Abstract
Complement is an essential component of innate immunity. Its activation results in the assembly of unstable protease complexes, denominated C3/C5 convertases, leading to inflammation and lysis. Regulatory proteins inactivate C3/C5 convertases on host surfaces to avoid collateral tissue damage. On pathogen surfaces, properdin stabilizes C3/C5 convertases to efficiently fight infection. How properdin performs this function is, however, unclear. Using electron microscopy we show that the N- and C-terminal ends of adjacent monomers in properdin oligomers conform a curly vertex that holds together the AP convertase, interacting with both the C345C and vWA domains of C3b and Bb, respectively. Properdin also promotes a large displacement of the TED (thioester-containing domain) and CUB (complement protein subcomponents C1r/C1s, urchin embryonic growth factor and bone morphogenetic protein 1) domains of C3b, which likely impairs C3-convertase inactivation by regulatory proteins. The combined effect of molecular cross-linking and structural reorganization increases stability of the C3 convertase and facilitates recruitment of fluid-phase C3 convertase to the cell surfaces. Our model explains how properdin mediates the assembly of stabilized C3/C5-convertase clusters, which helps to localize complement amplification to pathogen surfaces.
Complement is a crucial component of innate immunity. It is a first line defense mechanism against pathogens and it is essential in the modulation of adaptive immune responses and to remove apoptotic cell debris and immune complexes (1). Activation of complement results in the formation of unstable protease complexes, named C3 convertases (C3bBb in the alternative pathway) (AP), which catalyze the cleavage of C3 to generate the activated fragment, C3b. This exposes a reactive thioester that attaches covalently to the target surfaces (opsonization), initiating the terminal pathway that causes cell lysis and generates inflammation at the site of activation (1, 2).
The complement AP is exquisitely regulated and pathological conditions are associated with both loss-of-function variants of the regulatory molecules, as well as gain-of-function variants of propagating components of the pathway (2). Accelerated dissociation of the AP C3 convertase and inactivation of C3b are critical steps to maintain complement homeostasis and to prevent nonspecific damage to self-cellular components when complement is activated. These activities are performed primarily by factor H (FH), in collaboration with the plasma serine protease factor I (FI) (2). Self-tissues are also protected by membrane-bound proteins that restrict complement activation by acting as cofactor for proteolytic inactivation of C3b by FI or accelerating the dissociation of the C3bBb convertase. Thus, in health, spontaneous activation of C3 in plasma is kept at a low level and further complement activation and C3b deposition is restricted to targets lacking surface regulators. Recent advances in understanding the structural basis of the assembly, activation, and regulation of the AP C3 convertase have provided important insights into the regulation of the AP and the pathogenic consequences of its dysregulation (2–4).
Properdin is the only known complement regulator that enhances the stability of the C3bBb convertase and the activity of the AP. Properdin binds to C3bB and C3bBb more efficiently than to C3b alone, stabilizing preformed C3bBb convertase complexes (5). Properdin is also a pattern-recognition molecule that binds to negatively charged molecules on certain microbial surfaces, apoptotic and necrotic cells, as well as cells undergoing malignant transformation. Once bound to a surface, properdin can direct C3b deposition and C3bBb assembly, thus serving as a focal point for amplifying complement activation (6). Although the importance of properdin has been somehow neglected, it plays important roles in antibacterial defense and in inflammatory or autoimmune diseases, as illustrated by the high vulnerability of properdin-deficient individuals to Neisseria meningitides infections and the reported role of properdin in a number of pathological conditions (7, 8).
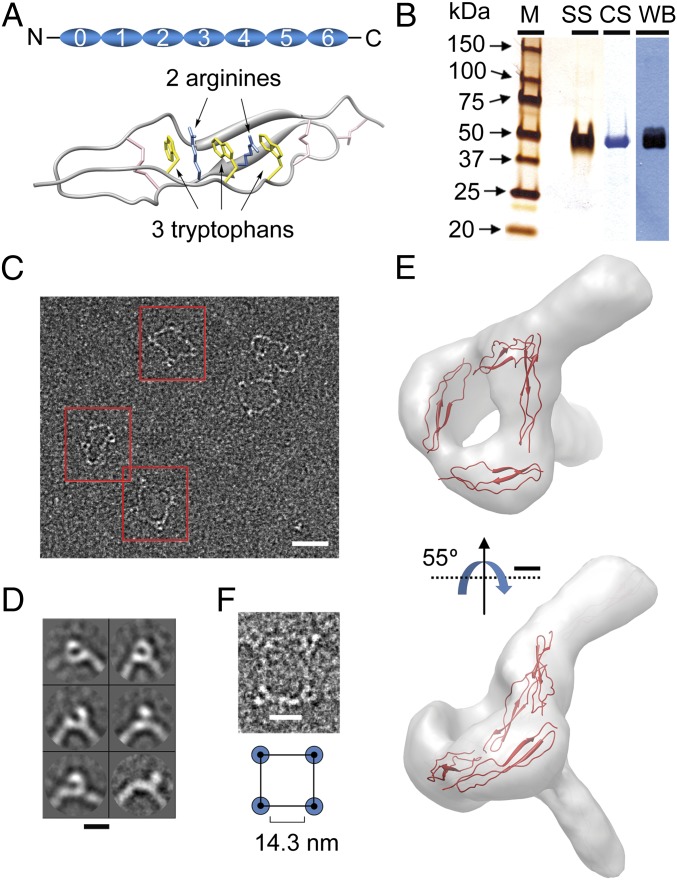
