Abstract
The objective of this study was to use next-generation sequencing technologies to dissect quantitative trait loci (QTL) for southern root-knot nematode (RKN) resistance into individual genes in soybean. Two hundred forty-six recombinant inbred lines (RIL) derived from a cross between Magellan (susceptible) and PI 438489B (resistant) were evaluated for RKN resistance in a greenhouse and sequenced at an average of 0.19× depth. A sequence analysis pipeline was developed to identify and validate single-nucleotide polymorphisms (SNPs), infer the parental source of each SNP allele, and genotype the RIL population. Based on 109,273 phased SNPs, recombination events in RILs were identified, and a total of 3,509 bins and 3,489 recombination intervals were defined. About 50.8% of bins contain 1 to 10 genes. A linkage map was subsequently constructed by using bins as molecular markers. Three QTL for RKN resistance were identified. Of these, one major QTL was mapped to bin 10 of chromosome 10, which is 29.7 kb in size and harbors three true genes and two pseudogenes. Based on sequence variations and gene-expression analysis, the candidate genes underlying the major QTL for RKN resistance were pinpointed. They are Glyma10g02150 and Glyma10g02160, encoding a pectin methylesterase inhibitor and a pectin methylesterase inhibitor -pectin methylesterase, respectively. This QTL mapping approach not only combines SNP discovery, SNP validation, and genotyping, but also solves the issues caused by genome duplication and repetitive sequences. Hence, it can be widely used in crops with a reference genome to enhance QTL mapping accuracy.
Keywords: high throughput genotyping, high resolution linkage map
Root-knot nematode [RKN, Meloidogyne incognita (Kofoid and White) Chitwood] is an important soybean pest that causes severe yield loss in the southern United States because of the region’s warm climate and sandy soil (1). Resistant cultivars, in combination with nonhost crop rotation, play an important role in reducing yield loss caused by RKN. Although enormous efforts have been directed toward screening resistant germplasm (1) and characterizing quantitative trait loci (QTL) for RKN resistance (2, 3), genes underlying these QTL are still elusive due to lack of high-resolution maps and the labor-intensive nature of map-based cloning. As a result, diagnostic markers are still not available for marker-assisted selection of genes/QTL for RKN resistance.
Mapping resolution is dependent on marker density and population size. The advent of array-based platforms allows for the simultaneous genotyping of a considerable number of SNPs in a large set of individuals in soybean. However, informative markers obtained in a particular mapping population are limited due to ascertainment bias, the sampling bias that arises from how SNPs are chosen for inclusion on SNP arrays. Recently, the Illumina Infinium iSelect SoySNP50K chip, which provides more than 50,000 genotypes in a single assay, was developed (4). However, the cost makes it prohibitive to genotype mapping populations for a majority of investigators.
Recent advances in sequencing technologies make it attractive to genotype mapping populations with next-generation sequencing (NGS) technologies. The reduced representation of sequencing strategy was used in biparental population mapping (5–7). In crops with complex genomes, such as maize and barley, this strategy is particular useful because one can exclude repetitive regions by choosing methylation-sensitive enzymes to avoid cutting methylated transposons (7). An alternative approach is whole-genome resequencing (WGR), which was used to genotype recombinant inbred lines (RIL) populations in rice (8, 9). Using the WGR approach, Huang et al. mapped QTL for plant height to a region where the “green revolution” gene resides (8), and Xie et al. localized a QTL for grain width to a bin harboring the cloned gene GW5 for grain width (9), lending credence to this approach.
The complexity of the soybean genome poses challenges to the applications of sequencing-based genotyping approaches. Soybean is a paleopolyploid with a genome size of 1,115 Mb (10). A striking feature of the soybean genome is that it experienced two genome duplications that occurred ∼59 and 13 Mya, respectively. As a result, about 75% of the genes are present in multiple copies (10). These paralogous sequences are substantive issues for sequencing-based genotyping because the short sequence reads, a typical feature of NGS technologies, may not be uniquely mapped to the reference genome, and allelic variations can’t be distinguished from differences among closely related paralogous sequences. Another challenge is the low polymorphism of the soybean genome, which was caused by the low diversity of its progenitor, Glycine soja, and genetic bottlenecks it has undergone in its evolutionary history (11). These include domestication bottleneck and introduction bottleneck, plus the intensive selection in modern soybean breeding (11). Besides, the abundant repetitive sequences, which dominate heterochromatic regions that account for 57% of the soybean genome (10), also present technical difficulties for sequence alignment. These repeats create ambiguities in alignment, which, in turn, produce errors when interpreting results.
