Abstract
Membrane trafficking is regulated by small Ras-like GDP/GTP binding proteins of the Rab subfamily (Rab GTPases) that cycle between membranes and cytosol depending on their nucleotide state. The GDP dissociation inhibitor (GDI) solubilizes prenylated Rab GTPases from and shuttles them between membranes in the form of a soluble cytosolic complex. We use attenuated total reflection–Fourier transform infrared spectroscopy to directly observe extraction of Rab GTPases from model membranes by GDI. In their native form, most Rab GTPases are doubly geranylgeranylated at the C terminus to achieve localization to the membrane. We find that monogeranylgeranylated Rab35 and Rab1b reversibly bind to a negatively charged model membrane. Correct folding and GTPase activity of the membrane-bound protein can be evaluated. The dissociation kinetics depends on the C-terminal sequence and charge of the GTPases. The attenuated total reflection experiments show that GDI genuinely accelerates the intrinsic Rab membrane dissociation. The extraction process is characterized and occurs in a nucleotide-dependent manner. Furthermore, we find that phosphocholination of Rab35, which is catalyzed by the Legionella pneumophila protein AnkX, interferes with the ability of GDI to extract Rab35 from the membrane. The attenuated total reflection–Fourier transform infrared spectroscopy approach enables label-free investigation of the interaction between GDI and Rab GTPases in a membrane environment. Thereby, GDI is revealed to actively extract monogeranylgeranylated membrane-bound Rab GTPases and, thus, is not merely a solubilization factor.
Keywords: lipid bilayers, vesicle spreading, Rab recycling, geranylgeranyl
Vesicular trafficking processes in eukaryotic cells are regulated by Rab GTPases that constitute the largest family of small GTPases (1). Rab GTPases cycle between the inactive GDP-bound and the active GTP-bound state. Interaction with various effectors takes place with the latter only. Exchange of GDP to GTP is catalyzed by guanine nucleotide exchange factors (GEFs) and hydrolysis of GTP to GDP is catalyzed by GTPase activating proteins (GAPs). Their function as regulators of membrane trafficking implies localization to and cycling between membranes of different cellular compartments. Membrane localization is mediated by geranylgeranylation of two C-terminal cysteine residues by Rab geranylgeranyl transferase (GGTase) with assistance of Rab escort protein (REP) (2, 3). Rab recycling is mediated by GDP dissociation inhibitor (GDI) that extracts membrane-bound Rab GTPases to form a soluble cytosolic complex. For over 60 Rab GTPases, there are five GDI isoforms in mammals (only two isoforms in humans), which are assumed to have a uniform mechanism (4). Certain mutations in GDI are responsible for a form of familial mental retardation (5). Recent evidence shows that the pathogenic bacterium Legionella pneumophila specifically disrupts the interaction between certain Rab GTPases and GDI by adenylylation or phosphocholination of Rab GTPases (6–8).
GDI has been studied by X-ray crystallography in its apo-form and in complex with Rab GTPases (9–11). Interactions with Rab GTPases are mediated by the Rab binding platform, the C terminus coordinating region, and the lipid binding pocket of GDI, which bind to the G domain, the C terminus, and the prenyl moieties of Rab, respectively. Nanomolar dissociation constants (KD) for the interaction of GDI with Rab GTPases have been determined by fluorescence titration experiments and by isothermal titration calorimetry (ITC) (11–13). A fluorescence readout was provided by using farnesyl–7-nitrobenzol[1,2,5]oxodiazol (NBD) as the lipid anchor, and the ITC measurements were performed using the soluble farnesylated form of Rab proteins. However, all biophysical studies of the interaction between GDI and Rab GTPases were conducted in the absence of a lipid membrane, and therefore the extraction of Rab from the membrane by GDI could not be characterized.
Attenuated total reflection–Fourier transform infrared spectroscopy (ATR-FTIR) is a valuable tool for detailed analysis of membrane-attached proteins (14–17). Model membranes can be formed by vesicle spreading on the internal reflection element, such as a germanium crystal (18, 19). Various techniques exist for the immobilization of proteins on the lipid bilayer, the most natural for small GTPases being via lipid anchors attached to the protein. The availability of lipidated Ras GTPase allowed investigation of the secondary structure and orientation of membrane-bound Ras with ATR-FTIR spectroscopy, and recent advances allow recording of difference spectra of protein activity, such as GTP hydrolysis (15, 19, 20).
Here, we have applied ATR-FTIR spectroscopy to investigate GDI-mediated membrane extraction of Rab35 and Rab1b. Rab35 is involved in phagosome–lysosome fusion, whereas Rab1b regulates endoplasmic reticulum to Golgi transport, and both GTPases are targeted by posttranslational modifications as the pathogen L. pneumophila hijacks or interferes with intracellular vesicle trafficking during infection (1, 6, 8, 21). We have improved existing preparation methods of monogeranylgeranylated Rab35 and Rab1b [prenylated via a CAAX-box (2)] to achieve binding to the membrane and verify the activity of bound protein. Finally, we have observed membrane extraction by GDI in a nucleotide-dependent manner, and have furthermore shown that phosphocholination of Rab35 directly interferes with membrane extraction by GDI.
Results
Lipid Bilayer Formation.
Solid-supported lipid bilayers were formed on germanium by in situ spreading of small unilamellar vesicles. Our model membrane consisted of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and negatively charged 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS) in a 9:1 ratio. We chose a negatively charged model membrane because Rab35 naturally resides on the plasma membrane and endosomes, which carry ∼10% negatively charged lipids (22, 23). Time-resolved infrared spectra of vesicle spreading were acquired with parallel and vertical polarized light, and spreading kinetics as well as orientational parameters of the bilayer were obtained.