Properdin is a 53-kDa glycoprotein comprising seven conserved domains with homology to thrombospondin repeats (TSRs) of type I, and numbered TSR0 to TSR6 from the N- to the C terminus (Fig. 1A) (9). Atomic structures for properdin have not been resolved yet, but the structure of a double-TSR domain from thrombospondin [Protein Data Bank (PDB) 3R6B] provides a reasonable model for TSR domains in properdin (10) (Fig. 1A). Each TSR comprises a folded core consisting of three antiparallel strands (A, B, and C) held together by three disulfides (11) (Fig. 1A). Human plasma contains a low concentration of properdin (0.02 mg/mL) in the form of a polydisperse mixture of oligomeric structures, mostly dimers, trimers, and tetramers (12). Examination of purified properdin using electron microscopy (EM) revealed that each monomer forms an elongated rod-like molecule, which associates into cyclic polymers (13). Despite early work identifying a potential region in C3b interacting with properdin (14), the structural basis for the AP C3 convertase stabilization by properdin is unknown. Using single-particle EM, image processing, 3D reconstruction techniques, and hybrid methods that combine electron microscopy and X-ray crystallography data (15), we propose a model for the 3D structure of the properdin–C3bBb complex.
Fig. 1.
Structure of properdin oligomers by electron microscopy. (A) Schematic cartoon of the arrangement of TSR domains in a properdin monomer (Upper) and a view of the atomic structure of one homolog TSR domain from thrombospondin (PDB 3R6B, Lower) (10). Side chains of the proposed key arginine and tryptophan residues are shown in blue and yellow, respectively. Disulfide bonds are shown in pink. (B) Final preparation of purified properdin analyzed by SDS/PAGE. SS, silver staining; CS, Coomassie staining; WB, Western blotting using polyclonal antibodies against properdin. (C) Typical micrograph of properdin. Selected single molecule images for several properdin oligomers are highlighted within a red square. (Scale bar, 26 nm.) (D) Reference-free 2D averages of properdin vertexes extracted from the micrographs reveal several views of the structure connecting two monomers. (Scale bar, 7 nm.) (E) 3D structure of the properdin vertex and pseudoatomic model obtained by fitting a crystal structure of a TSR domain from thrombospondin (PDB 3R6B) (10) into the EM density. (Scale bar, 1.2 nm.) (F) Carton representation of a properdin tetramer (Lower) and a raw image for a properdin tetramer (Upper). Vertexes are represented as a blue circle and the region whose distance was measured is indicated.
Results
Intricate Connections Between Properdin Monomers Assemble Large Oligomers.
Human properdin was purified to homogeneity from plasma by immunoaffinity followed by ionic exchange and size exclusion chromatography (Fig. 1B). The functional integrity of the purified properdin was verified using AP-dependent hemolytic assays with rabbit erythrocytes (Fig. S1). Properdin was observed in the electron microscope and found assembled into several oligomeric species in which the elongated monomers were connected at their ends. As previously reported (12, 13) the most common oligomers were triangle-shaped trimers, square-shaped tetramers, and pentagonal pentamers (Fig. 1C, and below). From these EM images we interpreted that the interaction between properdin monomers involves the N-terminal end of one monomer and the C-terminal end of another, permitting the assembly of a variety of oligomers with the only restriction of geometrical constraints. Interestingly, we found that the angle formed by two adjacent monomers ranged from 60° in the trimers to 108° in the pentamers, indicating a large flexibility in the interaction between monomers.
Analysis of the vertex of the properdin oligomers by single-particle image processing methods revealed a complex structure. A total of 6,425 images of vertexes from the tetramers were extracted from the micrographs to be sorted into homogenous groups and averaged to improve the signal/noise ratio (Fig. 1D). The 3D structure of the vertex at 23.4-Å resolution revealed a connectivity between monomers that was very different from that proposed from X-ray scattering data and modeling (16) (Fig. 1E and Fig. S1). We modeled the number of TSR domains composed of this vertex by manually fitting the atomic structure of one of the homologous TSR domains from thrombospondin (PDB 3R6B) (10) into the EM density. Each properdin monomer comprises seven TSR domains and we found that four of these units could be accommodated into the vertex. Thus, each properdin monomer contributes four TSR domains for the assembly of two vertexes, at the N- and C-terminal end of each monomer, leaving three TSR units for the elongated connection between vertexes. In agreement with this, we found that the average distance between vertexes, obtained from 150 images of complexes, measured 14.3 ± 1.2 nm (Fig. 1F), which fits the length spanned by three TSRs, assuming an averaged length of 5 nm per TSR domain based on the atomic structure (10).
Purification of the Properdin–C3bBb Convertase Complex.