The objective of this study was to dissect QTL for RKN resistance into genes by means of NGS technologies. Using a WGR approach that combines SNP discovery, SNP validation and genotyping, we genotyped a RIL population to construct a bin map. A high-resolution linkage map was subsequently constructed with bins serving as markers. This map enabled us to map QTL for RKN resistance with unprecedented accuracy and pinpoint the genes underlying the major QTL without a laborious and time-consuming fine-mapping process.
Results
Multiplexed Illumina Sequencing of Recombinant Inbred Lines.
We used a high throughput genotyping approach based on multiplexed Illumina sequencing to enhance QTL mapping resolution. In total, five DNA libraries were constructed and sequenced with standard Illumina sequencing protocols for 246 RILs derived from a cross between Magellan and PI 438489B. Using the Burrows–Wheeler Aligner (12) with default settings, a total of 780,201,692 reads, equivalent to 67.64-Gb sequences, were mapped to the Williams 82 reference genome. The 83-mer sequences were sorted based on the barcodes. About 141- to 458-Mb clean sequences were mapped for each RIL. Those mapped to multiple locations due to the duplicated nature of the soybean genome were excluded from analysis. The effective sequence depths for each RIL range from 0.10× to 0.31× with an average of 0.19×, and the corresponding sequence coverage ranges from 8.84% to 24.75% with a mean of 16.2%.
SNP Calling in Recombinant Inbred Lines.
To identify SNPs, sequences were aligned against the Williams 82 reference genome. SAMtools (13) was used to call SNPs in each RIL. Homozygous SNPs identified in each RIL ranged from 16,705–74,756 with an average of 42,577. Using the “mpileup” function of SAMtools, we merged all SNP datasets. Putative SNPs were identified by comparing SNPs at the same loci. Only biallelic SNPs were kept. In total, 1,464,938 SNPs were identified, yielding a SNP frequency of ∼1 SNP per 640 bp. In a RIL population, we expected that SNPs segregate in a 1:1 ratio. Thus, χ2 tests were conducted for all SNPs. There were 195,375 SNPs that did not deviate from the expected ratio (P > 0.01 for χ2 test), and these were selected as candidate SNPs. Candidate SNPs were not evenly distributed among chromosomes, ranging from 4,272 on chromosome 11–16,485 on chromosome 18 (Fig. S1). We used an approach based on the principle of maximum parsimony of recombination (MPR) to infer the parental genotypes of these candidate SNPs, followed by a refining procedure involving resampling and Bayesian inference (9). Finally, we identified a set of 109,237 phased SNPs to genotype the RIL population. The predicted parental alleles of these SNPs were in agreement with the actual alleles obtained from the deep sequencing of parents (see below). The phased SNPs on different chromosomes range from 2,229–11,087 (Fig. S1).
Sequencing of Parental Lines.
The two parental lines were sequenced with RILs at a low coverage. To confirm the inferred genotypes, 16.41- and 17.13-Gb additional sequences were generated from cultivars Magellan and PI 438489B, respectively. The depths of effective sequences are ∼13.48× (coverage: 95.2%) and 13.47× (coverage: 93.1%), respectively. Aligning these sequences against the Williams 82 reference genome, we identified 463,662 SNPs from Magellan and 1,004,361 SNPs from PI 438489B. Further analysis revealed 1,237,394 SNPs and 360,544 indels between Magellan and PI 438489B.
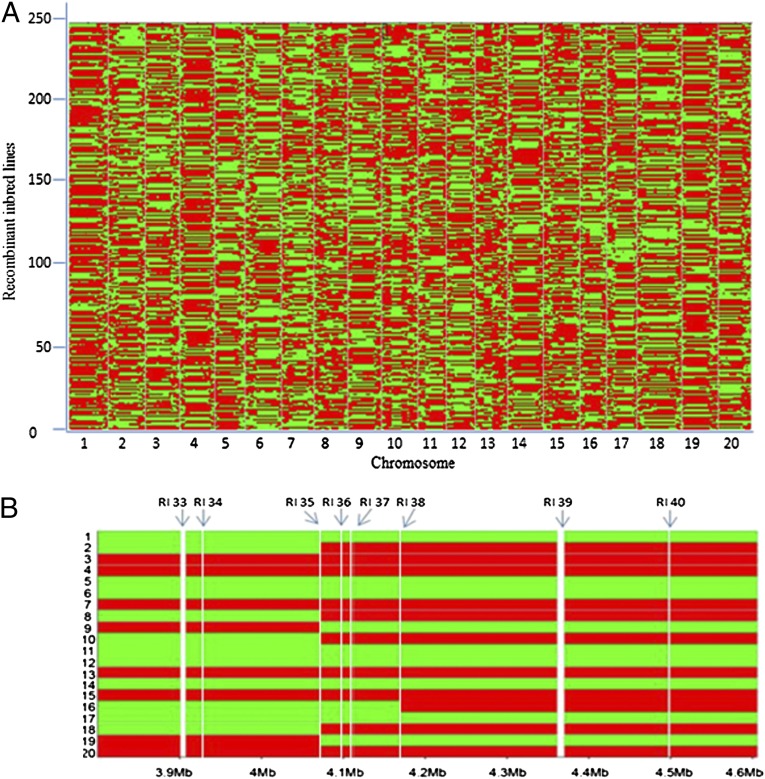
The sequencing error rate of the RIL population was estimated to be 3.84% by using an approach described by Xie et al. (9). This result was used to determine the transmission probabilities and emission probabilities of a Hidden Markov Model (HMM), which was used to compute a posterior probability that a genomic region is either homozygous for Magellan, homozygous for PI 438489B, or heterozygous. Based on HMM results, the genotypes of RILs at all SNP loci were imputed, and consecutive alleles were lumped into haplotype blocks (Fig. 1A). The average block size was 14.3 Mb.