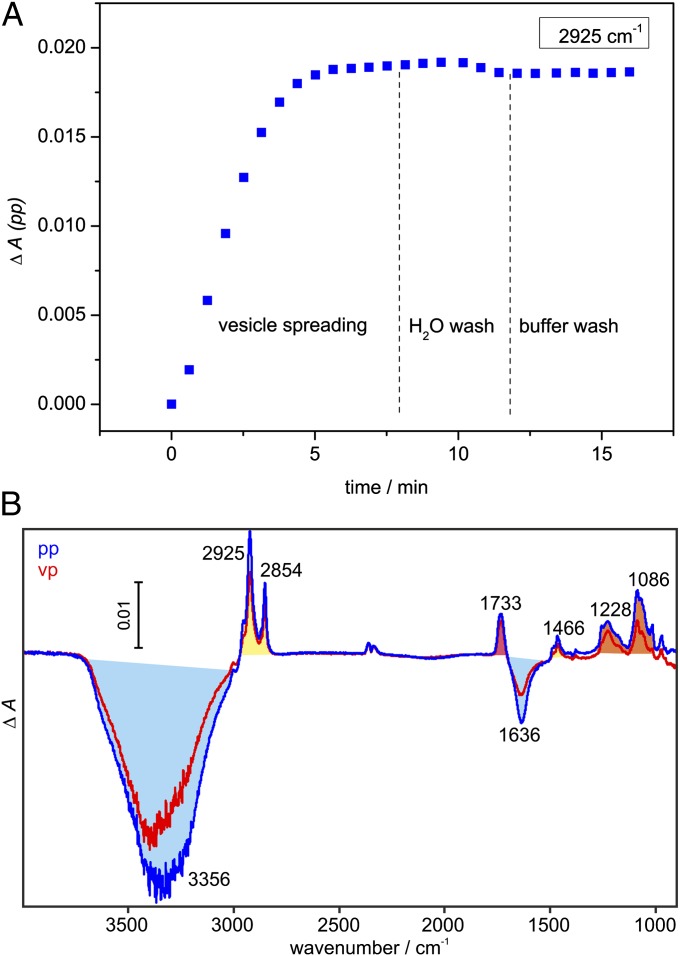
Fig. 1A shows the kinetics of vesicle spreading monitored at 2,925 cm−1 [νas(CH2)]. The spreading was complete after 10 min. The vesicles ruptured quickly upon adsorbing to the germanium crystal because no overshoot kinetics was observed, which is typical for the accumulation of intact vesicles at the surface (19). This strong interaction between vesicles and germanium is probably due to the negative charge introduced by DOPS because neutral vesicles show slower spreading kinetics with an overshoot (19). A short wash step with water led to a slight absorbance decrease, probably by rupturing any incompletely fused vesicles as has been observed on silica substrates as well (24). Afterward, the bilayer remained stable for the duration of the experiment. Layer completeness was verified with BSA as described for 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) layers (15).
Fig. 1.
Formation of the solid-supported lipid bilayer containing DOPC/DOPS in a 9:1 ratio. (A) A typical time course of vesicle spreading monitored at 2,925 cm−1 [νas(CH2)] with parallel polarized light. (B) Difference spectra of single bilayer formation with parallel (termed pp, blue) and vertical (termed vp, red) polarized light. Positive bands belong to the methylene (yellow), carbonyl (red), and phosphate (orange) groups of DOPC/DOPS. Negative bands belong to displaced water (light blue). A CO2 artifact is present at 2,361/2,341 cm−1.
Difference spectra of vesicle spreading (Fig. 1B) allow characterization of the model membrane. Negative bands belong to the displaced buffer layer; positive bands belong to the phosphate, carbonyl, and aliphatic groups of the lipids. The νas(CH2) and ν(CO) bands at 2,925 and 1,733 cm−1 were used to determine the surface concentration and molecular orientation of the lipids from dichroic measurements (SI Materials and Methods). The surface concentration of 339 ± 46 pmol/cm2 is similar to values for POPC films generated by vesicle spreading or the Langmuir–Blodgett technique [404–448 pmol/cm2 (15, 25)]. The lower value for DOPC/DOPS probably reflects a less dense packing of the unsaturated acyl chains compared with mixed acyl chains of POPC. Order parameters of the acyl chains (S = 0.23 ± 0.14) and the carbonyl group (S = −0.21 ± 0.08) are in good agreement with literature values for POPC bilayers [0.2 for the acyl chains, −0.18 to −0.26 for the carbonyl group (15, 26)], indicating that a planar single lipid bilayer is present. All relevant parameters are summarized in Table S1.
Membrane Binding of Geranylgeranylated Rab GTPases.
Next, we analyzed membrane-binding behavior of the used proteins and identified optimal conditions for the GDI assay. In controls, unprenylated Rab35 with a CAAX-box (Rab35CVIL) exhibited nonspecific binding to the model membrane (Fig. S1A). Increasing the salt concentration to 300 mM NaCl inhibited nonspecific binding and ensured that prenylated Rab35 would only be immobilized via its lipid anchor (Fig. S1 B and C). RabGGTase β used as a chaperone to solubilize prenylated Rab GTPases did not show any nonspecific binding even at low salt conditions (Fig. S1 B and C). GDI showed strong nonspecific binding that, however, was also eliminated with higher salt concentrations, at least for a maximum GDI concentration of 1 µM (Fig. S1 B and C).