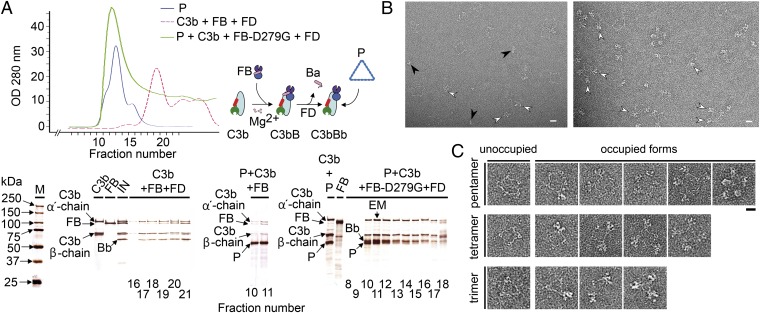
We assembled the complex between properdin and C3 convertase by incubating C3b, Factor B (FB), and Factor D (FD) in the presence of properdin. In these experiments, we used the FB-D279G mutant that increases the stability of C3 convertase (2). The mixture was resolved by gel filtration chromatography and the mobility of the complex compared with that of properdin alone. We observed comigration of C3b, the Bb fragment of FB and properdin in a large molecular weight species compared with the elution of C3bBb convertase alone. The purification of a stable complex containing C3bBb and properdin, which resisted purification, suggested properdin was contributing to stabilize the otherwise unstable C3bBb (17, 18) (Fig. 2A). The peak fraction was observed in the electron microscope, revealing properdin oligomers decorated by extra densities corresponding to C3bBb convertases (Fig. 2B). Interestingly, C3bBb convertase molecules were bound to properdin vertexes, revealing that the structure assembled by the oligomerization of two properdin monomers was essential for C3bBb convertase recognition. C3bBb molecules protruded outwards from these vertexes. We tested several concentrations of C3bBb convertase and found that the level of vertex occupancy was dependent on the amount of C3bBb convertase used in the experiment (Fig. 2B). This indicates that each properdin oligomer has the potential to use all its vertexes to bind C3bBb convertase (Fig. 2C).
Fig. 2.
Purification and electron microscopy of the properdin–C3bBb convertase complex. (A) Chromatograms (Upper) and silver-stained SDS/PAGE (Lower) for the fractions of size-exclusion chromatography experiments performed in a Superdex 200 gel-filtration column (GE Healthcare) using properdin, C3b, FD, and either wild-type FB or the FB-D279G mutant. Chromatograms show profiles for properdin injected alone (P, blue line), the incubation of C3b, FB, and FD to assemble a C3bBb convertase (C3bBb, magenta discontinuous line), and the incubation of properdin, C3b, FB-D279G, and FD to assemble a properdin–C3bBb convertase complex (PC3bBb, green line). Lower shows SDS/PAGE of selected fractions from the chromatographies above. (Left) Assembly of a C3bBb convertase (C3bBb, magenta discontinuous line). (Right) Properdin–C3bBb convertase complex (PC3bBb, green line). (Center) SDS/PAGE of a chromatography analyzing the interaction of properdin with the C3bB proconvertase. The input to the gel-filtration column is indicated as IN, and C3b, FB, and properdin are loaded as controls. Chains of C3b detected in the SDS/PAGE are indicated. The formation of the properdin–C3bBb convertase complex is revealed by the advanced elution of C3bBb (factions 10–15) in the presence of properdin compared with the elution of C3bBb convertase alone (C3bBb, fractions 17–21), as well as the appearance of a new band corresponding to the FB fragment Bb (labeled Bb) resulting from the proteolysis of the input FB (labeled FB). The fraction selected for EM analysis is labeled. (B) Representative micrograph corresponding to properdin–C3bBb convertase complexes collected at two experimental conditions generating partial (Left) or high occupancy (Right) of properdin by C3bBb convertase. Selected C3bBb convertase molecules bound to properdin have been labeled with an open arrow. Black arrows stand for unbound C3bBb convertase molecules. (Scale bar, 14 nm.) (C) Gallery of raw images of properdin–C3bBb convertase complexes selected from the micrographs and panelled according to the oligomeric state of properdin, and showing, from left to right, increased occupancy of properdin vertexes with C3bBb convertase molecules. (Scale bar, 14 nm.)
Properdin Cross-Links C3b and the Bb Fragment, Stabilizing the C3bBb Convertase.
