Significance
This study provides new insight into the requirements for observed silencing of RNA polymerase II transcription near tRNA genes. Mod5 is a conserved tRNA modification enzyme found in both the nucleus and cytoplasm, although it only modifies tRNAs in the cytoplasm. Mod5 is required for silencing near tRNA genes, and it is bound to both nuclear tRNA gene complexes and nuclear pre-tRNA transcripts. Possible mechanisms for this form of RNA-mediated transcriptional silencing are discussed.
Keywords: RNA silencing, Maf1
Abstract
The tRNA gene-mediated (tgm) silencing of RNA polymerase II promoters is dependent on subnuclear clustering of the tRNA genes, but genetic analysis shows that the silencing requires additional mechanisms. We have identified proteins that bind tRNA gene transcription complexes and are required for tgm silencing but not required for gene clustering. One of the proteins, Mod5, is a tRNA modifying enzyme that adds an N6-isopentenyl adenosine modification at position 37 on a small number of tRNAs in the cytoplasm, although a subpopulation of Mod5 is also found in the nucleus. Recent publications have also shown that Mod5 has tumor suppressor characteristics in humans as well as confers drug resistance through prion-like misfolding in yeast. Here, we show that a subpopulation of Mod5 associates with tRNA gene complexes in the nucleolus. This association occurs and is required for tgm silencing regardless of whether the pre-tRNA transcripts are substrates for Mod5 modification. In addition, Mod5 is bound to nuclear pre-tRNA transcripts, although they are not substrates for the A37 modification. Lastly, we show that truncation of the tRNA transcript to remove the normal tRNA structure also alleviates silencing, suggesting that synthesis of intact pre-tRNAs is required for the silencing mechanism. These results are discussed in light of recent results showing that silencing near tRNA genes also requires chromatin modification.
Transcription silencing can occur near yeast tRNA genes, which are distributed throughout the linear chromosomes (1, 2). The tendency of tRNA genes to suppress transcription from nearby RNA polymerase II (pol II) promoters is mechanistically distinct from other forms of silencing in yeast, because gene mutations and deletions that affect other silencing mechanisms fail to affect tRNA gene-mediated (tgm) silencing (3). This tgm silencing is also distinct from the ability of tRNA genes to serve as insulators or boundary elements, because it requires the tRNA gene to be transcribed (2) rather than requiring only the presence of the TFIIIC transcription factor (4–6).This form of silencing depends on the subnuclear localization of the tRNA loci to the nucleolus and is relieved when nucleolar architecture is compromised (3, 7). The correlation between the loss of silencing and the loss of subnuclear localization raised the question of whether nucleolar localization alone was sufficient to suppress nearby pol II transcription. The existence of additional mechanisms required for the silencing was suggested by the requirement for Maf1, which was originally shown to be a repressor of RNA polymerase III (pol III) transcription that acts through interactions with pol III and the upstream transcription factor, TFIIIB, although the precise mechanism of inhibition is not currently known (8–11).
Here, we show that another protein, Mod5, is also required for tgm silencing. Mod5 was originally tested for tgm silencing effects because of its genetic interaction with Maf1 (12, 13). Even so, it was not entirely expected that Mod5 would affect tgm silencing, because its previously known function is the isopentenylation of the A37 exocyclic amine in the tRNAs tRNATyr, tRNAPhe, and tRNASer (14–17). In addition, no other tRNA processing enzymes that we tested were required for the silencing, and therefore, it did not seem that tRNA maturation per se was needed (3). Here, we show that Mod5 binds to tRNA genes in vivo and the pre-tRNA transcripts, even if the RNA is not an appropriate substrate for Mod5. Possible explanations for the role of Mod5 and the pre-tRNA transcripts are discussed.
Results
Mod5 Is Required for Silencing near tRNA Genes.
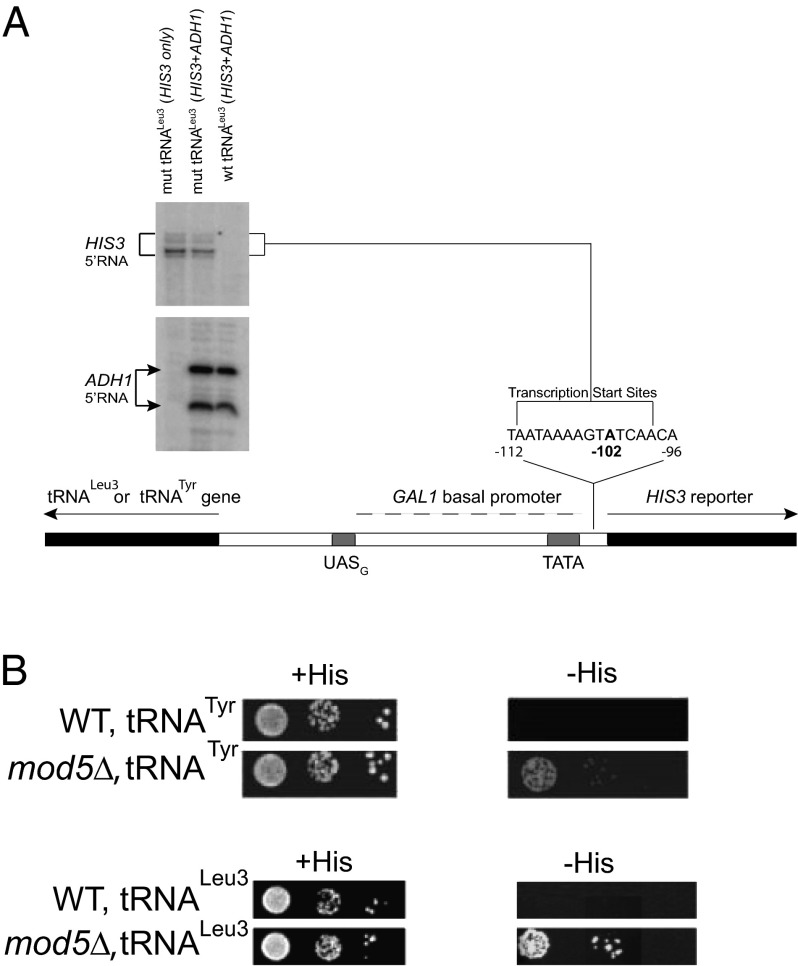
Over 200 gene deletions and mutations have previously been tested to determine which proteins are necessary for tgm silencing (3, 18). The plasmid reporter construct contains an active tRNA gene upstream of an HIS3 coding region in strains where the chromosomal HIS3 gene is deleted. HIS3 transcription is controlled by a modified GAL1 promoter with a single, consensus upstream activation sequence (UASG) binding site for Gal4 protein (Fig. 1A). WT cells are not able to grow in the absence of histidine, because the tRNA gene prevents pol II transcription (Fig. 1A), even when Gal4 is bound to the UASG (2, 18).
Fig. 1.
Deletion of MOD5 releases tgm silencing. (A) Plasmid construct used for testing for tgm silencing. A tRNALeu (SUP53) or tRNATyr (SUP4) gene in WT cells silences the HIS3 reporter gene under the control of a consensus Gal4 UAS and Gal1 basal promoter (2). In the mut tRNALeu version, point mutations in the A and B box promoters of tRNALeu (SUP53; G19C and C56G) eliminate association of all tRNA gene transcription components, and tgm silencing is lost. Primer extension analysis from the HIS3 coding region shows the level and 5′ termini of the HIS3 transcripts, with primer extension on ADH1 mRNA in the same reactions used as an internal quantitation control (2). In the first lane, the ADH1 primer is omitted to verify the identity of the HIS3 products. The major HIS3 transcript 5′ terminus at −102 relative to the translation start is indicated. (B) Cells in which tgm silencing is disrupted by deletion of MOD5 express HIS3 and are able to grow on media lacking histidine (SGR-ura-his). Silencing still requires MOD5 regardless of whether the SUP4 tRNATyr gene or the SUP53 tRNALeu3 gene is present.
Most mutations previously shown to alleviate tgm silencing cause a mislocalization of the tRNA genes, and therefore, they no longer cluster near the nucleolus (3). These mutations typically cause poor growth of the mutant yeast strains, because they usually disrupt nucleolar architecture. An exception is the deletion of MAF1, which both derepresses tRNA gene transcription and alleviates tgm silencing (9), but it does not have any detectable growth phenotype. Because MAF1 is known to affect both localization and function of another protein Mod5, a known tRNA modifying enzyme (12, 13, 19), we also tested mod5∆ and found that it alleviates tgm silencing (Fig. 1B) and has no negative growth phenotype under normal conditions. Because the tgm silencing assay uses a plasmid containing an active SUP4 tRNATyr gene, which produces a substrate for Mod5, we also used a second test construct having a tRNA gene that does not encode a substrate of Mod5, the SUP53 gene variant of the tRNALeu3 family (2). Silencing occurs in both constructs, and deletion of MOD5 caused alleviation of silencing in both constructs (Fig. 1B). Thus, Mod5 is required for local silencing, regardless of whether the tRNA gene product is a substrate for the enzyme.