We used geranylgeranylated Rab35CVIL (termed Rab35_G) chaperoned in complex with the β-subunit of RabGGTase (Rab35_G:β-su) for membrane-binding studies. Here, RabGGTase β functions solely as a chaperone to prevent aggregation of geranylgeranylated Rab and is washed away after membrane binding of Rab35_G. The chaperone was shown to be a weak binder and stabilizer of the lipid moiety of prenylated Rab GTPases and not to interfere significantly with other binding partners such as GDI or REP (12). ATR-FTIR enabled the evaluation of the secondary structure of membrane-bound Rab35, which proved to be a crucial advantage. Binding of Rab35_G to the DOPC/DOPS bilayer in control experiments at low salt conditions was affected by nonspecific interactions. Two fractions of Rab35 were identified, one being obviously denatured, as indicated by a broader amide I band (Fig. S2A). These two components were kinetically separated, the denatured fraction binding more slowly than the correctly folded protein (Fig. S1A). Raising the salt concentration in the buffer eliminated the slower binding phase, so that only the fast binding phase remained (Fig. S1A). Thus, Rab35_G bound to the DOPC/DOPS lipid bilayer specifically with an apparent rate constant of 1.7 min−1 at a Rab35_G:β-su concentration of 2 µM.
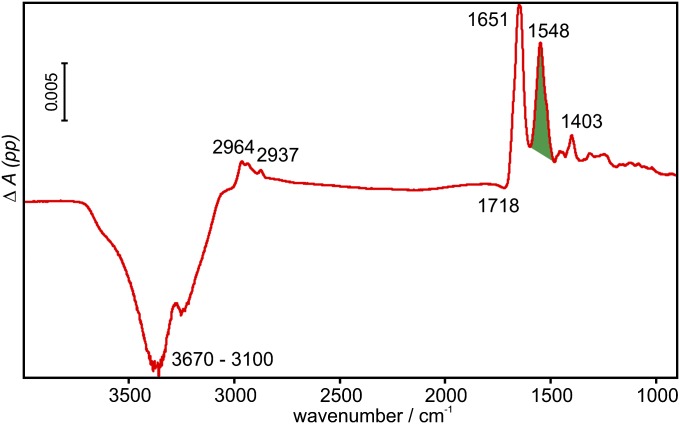
The difference spectrum of Rab35_G immobilization is shown in Fig. 2. Negative bands belong to the displaced buffer, whereas positive bands arise from the immobilized Rab35_G. The area of the amide II band (green) was used to monitor the kinetics of extraction by GDI in subsequent experiments. Also, the surface concentration of Rab35_G was calculated with the help of dichroic measurements to be 5 pmol/cm2. Compared with the reported surface concentration of 13 pmol/cm2 (78% surface coverage) for lipidated N-Ras monolayers on POPC (15), this shows that Rab35_G is present at the model membrane as a loosely packed monolayer, leaving sufficient space for interaction with binding partners such as GDI.
Fig. 2.
Difference spectrum of the immobilization of Rab35_G. For clarity, only the spectrum measured with parallel polarized light is shown. Positive bands are due to the immobilized protein, and negative bands are due to the displaced buffer. The area of the amide II band (green) was used to determine the surface concentration and also for the kinetics of protein immobilization and extraction.
To achieve membrane extraction of Rab proteins by GDI, we had to ensure that the membrane-bound proteins were in a native, GDI-binding competent state. The shape of the amide I band is determined by the secondary structure of the protein, and thus reflects its fold (14, 19). The amide I band of membrane-bound Rab35_G is very similar to that of lipidated N-Ras immobilized on a POPC bilayer (20) as demonstrated in Fig. S2B. Given that Ras and Rab proteins possess the same G-domain fold, this indicates that membrane-bound Rab35_G is correctly folded. Interestingly, the conditions in the in vitro prenylation assay had an impact on the folding state of membrane-bound protein. Our prenylation method relies on detergent for solubilization of the geranylgeranylated Rab protein, which necessitates subsequent detergent removal and introduction of RabGGTase β as a chaperone. In principle, prenylation in the presence of RabGGTase β instead of detergent leads to a seemingly identical product as judged by mass spectrometry. However, protein prenylated without detergent was found to be denatured in the ATR experiment as opposed to protein prenylated in presence of detergent, shown for the example of Rab1b_G in Fig. S2B. Notably, the protein found to be denatured in the ATR experiment was in the GDP-bound state after the prenylation, as judged by HPLC analysis, and could thus be assumed to have a partly functional G domain.
An additional advantage of ATR-FTIR is that the molecular switch function of membrane-bound GTPases can be probed by difference spectroscopy with beryllium trifluoride anion, which is a γ-phosphate analog (15, 27). Difference spectra of the BeF3−-induced “off” to “on” transition in membrane-bound Rab35_G was obtained and were comparable to difference spectra obtained with N-Ras elsewhere (28), further indicating that the protein bound to the membrane was active in the sense that the G domain was functional (Fig. S3). Only correctly folded, active protein, obtained by detergent-assisted prenylation, could be extracted from the membrane by GDI, as will be outlined below.
Membrane Extraction of Rab GTPases by GDI.