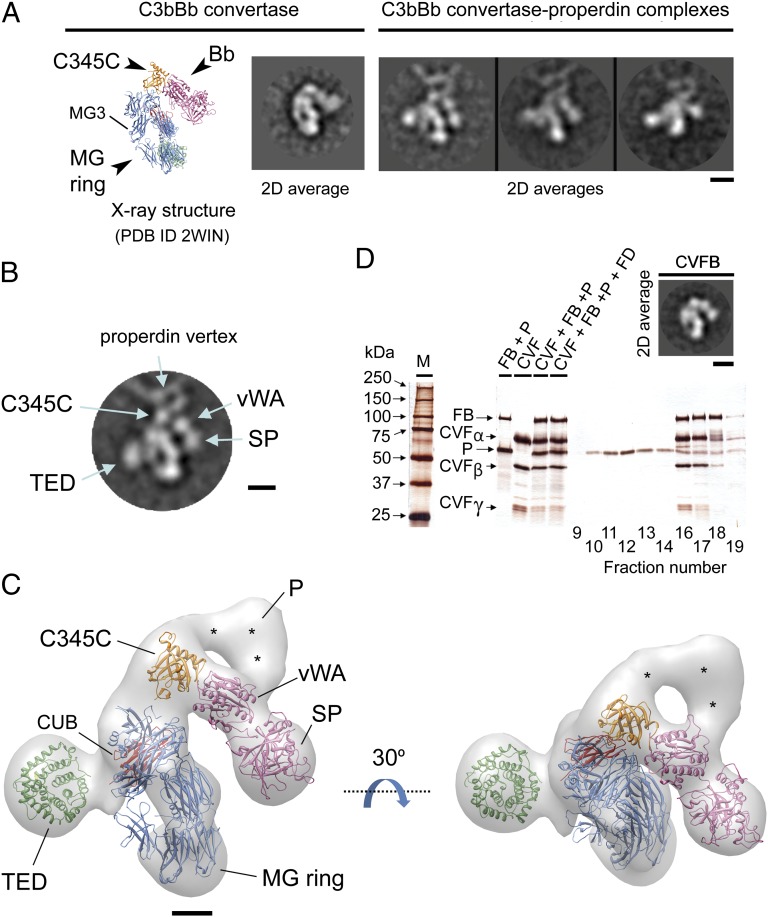
The basis for C3bBb convertase stabilization by properdin was explored by processing 21,891 images of C3bBb convertase molecules at properdin vertexes extracted from the micrographs. These images were computationally classified to find and average those corresponding to similar views of the complex (Fig. 3A). These averages were extremely revealing compared with the crystal and EM structure of C3bBb convertase (17, 18). The macroglobulin (MG) ring and C345C domain from C3b, and the von Willebrand factor type-A (vWA) and serine-protease (SP) domains from FB were clearly identified, as well as the properdin vertex contacting C345C and vWA (Fig. 3B). Other averages were interpreted as corresponding to additional views of the complex from a different angle and these were used for the 3D analysis of the complex (see below). Surprisingly, the most typical view of the properdin–C3bBb convertase complex was found in two distinct subtypes, either containing or not a strong globular density in the proximity of MG3 domain (see below).
Fig. 3.
Structure of the properdin–C3bBb convertase complex. (A) Representative reference free 2D averages of C3bBb convertase molecules bound to properdin vertexes (Right), compared with a view of the crystallographic and EM structures of C3bBb convertase (Left) (PDB 2WIN) (17). Each domain has been colored differently and labeled. (Scale bar, 5 nm.) (B) Selected average of the properdin–C3bBb convertase complex. Different domains and regions are labeled. (Scale bar, 5 nm.) (C) Two views of the structure of the properdin–C3 convertase complex at 29.3-Å resolution. A pseudoatomic model of the properdin–C3 convertase complex was obtained by fitting the atomic structure of C3bBb convertase (PDB 2WIN) (17) into the EM structure. The MG ring is displayed in blue. C345, CUB, and TED domains are colored in orange, red, and green, respectively. vWA and SP domains from the Bb fragment are colored in pink. Densities corresponding to properdin vertex are labeled with asterisks. (Scale bar, 2 nm.) (D) Fractions from a size-exclusion chromatography loaded with the incubation of CVF, FB-D279 mutant, properdin, and FD were analyzed by SDS/PAGE. Properdin does not interact with CVF-FB in the conditions tested, as revealed by the absence of comigration of the CVFB complex with properdin. Inset, Upper Right corner shows an average of images obtained for the purified CVFB complex using electron microscopy. (Scale bar, 5 nm.)
Images of the properdin–C3bBb convertase complex were then used to reconstruct its 3D structure. We found that two conformations were coexisting in the dataset, which corresponded to those images that either contained or not a globular density in the proximity of the MG3 domain. The dataset was consequently split in two subgroups (SI Materials and Methods). The 3D structure of the major population solved at 29.3-Å resolution, corresponding to 66.6% of the particles, was interpreted by fitting the atomic structure of C3bBb convertase (PDB 2WIN) (17) into the EM map to generate a hybrid pseudoatomic model of the complex (Fig. 3C). The Bb fragment together with the C345C domain had to be moved backward by ∼30° to fit into the density of the EM map, indicating a displacement compared with the crystal structure. This is conceivable by the flexible linker connecting C345C to the MG ring, as observed in the EM images of C3bBb convertase (17, 18). The fitting revealed that a small segment of the MG ring corresponding to MG4 was not well resolved in the reconstruction, which we interpret as an effect of the accumulation of staining agent in the center of the MG ring by the proximity of the globular density in MG3. Remarkably, we found that the TED domain was missing at its expected location in C3b, strongly suggesting that the globular density in the vicinity of the MG ring corresponds to the TED domain. The CUB domain was also not found in properdin–C3bBb at the expected location but a density nearby the TED domain was interpreted as the CUB, further supporting the repositioning of the TED domain (Fig. 3C).