Mod5 Is Not Required for Subnuclear Clustering of tRNA Genes.
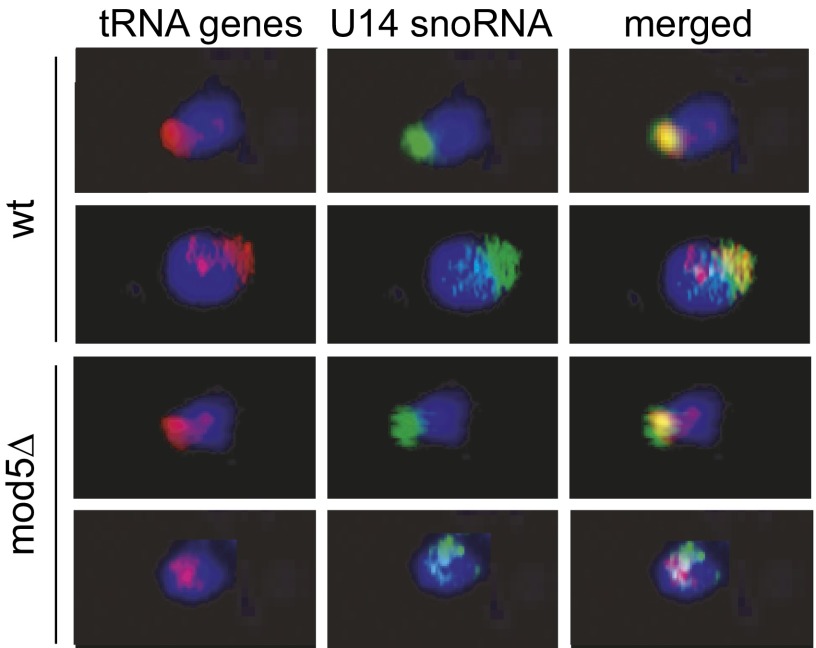
Previous deletions and mutations that caused alleviation of tgm silencing had been in proteins affecting nucleolar architecture and chromosome condensation, and they had dramatic effects on the subnuclear clustering of the tRNA genes (3, 18, 20). It is possible that mislocalization of tRNA genes alleviated tgm silencing in these cases, simply because the removal of loci from the nucleolus made them more accessible to pol II and pol III transcription factors (3) or the mislocalization disrupted spatial relationships needed for organizing active silencing events. To test whether subnuclear clustering alleviation is the case for the MOD5 gene deletion, this strain was tested for localization of the tRNA genes. The nucleolar clustering of 10 linearly dispersed tRNALeu3(CAA) genes was tested in these deletion strains using FISH as described previously (3, 7). In both the mod5∆ and the parental WT strains, the tRNALeu3 genes colocalized approximately with the nucleolar marker U14 in more than 90% of the cells imaged with probes to both (Fig. 2). This colocalization is quite different from results obtained with mutants that abrogate clustering and nucleolar localization of the tRNA genes (3, 18, 20), where there is little or no overlap between nucleolar markers and the tRNA gene signals. This result is also consistent with the normal growth rate of the mod5∆ strain, because strains with dispersed tRNA gene localization routinely grow slowly (3, 18, 20). Although we are unable to rule out more subtle disturbances of tRNA gene positions, other mutations giving loss of tgm silencing had displayed obvious mislocalization phenotypes (3). These data suggest that the alleviation of tgm silencing caused by the mod5∆ deletion is not caused by gross redistribution of the tRNA genes.
Fig. 2.
Deletion of MOD5 does not cause mislocalization of tRNA genes. The 10 tRNALeu(CAA) genes (red) and the U14 snoRNA nucleolar marker (green) were detected in fixed nuclei by in situ hybridization with fluorescent oligonucleotides, and representative cells are shown. Blue represents DAPI staining of nucleoplasmic DNA. As shown by these representative cells, the mod5∆ mutant strain maintains the WT localization of the tRNA genes as shown previously (18, 51).
Mod5 Is Not Required for Repression of tRNA Gene Transcription by Target of Rapamycin.
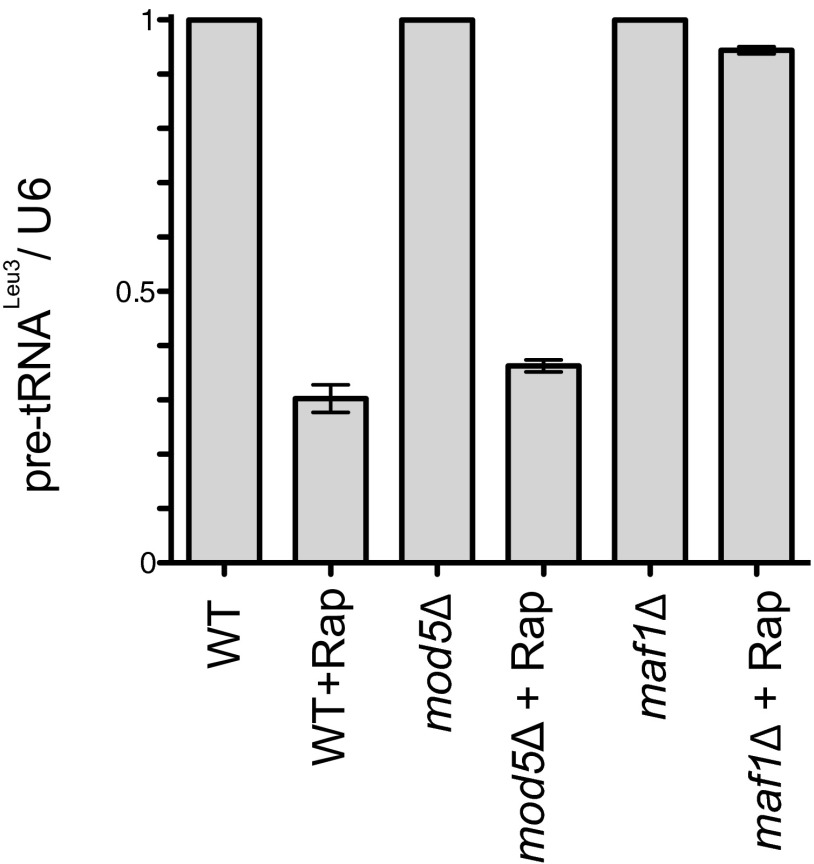
Treatment of yeast with rapamycin causes inhibition of pol III transcription of tRNA genes through action of the target of rapamycin pathway on Maf1 (10, 21, 22). We tested whether Mod5 might be required for Maf1 to repress tRNA transcription, thus suggesting that Maf1 was the target of Mod5 in the tgm silencing mechanism. Target of rapamycin inhibition of tRNA biosynthesis was assessed by Northern blot analysis of pre-tRNA:tRNA ratios as a measure of newly synthesized RNA (7–9). As expected, WT cells treated with rapamycin showed more than a threefold reduction in new tRNA synthesis, whereas maf1∆ cells showed no dramatic loss of transcription when treated with rapamycin (Fig. 3). In contrast, rapamycin treatment of mod5∆ cells caused repression of new pre-tRNA synthesis at a level similar to WT (Fig. 3). These results suggest that the actions of Mod5 in tgm silencing are either downstream or independent of the actions of Maf1 in repressing tRNA gene transcription.
Fig. 3.
Deletion of MOD5 does not alleviate repression of tRNA gene transcription mediated by Maf1. Three strains (WT, mod5∆, and maf1∆) were treated with 0.2 µg/mL rapamycin for 40 min. Denaturing polyacrylamide gel analysis of whole-cell RNA was analyzed by Northern blot from duplicate samples. The level of pre-tRNAs before endonucleolytic processing can be used as a measure of new synthesis when measured relative to a more stable pool of cellular U6 snRNA (22). Blots were probed for pre-tRNALeu3 and U6 snRNA as the internal normalization control. For each duplicate sample, the ratio of precursor tRNA to U6 signal is shown, and it is normalized to 1.0 for the ratio in that strain in the same experiment without rapamycin treatment. Deletion of MAF1 but not MOD5 reduces inhibition pre-tRNA synthesis by rapamycin.
Mod5 Modification of tRNAs Does Not Seem to Be Required for tgm Silencing.
