4A and Movie S1). This is evidenced by the wrinkles in the zone surrounding the neck of the drop (Fig. 4A) that can be more clearly observed following retraction of more liquid (Fig. S2C). Analysis of the wrinkles in the protein film revealed that no relaxation occurred over a period of 10 min after compression (Fig. 4B, Movie S1, and Fig. S2), suggesting this surface layer is stable. These findings demonstrate that BslA is capable of self-assembly into a stable, complex higher-order film without the aid of a protein or carbohydrate partner.
Fig. 4.
In vitro analysis of BslA self-assembly into an elastic protein film. Pendant droplet analysis of purified BslA42–181 protein shows elastic film formation at the protein–oil interface. (A) A 40-µL droplet of BslA42–181 (0.2 mg/mL in 25 mM phosphate buffer, pH 7) was expelled into glyceryl trioctanoate, and following 20 min of equilibration compressed by retraction of 5 µL. Wrinkles formed in the neck of the drop, indicative of film formation. (B) Film relaxation after droplet compression, as measured by loss of surface wrinkles (expressed as the reduction in normalized grayscale values). Wild-type BslA42–181 shown by black circles; also shown are BslA42–181 containing the amino acid substitutions L76K (red circles), L77K (green triangles), and L79K (yellow triangles).
Structural Analysis of the BslA Protein Reveals High Levels of Surface Hydrophobicity.
To understand the function of BslA at the molecular level the crystal structure of BslA was determined. Residues 48–172 of BslA [corresponding to the ordered region of BslA as assessed by JPred (30)], was overexpressed as a GST fusion in E. coli. Mutation of leucine 98 to methionine was introduced to aid structure determination by incorporation of additional selenomethionine. This mutation does not affect protein function in vivo (Fig. S3). To reduce self-assembly and aid protein crystallization, surface lysine residues were reductively methylated after cleavage of BslA from GST (31). Methylated BslA48–172-L98M formed crystals in lithium sulfate solutions. Crystals of BslA formed in space group P212121 with 10 molecules in the asymmetric unit (Table S1). The structure was determined by a selenomethionine single-wavelength anomalous dispersion experiment to 1.9 Å resolution, and refined to an R factor of 16.5 (Rfree = 19.9) with good stereochemistry (Table S1).
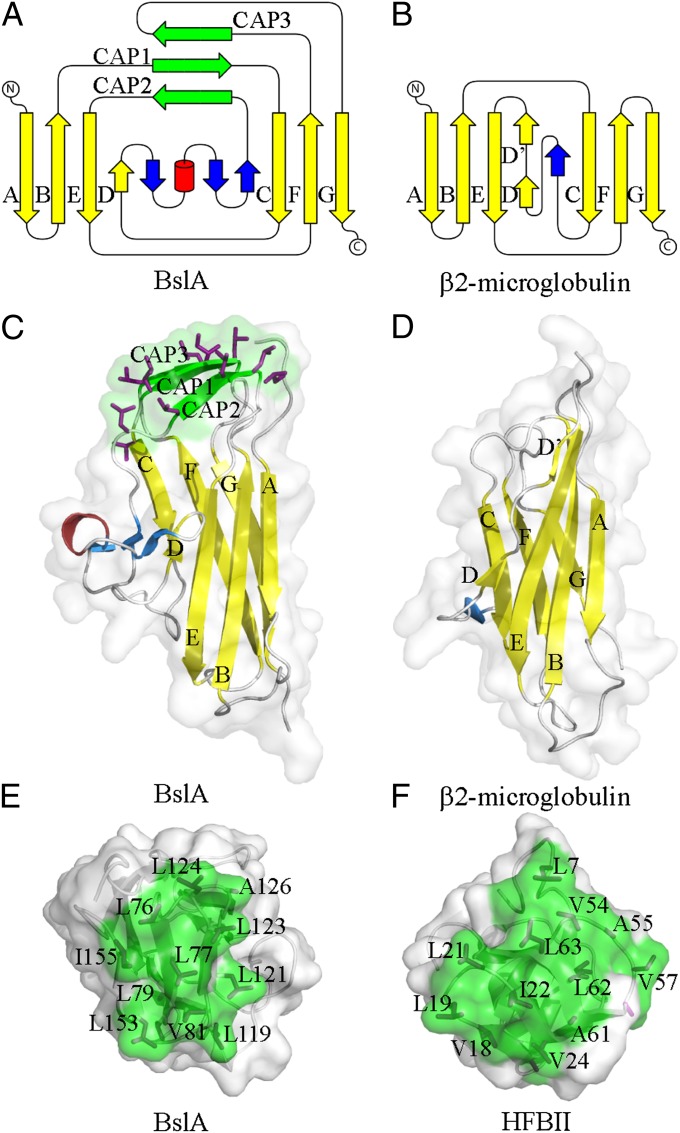