The structure revealed that properdin contacts both the C345C domain in C3b and the vWa domain in Bb (Fig. 3C), indicating that properdin stabilizes the C3bBb convertase by holding together the two components of this enzymatic complex. The structure also suggested that properdin would interact with these two domains more efficiently when the Bb fragment is in the conformation found in C3bBb convertase than the closed conformation of the C3bB proconvertase. We confirmed this hypothesis after observing that properdin did not interact with the complex between C3b homolog cobra venom factor (CVF) and FB, because CVF and FB form a tight complex, but FB is maintained in its closed conformation (19) (Fig. 3D).
Properdin Promotes a Relocation of the TED Domain.
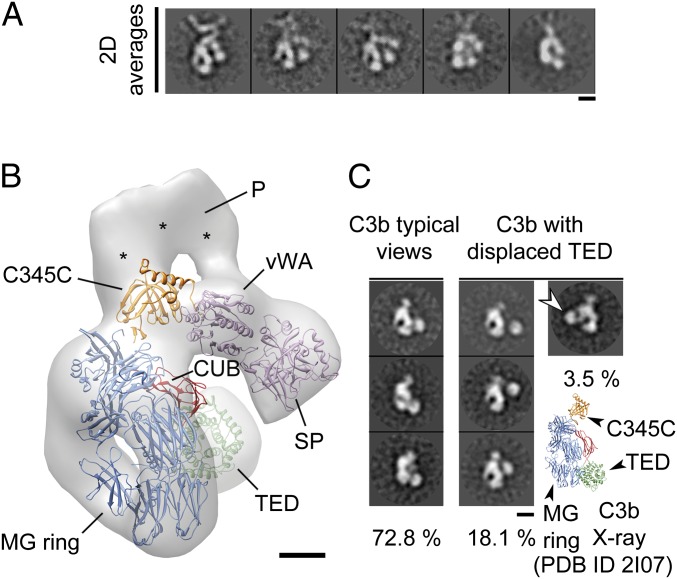
The 3D structure of the minor population of properdin–C3bBb convertase complexes revealed essentially the same structural features of the most abundant conformation albeit at lower resolution, but the TED domain was at its expected location in C3b (Fig. 4 A and B). Thus, the TED domain was found in two positions in the context of the properdin–C3bBb convertase complex, at an approximate 1:2 ratio between the two species (33.3 vs. 66.6%). We searched for this conformation of the TED domain in the C3b preparation used for these studies by collecting single molecule images, which were classified and averaged as before (Fig. 4C). We found that only 3.5% of 5,000 images of C3b showed this unusual conformation, whereas ∼73% corresponded to the TED position described in the crystal structure (2–4). Interestingly, ∼18% of the images revealed alternative locations for the TED domain. As a whole, these results indicate that properdin stimulates the rearrangement of the TED domain in C3b.
Fig. 4.
Positioning of the TED domain in the properdin–C3bBb convertase complex. (A) Representative 2D averages of the minor conformation of the properdin–C3bBb convertase complex. These show that the TED domain is not in the proximities of the MG3 domain, but in the location found in the crystal structure of C3b. (Scale bar, 5 nm.) (B) One view of the structure of the minor conformation of the properdin–C3bBb convertase complex at 33.0-Å resolution. Densities corresponding to properdin vertex are labeled with asterisks. (Scale bar, 2 nm.) (C) Processing and classification of images of C3b revealed that most molecules show the TED domain in the classical conformation, whereas a small percentage of molecules display the TED domain in alternative conformations. An arrow points to the TED domain placed close to the MG3 domain, found in 3.5% of the images analyzed. A view from the crystal structure of C3b (PDB 2I07) is shown to help comparison with the EM images. Each domain has been colored differently and labeled. (Scale bar, 5 nm.)
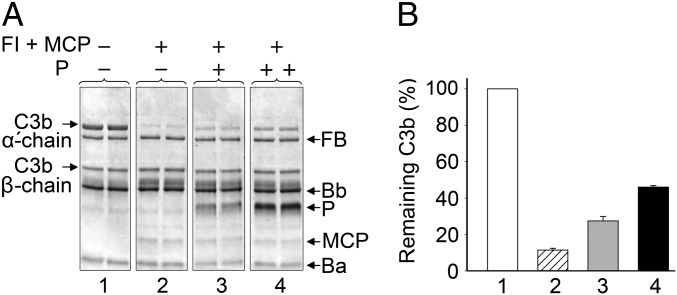
Properdin Interferes with C3bBb Convertase Decay.
The repositioning of the TED domain, and presumably also the CUB domain to which it is tethered, is predicted to remove essential structural determinants for the interaction of C3b with the complement regulators FH, decay-accelerating factor (DAF), and complement receptor 1 (CR1), turning the properdin–C3bBb complex less prone to accelerated decay compared with C3bBb alone (4, 20). Similarly, the repositioning of the TED domain should impair the interaction between C3b and membrane cofactor protein (MCP or CD46), whereas changes in the CUB domain could affect Factor I (FI) binding. The combined effect of these changes should slow down the FI-dependent proteolytic inactivation of C3b. We tested this hypothesis by assembling a C3bBb convertase through the incubation of C3b, FB, and FD in the absence or presence of properdin (Fig. 5A). Next, soluble versions of MCP and FI were added and incubated for 15 min in all cases, to allow for cofactor activity, and the intact C3b was quantified by measuring the ratio between α′ chain/β chain of C3b. We observed that the amount of intact C3b remaining after the incubation was significantly increased in the presence of properdin in a dose-dependent manner (Fig. 5B). These data show that properdin also contributes to stabilize the C3bBb convertase by interfering the interaction of C3b with the complement regulators, which should impact both their accelerated decay and factor-I mediated cofactor activities.