Mod5 is an isopentenyl transferase that catalyzes the transfer of a dimethylallyl group from dimethylallyl pyrophosphate (DMAPP) to the exocyclic amine on position 37 of a few tRNAs in yeast (14–17). Although not required for life under most conditions, these modifications enhance the function of these tRNAs, which has allowed the development of a tRNA suppressor assay for translation efficiency (17). Previous studies of the enzymatic activity of Mod5 have used a strain (MT-8) with an ochre termination mutation in ade2 and an SUP7 ochre (14) tRNA gene that can suppress the ochre mutations only when the tRNA is modified by Mod5. Deletion of MOD5 renders the SUP7 ochre suppressor ineffective (16). Thus, the strain requires adenine in the media for growth, unless Mod5 is expressed from genes supplied on plasmids (14) (Fig. 4).
Fig. 4.
DMAPP depletion does not affect tgm silencing. (A) Atorvastatin, which prevents DMAPP formation by inhibiting HMG-CoA reductase, inhibits Mod5 sufficiently at 20 µg/mL to inhibit growth in the MT-8 strain, which tests for SUP7 tRNA modification by suppression of ade2 and lys2 ochre mutations. (B) Atorvastatin could not alleviate tgm silencing (test construct like in Fig. 1), even at 100 µg/mL of the drug. On Atorvastatin plates lacking histidine (−His), the mod5∆ strain allows growth, but in the WT strain, tgm silencing remains in force, suggesting that tRNA modification by Mod5 is not required. (C) A high-copy plasmid expressing ERG20 (YepERG20) was inserted into both WT and mod5∆ strains. Overexpression of Erg20p from this plasmid limits the ability of Mod5 to modify tRNAs by depleting cellular pools of DMAPP into a competing pathway for protein prenylation (13). Dilutions of these strains were plated in duplicate to selective (−His) and nonselective (+His) plates to test for tgm silencing from the reporter plasmid. Overexpression of ERG20 does not alleviate tgm silencing in the WT strain.
We tested whether the previously defined catalytic activity of Mod5 is required for tgm silencing by depleting its DMAPP substrate in two different ways (23–25). The first method was to treat cells with the drug Atorvastatin, which inhibits an early enzyme in DMAPP biosynthesis, 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) reductase (24). Atorvastatin at 20 µg/mL was sufficient in this assay to inhibit Mod5 tRNA modification activity and eliminate growth on solid medium lacking adenine (−Ade in Fig. 4A; but containing lysine) in strains containing MOD5 (Fig. 4A) as previously shown. We then tested the ability of Atorvastatin to limit tgm silencing. There was no loss of silencing up to 100 µg/mL Atorvastatin (higher concentrations are toxic), which is five times the concentration needed to effectively inhibit in the tRNA modification assay (Fig. 4B). The second method for depleting the DMAPP substrate was to overexpress the enzyme Erg20p from a high-copy plasmid. Erg20p uses DMAPP to create precursors for sterol biogenesis, and its overexpression depletes the DMAPP pool and decreases N6-isopentenyladenosine (i6A) tRNA modification. Although decrease of i6A tRNA modification was previously shown to cause a loss of ochre suppression in the SUP7 test system (23), overexpression of Erg20p did not cause an alleviation of tgm silencing (Fig. 4C). In combination, these data suggest that the tRNA modifying activity of Mod5 is not required for tgm silencing. The observation that Mod5 modification of tRNAs is not required for tgm silencing is consistent with the observation that the tRNA gene causing the silencing of nearby pol II transcription does not have to encode a pre-tRNA that can be modified by Mod5. Not only do the SUP4 and SUP53 pre-tRNA in the nucleus have introns that preclude their use as substrates, but mature SUP53 tRNA never becomes an Mod5 substrate because of an incompatible sequence at the site of modification.
We attempted to also identify mutations in MOD5 that would compromise the tgm silencing functions without affecting tRNA modification activity (SI Materials and Methods and Fig. S1). All mutations that affected one activity also affected the other activity, although most of these seemed to destabilize the expressed protein. One mutation in the putative tRNA modification site compromised both activities and seemed to express well (Fig. S1), but we are not currently able to differentiate between effects on catalysis vs. tRNA binding.
Mod5 Function in tgm Silencing Is Conserved in Eukaryotic Homologs.
The Mod5 protein is highly conserved from bacteria to mammals as a tRNA modification enzyme, with two highly conserved domains being an ATP/GTP binding domain and the DMAPP binding site (26) (Fig. 5A). One notable feature of the eukaryotic versions is that they have a longer C terminus, and this region has a zinc finger motif (26). Homologs of Mod5 have been cloned from both Arabidopsis thaliana and humans, and they are capable of performing the appropriate yeast tRNA modification as assessed by the SUP7 suppression assay (26–28). To test whether the ability to confer active tgm silencing in an mod5∆ strain is also conserved, we expressed ORFs of either the human or Arabidopsis homologs in constructs previously shown to confer tRNA modification along with a plasmid containing the tgm silencing test construct. The human Mod5 homolog (TRIT1) was able to fully restore tgm silencing in yeast, and the A. thaliana homolog partially restored tgm silencing (Fig. 5B). These results show that the participation of Mod5 in tgm silencing is a conserved property of the eukaryotic enzyme.
Fig. 5.
Conservation of the Mod5 protein and silencing function in eukaryotes. (A) The alignment of three Mod5 proteins from S. cerevisiae, A. thaliana, and H. sapiens is depicted, with areas of conservation indicated by shaded boxes. All variants contain motifs for ATP/GTP binding and DMAPP binding, including E. coli (14, 40). Eukaryotic homologs also have a zinc finger motif in the C-terminal extensions. (B) tgm silencing functions of yeast Mod5 are conserved in plant and human proteins. It was previously shown that human and Arabidopsis Mod5 homologs are able to restore i6A modification of tRNAs in yeast in an mod5∆ strain (14, 24). Plasmids expressing these ORFs were tested for supporting tgm silencing in an mod5∆ strain containing a tgm silencing reporter plasmid, with loss of silencing shown by growth on media lacking histidine (−His). Although both homologs restore silencing, the H. sapien version of Mod5 (TRIT1) is consistently more robust in preventing growth without histidine.
Mod5 Is Present at tRNA Genes.
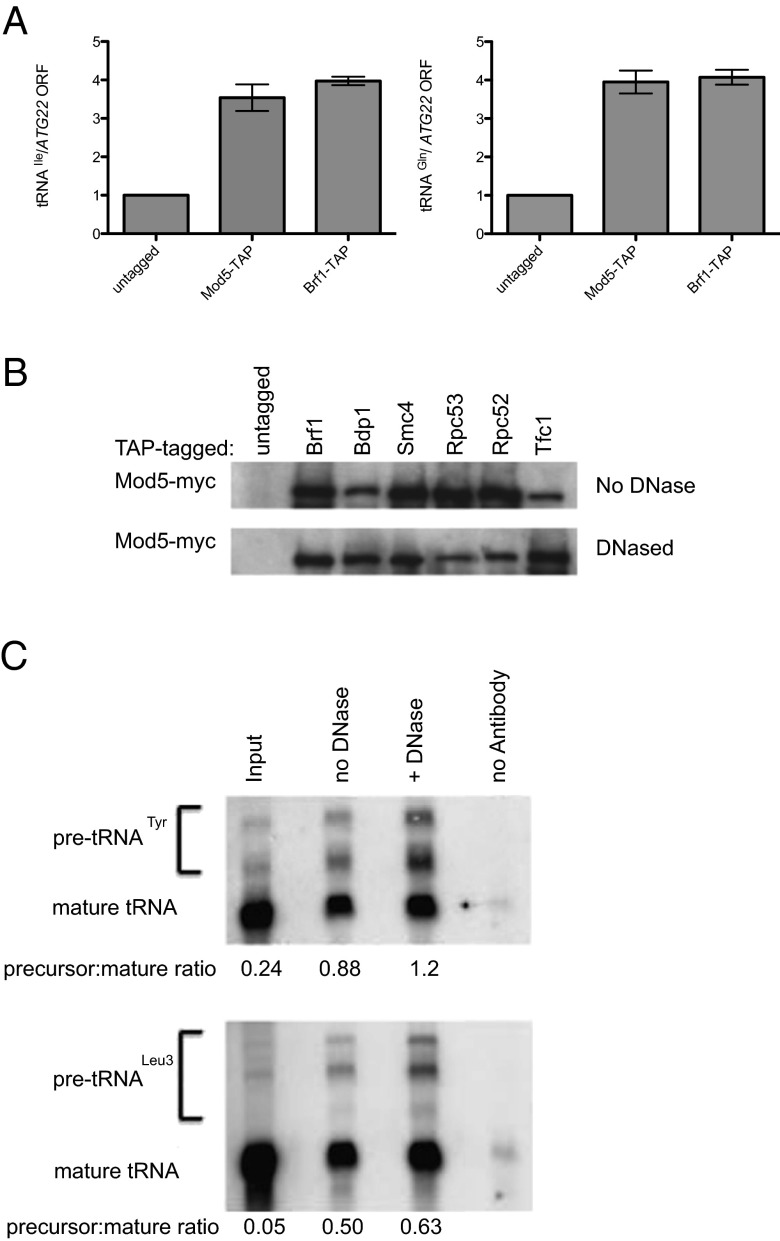
Because some Mod5 and most tRNA genes are located in the same subnuclear compartment, the nucleolus (29, 30), the hypothesis was suggested that Mod5 might be directly associated with tRNA genes and acting in cis to exert its effect on tgm silencing. To determine if Mod5 is present at tRNA genes, we used ChIP using endogenous Mod5 fused to a C-terminal tandem affinity purification (TAP) tag, where the gene is transcribed from its endogenous promoter. We performed ChIP using Mod5-TAP and the pol III transcription factor TFIIIB subunit, Brf1p, as a control, because it is stably bound to tRNA genes (11). Occupancy at two different chromosomal tRNA genes tRNAIle (tI[AUU]D on chromosome IV) and tRNAGln (tQ[UUG]E1 on chromosome V) was detected by semiquantitative PCR amplification (Fig. 6A) relative to an internal amplification control—the coding region of ATG22 that has no nearby tRNA genes. These loci had previously been shown to provide ChIP results comparable with genome-wide association studies for pol III complex components and condensin (20). As shown in Fig. 6A, Mod5 is enriched three- to fourfold at each tRNA gene relative to an internal control, which is similar to the results with Brf1p, although we are unable to determine the degree of occupancy from this type of experiment. It is notable that neither of these two tRNA genes produces a substrate for Mod5 (17), indicating that Mod5 is present at tRNA genes, even in the absence of synthesis in cis of an i6A modification substrate. The observation of Mod5 at tRNA genes that do not code for a Mod5 substrate is in agreement with our results that showed that both SUP4 and SUP53 cause tgm silencing, although neither pre-tRNA can be modified by Mod5.