Fig. 1.
Recombination bin map of 246 RILs. (A) Genetic constituents of 246 soybean RILs. Chromosomes are separated by vertical lines. Red and green represent Magellan and PI 438489B genotype, respectively. (B) An enlarged bin map showing part of chromosome 10, ranging from 3.8 Mb to 4.6 Mb, in 20 RILs. The white vertical line shows recombination interval (RI), which was defined as the transition region between two haplotype blocks in at least one of the 246 RILs. In RI 35, recombination took place in RIL 2, 8, 9, 10, 18, and 19. In RI 38, recombination occurred only in RIL16. The widths of these lines were based on the sizes of RIs. The chromosome fragment between two adjacent RIs was defined as a bin, which was used as a molecular marker.
Construction of a Bin Map.
Based on genotypic data, a total of 3,489 recombination intervals, which were defined as the transition regions between two different genotypic blocks, were identified in the 246 RILs. The chromosome fragment between two adjacent recombination intervals was defined as a bin. A total of 3,509 bins were identified (Fig. 1B). Among them, 2,566 resided in euchromatin regions (hereafter, chromosome arms) occupying ∼43% of the soybean genome, and 943 were harbored in the pericentromeric regions, which account for ∼57% of the soybean genome and are mainly heterochromatin characterized by repetitive sequences and low recombination rate (10). The mean and median bin size on chromosome arms were 136.9 and 80.7 kb, respectively, whereas in pericentromeric regions, they were 522.4 and 259.9 kb, respectively (Fig. S2). In total, 165 bins (4.7% of the total) had a bin size bigger than 1 Mb, and 150 of them localized in pericentromeric regions. On chromosome arms, 15 bins were longer than 1 Mb, ranging from 1.02 to 3.11 Mb, with six of them being terminal bins. The average size of terminal bins was 496.3 kb, 3.6 times as large as the average bin size in the euchromatin region. This finding suggests that recombination in the telomere regions may be suppressed. Similar results were reported in Caenorhabditis elegans genome, in which no recombination occurred in all telomere regions (14).
The size of recombination interval is mainly dependent on the marker density. In SNP-rich regions, the recombination interval can be narrowed down to one or a few base pairs. The total length of 2,546 recombination intervals on the chromosome arm was 52.95 Mb, ranging from 3 bp to 1.89 Mb with a mean of 20.8 kb for each recombination interval. However, in SNP-sparse regions, mainly pericentromeric regions, the recombination interval is large. The total length of 943 recombination intervals in the pericentromeric regions was 53.11Mb, ranging from 1 bp to 1.27 Mb with an average of 56.3 kb for each recombination interval. The distribution of recombination interval sizes is shown in Fig. S3.
The mean number of recombination events in a single RIL were 46.3, ranging from 1.82 to 3.05 in each chromosome. The χ2 test suggested that the average number of recombination events in each chromosome was determined by chromosome length (null hypothesis: recombination events on a chromosome are proportional to its length; χ2 = 1.17; df = 19; not significant).
Resolution of Genes.