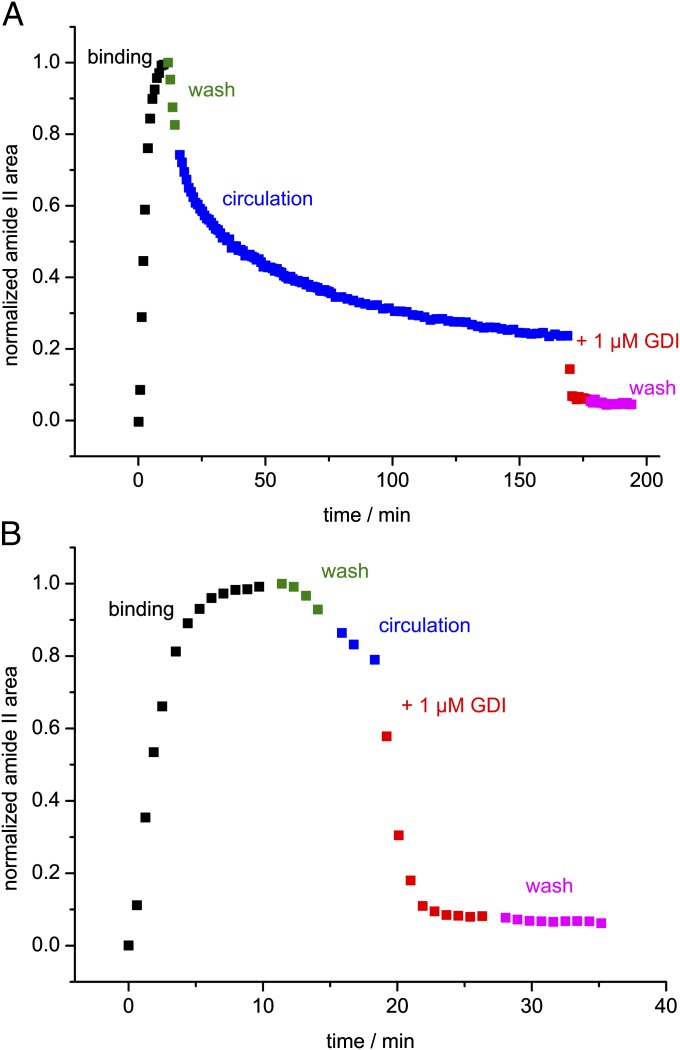
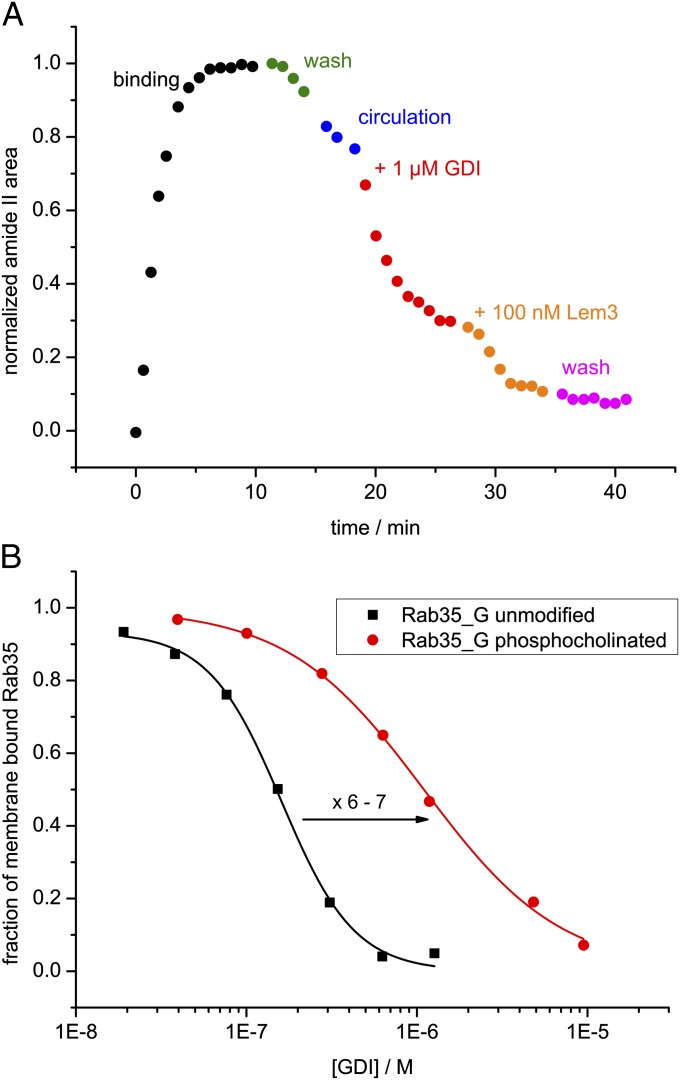
After establishing binding of Rab35_G to the lipid bilayer and confirming protein activity, we studied membrane dissociation of Rab35_G in the absence and presence of GDI. A washing step was performed to remove excess Rab35_G:β-su from the sample cell. We found that Rab35_G quickly dissociated from the membrane during the washing step with an apparent rate constant of 0.042 min−1 at a flow rate of 1 mL/min. Therefore, the washing step was kept as short as possible and the system was switched into a circulating mode, in which the solution with dissociated Rab35_G was retained in the sample cell. Under these conditions, bound Rab35_G dissociated until equilibrium between the circulating solution and the membrane was reached (Fig. 3A). The observed fast membrane binding and dissociation is in agreement with the behavior of monoprenylated peptides studied by fluorescence (29).
Fig. 3.
Time course of the immobilization of Rab35_G on a lipid bilayer and subsequent extraction by GDI. The experiment consisted of protein binding in a circulating system (black), wash (green), a circulation phase (blue), addition of GDI to the system (red), and a final wash step (magenta). The data points represent the normalized area of the amide II band. (A) An experiment with a prolonged circulation phase. Bound Rab35_G dissociates until equilibrium between the circulating solution and the membrane is reached. Addition of GDI leads to a rapid extraction of the remaining membrane-bound Rab35_G. (B) The same experiment with a short circulation phase. The extraction by GDI is seen more clearly by keeping the circulation phase short.
Addition of 1 µM GDI to the system led to rapid extraction of membrane-bound Rab35_G (Fig. 3A). This effect was seen more clearly by keeping the circulation phase short (Fig. 3B). The kinetics of GDI-mediated membrane extraction could not be determined because the rate of the process was limited by mixing kinetics of the sample cell, so that the rate constant was at least 0.5 min−1. For comparison with the spontaneous intrinsic membrane dissociation of Rab35_G, rebinding effects need to be considered. The plot of the apparent dissociation rate koff against the flow rate shows that koff moves toward saturation at higher flow rates (Fig. S4), as rebinding (occurring with the apparent association rate constant kon) is reduced. However, the observed extraction rate constant of GDI is approximately one order of magnitude greater (Fig. S4). Therefore, the effect of GDI is not due to capturing of free Rab, but to an increase of koff.