Fig. 6.
Mod5 is physically associated with tRNA gene complexes and pre-tRNAs. (A) ChIPs were performed using TAP-tagged versions of Mod5 and a positive control (the Brf1p subunit of the continuously bound TFIIIB transcription factor). PCR detection used primers flanking tRNAIle and tRNAGln genes as previously determined (20), with the coding region of ATG22 as an internal negative control, because the nearest tRNA gene is >5,000 bp away. Triplicate reactions from duplicate experiments are expressed as ratios of linear range PCR signal at the tRNA gene/ATG22 control. Ratios are normalized to parallel controls using an untagged strain. (B) Mod5-myc is coimmunoprecipitated with proteins bound at tRNA genes. The chromosomal MOD5 ORF was C-terminally tagged with 13xmyc in strains where the other indicated proteins are TAP-tagged as well as a strain with no secondary TAP tag. Cell lysates from 1 L midlog cultures were divided, and one-half of the samples were treated with excess DNase I (as assessed by PCR) to eliminate interactions that are mediated by co-occupation at a distance on the same DNA fragments. TAP-tagged proteins were isolated in single affinity steps, and Western blots were probed for coisolation of Myc-tagged Mod5. These results indicate that Mod5-myc is reproducibly present in affinity pull-downs of TAP-tagged Brf1p, Bdp1, Smc4, Rpc53, Rpc82, and Tfc1 but not when a TAP tag is absent. Although formation of pol III transcription complexes, including condensin, is likely to be DNA-mediated on the tRNA genes, the DNase insensitivity indicates that Mod5 is likely in close contact with the pol III transcription complex. (C) Mod5-myc is associated with pre-tRNAs. After cross-linking in culture, Mod5-myc was affinity isolated, cross-linking was reversed, and the associated RNA was analyzed by Northern blot with probes to either tRNALeu3 (not an Mod5 substrate) or tRNATyr (only the spliced form is an Mod5 substrate). In both cases, antibody to the myc tag isolated both the nonsubstrate precursors and the mature forms, with substantial enrichment for precursors (ratios given below). RNA coisolations and precursor enrichments were not sensitive to DNase treatment.
Mod5 Associates with RNA Pol III Transcription Components.
Proteomic analysis of proteins that coisolate with TAP-tagged Mod5 by Multidimensional Protein Identification Technology (31) showed a large number of polypeptides, possibly because of chromatin association as well as cytoplasmic associations (SI Materials and Methods and Table S1). Although it was not possible to form confident hypotheses from results of this complexity, there were preliminary indications that Mod5 might interact with protein components expected to be found at the tRNA genes. In addition to multiple nucleolar proteins, there were subunits of RNA pol III and condensin, which are found preferentially associated with tRNA genes and required for their nucleolar clustering and silencing (20). Using coimmunoprecipitation, we tested directly whether Mod5-myc could be pulled down with TAP-tagged polypeptides that are part of tRNA gene transcription complexes, including subunits of condensin (Smc4), TFIIIB (Brf1 and Bdp1), TFIIIC (Tfc1), and RNA pol III (Rpc53 and Rpc82). Affinity isolation of each of the tested TAP-tagged proteins was able to coisolate the myc-tagged Mod5 compared with a negative control strain without any TAP tag (Fig. 6B). Because these components are not thought to associate with each other away from the pol III-transcribed genes, these results are consistent with the data that indicated that Mod5 is located in the nucleolus (30) and at the tRNA genes (Fig. 6A). To determine whether the interactions of Mod5 with these proteins required the continued presence of DNA, we repeated the coimmunoprecipitations in the presence of sufficient DNase I to completely remove any exposed DNA from the immunoprecipitated fractions, which were assayed by exhaustive PCR. DNase treatment did not cause the total loss of any interaction, although a mild decrease in signal was seen in RNA pol III TAP-tagged samples (Fig. 6B) (Mod5-myc signals from the Rpc53 and Rpc82 are decreased 45–50% in the data shown). Although the pull-down efficiencies are too variable between experiments for quantitative interpretation, this finding is consistent with previous observations that pol III is the most DNase-unstable member of the tRNA gene complexes (20). These results were repeated for selected proteins in a reciprocal manner, where Tfc1-myc, Smc2-myc, and Smc4-myc were all able to coimmunoprecipitate Mod5-TAP (SI Materials and Methods and Fig. S2).
Mod5 Binds to pre-tRNAs.
One possible explanation of why Mod5 is involved in the silencing mechanism is that Mod5 binds the pre-tRNA transcript, and the resulting ribonucleoprotein complex contributes to antagonizing nearby pol II transcription. Such dual use of an ancient RNA processing enzyme is not without precedent in eukaryotes, and the presence of Mod5 in the nucleus as well as the cytoplasm, where it modifies a limited selection of tRNAs, would suggest a possible alternative function. The facts that it binds to genes regardless of whether they produce substrates and is required for silencing are consistent with a function independent of the catalytic activity requiring DMAPP, but does not preclude binding of the protein to the nascent pre-tRNA transcripts.
To test if Mod5 binds to the nascent pre-tRNA transcripts, we performed in vivo cross-linking to stabilize RNA–protein complexes and isolated Mod5 using a C-terminal myc affinity tag. The results (Fig. 6C) show that a portion of the affinity-isolated Mod5 was coisolated with both Tyr and Leu3 pre-tRNA and tRNA species. Importantly, the precursors, including primary transcripts, were significantly enriched compared with the precursor:mature ratios in the cellular extracts. The multiple precursor bands seen for these two tRNA types have been characterized previously, and they correspond to the size of both primary transcripts (highest band) and intron-containing but end-matured pre-tRNAs, which suggests that Mod5 binds pre-tRNAs soon after synthesis and can remain bound (or rebind) for some time after synthesis and release from the gene. Because neither the intron-containing precursors nor the mature tRNALeu3 can be modified by Mod5, finding in the absence of i6A modification is sufficient for silencing. Although the integral association of Mod5 with the large polypeptide assembly on tRNA genes precludes assigning a direct Mod5–pre-tRNA association, a tentative hypothesis consistent with these observations is that Mod5 captures nascent transcripts and acts downstream to affect pol II transcription.