Fig. 5.
C3bBb convertase decay in the presence of MCP, FI, and properdin. (A) C3bBb convertase was formed by incubating C3b, FB, and FD in the absence (P) (−) or the presence of two amounts of properdin (P) (0.9 µg + and 1.8 µg ++) and incubated with MCP and FI. SDS/PAGE shows the result of this reaction after incubating for 15 min. Each experiment was performed in duplicate. (B) The amount of C3b remaining after incubation was estimated by quantifying the ratio between α′ chain/β chain of C3b. Experiments labeled as 1–4 correspond to the matching experiment in A (0.9 µg of P, gray and 1.8 µg of P, black). Error bars indicate the mean ± SD of two independent experiments.
Discussion
A major point of regulation in the activation of complement is altering the stability of the alternative pathway C3bBb convertase. Down-regulation to control homeostasis and prevent tissue damage is provided by a number of plasma and membrane-associated regulators that accelerate the dissociation of the C3bBb complex (3). In contrast, properdin is the only complement regulator that stabilizes the C3bBb convertase, which may be critical to tip the balance in favor of amplification on microbial pathogens (9). The molecular bases of the properdin functions are still poorly understood. Using EM single-particle image processing methods we describe here a structural model for the properdin–C3bBb complex supporting that properdin stabilizes the C3bBb convertase by holding together the two components of the AP C3 convertase, C3b and Bb, and by promoting a large displacement of the TED domain that likely interferes with the decay of C3 convertase by complement down-regulators.
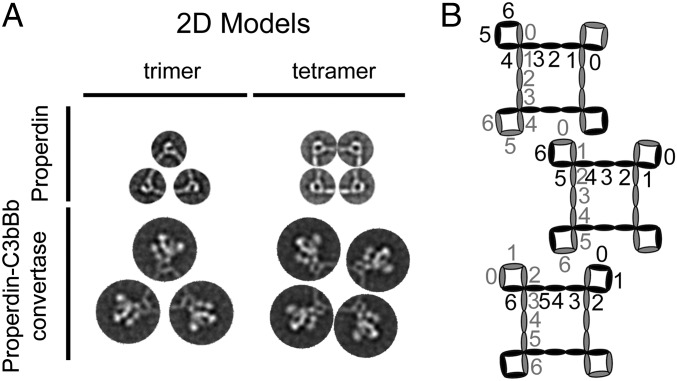
Properdin oligomerizes by a complex interplay between N- and C-terminal ends of two elongated monomers forming a curly vertex structure (Fig. 6A). This interaction allows the assembly of oligomers containing a variable number of monomers, and the maximum number of units that could be accommodated per oligomer is probably only limited by conformational and geometrical restrains. Our structural model for properdin is different from that proposed by Sun et al. based on X-ray scattering and modeling data (16). Their model showed connections between the N- and C-terminal ends of properdin but the 3D structure of these contact points is very different from the structure of the vertexes that we have resolved using EM (Fig. S1). In addition, their model proposed that the properdin oligomers were partially collapsed, whereas we find well-defined triangular and squared-shaped molecules supporting full extension of the properdin monomers. Although we cannot rule out an effect on the conformation of properdin by the carbon surface used as support film in electron microscopy, compared with the structure described from measurements in solution, we believe the EM conformation could reflect closer the situation on cellular surfaces.
Fig. 6.
Model for complement activation by properdin. (A) Idealized images of properdin and properdin–C3bBb convertase complexes generated by combining the averages and the dimensions of experimental single molecule images. (B) Cartoon representing the three alternative models for the arrangement of subunits at the vertex of properdin oligomers (see Discussion). Alternating oligomers are shown in black and gray. The TSR domains are numbered from 0 to 6.
Each vertex is the structural unit of recognition and stabilization of C3bBb convertase by holding together the C345C and the vWA domains from C3b and Bb, respectively (Fig. 6A). Thus, oligomerization, which requires the involvement of the N- and the C-terminal TSRs, is essential for interaction with the C3bBb convertase. This model is consistent with previous evidence showing that properdin binds equivalently to C3b, iC3b, and C3c (21), which we now justify by the interaction of properdin with the C345C domain, and with experiments showing that properdin interacts with FB (22). Early studies using a combination of cyanogen bromide (CNBr) digestions and synthetic peptide synthesis, have proposed that the properdin binding site in C3b is located within a 34-amino acid peptide region in the MG8 domain of the α′-chain, spanning residues 1402–1435 (14). Our model of the structure of the properdin–C3bBb complex does not show contacts compatible with that region in C3b. To explain these apparently conflicting data we like to suggest that the interactions between C3b, FB, and properdin transit through several intermediates so that properdin may initially recognize sites in C3b that are distinct from those conforming the fully assembled properdin–C3bBb complex.