A subsequent wash led to no further dissociation, indicating that Rab35_G had been completely extracted from the membrane by 1 µM GDI. A small amount of protein (6–8%) remained irreversibly bound to the membrane and might represent the fraction of inactive or nonspecifically bound Rab35_G.
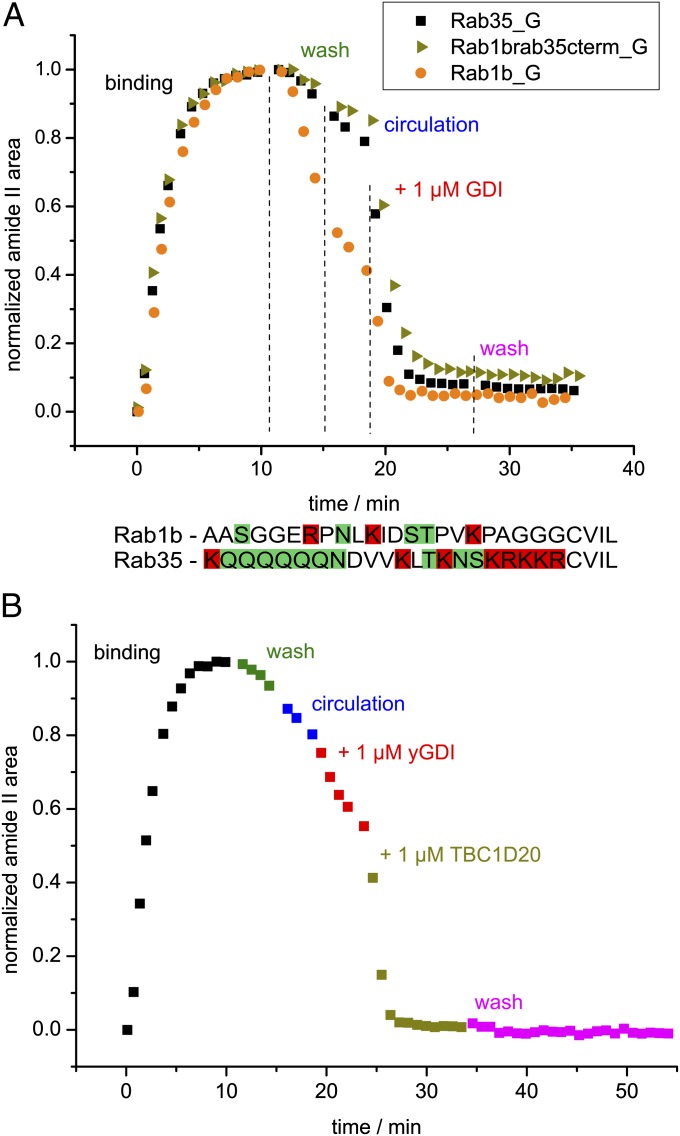
We also examined membrane extraction of Rab1b_G by GDI (Fig. 4A). Notably, spontaneous membrane dissociation of Rab1b_G was faster than that of Rab35_G. This can be attributed to the C terminus of Rab35 that possesses a polybasic stretch and a polar polyglutamine stretch, leading to a higher affinity toward the negatively charged membrane. Chimeric Rab1b with the C terminus of Rab35 behaves exactly like Rab35, proving that the nature of the C terminus is responsible for the different dissociation kinetics (Fig. 4A). Also, the amount of irreversibly bound protein was smallest with Rab1b, indicating that the charged C terminus of Rab35 contributes to nonspecific binding. However, all proteins were effectively extracted from the membrane by GDI in the same manner.
Fig. 4.
Influence of the C terminus on the dissociation rate and of the nucleotide state on the extraction by GDI. (A) Time course of the immobilization of Rab35_G (black), Rab1b_G (orange), and chimeric Rab1b with the C terminus of Rab35 (Rab1brab35cterm_G, ochre) on a lipid bilayer and subsequent extraction by GDI. The phases of the experiment correspond to those in Fig. 3 and are labeled accordingly. The sequences of the C termini of Rab1b and Rab35 are compared below the diagram (positively charged residues marked in red, polar uncharged residues marked in green). The positively charged C terminus of Rab35 results in slower dissociation from the negatively charged membrane compared with Rab1b. The chimeric protein behaves similarly to Rab35 as the electrostatic interaction between the C terminus and the membrane is enabled in the construct. Notably, Rab1b exhibits the smallest degree of nonspecific binding to the membrane, which can be due to the less charged C terminus. (B) Time course of a control experiment with Rab35_G in the GTP-bound state. The data points represent the normalized area of the amide II band with color coding according to Fig. 3. Additionally, the GAP TBC1D20 was added after GDI (ochre). Dissociation of Rab35·GTP_G is only insignificantly accelerated by GDI. The slight effect may be due to a fraction of Rab35_G being in the GDP-bound state. TBC1D20 catalyzes GTP hydrolysis and enables interaction of Rab35_G with GDI.
Because the affinity of GDI to Rab GTPases is significantly lowered in the GTP-state (13), we performed an experiment with Rab35·GTP_G. For this, nucleotide exchange was performed using the GEF Connecdenn before the ATR-FTIR experiment, which was then done in the same manner as before. Indeed, addition of GDI did not lead to a significant acceleration of membrane dissociation as opposed to previous experiments with Rab35·GDP_G (Fig. 4B). A minor effect was still observed, which may be due to incomplete nucleotide exchange. Addition of the GAP TBC1D20 initiated GTP hydrolysis, and the typical rapid extraction of membrane-bound Rab35_G was observed (Fig. 4B), proving the specificity of the interaction between Rab35 and GDI. Titration of membrane-bound Rab35_G with GDI revealed an EC50 value of 165 ± 5 nM for the membrane extraction of Rab35_G in the GDP-bound state (Fig. S5). For the GTP-bound state, the titration confirms that a fraction of Rab35_G is present in the GDP-bound state, whereas the EC50 value for the GTP-bound state is >5 µM, but could not be precisely determined due to nonspecific binding of GDI to the membrane at high concentrations (Fig. S5).