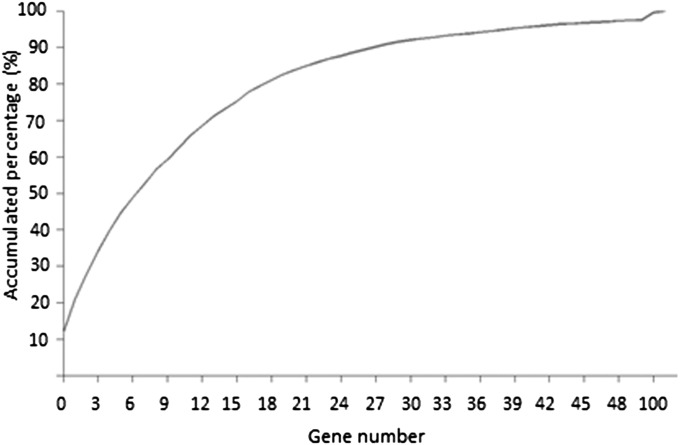
Mapping resolution, the size of a physical region that is confidently associated with a trait, is a function of population size and marker density. The distribution of genes in bins and recombination intervals indicates the mapping resolution of a given population. In the soybean genome sequence assembly version 7.0 (Gmax_109_gene.gff3.gz”; ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Gmax/annotation/), a total of 47,245 genes were predicted. We counted the gene number in each bin, as well as each recombination interval. A total of 40,832 genes were found in 3,100 bins. Among these bins, 318 (9.1% of the total) contained a single gene, and 1,456 (40.7% of the total) contained two to ten genes (Fig. 2). Thus, 50.8% of the total bins contained no more than 10 genes, suggesting that reasonable mapping resolution would be achieved without a fine mapping process for a considerable number of QTL/genes. Four hundred and nine bins did not harbor any gene. Another 6,413 genes were localized in 1,661 recombination intervals.
Fig. 2.
Plot of gene resolution. Gene number in each bin is on the horizontal axis, and the accumulated percentage of bins is on the vertical axis.
Construction of a Genetic Map with Bins Serving as Markers.
We used 3,509 bins as molecular markers to construct a linkage map. On average, a bin consisted of 31 phased SNPs. Using the program R/QTL package (15), we obtained a linkage map which covered a total genetic distance of 2,314 centiMorgans (cM) on chromosomes ranging from 91.0 cM to 155.6 cM in size (Fig. S4). Thus, the physical-to-genetic ratio was 411 kb/cM. We have compared the orders of bins on the linkage map and the soybean genome assembly 7.0 physical map, and observed an excellent concordance. The genetic distance of 2,566 bins on the arms, which account for 43% of soybean genome, was 1,747.5 cM, yielding a physical-to-genetic ratio of 233.8 kb/cM. In the 57% pericentromeric regions, the genetic distance of 943 bins was 566.5 cM, and the physical-to-genetic ratio was 888.8 kb/cM.
Mapping QTL for RKN Resistance.
The RIL population was evaluated for resistance to RKN (M. incognita Chitwood) by rating root galling in a greenhouse. Resistance to RKN was mapped using a gall index. The distribution of gall index in the RIL population is shown in Fig. S5. Three QTL for RKN resistance were identified by both interval mapping and multiple-QTL model (MQM) mapping (16). A major QTL explaining 23.6% of total genetic variance was mapped to bin 10 of chromosome 10 with a logarithm of odds (LOD) score of 15.4 (Fig. S6). The additive effect of this QTL was 0.75. The second QTL was mapped to bin 113 of chromosome 8 with an LOD score of 5.3, and explained 7.4% of the total phenotypic variance. In addition, bin 131 of chromosome 13 also harbored a minor QTL explaining 5.6% of phenotypic variance (Table 1).
Table 1.
A comparison of mapping resolutions of WGR approach and the traditional method based on GoldenGate SNP array and SSR markers
| Location | SNP array and SSR |
WGR |
||||||||
| Flanking markers | LOD | R2, % | A | Interval, kb | bin | LOD | R2, % | A | Bin size, kb | |
| Chr 8 | BARC-051847–11270; BARC-039273–07476 | 4.5 | 6.4 | 0.4 | 13,808.20 | 113 | 5.3 | 7.4 | 0.4 | 340 |
| Chr 10 | BARC-065469–11494; BARC-018101–02517 | 15.4 | 23.6 | 0.8 | 495.9 | 10 | 15 | 23.2 | 0.75 | 29.7 |
| Chr 13 | BARC-010501–00676; Sct-033 | 3.4 | 4.8 | 0.3 | 1,141.6 | 131 | 5.4 | 5.6 | 0.4 | 7.9 |
GoldenGate (universal soybean linkage panel 1.0) SNP array contains 1,536 SNPs. A set of 204 informative SSR markers were also used to genotype the RIL population. A, additive effects; Chr, chromosome.