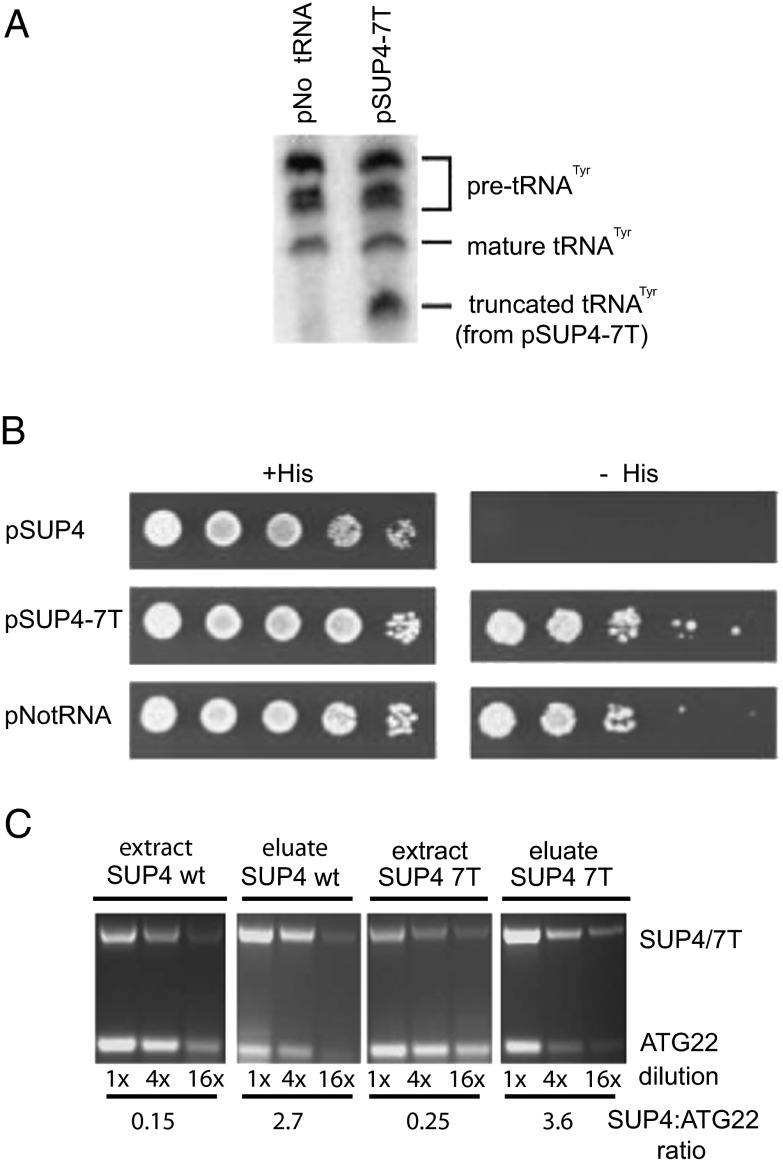
If Mod5 needs to bind the pre-tRNA for silencing to occur, one predicts that nontRNA transcripts, which do not conform to the general structure recognized by the enzyme, might not be able to confer silencing, although the pol III transcription complex is active on the gene. We created a modified tRNATyr (SUP4) gene-silencing reporter construct, in which the tRNA gene promoters were intact and correctly spaced, but transcripts were truncated immediately after the anticodon by inserting additional T residues in the intron sequence (to give seven consecutive T residues), which efficiently terminates pol III transcription. After confirming that the truncated tRNATyr transcript was made from this construct by Northern blot (Fig. 7A), we tested for tgm silencing of the neighboring nat1 gene and found that silencing had been completely removed to the level of having no tRNA gene present (Fig. 7B). This result is consistent with a need for a bona fide pre-tRNA transcript to activate the silencing mechanism. To test whether the full-length pre-tRNA transcript was necessary for Mod5 to associate with the tRNA gene complex, we performed ChIP in a strain with a chromosomal TAP tag on Mod5 containing either the normal SUP4 silencing test plasmid or the 7T construct that produces truncated transcripts. The Mod5 remains associated with the 7T SUP4 gene, consistent with Mod5 associating with the tRNA gene complex in the absence of a full-length pre-tRNA.
Fig. 7.
tgm silencing is abolished by truncation of the tRNA transcript. A modified SUP4 tRNA gene was used in the silencing test construct (Fig. 1), in which a TTTTTTT transcription termination sequence was substituted for the tRNA intron sequence starting immediately after the anticodon. The internal promoters are intact and spaced identically, but the truncated transcripts lack both tRNA structure and an anticodon stem, and they are not predicted to bind to Mod5. (A) Northern blots of total cell RNA were probed with an oligo complimentary to the 5′ half all tRNATyr transcripts. The upper pre-tRNA and mature tRNA bands are from the eight chromosomal tRNATyr genes, with the lowest band being the size expected for the pSUP4 transcript truncated by termination at the 7T sequence after the anticodon. The truncated 7T primary transcript is ∼40% (±10%) of the combined pre-tRNA signal from the other eight genes in triplicate experiments, although quantitative comparison of synthesis rate is not possible, because the RNA turnover is by different pathways. The oligonucleotide probe used here hybridizes preferentially to pre-tRNAs rather than mature tRNAs (Fig. 6 shows comparison with WT precursor to mature signals). (B) tRNA truncation releases tgm silencing as well as deletes the tRNA gene entirely, allowing growth on selective media (−His). (C) ChIP of the test SUP4 WT and truncated transcript (7T) genes was performed in a strain containing TAP-tagged Mod5. PCR of serial dilutions (1×, 4×, and 16×) of DNA from 1% of the input extract or calmodulin affinity isolation of chromatin fragments shows substantial enrichment relative to the starting extract of both test SUP4 tRNA genes relative to the control ATG22 locus.
Discussion
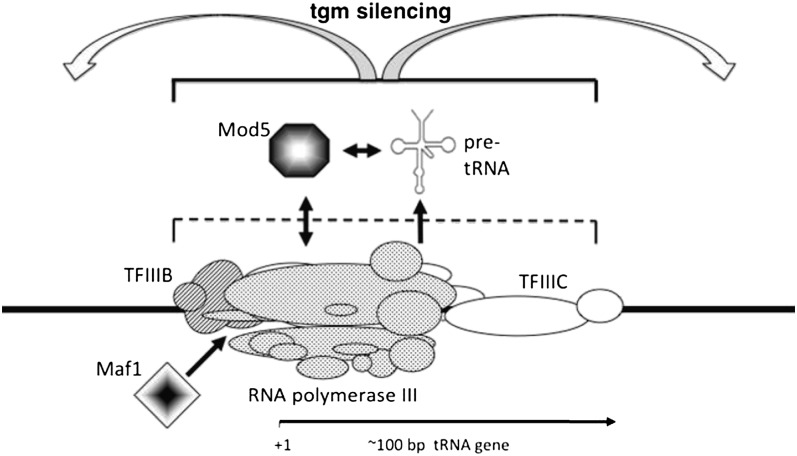
Previous genetic analysis has shown that the mechanism of tgm silencing is distinct from other forms of transcriptional silencing. Our earlier investigations of this mechanism revealed that condensin-dependent clustering of tRNA genes and microtubule-dependent localization to nucleoli were required, because mutations that mislocalized tRNA genes in the cell also relieved tgm silencing (3, 18). Additional evidence now shows that, although localization of tRNA genes in the nucleolus is necessary for silencing near tRNA genes, it is not sufficient, and additional mechanisms come into play. We previously showed that Maf1, a pol III repressor that binds to tRNA genes, is also required for nearby silencing, although it has no strong effect on nuclear positioning of the genes (9). Here, we show that Mod5, a protein with genetic ties to Maf1, is also required for tgm and does not seriously disrupt tRNA gene clustering and nucleolar localization. Although these proteins have previously been shown to interact genetically (9), the current demonstration that Mod5 is also present at the tRNA genes is the first indication that they are both present at tRNA genes and may act in concert to affect local transcription by RNA pol II. A schematic representation of the pol III transcription complex interactions with Maf1, Mod5, and the pre-tRNA transcripts that are required for tgm silencing is shown in Fig. 8. Although condensin also associates with these complexes, it is not represented here, because it is thought to exert its effects through clustering of the tRNA genes.
Fig. 8.
Schematic representation of tRNA gene complex components that are required for tgm silencing. RNA pol III, TFIIIB, and TFIIIC are components of the chromosomal tRNA gene transcription complexes (11). Involvement of Maf1, Mod5, and the pre-tRNA transcript are described in the text. Another required component bound to the tRNA genes through TFIIIC, the condensin complex (20), is not depicted, because it likely acts separately by spatially clustering the tRNA genes.
The isopentenyl transferase enzyme encoded by MOD5 in yeast is conserved from Escherichia coli to Homo sapiens (32–34). Other than the universal conservation of the ATP/GTP binding domain and the DMAPP substrate binding domain, the eukaryotic homologs possess an ∼100-aa C-terminal extension not found in the prokaryotic version. This extension has a conserved zinc finger motif (26), and although the function of this region is currently unknown (26), it has been implicated in nucleolar localization (30). It seems possible that the C-terminal domain might also allow additional functions that contribute to tgm silencing.