Influence of Phosphocholination on the GDI-Mediated Membrane Extraction.
Phosphocholination is used by the pathogenic bacterium L. pneumophila during infection to interfere with vesicular transport (8, 30). We used the ATR assay to directly observe the effect of phosphocholination on the ability of GDI to extract modified Rab35 from the membrane. Phosphocholinated wild-type Rab35 was used for titration experiments, but the Rab1b-like mutant Rab35T76S was used for the dephosphocholination with Lem3 because Lem3 naturally acts upon Rab1b, but not Rab35. Rab35 was first phosphocholinated, and then geranylgeranylated (Rab35-PC_G) and bound to the model membrane in the same manner as unmodified Rab35. However, 1 µM GDI was unable to completely extract Rab35-PC_G from the membrane, indicating an affinity decrease compared with unmodified Rab35 (Fig. 5A). Because GDI is circulating over the membrane, phosphocholinated Rab is only removed from the membrane until equilibrium between membrane-bound and GDI-bound Rab is reached. The dephosphocholinase Lem3 was able to restore the interaction between GDI and Rab35T76S, showing that the effect was specifically due to phosphocholination. Then, we quantified this effect by titrating membrane-bound Rab35_G and Rab35-PC-_G with GDI and determining the respective EC50 values for the extraction (Fig. 5B). Phosphocholination led to an increase of the EC50 value from 165 ± 5 nM to 1.05 ± 0.05 µM and thus markedly inhibited extraction of Rab35 from the membrane. The phosphocholinated residue Thr76 is situated in the interface between GDI and Rab35 (31), and therefore phosphocholination is expected to disturb this interaction. The relevant kinetic and thermodynamic parameters are summarized in Table S2.
Fig. 5.
Effect of phosphocholination of Rab35_G on the GDI-mediated membrane extraction. (A) Time course of an extraction experiment analogous to that in Fig. 3. GDI is unable to extract Rab35_PC_G completely. The dephosphocholinase Lem3 specifically restores the interaction (orange data points, the Rab1b-like mutant Rab35T76S was used because Lem3 naturally acts upon Rab1b, but not Rab35). (B) Titration of membrane-bound Rab35_G with GDI. The data were fitted with a logistic hyperbolic function (SI Materials and Methods) and yielded EC50 values of 165 nM and 1.05 µM for wild-type (black) and phosphocholinated (red) Rab35, respectively.
Discussion
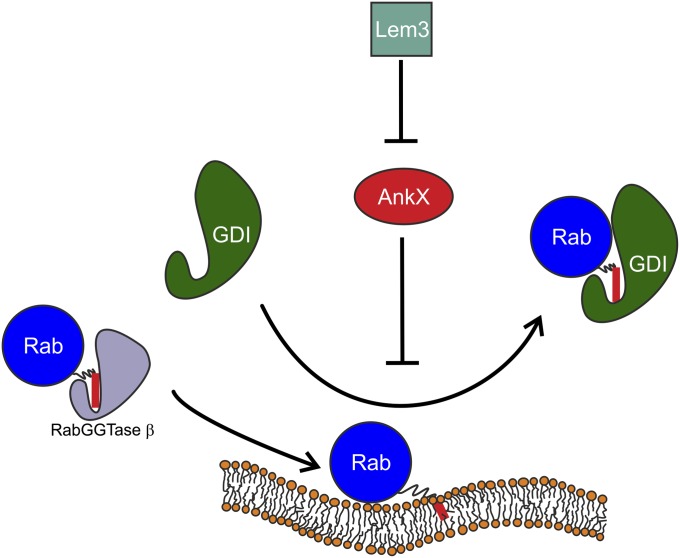
The interaction between GDI and Rab GTPases occurs on cellular membranes and involves many components, such as GDI and the GTPase themselves, the membrane, nucleotide, and the lipid anchor. This complexity poses challenges for the analysis and understanding of this process in vitro using biophysical methods. Insight into the interaction has been gained from studies performed in solution without a membrane involved (11–13). In many cases farnesyl-NBD, an artificial fluorescent probe, was used to provide a readout. Our advance allows investigation of the interaction with a label-free geranylgeranyl moiety and, most importantly, on a membrane instead of unnaturally in solution. Fig. 6 summarizes the processes and interactions that we were able to observe with the ATR-FTIR setup. Geranylgeranylated Rab35 solubilized with RabGGTase β was bound to a solid-supported model lipid bilayer. Although a label-free detection of membrane binding is also possible with surface plasmon resonance (SPR), ATR-FTIR additionally provides full spectral information. This crucial advantage allows confirmation of the GTPase activity of membrane-bound protein. The protein folding and the GTPase activity can be assessed by means of difference spectroscopy. For Rab GTPases studied here, this proved to be an invaluable tool because the GTPase activity of membrane-bound Rabs and their ability to be extracted by GDI strongly depended on sample preparation and experimental conditions, i.e., Rab proteins without GTPase activity could not be extracted. Thus, lack of information on correct folding and activity of a protein might lead to wrong conclusions especially when dealing with protein–protein interactions.
Fig. 6.
A scheme of the interactions observed in the ATR experiments. Monogeranylgeranylated Rab35 solubilized with RabGGTase β is reversibly bound to a lipid bilayer (the red bar represents the lipid anchor). GDI mediates membrane extraction of Rab35, which can be inhibited by phosphocholination of Rab35 by AnkX. This effect is reversed by action of the dephosphocholinase Lem3.