The structure of BslA consists of one 310 helix and 13 β-strands that form two distinct faces on opposite sides of the molecule, with strands A, B, E, and D forming one β-sheet and strands C, F, and G creating the opposing β-sheet face (Fig. 5 A and C). Strikingly, despite low primary sequence homology, the overall fold clearly identifies BslA as a member of the Ig superfamily (Fig. 5 B and D) (32). Structural homology searches using secondary-structure matching (33) reveal several members of the Ig family that share structural features with BslA. One of the most similar structures is the β-2 microglobulin (34) with a Z-score of 4.9 and a root mean square deviation of 2.7 Å over 84 equivalent Cα atoms. The structure and topological organization of BslA and β-2 microglobulin are compared in Fig. 5.
Fig. 5.
Structural analysis of BslA. (A and B) Topological representation of (A) BslA and (B) the structurally similar β2-microglobulin (34) constructed using TopDraw (44). Yellow and green β-strands represent conservation with the canonical Ig fold and the hydrophobic cap, respectively. Blue β-strands and the red α−helix represent secondary structure not part of the classical Ig fold. (C and D) Ribbon representation of the structure of (C) BslA and (D) β2-microglobulin using the same color scheme as in A and B. The surface-exposed leucine, isoleucine, and valines are represented as sticks with magenta carbon atoms. (E and F) The hydrophobic regions of (E) BslA and (F) HFBII (35). The hydrophobic region is shown in green, and surface-exposed leucine, isoleucine, and valines are annotated.
An additional, albeit smaller, three-stranded β-sheet (β-strands CAP1, CAP2, and CAP3) straddles the two main β-sheet faces and is positioned above the Ig fold (Fig. 5 A and C), forming an almost flat surface. The third β-strand (CAP3) is a transient β-strand because it only forms part of the β-sheet in some of the monomers in the asymmetric unit (PDB ID code 4bhu). Comparison with other Ig proteins reveals that this β-sheet cap is unique to BslA. Strikingly, this region displays numerous solvent-exposed hydrophobic amino acids (Fig. 5 C and E). These unusual exposed hydrophobic amino acids are masked from the solvent in the crystal structure by virtue of the crystal packing with the other nine molecules of BslA that form the asymmetric unit (Fig. S4). This protein packing arrangement is reminiscent of a micelle, with the hydrophobic cap orientated toward the center of the decamer and thus excluding solvent molecules (Fig. S4). The total surface area of BslA is ∼6,670 Å2, and the estimated area of the hydrophobic patch is about 1,620 Å2 (24% of the total surface area) (Fig. 5E). Although the BslA fold is not similar to that of the fungal hydrophobins, the hydrophobic cap bears remarkable physiochemical similarity with the hydrophobic patch observed in the crystal structure of the hydrophobin HFBII (35) (Fig. 5F). Fungal hydrophobins are known for their high surface activity (36), with exposed hydrophobic amino acids forming 12% of the surface of the HFBII protein (e.g., leucines, valines, and isoleucines, as shown in Fig. 5F).
Analysis of the Hydrophobic Cap.