The quality and accuracy of this map was evaluated by comparing it to a map derived from the same population using the universal soybean linkage panel (USLP) 1.0 SNP array, which contains 1,536 SNPs, and a set of SSR markers. A total of 700 SNPs and 204 SSR markers were mapped in this mapping population. All three QTL identified in this study were mapped in the same regions with similar R2 values, LOD scores, and additive effect values (Table 1). The intervals harboring the QTL on chromosomes 8, 10, and 13 were 13,808.2, 495.9, and 1,141.6 kb, respectively, whereas the corresponding sizes of bins harboring these QTL were 340, 29.7, and 7.9 kb. Thus, the mapping resolution of the bin-based map was 16.7–144.5 times higher than the map based on SNP and SSR markers at these loci.
Identification and Confirmation of the Candidate Genes Underlying the QTL for RKN Resistance.
The current annotation of the Williams 82 reference genome (10) predicts five genes in bin 10 of chromosome 10, including Glyma10g02140, Glyma10g02150, Glyma10g02160, Glyma10g02170, and Glyma10g02180. We examined these gene sequences in Williams 82, Magellan, and PI 438489B, and conducted gene expression experiments after inoculation with RKN eggs. We found that two of them, Glyma10g02140 and Glyma10g02170, are likely pseudogenes because of the absence of start codon (Glyma10g02140 and Glyma10g02170) or stop codon (Glyma10g02140). Glyma10g02180 appears to be a true gene that is predicted to encode a protein of unknown function (DUF538). The gene sequences in both Magellan and PI 438489B are identical to that of the Williams 82 reference genome.
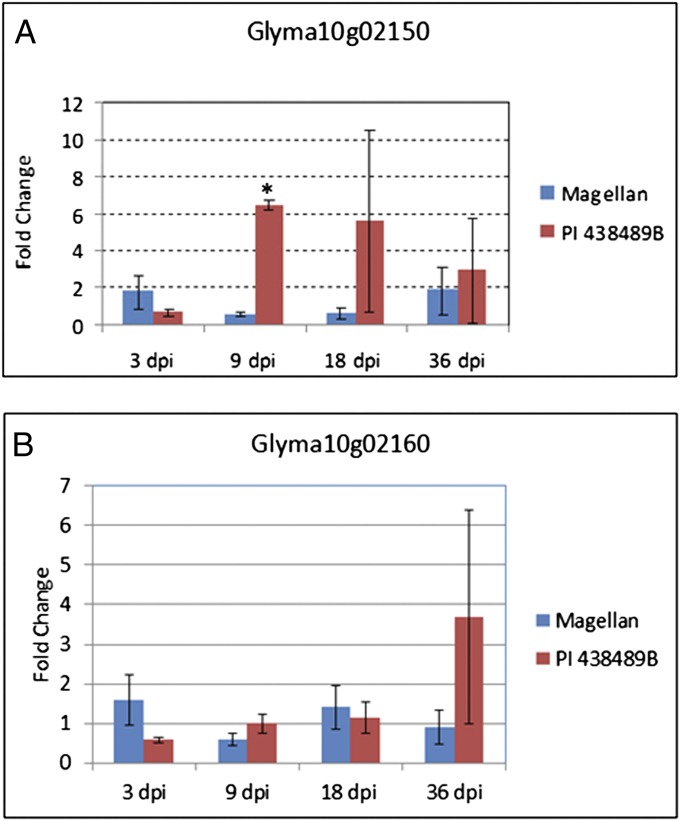
Glyma10g02150 and Glyma10g02160 also appear to be true genes predicted to encode a pectin methylesterase inhibitor (PMEI) and PMEI-pectin methylesterase, respectively. Comparing sequences of Magellan and PI 438489B, we found five SNPs, including four nonsynonymous SNPs, and one indel in Glyma10g02150. The 1-bp insertion in the exon in Magellan and Williams 82 shift the ORF of this gene, resulting in a different and much longer protein compared with that in PI 438489B. Gene-expression studies revealed that this gene was induced by RKN inoculation in PI 438489B over time, but no significant induction was observed in Magellan (Fig. 3A). The gene Glyma10g02160 was identical in Magellan and the Williams 82 reference genome. However, a total of 12 SNPs and two 1-bp deletions were found in PI 438489B. Among them, six were harbored in coding regions, and three were nonsynonymous SNPs. The quantitative RT-PCR results showed that Glyma10g02160 was not significantly induced by RKN inoculation in either PI 438489B or Magellan except at 36 d postinoculation (Fig. 3B). At this time point, certain inductions appeared to occur in PI 438489B, although such induction was not statistically significant compared with that in Magellan, likely due to large experimental variations between the replicates. Interestingly, the protein encoded by Glyma10g02160 is similar to the Arabidopsis protein pectin methylesterase, PME3, which was shown to be important in nematode parasitism (17). Taking these together, we conclude that Glyma10g02150 and Glyma10g02160 are good candidates for the genes underlying the major QTL for RKN resistance.