The structure of each of the properdin vertexes could be interpreted as a combination of TSR0 from one monomer and TSR4-5-6 from another (TSR0/TSR4-5-6), or the alternative options TSR0-1/TSR5-6 and TSR0-1-2/TSR6, but we could not discriminate between these three options at the resolution of these studies (Fig. 6B). The studies by Higgins et al. (23) proposed functions for several TSR domains in properdin by characterizing recombinant properdin lacking single specific TSRs. TSR0, TSR1, and TSR2 were not included in those studies. They suggested that TSR4 and TSR5 were required for C3b binding and C3bBb convertase stabilization, respectively. These results are not in conflict with our findings but they need to be reanalyzed, as any mutation affecting oligomerization would indirectly affect C3bBb binding. A vertex comprising TSR0 from one monomer and TSR4-5-6 (TSR0/TSR4-5-6) from another monomer would agree with the published results. Importantly, the TSR4 and TSR5 mutants were also affected in oligomerization, not assembling as trimers and tetramers. This could be interpreted as TSR4 and TSR5 being part of the vertex, but alternatively, if the mutations affect oligomerization indirectly, they could fail to stabilize the C3bBb convertase as a consequence of the oligomerization defect rather than by being involved in C3bBb binding. The involvement of TSR6 at the vertex in stabilization of C3bBb is supported by the disease-associated Y387D mutation in TSR6, which produces normal plasma levels of properdin, which assembles oligomer lacking the capability to stabilize the C3bBb convertase (24).
A remarkable finding is the positioning of the TED domain in two alternative locations, one in agreement with the crystal structure of C3b and an alternative location in the vicinity of the MG3 domain, and the accompanying relocation of the CUB domain attached to the TED. Such movements appear to be an intrinsic property of C3b, as we find a similar conformation in a small percentage of C3b molecules, but certainly properdin turns this alternative conformation into the major species. In agreement with our findings, previous EM studies by Nishida et al. found a proportion of C3b molecules in this alternative conformation (25). In addition, recent FRET data obtained for C3b in complex with SCR1–4 from FH suggested some degree of mobility of the TED domain (26). Large displacements of the CUB-TED region seem feasible as these take place during the structural transition from C3 to C3b (4) and also from C3b to iC3b (27).
The rearrangement of the TED removes essential structural determinants for recognition of C3b by some regulators such as FH and MCP (4). SCR1–4 of FH interacts with C3b as an elongated string of monomers and this causes decay of C3 convertase, as revealed in the crystal structure of this complex (20). Movements of the TED domain in iC3b, a proteolytic fragment of C3b, have been shown to disrupt FH binding and consequentially iC3b is not regulated by FH (27). Similarly, the rearrangements of the TED domain that we find in the properdin–C3bBb convertase complex would limit C3 convertase regulation by FH, MCP, and other regulators that use similar mechanisms, such as DAF. These effects probably combine with the consequences of the changes observed in the CUB domain that are predicted to affect the interaction of C3b and FI (4). In agreement with this interpretation, we observed a reduced FI-dependent cofactor activity of MCP for the proteolysis of C3b in the presence of properdin. We speculate this will contribute, in addition to the holding of C3b and Bb together, to enhance the complement responses in vivo.
Materials and Methods
Generation and Purification of Properdin–C3bBb Convertase Complexes.
C3b and properdin were purified from plasma and FB from the supernatant of CHO cells. C3b (5 μg), FB (10 μg, FB or FB-D279G), and properdin (P, 1 μg) were incubated for 35 min at room temperature in 20 mM Hepes (pH 7.5), 75 mM NaCl and 5 mM MgCl2 at a final molar ratio P:C3b:FB 0.7:1:4. Subsequently, 100 ng of FD (Calbiochem) was added and the mixture was injected in a Superdex 200 column (GE Healthcare). Fractions were analyzed using 10% (wt/vol) SDS/PAGE.
Electron Microscopy and Image Processing.
A few microliters of freshly purified complexes were adsorbed onto carbon-coated grids and negatively stained with 2% (wt/vol) uranyl formate. Micrographs were recorded using a JEOL 1230 transmission electron microscope and a TemCam-F416 detector from Tietz Video and Image Processing Systems (TVIPS) using EM-TOOLS (TVIPS). Images were collected at a final magnification of 54,926×. Using EMAN (28), 6,425 images for properdin vertexes and 21,891 images for C3bBb convertases bound to a propedin vertex were selected and processed. Ab initio templates for refinement were obtained using the command “e2initial model” in EMAN2 (28) and the random conical tilt (RCT) method.