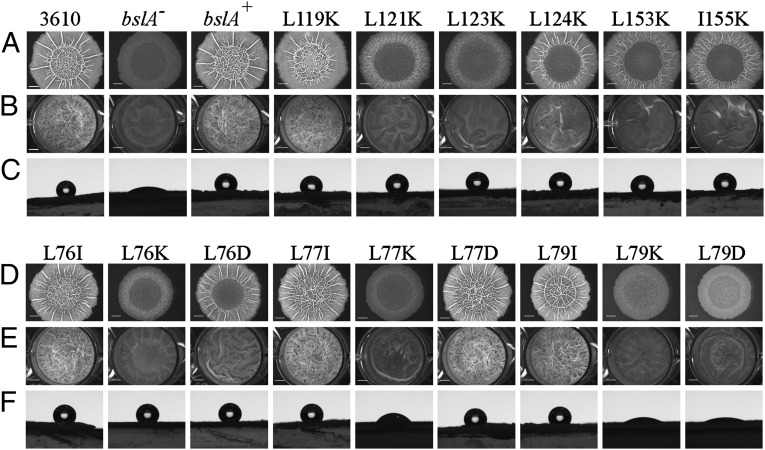
To investigate the role of the extensive BslA hydrophobic cap, mutations were introduced in the coding sequence that changed each of the exposed leucines and isoleucines in the three β-strands (CAP1, CAP2, and CAP3) in turn to lysine. These mutations should disrupt the hydrophobic face at each position (Fig. 6 and Table S2). In vivo analyses of the effects of these BslA mutations on biofilm formation showed that the three leucines in the middle strand (β-strand CAP1: L76, L77, and L79) individually had the greatest effect on BslA function in the biofilm. Heterologous expression of these mutant alleles could not restore wild-type biofilm formation to the bslA mutant (Fig. 6D), as evidenced by the flat, unwrinkled biofilms that formed. In the presence of the L77K and L79K BslA mutant proteins the colony biofilm remained wettable by water (Fig. 6F and Table S2). The presence of BslA protein was confirmed by Western blots of all of the complex colonies (Fig. S5 A and B) and of pellicles formed by the CAP1 mutants (Fig. S5C). The loss of colony hydrophobicity was not merely due to lack of colony complexity, because the strain possessing the L76K mutation exhibited a morphology near that of the bslA null mutant yet retained the nonwetting, hydrophobic nature of the wild-type colony (Fig. 6 D and F). Of the hydrophobic residues on the outer strands (β-strands CAP2 and CAP3), only mutating the amino acids in the central parts of the strands (L121, L123, L153, and I155) had any effect on BslA function. Those at the outer edges of the strands (L119 and L124) had no observable effect on the biofilm structure or hydrophobicity (Fig. 6 A–C). Thus, the three hydrophobic residues along the middle CAP1 strand (L76, L77, and L79) were selected for further analyses after recombinant proteins containing these mutations were shown by CD spectroscopy to fold correctly (Fig. S6).
Fig. 6.
In vivo biofilm analysis of the hydrophobic cap of BslA. (A) Complex colony morphologies of strains containing leucine/isoleucine-to-lysine mutations in the β-sheets CAP2 and CAP3 alongside wild-type (NCIB3610), bslA− (NRS2097), and bslA+ (NRS2299) controls. (B) Pellicle morphology of the CAP2 and CAP3 mutants shown in A. (C) Sessile water-drop analysis of colony hydrophobicity of CAP2 and CAP3 mutants. Colonies were grown as for morphology analysis and 5-µL water drops placed on top. (D) Complex colony morphologies of strains containing mutations in the central CAP1 β-sheet. (E) Pellicle morphology of the CAP1 mutants shown in D. (F) Water-droplet analysis of colony hydrophobicity of CAP1 mutants. Table S3 gives strain details.