Fig. 3.
Analysis of the genes in the major QTL region on chromosome 10 in response to RKN inoculation using qPCR. Based on sequence variations, Glyma10g02150 (A) and Glyma10g02160 (B) were selected as candidate genes. Both PI 438489B and Magellan were inoculated with RKN for 3, 9, 18, and 36 d. The relative fold change of a target gene in each of these lines was obtained using qPCR from the comparison between the RKN-treated plants and the similarly water-treated plants after normalization with the reference gene Actin. The value at each time point was the average of three biological replicates. Blue and red bar represent Magellan and PI 438489B, respectively. The asterisk indicates the difference between PI 438489B and Magellan plants at a particular time point (dpi, days postinoculation) was statistically significant via Student t test (P < 0.05).
The second QTL was mapped to the bin 113 of chromosome 8, which was in the pericentromeric region. The current gene annotation of this 340-kb region predicts 18 high-confidence gene models and 12 low-confidence gene models. The functions of these genes are not clear yet. The third QTL region, bin 131 of chromosome 13, is 7.9 kb in size. No gene model is predicted in this region. However, there are three resistance genes (R genes) located about 10 kb downstream. Further study of sequences in this region and its relationship with these genes may reveal the nature of this QTL.
Discussion
The WGR Approach Combines SNP Discovery, SNP Validation, and Genotyping.
The bottlenecks for soybean map-based cloning include the insufficiency of molecular markers and the lack of highly efficient genotyping approaches. Currently, NGS technologies have been widely used to identify SNPs. A typical SNP discovery project usually sequences and compares the low copy regions of a few genotypes (cultivars) by using restriction enzymes (18). However, when they are applied to crops with a complex genome, challenges arise due to the short reads produced by NGS technologies. It is difficult to discriminate orthologous sequences from paralogous sequences. Also, these massively parallel sequencing technologies are error-prone. As a result, only 50–85% of identified SNPs were validated in crop plants (19), and the validation of SNP markers is costly and labor intensive. The recent application of array-based genotyping platforms, such as Affymetrix array and Illumina GoldenGate array, greatly improves the efficiency of genotyping. However, these markers were developed from a small panel of germplasm. Thus, the ascertainment bias may compromise the power of these platforms.
A striking feature of the WGR QTL mapping approach used in this study was that it combined SNP discovery, SNP validation, and genotyping. Using this method, we defined a set of unbiased, genome-wide SNPs. Different from traditional SNP discovery approaches, we not only sequenced both parental lines at a higher coverage, but also sequenced their progenies at a lower coverage. SNPs identified in parental lines were validated by their segregations in their progenies. With stringent criteria, we selected a set of 109,237 phased SNPs to genotype the RIL population. Although we excluded a considerable number of true SNPs that did not pass the χ2 test, this set of SNPs still represent the largest set of markers used in soybean QTL mapping. The linkage map constructed with bins, which were based on these SNPs, further validated them by the excellent concordance between the genetic map and the soybean sequence assembly 7.0.
Application of the WGR Approach in Soybean Genomics Studies.
The central task of the WGR QTL mapping approach is to infer recombination breakpoints in each RIL. Given that recombination events in each RIL are limited, ranging from 27 to 76 with a mean of 46.5 in the population used in this study, sequencing each RIL with 0.2× depth is sufficient to capture these recombination breakpoints. In this study, the total genetic distance of the linkage map was 2,314 cM, similar to the consensus genetic linkage map 4.0 with a total length of 2,296.4 cM (20). This result suggested that all or most of the recombinant events in the RIL population were identified. In this study, 50.8% of bins contained less than 10 genes. Given that genome-wide sequence variations between the two parents were detected and verified, we have a good chance to pinpoint the genes underlying a considerable number of QTL without a laborious fine mapping process.
Due to recombination suppression, the bin size in the pericentromeric regions was bigger than that on the chromosome arms (Fig. S2). However, gene density in the pericentromeric regions was lower (10). Thus, the mapping resolution in pericentromeric regions, in terms of gene number in each bin, was not significantly different from that on chromosome arms. On the contrary, the mapping resolution at the chromosome ends was relatively low because of the high gene density and large bin size caused by recombination suppression. The average gene number in the 40 terminal bins was 52.3, much greater than the genome-wide average of 11.6. A larger RIL population would be necessary to precisely map genes/QTL in these regions.
The WGR Approach Can Be Widely Used in Crops with a Reference Genome.