To test whether the catalytic activity of Mod5 is required for silencing, we used two different methods to deplete the known substrate of Mod5 for tRNA modification, DMAPP. Neither of these methods alleviated tgm silencing (Fig. 4 B and C), although they do decrease the tRNA modification of Mod5. This apparent continuation of silencing in the face of reduced tRNA modification is consistent with the observation that the pre-tRNAs synthesized at the site of silencing do not need to be substrates for modification. Thus, we hypothesize that Mod5 acts in silencing through binding to nascent transcripts, although at this time, we cannot definitively rule out participation of the enzyme’s catalytic center. We have not yet been able to identify mutations that inactivate tRNA modification without inactivating tgm silencing (SI Materials and Methods and Fig. S1), but it is possible that more detailed knowledge of the tRNA modification mechanism will allow the two activities to be differentiated. The lack of silencing by the truncated tRNA transcript extends our previous observation that active pol III transcription is required by suggesting that a recognizable pre-tRNA needs to be formed, consistent with a requirement for a protein that recognizes tRNAs (2), which is consistent with the association of Mod5 with pre-tRNA transcripts (Fig. 7). A detailed future analysis of pre-tRNA requirements in parallel with in vitro binding requirements for the purified enzyme will be of interest. Although Mod5 associates with the active tRNA transcription complex in the absence of a full-length transcript, these results suggest that the pre-tRNA is required for a subsequent event.
Mod5 exists in both the cytoplasm and nucleus, with a concentration in the nucleolus (29, 30). The results presented here indicate that Mod5 possesses a previously unsuspected influence on nuclear gene expression that does not require modification of tRNAs. This dual functionality has been observed previously for other tRNA processing enzymes as well. One example is the yeast protein Nam2p, a tRNA synthetase, that is involved in tRNA aminoacylation and also participates in the excision of an intron in bI4 maturase premRNA (35, 36). A second example is the Pus1 protein, which modifies uridine to pseudouridurine in tRNAs but also modifies steroid receptor RNA activator and cooperates with retinoic acid receptor to enhance transcription at target promoters (37). The modification of dissimilar substrates with different chemical groups provides an intriguing set of possibilities for Mod5, however unlikely. The characterized enzymatic reaction catalyzes the transfer of an isopentenyl group to the exocyclic amine of adenosine, with a pyrophosphate leaving group from the DMAPP donor. Although the DMAPP depletion studies suggest that it is not a substrate for any alternative reactions, it is not impossible that entirely different donor and acceptor substrates could have evolved in the context of the tRNA gene complex. For example, a previously uncharacterized activity might transfer a different small substrate to an amino group on either a protein or the pre-tRNA.
An unanswered question to date is what regulatory event is being affected in the immediate vicinity of the tRNA genes. The lack of Mod5 does not seem to affect overall tRNA gene localization and it is not essential for Maf1 regulation of pol III transcription, suggesting that it is either downstream of the effect of Maf1 or independent of it. One possibility is that chromatin structure is involved in tgm silencing after all, although it is regulated by different nucleosome modification and remodeling activities than were tested earlier, because they are used for other silencing forms. It is possible that a complex between Mod5 and pre-tRNAs helps to recruit activities that modify local chromatin, and this hypothesis is currently under investigation. Other known forms of RNA-mediated chromatin silencing involving siRNAs do not seem to be present in Saccharomyces.
Another interesting possibility is that the pre-tRNA transcripts are directly inhibiting RNA pol II in the vicinity, with the interaction mediated by Mod5 and would be compatible with earlier observations that tRNAs can inhibit purified pol II (32, 33) and that pol III-synthesized transcripts from small interspersed elements can directly inhibit mammalian pol II in vitro (34) and repress pol II transcription in vivo (38, 39). It is not immediately clear for either this hypothesis or possible chromatin modification why pre-tRNAs are functional and truncated half-tRNAs are not, but different small interspersed elements have differential abilities to inhibit pol II; also, it seems plausible that there are specific constraints on the RNA effector beyond binding to Mod5, such that Mod5 binding might or might not be the descriminator of function.
The complementation of tgm silencing by both the human and Arabidopsis Mod5 homologs suggests that it is likely to be broadly applicable in eukaryotes. The human homolog of Mod5, TRIT1, is of particular interest, because previous studies have suggested that TRIT1 might be a tumor suppressor in specific types of human cancers. It is down-regulated or alternatively spliced in lung adenocarcinoma, and when transfected into a human lung carcinoma line that had low expression of TRIT1, it is able to significantly reduce tumor development in a nude mouse test system (40). In addition, the yeast Mod5 has recently been shown to have prion-like misfolding properties that help confer resistance to certain antifungal agents (41). Although this activity, like the tumor suppressor properties of TRIT1, was attributed to the lack of tRNA modification by DMAPP, it will be interesting to examine these properties in light of possible effects on gene expression in the nucleus.
Materials and Methods
Yeast Strains.
Strains were derived from S. cerevisiae strains W3031 (MATα leu2-3, 112 his3-11, 15 ade2-1 trp1-1 ura3-1 can1-100), MT-8 (MATα SUP7, ura3-1, his5-2, leu2-3,112, ade2-1, trp1, lys1-1, lys2-1, can1-100, mod5:TRP1), BY4741 (MATα his3∆1, leu2Δ, met15∆, ura3∆), and YPH500α (MATα ade2-101, his3∆200, leu2∆1, lys2-801, trp1∆63,ura3-52). Strains with TAP-tagged protein ORFs were purchased from Open Biosystems (42). Additional affinity-tagged strains were created by tagging chromosomal ORFs with 13xmyc-epitope by direct recombination at the C termini as described in ref. 43. Descriptions of all of the strains and plasmids used are listed in Table 1.
Table 1.
Strains, plasmids, and oligonucleotides
| Name | Genotype, description, or sequence | Source |
| BY4741 | MATα, his3∆1, leu2∆0, met15∆0, ura3∆0, GAL4, GAL80 | 52 |
| BY4741 mod5∆ | MATα, his3∆1, leu2∆0, met15∆0, ura3∆0, GAL4, GAL80, mod5∆ | 53 |
| BY4741 MOD5-TAP | MATα, his3∆1, leu2∆0, met15∆0, ura3∆0, GAL4, GAL80, MOD5-TAP:HIS5 | 42 |
| W3031A | MATα, ura3-1, ade2-1, his3-11,15 trp1-1, can1-100 GAL4, GAL80 | 54 |
| MT-8 | MATα, SUP7, ura3-1, his5-2, leu2-3,112, ade2-1, trp1, lys1-1, lys2-1, can1-100, mod5:TRP1 | 16 |
| YPH500α | MATα, ade2-101, his3- ∆200, leu2- ∆1, lys2-801, trp1- ∆63, ura3-52 | 44 |
| yOH1 | MATα, ade2-101, his3- ∆200, leu2- ∆1, lys2-801, trp1-∆63, ura3-52, TFC1-13Myc:kanMX6 | 21 |
| pSUP4o | tgm silencing construct | 2 |
| pSUP53o | tgm silencing construct | 2 |
| ARAB-tRNA-IPT:pFL61 | A. thaliana tRNA isopentenyltransferase gene cloned into pFL61 plasmid | 27 |
| TRIT1:pFL61 | TRIT1 cloned into pFL61 plasmid | 26 |
| pMPH1 | MOD5 in pRS316 | This work |
| pMPH2 | tgm silencing construct in pRS315 | This work |
| pMPH3 | MOD5-TAP in pRS316 | This work |
| YEP-ERG20 | ERG20 in Yep13 | 23 |
Cloning of MOD5.
MOD5 was PCR-amplified from BY4741 from 500 bp upstream of the AUG start codon to 142 bp after the stop codon, with BamHI sites added to the ends. This product was ligated into the BamHI site of plasmid pRS316 (44), creating plasmid pMPH1. The TAP tag was PCR-amplified from the MOD5-TAP strain from Open Biosystems using primers containing 100-nt pairs of the MOD5 ORF C terminus and 50 nt corresponding to the pRS316 plasmid. The amplified TAP tag was added to the pMPH1 clone by gap repair by transforming the amplified product and pMPH1-digested with XhoI and HindIII into an mod5∆ strain, creating pMPH3. This construct contains the MOD5 promoter starting 10 bp after the Spe I site and the insert ends at 10 bp before the SmaI site.
In Situ Hybridization and Microscopy.