Mutant forms of BslA containing either a conservative change (to isoleucine) or the introduction of an isosteric negative charge (aspartic acid) were constructed for L76, L77, and L79 and the colony and pellicle biofilms analyzed in vivo (Fig. 6 D–F, Fig. S4 B and C, and Table S2). Of these, only two had an effect on the function of BslA: L76D resulted in partial loss of morphological complexity and L79D had a BslA-null morphology and was wettable by water. Analysis of the localization of BslA in the floating pellicle biofilms (comparing pellicle and media fractions) revealed that in wild-type samples and those with wild-type morphology (L76I, L77I, L77D, and L79I) BslA is predominantly found in the pellicle (cell and matrix) fraction. In contrast, in pellicles that are morphologically BslA-null, the mutant BslA proteins were found in both the pellicle and media fractions, but predominantly within the media fractions (Fig. S5C). The loss of association of BslA from the cells in the biofilm has previously been reported for a G80D BslA mutation (22). Analysis of the G80D mutation with regard to the crystal structure indicates that an aspartic acid at this position would perturb the local protein structure and thus disrupt the hydrophobic cap β-sheet, because this side chain would point toward the core of the cap domain rather than the solvent. Together, these in vivo analyses have shown that the hydrophobic cap of BslA is vital for full BslA functionality within the biofilm, being required for both the complexity of the biofilm morphology and for the production of the hydrophobic surface layer. Moreover, it is the hydrophobic nature of the amino acids within the cap that is most important, not the specific amino acids themselves (because the leucine-to-isoleucine mutations had no effect on BslA function), and the central hydrophobic residues are of greatest importance for BslA function.
In Vitro Analyses of the Hydrophobic Residues on Polymerization and Film Formation.
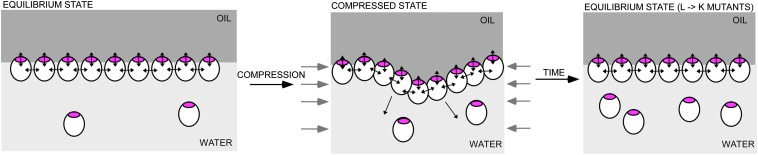
Having shown that the hydrophobic residues L76, L77, and L79 are important for the function of BslA in the B. subtilis biofilm, BslA42–181 protein containing each of the mutations L76K, L77K, and L79K was purified from E. coli to determine the impact of mutation on protein film formation in vitro. The pendant drop experiments were repeated for each of the mutant proteins and both film formation and relaxation of the elastic film after compression were compared with wild-type protein (Fig. S2, Fig. 4B, and Movies S2–S4). After 20 min of equilibration the 40-µL droplet of protein was compressed by removal of 5 µL. The observation of wrinkles on the drop surface indicates the formation of an elastic film; any relaxation of these wrinkles over time suggests the film is not stable and that protein can be released back into the aqueous reservoir (Fig. S2 and Fig. 7). During the measurement period there was no relaxation in the wrinkles formed by the wild-type protein (Fig. 4B). By comparison, the wrinkles in the film formed by the L77K protein immediately began to relax, and were fully relaxed after 15 s. The wrinkles in the film formed by the L79K protein upon droplet compression had an ∼45-s delay before relaxation started but were fully relaxed within 2 min, whereas the wrinkles formed in the film produced by the L76K protein upon droplet compression had an extended delay of ∼2 min before the start of relaxation but were almost fully relaxed after 5 min (Fig. 4B). This suggests that the surface activity of the monomers is decreased in each of the three mutant proteins, such that under compression of the droplet the decrease in surface activity allows release of the monomers from the interface and subsequent relaxation of the wrinkles in the protein film, with the greatest decrease in surface activity seen in the L77K protein (Fig. 7 shows a model).
Fig. 7.
Model of BslA film formation and relaxation after compression. In the equilibrium state, BslA will form a film at the water–oil interface, with both lateral protein–protein interactions between BslA monomers and interactions between the hydrophobic cap (shown in magenta) and the oil–water interface. After compression (by removal of some of the water) the monomers are moved closer together, creating the visible wrinkles. For the wild-type proteins the surface activity of the hydrophobic cap prevents monomers from being released from the BslA film, causing long-lasting wrinkles. However, in the proteins containing mutations in the CAP1 β-sheet the surface activity is lowered enough to allow release of some of the BslA monomers in the film, allowing the film to return to an equilibrium state and the relaxation of the wrinkles.