Genome duplication and abundant repetitive sequences, which are common in crop genomes, are major issues for sequencing-based genotyping. These make it difficult to distinguish allelic variation from differences between paralogous sequences, and the approaches developed in species with a relatively small and simple genome can't be used in crops with a complex genome, such as soybean, maize and barley. To solve these issues, we used unique sequences to genotype the population and conducted χ2 tests. All reads mapped to multiple locations of the reference genome were excluded from analysis. The χ2 tests efficiently eliminated potential false SNPs caused by misalignment of paralogous sequences or sequencing errors. However, the χ2 tests also excluded a considerable number of true SNPs which did not pass the tests from analysis. In this study, only 195,375 SNPs (13.3%) passed χ2 tests. Although this was still a decent number for genotyping the RIL population, we did identify several genomic regions where a much lower portion of SNPs passed χ2 tests on chromosome 1, 10, 11, 15, and 19. All of them resided in pericentromeric regions (Fig. S7). Because phased SNPs were relatively rare in these repeat-rich regions, some recombination intervals were large, and 13 of them were larger than 1 Mb. The mapping resolution in these regions was relatively low. Thus, it is necessary to find a more effective approach to filter false SNPs. However, the dilemma is that no satisfactory results could be achieved without χ2 tests, even when much more stringent criteria were applied to SNP calling. A major factor contributing to this issue is the short sequence reads produced by NGS technologies (83 bp per read in this study), which created ambiguities in alignment in duplicated or repetitive genomic regions. Given that the third generation sequencing technologies can yield multikilobase sequence reads (21), the application of these sequencing platforms plus the development of more sophisticated bioinformatics tools should be able to completely solve this issue in the near future. Also, we sequenced the RILs at an average of ∼0.19× depth, and the SNP copy numbers in the RIL population were generally low, ranging from 2 to 236 with a mean of 44. A higher sequence coverage may localize recombination intervals more precisely and improve the mapping accuracy in these repeat-rich regions.
Different from previous studies in which genotypic data in recombination intervals were imputed (6, 8, 9), we excluded these regions from analysis and only used bins, which were based on accurate genotypic data, as molecular markers to construct the linkage map. The rational was that 13.6% of the total genes were harbored in recombination intervals, and the imputed data may seriously compromise the power of QTL mapping. Moreover, in the recombination hot spots, recombination events occurred in many RILs in the same recombination interval. One example was that we detected recombination events in 95 RILs (38.4% of the total) in an interval on chromosome 13, which was 67,342 bp in size and localized between bin 100 and bin 101. Accurate genotypic data are crucial for mapping QTL/genes in these regions. Our results suggested that our approach successfully solved issues caused by duplicated genomes and repetitive sequences. Thus, we expect that this method can be widely used in other crops with a reference genome.
RKN Resistance Genes in PI 438489B.
The mapping results of RKN resistance exemplified the advantages of the WGR approach. In earlier studies, the QTL on chromosome 10 was mapped to an interval of 13.1 cM with RFLP markers (2) and 9.1 cM (0.85 Mb) with SSR markers in PI 96354 (3). Tamulonis et al. indicated that this QTL might be the Rmi1 gene identified in cultivar Forrest by Luzzi et al. (2, 22). Using the WGR approach, we were able to map a major QTL in the same region in PI 438489B, pinpoint the candidate genes, and identify the putative causative indel maker in Glyma10g02150. Subsequently, we sequenced the target region in a set of cultivars and germplasm. We found that two cultivars known to contain gene Rmi1, Forrest and S99-2281, also have the same allele at this indel locus, whereas all other accessions susceptible to RKN have the alternative allele. This finding suggested that the QTL identified in PI 438489B may be the Rmi1 gene.
RKN is an economically important plant parasitic nematode. The preferred strategy to control RKN parasitism is to use RKN resistance genes as an environmentally benign and cost-effective alternative to chemical controls. Currently, the most understood RKN resistance gene is Mi-1.2, which came from a wild tomato species, Lycopersicon peruvianum. Mi-1.2 is a typical R gene that encodes a protein with a nucleotide binding site (NBS) domain and a carboxyl-terminal leucine rich repeat (LRR) domain. A complementation study suggested that a single copy of Mi-1.2 is sufficient to confer full resistance to RKN (23). In pepper, an RKN resistance gene that shares 99% of deduced amino acid with Mi-1.2, CaMi, was isolated (24). These genes play an important role in reducing yield losses caused by RKN.