Following membrane binding, spontaneous membrane dissociation and the process of membrane extraction by GDI were directly observed in a label-free manner. We showed dependency of the dissociation on the properties of the C terminus, and GTP/GDP-state dependency of the GDI-mediated membrane extraction. The ability of Rab35_G to spontaneously dissociate from the membrane raises the question of the mechanism of GDI. Possibly, GDI could tightly bind spontaneously dissociated Rab and thereby passively remove Rab from the membrane. The alternative would be that GDI actively extracts membrane-bound Rab. Notably, GDI was able to significantly accelerate membrane dissociation of Rab GTPases. This means that GDI not only shifts the equilibrium through its thermodynamically favorable interaction with prenylated Rab GTPases, but also genuinely accelerates dissociation of the GTPase from the membrane. Previous indirect evidence suggested that Hsp90 plays a role in GDI-mediated membrane extraction (32). However, we show here that GDI does not need additional factors for the extraction process. Possible intermediate states of the extraction process, such as GDI interacting with membrane-bound Rab, would probably not be observed in this flow cell because of their transient nature. The highly dynamic interaction of monogeranylgeranylated Rab GTPases with the membrane is in agreement with results obtained with monogeranylgeranylated peptides (29). It has to be considered that most Rab GTPases are doubly geranylgeranylated in vivo. Doubly geranylgeranylated peptides were found to bind to membranes more tightly (33). Therefore, monogeranylgeranylated Rab GTPases are likely to be more readily extracted than the doubly prenylated form. Although a number of Rab GTPases such as Rab8 or Rab23 are naturally monogeranylgeranylated (31), it remains to be seen whether GDI is able to accelerate membrane dissociation of doubly geranylgeranylated Rab GTPases in the same manner.
Finally, we used our method to study the effect of phosphocholination, a posttranslational modification introduced by the pathogen L. pneumophila. We observed that phosphocholination directly interferes with membrane extraction by disturbing the interaction with GDI. This result is in agreement with previous works that used a fluorescent farnesyl-NBD lipid anchor instead of geranylgeranyl for interaction studies with GDI, but were conducted in solution only, lacking the Rab-membrane interaction (6, 7). It has been recently proposed that phosphocholination passively displaces GDI from the cytosolic Rab:GDI complex by inhibiting rebinding of GDI, and thus recruits Rab proteins to membranes and traps them in the inactive GDP state (6, 7). Evidence from in vivo experiments (34) suggests that the same principle is used to tackle the natural problem of Rab membrane targeting: GEF-mediated nucleotide exchange displaces GDI and leads to recruitment of Rab proteins to specific membranes. Therefore, not only the mechanism of membrane extraction but also that of GDI displacement and membrane delivery of Rab GTPases presents an interesting target for future biochemical and biophysical investigations. ATR-FTIR is an excellent method for the investigation of such membrane-dependent processes, providing detailed chemical information invaluable for the interpretation of results, which other methods, such as SPR, cannot provide.
Materials and Methods
Protein Expression and Purification.
The procedures for the preparation of GDI-1, Connecdenn 1, Rab35, Rab1b, AnkX, GGTase I, RabGGTase β, TBC1D20, and yeast GDI are described elsewhere (6, 7, 12, 31, 35) and in detail in SI Materials and Methods.
Preparative Geranylgeranylation.
Five milligrams of Rab with 5 mol% of GGTase I and 165 µM geranylgeranyl pyrophosphate were incubated overnight at 8 °C in 50 mM Hepes, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 2.5 mM dithioerythreitol, 20 µM GDP, 2% (wt/vol) CHAPS, 10% (wt/vol) glycerol, and 10 µM ZnSO4. Details of purification are given in SI Materials and Methods. The prenylated protein was analyzed by MALDI-MS to verify the prenylation (Fig. S6A).
Preparative Phosphocholination.
Seven milligrams of Rab35 were incubated with 2 mol% AnkX and 2 mM CDP-choline (Sigma-Aldrich) for 3 h at room temperature in 50 mM Hepes, pH 7.5, 200 mM NaCl, 2 mM MgCl2, 2.5 mM dithioerythreitol, 20 µM GDP, and 5% (vol/vol) glycerol. Products were separated by gel filtration and phosphocholination was verified by electrospray ionization-MS (Fig. S6B).
ATR-FTIR Measurements.
ATR-FTIR measurements were performed with a Vertex 80V spectrometer (Bruker Optik), using a vertical ATR multireflection unit (Specac) and trapezoidal germanium plates (ACM or Korth Kristalle) in a home-built flow cell. The preparation of the lipid bilayer (19, 20) is described in SI Materials and Methods. For protein binding, 2.5 µM geranylgeranylated Rab in complex with RabGGTase β (Rab_G:β-su) were added in a total volume of 2 mL. Unbound protein was washed out of the system. For titrations with GDI, the system was switched from a flow-through to a circulating mode and allowed to reach equilibrium between membrane-bound and dissociated Rab. Then, GDI was added at increasing concentrations. We used bovine GDI-1 for all titrations (Fig. 5 and Figs. S4 and S7), but yeast GDI from Saccharomyces cerevisiae was found to have an even smaller EC50 value for membrane extraction of Rab35 (Fig. S7) and therefore also is a good model. Experiments shown in Figs. 3, 4, and 5A were performed with yeast GDI. Instrumental parameters, data analysis, and further experimental details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Lena Oesterlin and Nathalie Bleimling for GDI-1 and Connecdenn protein samples, Katja Kuhlmann and Xi Liu for the mass-spectrometric analysis of prenylated proteins, Jörn Güldenhaupt for helpful discussions, and Harald Chorongiewski for technical assistance. This work was supported by the Ruhr University Research School funded by the Deutsche Forschungsgemeinschaft in the framework of the Excellence Initiative, by the Deutsche Forschungsgemeinschaft within the Sonderforschungsbereich 642, and by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.R.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307655110/-/DCSupplemental.