Discussion
The unique properties of BslA reported herein allow BslA to be defined as a surface-active protein and, moreover, a hydrophobin. Surface-active proteins modify the chemical and physical properties of an interface and are diverse in nature and function (36). Other examples of surface-active proteins include latherin from horse sweat (37), various amyloid proteins including bacterial curli (38), SapB, the chaplins and the rodlins from Streptomyces coelicolor (36, 39, 40), and the hydrophobins and repellents from fungi (36). Hydrophobins are small proteins found in fungi that confer water resistance to spores and prevent wetting after assembly (41). Hydrophobins typically contain a surface-exposed hydrophobic patch that is stabilized by the presence of disulphide bonds and can be distinguished into two classes that are defined by the solubility of the monolayer films (36). Comparative analysis of the crystal structure of HFBII from Trichoderma reesei with that of BslA identified physiochemical conservation of the hydrophobic domains (Fig. 5 E and F) (35), although there are no disulphide bonds present within the BslA protein for stabilization of its hydrophobic cap and the overall structures are entirely divergent. This is suggestive of parallel evolution of functional homologs in these two distantly related organisms. Based on both the physiochemical and functional homology with the hydrophobins of fungi, BslA is defined here as a “bacterial hydrophobin.”
Using in situ immunofluorescence microscopy we have revealed that natively produced and secreted BslA localizes to the interfaces of the B. subtilis biofilm, conferring hydrophobic properties (Figs. 1 and 2). Consistent with this, higher-order self-assembly of BslA in the absence of any biofilm matrix partner was shown by in vitro biophysical analysis (Fig. 4). Purified BslA exhibits surface activity and forms an elastic skin at the interface. Atomic-level analysis of the BslA monomer elucidated a unique combination of an Ig-like fold and a hydrophobic cap (Fig. 5), the latter component being reminiscent of the physiochemical properties of the fungal hydrophobins (35). The BslA crystal structure allows a molecular-level interpretation of the findings of Kobayashi and Iwano (22), who reported that mutation of serine 63 to proline disrupted BslA function and protein stability in vivo. The presence of this mutation would significantly disrupt BslA folding because the backbone nitrogen of serine 63 donates a β-sheet–type hydrogen bond to threonine 60. Here, using a targeted approach the importance of the hydrophobic cap of BslA was demonstrated by in vivo and in vitro analyses of site-directed mutants, with leucine 77 identified as critical for biological function, both for morphological complexity and surface hydrophobicity (Fig. 6). Leucine 77 is located at the center of the hydrophobic cap of the BslA monomer, and analysis demonstrated that it influences surface activity of the protein and not the polymerization process per se. Selective localization of BslA to the biofilm interface and self-assembly into a higher-order structure is consistent with BslA forming a hydrophobic barrier or integument of the biofilm that may serve a protective role (19, 22, 24).
The biofilm extracellular matrix is critical for stability, protection, and hydration of the community, but little is known about its structure and organization (1, 2, 8, 14). The high-resolution in situ microscopy used here provides information regarding the localization of a natively produced matrix associated protein in biofilms formed by B. subtilis. BslA was previously defined as a component of the biofilm matrix and it was shown that heterologous addition of purified BslA to the developing biofilm generated a surface layer (22). Here, using immunofluorescence microscopy analysis the localization of natively produced BslA within mature pellicle and complex colony biofilms was visualized and it was seen to form a protein shield surrounding the biofilm community (Figs. 1 and 2). Therefore, we conclude that BslA is not a core biofilm matrix component but a protein integument of the biofilm. The formation of an outer layer by natively produced BslA is reminiscent of the localized accumulation of curli-producing cells at the air–biofilm interface of E. coli rugose biofilms, which is known to serve a protective function (15).