Yeast was grown to midlog in synthetic dextrose complete (SDC) media and fixed for 30 min by adding 3.6% (vol/vol) paraformaldehyde directly to the growth media to retain native cellular and nuclear architecture before fixation (3, 7, 45). After fixation, cells were harvested and treated as previously described (45, 46), except that spheroplasting was performed by treating with 0.38 mg/mL Zymolyase 20T (Seikagaku) for 45 min at 37 °C. Oligonucleotide probes, labeled with Oregon Green 488 (Molecular Probes) or Cy3 (GE Healthcare), to U14 snoRNA and tRNALeu(CAA) were previously described, and in situ hybridization and deconvolution fluorescence microscopy were also previously described (3, 7, 45).
Rapamycin Treatment and RNA Analysis.
Cells grown to an OD at 600 nm of 0.6–0.8 were treated with a final concentration of 0.2 µg/mL rapamycin (LKT Laboratories) for 40 min (22). Cell pellets were harvested and stored at −80 °C before RNA extraction. Total yeast RNAs (20 μg/lane) were electrophoresed on a denaturing 8% polyacrylamide gel and electrotransferred to a Nytran SuperCharge membrane (Schleicher & Schuell Bioscience). The blots were probed with [32P]-labeled oligonucleotides as described previously (47). Signals on the Northern blots were detected with a Typhoon Trio+ (GE/Amersham) and quantified with IPlab Gel software (Signal Analytics). For each duplicate sample, the ratio of precursor tRNA to U6 signal is shown, and it is normalized to 1.0 in that strain in the same experiment without rapamycin treatment.
Atorvastatin Treatment.
Solid media of SDC-trp, SDC-ade, synthetic medica with galactose and raffinose (SGR)-ura, or SGR-ura-his contained varied concentrations of Atorvastatin (Toronto Research Chemicals): 5, 10, 20, 50, 100, and 150 µg/mL.
Testing for Silencing.
Testing for tgm silencing was done as described (3) using plasmids pSUP40, pSUP530, and pMPH2 (2). pSUP4-7T and pNotRNA are derivates of pSUP40, in which the intron sequence after the anticodon of SUP4 has been converted from TTTATCAC to TTTTTTTATCAC (pSUP4-7T) or the entire BamH1 fragment encoding SUP4 has been removed from the plasmid by digestion and relegation. To analyze the level and start site of the HIS3 transcript relative to ADH1 mRNA as an internal control, primer extension was performed on whole-cell RNA essentially as described previously (2), with radiolabeled primers complementary to the 5′ ends of the HIS3 (5′-TCGAGTGCTCTATCGCTAG) and ADH1 coding regions used simultaneously in the same extension reactions as products analyzed after separation on denaturing polyacrylamide sequencing gels (2).
Alignment of Mod5 Homologs.
Protein sequence alignments were obtained with the STRAP sequence alignment program (48).
Testing for i6A Modification of tRNAs by Mod5.
To test for i6A modification of tRNAs, the strain MT-8 was used to monitor modification-dependent translational ochre suppression by an SUP7 r ochre suppressor tRNA (16). Plasmids were transformed into the MT-8 strain and then plated onto media lacking adenine. The i6A modification of the SUP7 tRNA was indicated by growth on media lacking adenine, indicating suppression of an ade2 ochre mutation.
ChIP and RNA Immunoprecipitation.
ChIPs were performed as described (49) except for the following adaptations. Cells were grown in 100 mL yeast extract/peptone/dextrose (YPD) media to OD at 600 nm of 0.5–0.7 and fixed with 1% formaldehyde for 1 h at room temperature. Fixation was stopped by the addition of glycine (final concentration of 400 mM) for 10 min. Cell pellets were resuspended in 800 µL lysis buffer (50 mM Hepes-KOH, pH 7.5, 100 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% deoxycholate, complete EDTA-free protease inhibitors; Roche) and transferred to 2-mL screw cap tubes. Zymolyase 20T was added to the cells (20 µL 12 µg/µL stock) and incubated at 30 °C for 30 min; 1-mL glass beads (425–600 µm; Sigma) were added, and chilled samples were lysed in a Bead Beater at 4 °C (Biospec Products) four times at a setting of five. Tubes were pierced at the bottom to collect lysate. The extracts were spun for 25 min at 20,000 × g, and cell pellets were resuspended in 800 µL lysis buffer. The resuspensions were sonicated eight times for 20 s at power 6 on Hold and Constant settings in a Branson Sonifer 250 and spun at 24,000 × g for 20 min. Supernatants were bound to 40 µL IgG Separose (GE Healthcare) for 2 h at 4 °C. The beads were washed five times with 15 mL lysis buffer, and samples were eluted as described (49). The ChIPs in Fig. 7 used altered conditions to maximize yield of plasmid and chromosomal chromatin. Lysis after Zymolyase digestion was done in calmodulin binding buffer (20 mM Tris, pH 7.2, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2) by sonication 10 × 10 s at 4 °C. Lysate was spun 10 min at 4 °C in a microfuge, and the chromatin in the soluble extract was sheared by a 25-min cycle (30 s on, 30 s off, 0 °C) in a Diagenode Bioruptor XL on high power. Lysate (2.4 mL from 50 mL culture) was bound to 0.2 mL Calmodulin Affinity Resin (Agilent Technologies) for 30 min at 4 °C, the resin washed three times with 1.2 mL binding buffer, and bound chromatin was eluted for 20 min with 2 × 0.2 mL buffer containing 5 mM EGTA instead of 2 mM CaCl2. Nucleic acids were precipitated with 2.5 volumes ethanol and resuspended in 50 μL 50 mM Tris (pH 8.3) at 65 °C for 4 h.
RNA immunoprecipitations were performed on BY4741 with a C-terminal myc tag on MOD5. Cultures were grown in 250 mL YPD to OD at 600 nm of 0.8 and fixed in 1% formaldehyde for 30 min at 23 °C. After glycine-quenching, cell pellets were resuspended in lysis buffer supplemented with 40 units/µL RNasin (Promega). Cells were broken by glass bead lysis in a FastPrep 24 (MP Biomedicals), and cell suspension was recovered from the glass beads and sonicated as above three times for 10 s each. The solubilized fraction was applied to Magnabind Protein A magnetic beads (Pierce) prebound to myc antibody (9E10; Santa Cruz Biotechnology) and incubated for 3 h at 4 °C. For DNase-treated samples, before the immunoprecipitation, 40 units TURBO DNase (Ambion) were added to the sonicated extracts and incubated for 15 min at 37 °C. Beads were washed successively with lysis buffer, lysis buffer containing 500 mM NaCl, wash buffer, 10 mM Tris, and 1 mM EDTA, and then eluted, and cross-links were reversed as described (49). Samples were treated with 150 µg Proteinase K (Roche), extracted, precipitated, and loaded onto a 6% denaturing gel. Samples were electrotransferred and immobilized onto Immobilon Ny+ nylon membrane (Millipore) and blotted with a radiolabeled probe either to the tRNATyr (5′-CTT GCG CCT TAA ACC AAC TTG GCT ACC-3′) or the tRNALeu3 (5′-TCA GGC GCC TTA GAC CGC TCG GCC AAA-3′). Images were visualized on a Typhoon Trio+ Imager (Amersham) and quantitated with Image J (National Institutes of Health).
PCR.
DNA from ChIPs (2.0 µL from 100 µL) was used as template in 50-µL reactions containing 0.20 mM dNTPs and 0.4 µg each primer using an Applied Biosystems GeneAmp PCR system 9700. Cycling parameters for the amplications were 94 °C for 30 s, 63 °C for 30 s, and 72 °C for 60 s for 21, 24, and 27 cycles. For the experiment in Fig. 7C, 34 cycles were used with fourfold serial dilutions of DNA samples. The primer sequences were 5′-TTATTAGCACGGTGCCTTAACCAACT and 5′-GCGCTTCCACCACTTAGTATGATTC for tRNAIle [tI(AUU)D on Chr IV], 5′-GAAAGCGGGTGTTTCTCCAATAAAT and 5′-GTGGTTATCACTTTCGGTTTTGATCC for tRNAGln [tQ(UUG)E1 on Chr V], 5′-GGCGACCACACCCGTCCTGTGGATC and 5′-CGTTCATTTGAAGGTTTGTGGGGCCA for plasmid SUP4 WT and 7T genes, and 5′-CAAAGTTTCGGTGGACTTCTAGTCAAA and 5′-GCTTTAAACCGAACGCATTGAAGAAAA for ATG22 coding region.
PCR products were analyzed on a 2.5% agarose gel stained with ethidium bromide and imaged on a Syngene Bioimaging System with GeneSnap (Syngene), with products quantified with GeneTools (Syngene). To determine enrichment, tRNA gene products are expressed as a ratio to the control ORF product in the linear range at 24 cycles.
Preparation of Yeast Soluble Extracts and Coimmunoprecipitations.