For further details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank our colleagues at the laboratory of S.R.d.C. for help in the purification of C3b, FB, and properdin. This work was funded by the Autonomous Region of Madrid (S2010/BMD-2316 to S.R.d.C. and O.L.), the Ramón Areces Foundation (O.L.), and the Spanish government (SAF2011-22988 to O.L. and SAF2011-26583 to S.R.d.C.). O.L. is additionally supported by Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/1001), and S.R.d.C. is also supported by the Fundación Renal Iñigo Alvarez de Toledo and the Seventh Framework Programme European Union Project EURenOmics (European Consortium for High-Throughput Research in Rare Kidney Diseases). M.A. is a Sara Borrell Fellow from the Instituto de Salud Carlos III (CD09/00282).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The 3D-EM maps have been deposited in the Electron Microscopy Data Bank database, www.emdatabank.org and www.ebi.ac.uk/pdbe (EMD-2402 and EMD-2403).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309618110/-/DCSupplemental.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez de Córdoba S, Harris CL, Morgan BP, Llorca O. Lessons from functional and structural analyses of disease-associated genetic variants in the complement alternative pathway. Biochim Biophys Acta. 2011;1812(1):12–22. doi: 10.1016/j.bbadis.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8(1):48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 4.Lea SM, Johnson S. Putting the structure into complement. Immunobiology. 2012;217(11):1117–1121. doi: 10.1016/j.imbio.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hourcade DE. The role of properdin in the assembly of the alternative pathway C3 convertases of complement. J Biol Chem. 2006;281(4):2128–2132. doi: 10.1074/jbc.M508928200. [DOI] [PubMed] [Google Scholar]
- 6.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179(4):2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 7.Densen P. Interaction of complement with Neisseria meningitidis and Neisseria gonorrhoeae. Clin Microbiol Rev. 1989;2(Suppl):S11–S17. doi: 10.1128/cmr.2.suppl.s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesher AM, et al. Combination of factor H mutation and properdin deficiency causes severe C3 glomerulonephritis. J Am Soc Nephrol. 2013;24(1):53–65. doi: 10.1681/ASN.2012060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kemper C, Atkinson JP, Hourcade DE. Properdin: Emerging roles of a pattern-recognition molecule. Annu Rev Immunol. 2010;28:131–155. doi: 10.1146/annurev-immunol-030409-101250. [DOI] [PubMed] [Google Scholar]
- 10.Klenotic PA, Page RC, Misra S, Silverstein RL. Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system. Protein Expr Purif. 2011;80(2):253–259. doi: 10.1016/j.pep.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chondrou M, Papanastasiou AD, Spyroulias GA, Zarkadis IK. Three isoforms of complement properdin factor P in trout: Cloning, expression, gene organization and constrained modeling. Dev Comp Immunol. 2008;32(12):1454–1466. doi: 10.1016/j.dci.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Pangburn MK. Analysis of the natural polymeric forms of human properdin and their functions in complement activation. J Immunol. 1989;142(1):202–207. [PubMed] [Google Scholar]
- 13.Smith CA, Pangburn MK, Vogel CW, Müller-Eberhard HJ. Molecular architecture of human properdin, a positive regulator of the alternative pathway of complement. J Biol Chem. 1984;259(7):4582–4588. [PubMed] [Google Scholar]
- 14.Daoudaki ME, Becherer JD, Lambris JD. A 34-amino acid peptide of the third component of complement mediates properdin binding. J Immunol. 1988;140(5):1577–1580. [PubMed] [Google Scholar]
- 15.Orlova EV, Saibil HR. Structural analysis of macromolecular assemblies by electron microscopy. Chem Rev. 2011;111(12):7710–7748. doi: 10.1021/cr100353t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Z, Reid KB, Perkins SJ. The dimeric and trimeric solution structures of the multidomain complement protein properdin by X-ray scattering, analytical ultracentrifugation and constrained modelling. J Mol Biol. 2004;343(5):1327–1343. doi: 10.1016/j.jmb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Rooijakkers SH, et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10(7):721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torreira E, Tortajada A, Montes T, Rodríguez de Córdoba S, Llorca O. 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc Natl Acad Sci USA. 2009;106(3):882–887. doi: 10.1073/pnas.0810860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janssen BJ, et al. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28(16):2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, et al. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10(7):728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farries TC, Lachmann PJ, Harrison RA. Analysis of the interactions between properdin, the third component of complement (C3), and its physiological activation products. Biochem J. 1988;252(1):47–54. doi: 10.1042/bj2520047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farries TC, Lachmann PJ, Harrison RA. Analysis of the interaction between properdin and factor B, components of the alternative-pathway C3 convertase of complement. Biochem J. 1988;253(3):667–675. doi: 10.1042/bj2530667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JM, Wiedemann H, Timpl R, Reid KB. Characterization of mutant forms of recombinant human properdin lacking single thrombospondin type I repeats. Identification of modules important for function. J Immunol. 1995;155(12):5777–5785. [PubMed] [Google Scholar]
- 24.Fredrikson GN, et al. Molecular characterization of properdin deficiency type III: Dysfunction produced by a single point mutation in exon 9 of the structural gene causing a tyrosine to aspartic acid interchange. J Immunol. 1996;157(8):3666–3671. [PubMed] [Google Scholar]
- 25.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc Natl Acad Sci USA. 2006;103(52):19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pechtl IC, Neely RK, Dryden DT, Jones AC, Barlow PN. Use of time-resolved FRET to validate crystal structure of complement regulatory complex between C3b and factor H (N terminus) Protein Sci. 2011;20(12):2102–2112. doi: 10.1002/pro.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcorlo M, et al. Unique structure of iC3b resolved at a resolution of 24 Å by 3D-electron microscopy. Proc Natl Acad Sci USA. 2011;108(32):13236–13240. doi: 10.1073/pnas.1106746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.