There are several outstanding questions that follow the identification of the BslA as a surface active protein, the first being how the monomers of BslA interact during self-assembly into an elastic skin. This can be split into two distinct processes: partitioning to the interface and self-assembling into a film that can sustain elastic deformation. At this point we cannot conclusively determine which process the BslA point mutations are perturbing. However, we can speculate based on the crystal packing arrangements of the BslA decamer and the in vitro biophysical analysis of BslA self-assembly that BslA function in vivo is dependent on the hydrophobic cap and that disruption of this cap decreases the surface activity (Fig. 7). We predict that in the colony biofilm the hydrophobic cap of BslA will face out to the environment, conferring the hydrophobic properties of the mature biofilm (Fig. 7) (22, 24). In contrast, it is difficult to predict how BslA will orientate at the aqueous interface of the pellicle biofilm. In this location, the hydrophobic face of BslA will not be exposed to the aqueous environment, thus it is possible that BslA forms multiple protein layers in vivo. Such multilayers may allow either the hydrophobic or hydrophilic side of BslA to be exposed depending on the environmental conditions and would be entirely consistent with the thickness of the BslA layer observed by microscopy. It will be of interest to identify the nature of the BslA film in the biofilm structure and identify whether the TasA amyloid fibers and the exopolysaccharide in the biofilm matrix interact with the BslA hydrophobic surface layer to facilitate matrix assembly.
In the context of the natural environment the bacterial cells may benefit from the formation of the BslA surface layer by being buffered from the changing environment of the soil by excluding water, but there would be negative implications associated with this function, including desiccation and lack of nutrient uptake. Selective permeability of the BslA barrier would control the diffusion of molecules into the interior of the biofilm and therefore influence the perception of extracellular signaling molecules from other members of the microbial community and also nutrient uptake.
Materials and Methods
Full details of all methods used are provided in SI Materials and Methods.
Growth Conditions.
The B. subtilis strains used and constructed in this study are detailed in Table S3. The full details of growth conditions are provided in SI Materials and Methods. Biofilm pellicles were grown in MSgg medium (42) at 25 °C for 72 h, and complex colonies were grown on MSgg solidified with 1.5% Select Agar (Invitrogen) at 30 °C for 48 h.
Strain Construction.
All strains, plasmids, and primers used are presented in Table S3 and were constructed using standard techniques. SI Materials and Methods gives full details.
Quantification of the Abundance of Fluorescence.
To assess the abundance of fluorescence throughout the depth of the pellicle, the images acquired by confocal microscopy were stored and annotated with regions of interest in OMERO (43). Following this, automated batch image analysis was performed with bespoke software written in Matlab (MathWorks) via the OMERO API (code available upon request). SI Materials and Methods gives full details.
Supplementary Material
Acknowledgments
We thank the Diamond LightSource Beamline I-02 for data collection time and assistance, Dr. R. Clarke for assistance with flow cytometry, and Drs. G. Penman and V. Rao for purification advice and assistance with structure solution, respectively. We thank Dr. M. Elliot for helpful discussions. This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) Grants BB/C520404/1 and BB/I019464/1 and BBSRC Doctoral Training Account Grant BB/D526161/1 (to A.O.). F.V.R. is supported by a Federation of European Biochemical Societies (FEBS) Return-to-Europe Fellowship and Marie Curie European Research Council International Re-Integration Grant (ERC-IRG) IRG26838. D.M.F.v.A. is funded by a Medical Research Council Programme Grant G0900138 and a Wellcome Trust Senior Research Fellowship WT087590MA. The microscopy facilities are supported by Wellcome Trust Strategic Award Grant 083524/Z/07/Z.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4bhu).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306390110/-/DCSupplemental.