The purification protocol was modified from previous work (50). Cells were grown in 500 mL YPD to an OD at 600 nm of 0.8–1.2, and cell pellets were stored at −80 °C. Pellets were resuspended in 1 mL buffer [10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Nonidet (Nonidet P-40), Complete protease inhibitors]. Zymolyase 20T was added to the cells (20 µL 12 µg/µL stock) and incubated for 1 h, and the cells were lysed by mechanical bead disruption. Lysates were spun for 30 min at 20,000 × g and the supernatant was collected. Extracts (3 mL) were added to 100 µL IgG Sepharose beads (GE Healthcare) and incubated with mixing at 4 °C for 3 h. Beads were washed four times with 10 mL lysis buffer and one time with cleavage buffer of 10 mM Tris, 150 mM NaCl, 01% Nonidet P-40, 0.5 mM EDTA, and 1 mM DTT. Beads were resuspended with 300 µL tobacco etch virus (TEV) cleavage buffer plus 50 µg TEV protease. Elutions were precipitated by trichloroacetic acid and resolved using SDS/PAGE gel 10% (Bio-Rad). Proteins were transferred to PVDF membrane and probed with anti-myc antibody (Santa Cruz) followed by anti-mouse from sheep (GE Healthcare), with signals detected by ELC Plus (GE Healthcare). In DNase-treated samples, 50 units DNase I (Cooper Biomedical) were added and incubated at 4 °C for 30 min before adding the lysate to the IgG Sepharose to bind TAP-tagged proteins. PCR analysis of parallel samples determined that this level of DNase eliminates detectable DNA fragments at the tRNA genes using 35 rounds of amplification. For qualitative analysis of the pull-down efficiency using the various proteins ± DNase I treatment, film exposures were digitized with an Epson 1680 scanner and analyzed using ImageJ software (http://rsbweb.nih.gov/ij/).
Northern Blot of Total Cell RNA.
BY4741 transformed with either pSUP4-7T or pNotRNA was grown in SD-ura to OD at 600 nm of 0.8, and cell pellets were resuspended in RNA extraction buffer (50 mM sodium acetate, pH 5.2, 10 mM EDTA, 1% SDS). RNA was extracted and precipitated. To verify synthesis of the truncated 7T transcripts compared with pre-tRNAs from the eight chromosomal tRNATyr genes, samples were run on 6% denaturing polyacrylamide gels with 10-nt increment size markers, blotted as for the RNA immunoprecipitation above, and probed with a radiolabeled oligo complementary to the 5′ half of tRNATyr (5′-CTTGCGCCTTAAACCAACTTGGCTACC-3′). This probe hybridizes preferentially to pre-tRNAs rather than mature tRNAs, likely because of base modifications or tightened structure in the mature, spliced tRNA. The steady state 7T truncated transcript signal was reproducibly over one-half the total signal of precursors from the eight chromosomal pre-tRNATyr genes combined, although this finding cannot be interpreted in terms of transcription rate, because the truncated transcript would not be recognized as a pre-tRNA and would be subject to completely different turnover paths.
Supplementary Material
Acknowledgments
We thank Olivier Lefebvre for strains, Bjorn Nicander for plasmids, May Tsoi and Sara Neill for their technical assistance, and Dan Bochar for advice on ChIP. This work was supported by National Institutes of Health Predoctoral Training Grants T32 GM007315 (to M.P.-H.), T32 GM07544 (to D.A.P), P41 RR011823 (to J.R.Y. for the support of I.X.M.), R01 GM27930 (to A.K.H.), and R01 GM063142 (to D.R.E.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219946110/-/DCSupplemental.
References
- 1.Goffeau A, et al. Life with 6000 genes. Science. 1996;274(5287):546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 2.Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14(2):1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, et al. Silencing near tRNA genes requires nucleolar localization. J Biol Chem. 2005;280(10):8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125(5):859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 5.Simms TA, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7(12):2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183(1):131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302(5649):1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai N, et al. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J Biol Chem. 2005;280(8):6455–6462. doi: 10.1074/jbc.M412375200. [DOI] [PubMed] [Google Scholar]
- 9.Moir RD, et al. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci USA. 2006;103(41):15044–15049. doi: 10.1073/pnas.0607129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pluta K, et al. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(15):5031–5040. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100(25):14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boguta M, Czerska K, Zoładek T. Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene. 1997;185(2):291–296. doi: 10.1016/s0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- 13.Murawski M, et al. maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim Pol. 1994;41(4):441–448. [PubMed] [Google Scholar]
- 14.Dihanich ME, et al. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7(1):177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gefter ML, Bikoff E. Studies on synthesis and modification of transfer RNA. Cancer Res. 1971;31(5):667–670. [PubMed] [Google Scholar]
- 16.Gillman EC, Slusher LB, Martin NC, Hopper AK. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol Cell Biol. 1991;11(5):2382–2390. doi: 10.1128/mcb.11.5.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laten H, Gorman J, Bock RM. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1978;5(11):4329–4342. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendall A, et al. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci USA. 2000;97(24):13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamińska J, et al. The isoprenoid biosynthetic pathway in Saccharomyces cerevisiae is affected in a maf1-1 mutant with altered tRNA synthesis. FEMS Yeast Res. 2002;2(1):31–37. doi: 10.1111/j.1567-1364.2002.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 20.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22(16):2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oficjalska-Pham D, et al. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22(5):623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10(6):1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 23.Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc Natl Acad Sci USA. 2000;97(1):61–66. doi: 10.1073/pnas.97.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimster-Denk D, et al. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J Lipid Res. 1999;40(5):850–860. [PubMed] [Google Scholar]
- 25.Warner GJ, et al. Inhibition of selenoprotein synthesis by selenocysteine tRNA[Ser]Sec lacking isopentenyladenosine. J Biol Chem. 2000;275(36):28110–28119. doi: 10.1074/jbc.M001280200. [DOI] [PubMed] [Google Scholar]
- 26.Golovko A, Hjälm G, Sitbon F, Nicander B. Cloning of a human tRNA isopentenyl transferase. Gene. 2000;258(1–2):85–93. doi: 10.1016/s0378-1119(00)00421-2. [DOI] [PubMed] [Google Scholar]
- 27.Golovko A, Sitbon F, Tillberg E, Nicander B. Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol Biol. 2002;49(2):161–169. doi: 10.1023/a:1014958816241. [DOI] [PubMed] [Google Scholar]
- 28.Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J Biol Chem. 2001;276(28):26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- 29.Boguta M, et al. Subcellular locations of MOD5 proteins: Mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol. 1994;14(4):2298–2306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolerico LH, et al. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151(1):57–75. doi: 10.1093/genetics/151.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Washburn MP. Utilisation of proteomics datasets generated via multidimensional protein identification technology (MudPIT) Brief Funct Genomics Proteomics. 2004;3(3):280–286. doi: 10.1093/bfgp/3.3.280. [DOI] [PubMed] [Google Scholar]
- 32.Johnson TL, Chamberlin MJ. Complexes of yeast RNA polymerase II and RNA are substrates for TFIIS-induced RNA cleavage. Cell. 1994;77(2):217–224. doi: 10.1016/0092-8674(94)90314-x. [DOI] [PubMed] [Google Scholar]
- 33.Sawadogo M. On the inhibition of yeast RNA polymerases A and B by tRNA and alpha-amanitin. Biochem Biophys Res Commun. 1981;98(1):261–267. doi: 10.1016/0006-291x(81)91897-0. [DOI] [PubMed] [Google Scholar]
- 34.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11(9):822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 35.Herbert CJ, Labouesse M, Dujardin G, Slonimski PP. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988;7(2):473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labouesse M. The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol Gen Genet. 1990;224(2):209–221. doi: 10.1007/BF00271554. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, et al. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell. 2004;15(4):549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 38.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11(9):816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 39.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29(4):499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Spinola M, et al. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene. 2005;24(35):5502–5509. doi: 10.1038/sj.onc.1208687. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336(6079):355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 42.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 43.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12(16):2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long RM, Elliott DJ, Stutz F, Rosbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1(10):1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Moir RD, Sethy-Coraci IK, Warner JR, Willis IM. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol Cell Biol. 2000;20(11):3843–3851. doi: 10.1128/mcb.20.11.3843-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gille C, Goede A, Preissner R, Rother K, Frömmel C. Conservation of substructures in proteins: Interfaces of secondary structural elements in proteasomal subunits. J Mol Biol. 2000;299(4):1147–1154. doi: 10.1006/jmbi.2000.3763. [DOI] [PubMed] [Google Scholar]
- 49.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 50.Puig O, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24(3):218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 51.Alberts AW, et al. Mevinolin: A highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci USA. 1980;77(7):3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 53.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 54.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.