References
- 1.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 2.Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271(10):5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y-W, Goody RS, Abagyan R, Alexandrov K. Structure of the disordered C terminus of Rab7 GTPase induced by binding to the Rab geranylgeranyl transferase catalytic complex reveals the mechanism of Rab prenylation. J Biol Chem. 2009;284(19):13185–13192. doi: 10.1074/jbc.M900579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alory C, Balch WE. Organization of the Rab-GDI/CHM superfamily: The functional basis for choroideremia disease. Traffic. 2001;2(8):532–543. doi: 10.1034/j.1600-0854.2001.20803.x. [DOI] [PubMed] [Google Scholar]
- 5.D’Adamo P, et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet. 1998;19(2):134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- 6.Goody PR, et al. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 2012;31(7):1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oesterlin LK, Goody RS, Itzen A. Posttranslational modifications of Rab proteins cause effective displacement of GDP dissociation inhibitor. Proc Natl Acad Sci USA. 2012;109(15):5621–5626. doi: 10.1073/pnas.1121161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, et al. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schalk I, et al. Structure and mutational analysis of Rab GDP-dissociation inhibitor. Nature. 1996;381(6577):42–48. doi: 10.1038/381042a0. [DOI] [PubMed] [Google Scholar]
- 10.Rak A, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302(5645):646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 11.Ignatev A, Kravchenko S, Rak A, Goody RS, Pylypenko O. A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism. J Biol Chem. 2008;283(26):18377–18384. doi: 10.1074/jbc.M709718200. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y-W, Tan K-T, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci USA. 2007;104(30):12294–12299. doi: 10.1073/pnas.0701817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y-W, et al. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol. 2010;6(7):534–540. doi: 10.1038/nchembio.386. [DOI] [PubMed] [Google Scholar]
- 14.Goormaghtigh E, Raussens V, Ruysschaert JM. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim Biophys Acta. 1999;1422(2):105–185. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]
- 15.Kötting C, Güldenhaupt J, Gerwert K. Time-resolved FTIR spectroscopy for monitoring protein dynamics exemplified by functional studies of Ras protein bound to a lipid bilayer. Chem Phys. 2012;396:72–83. [Google Scholar]
- 16.Tamm LK, Tatulian SA. Infrared spectroscopy of proteins and peptides in lipid bilayers. Q Rev Biophys. 1997;30(4):365–429. doi: 10.1017/s0033583597003375. [DOI] [PubMed] [Google Scholar]
- 17.Ataka K, Kottke T, Heberle J. Thinner, smaller, faster: IR techniques to probe the functionality of biological and biomimetic systems. Angew Chem Int Ed Engl. 2010;49(32):5416–5424. doi: 10.1002/anie.200907114. [DOI] [PubMed] [Google Scholar]
- 18.Richter RP, Bérat R, Brisson AR. Formation of solid-supported lipid bilayers: An integrated view. Langmuir. 2006;22(8):3497–3505. doi: 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- 19.Güldenhaupt J, et al. Secondary structure of lipidated Ras bound to a lipid bilayer. FEBS J. 2008;275(23):5910–5918. doi: 10.1111/j.1742-4658.2008.06720.x. [DOI] [PubMed] [Google Scholar]
- 20.Güldenhaupt J, et al. N-Ras forms dimers at POPC membranes. Biophys J. 2012;103(7):1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329(5994):946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 22.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16(17):1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 23.Holthuis JCM, Levine TP. Lipid traffic: Floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6(3):209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 24.Morigaki K, Tawa K. Vesicle fusion studied by surface plasmon resonance and surface plasmon fluorescence spectroscopy. Biophys J. 2006;91(4):1380–1387. doi: 10.1529/biophysj.106.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiter G, et al. Interaction of a bacterial endotoxin with different surfaces investigated by in situ Fourier transform infrared attenuated total reflection spectroscopy. Langmuir. 2002;18(15):5761–5771. [Google Scholar]
- 26.Hübner W, Mantsch HH. Orientation of specifically 13C=O labeled phosphatidylcholine multilayers from polarized attenuated total reflection FT-IR spectroscopy. Biophys J. 1991;59(6):1261–1272. doi: 10.1016/S0006-3495(91)82341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Díaz JF, Sillen A, Engelborghs Y. Equilibrium and kinetic study of the conformational transition toward the active state of p21Ha-ras, induced by the binding of BeF3- to the GDP-bound state, in the absence of GTPase-activating proteins. J Biol Chem. 1997;272(37):23138–23143. doi: 10.1074/jbc.272.37.23138. [DOI] [PubMed] [Google Scholar]
- 28.Schartner J, et al. Universal method for protein immobilization on chemically functionalized germanium investigated by ATR-FTIR difference spectroscopy. J Am Chem Soc. 2013;135(10):4079–4087. doi: 10.1021/ja400253p. [DOI] [PubMed] [Google Scholar]
- 29.Silvius JR, l’Heureux F. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry. 1994;33(10):3014–3022. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- 30.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320(5883):1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pylypenko O, et al. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J. 2006;25(1):13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, Balch WE. The Hsp90 chaperone complex regulates GDI-dependent Rab recycling. Mol Biol Cell. 2006;17(8):3494–3507. doi: 10.1091/mbc.E05-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahinian S, Silvius JR. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34(11):3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 34.Blümer J, et al. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200(3):287–300. doi: 10.1083/jcb.201209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavriljuk K, et al. Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. Proc Natl Acad Sci USA. 2012;109(52):21348–21353. doi: 10.1073/pnas.1214431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.