References
- 1.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 3.Davey ME, O’toole GA. Microbial biofilms: From ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Singh R, Paul D, Jain RK. Biofilms: Implications in bioremediation. Trends Microbiol. 2006;14(9):389–397. doi: 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Emmert EA, Handelsman J. Biocontrol of plant disease: A (gram-) positive perspective. FEMS Microbiol Lett. 1999;171(1):1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 7.Nagórska K, Bikowski M, Obuchowski M. Multicellular behaviour and production of a wide variety of toxic substances support usage of Bacillus subtilis as a powerful biocontrol agent. Acta Biochim Pol. 2007;54(3):495–508. [PubMed] [Google Scholar]
- 8.Costerton JW, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 9.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125(2):237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 10.López D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010;34(2):134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 11.Murray EJ, Kiley TB, Stanley-Wall NR. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology. 2009;155(Pt 1):1–8. doi: 10.1099/mic.0.023903-0. [DOI] [PubMed] [Google Scholar]
- 12.Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13(7):1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- 13.Petrova OE, Sauer K. Sticky situations: Key components that control bacterial surface attachment. J Bacteriol. 2012;194(10):2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.DePas WH, Chapman MR. Microbial manipulation of the amyloid fold. Res Microbiol. 2012;163(9-10):592–606. doi: 10.1016/j.resmic.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Serra DO, Richter AM, Klauck G, Mika F, Hengge R (2013) Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4(2):e00103-13. [DOI] [PMC free article] [PubMed]
- 17.Ongena M, Jacques P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16(3):115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Morikawa M. Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J Biosci Bioeng. 2006;101(1):1–8. doi: 10.1263/jbb.101.1. [DOI] [PubMed] [Google Scholar]
- 19.Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134(1):307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59(4):1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 21.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55(3):739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85(1):51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 23.Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol. 2011;193(18):4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epstein AK, Pokroy B, Seminara A, Aizenberg J. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci USA. 2011;108(3):995–1000. doi: 10.1073/pnas.1011033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R (2012) Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 3(4):e00184-12. [DOI] [PMC free article] [PubMed]
- 26.Kovács AT, Kuipers OP. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J Bacteriol. 2011;193(4):998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22(7):945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexandrov N, Marinova KG, Danov KD, Ivanov IB. Surface dilatational rheology measurements for oil/water systems with viscous oils. J Colloid Interface Sci. 2009;339(2):545–550. doi: 10.1016/j.jcis.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Russev SC, et al. Instrument and methods for surface dilatational rheology measurements. Rev Sci Instrum. 2008;79(10):104102. doi: 10.1063/1.3000569. [DOI] [PubMed] [Google Scholar]
- 30. Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36(Web Server issue):W197–201. [DOI] [PMC free article] [PubMed]
- 31.Walter TS, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14(11):1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238(4):528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 33. Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2256–2268. [DOI] [PubMed]
- 34.Calabrese MF, Eakin CM, Wang JM, Miranker AD. A regulatable switch mediates self-association in an immunoglobulin fold. Nat Struct Mol Biol. 2008;15(9):965–971. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakanpää J, Linder M, Popov A, Schmidt A, Rouvinen J. Hydrophobin HFBII in detail: Ultrahigh-resolution structure at 0.75 A. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):356–367. doi: 10.1107/S0907444906000862. [DOI] [PubMed] [Google Scholar]
- 36.Elliot MA, Talbot NJ. Building filaments in the air: Aerial morphogenesis in bacteria and fungi. Curr Opin Microbiol. 2004;7(6):594–601. doi: 10.1016/j.mib.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Beeley JG, Eason R, Snow DH. Isolation and characterization of latherin, a surface-active protein from horse sweat. Biochem J. 1986;235(3):645–650. doi: 10.1042/bj2350645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capstick DS, Jomaa A, Hanke C, Ortega J, Elliot MA. Dual amyloid domains promote differential functioning of the chaplin proteins during Streptomyces aerial morphogenesis. Proc Natl Acad Sci USA. 2011;108(24):9821–9826. doi: 10.1073/pnas.1018715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliot MA, et al. The chaplins: A family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003;17(14):1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris VK, et al. Recruitment of class I hydrophobins to the air:water interface initiates a multi-step process of functional amyloid formation. J Biol Chem. 2011;286(18):15955–15963. doi: 10.1074/jbc.M110.214197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98(20):11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allan C, et al. OMERO: Flexible, model-driven data management for experimental biology. Nat Methods. 2012;9(3):245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bond CS. TopDraw: A sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19(2):311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.