The candidate genes identified in this study were predicted to encode PMEI and PMEI-PME, respectively, and therefore represent a different type of RKN resistance. Previous studies revealed that bacterial PME makes pectin, a structurally complex polysaccharide that accounts for about 35% of the dicot and nongraminaceous monocot cell wall, susceptible to hydrolysis by enzymes such as endopolygalacturonases, contributing to the softening of the cell wall (25). In Erwinia chrysanthemi, PME catalyzes the important first step in the bacterial invasion of plant tissues, the deesterification of O6 methylesterified d-galacturonate (GalA) residues in pectic polysaccharides of the plant cell wall, which causes the subsequent degradation of the cell wall and allows the pathogen to invade and spread diseases (26). Pathogens, such as tobacco mosaic virus, also exploit endogenous plant PME as a vehicle for cell-to-cell movement (27). In Arabidopsis, PME3 is a potential target of a cellulose binding protein (CBP) produced by cyst nematode and may aid cyst nematode parasitism (17). Overexpression of PMEI resulted in the decrease of PME activity in transgenic Arabidopsis (28). We don’t know how the candidate genes identified in this study are involved in soybean RKN resistance. However, we speculate that they are likely involved in regulating plant cell modification either by plant or RKN enzymes. Further cloning and characterizing these two genes will be a starting point in understanding the property of this type of resistance in soybean. We are now in the process of making transient, as well as stable, transgenic plants to test their functionality in resistance to RKN.
Materials and Methods
Materials.
A RIL population derived from a Magellan × PI 438489B cross was used in this study. Two hundred forty-six F8-derived RILs were grown in a greenhouse. Two-week-old plantlet leaves were collected to extract genomic DNA with the CTAB method.
Construction of Sequencing Libraries and Illumina Sequencing
About one microgram of DNA for each sample was sheared into 300- to 500-bp DNA fragments using Covaris S2/E210. Sheared DNA was end-repaired, tailed with “A” nucleotides, and ligated to customized 7-mer pooling barcodes and Illumina paired-end sequencing adapters. DNA from 48 to 50 samples was pooled into a library. All libraries were sequenced using standard protocol on Illumina Hiseq2000, and 83-mer paired-end reads were generated. The sequence depth of 246 RILs and two parental lines was about 0.2×. Additional sequences were generated from two parents to achieve >10× sequence depth.
SNP Calling and Genotyping.
A pipeline combining Burrows–Wheeler Aligner (16) and Sequence Alignment/Map tools (SAMtools) (13) was used to call SNPs in RILs. The RIL sequences were aligned against the Williams 82 reference genome using the Burrows–Wheeler Aligner with default parameters to identify SNPs. The “pileup” function of SAMtools was used to merge the SNP dataset. Only biallelic SNPs were kept in the dataset. A χ2 test was conducted for each SNP with a null hypothesis that the two alleles at a locus segregate with a ratio of 1:1 in the RIL population. All SNPs that significantly deviated from this ratio (P < 0.01) were excluded from the SNP dataset. An approach described by Xie et al. was adopted to infer and refine parental sources of each SNP allele and genotype the RIL population (9).
The RIL population was previously genotyped by the universal soybean linkage panel one (USLP 1.0), which contains 1,536 mapped SNPs (20), and a set of SSR markers (29).
QTL Mapping.
The switch from one genotype to another within a chromosome represents a recombination event, and the transition region was defined as recombination interval. To conduct genetic analysis, we aligned the 246 RILs and defined the genomic region between any two adjacent recombination intervals within a chromosome as a bin with terminal bins exceptions, which are between a recombination interval and a chromosome end. We used bins as molecular markers to construct a linkage map using the program R/QTL package (15), and QTL was mapped using the MQM method of the program MapQTL 5.0 (16).
Phenotyping.
The 246 RILs were evaluated for RKN resistance in a greenhouse at the University of Georgia. A randomized complete block experimental design with four replications was used in this study. The 7- to 10-d-old plantlets were inoculated with ∼3,000 M. incognita Chitwood eggs as described (3). The experiment was terminated at 36 d after the inoculation when galls had developed on the susceptible controls, and gall numbers in each plant were counted and gall index was calculated as described by Hussey and Boerma (30).
Gene Expression.
PI 438489B and Magellan were grown in a greenhouse at the University of Georgia. A randomized complete block design with three biological replications was used. Each 7- to 10-d-old plant was inoculated with either 3,000 M. incognita Chitwood eggs (treatment) or water (control). Root tissues were collected at 3, 9, 18, and 36 d postinoculation. RNA extraction, cDNA synthesis, primer design, and quantitative RT-PCR were performed as described by Wan et al. (31). The relative fold change of each target gene was calculated by first normalizing its expression to that of the reference gene, Actin, and then comparing it with the corresponding water-treated sample as described (32).
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their insightful comments and Dr. Steven B. Cannon (US Department of Agriculture–Agricultural Research Service, Crop Insects and Crop Genetics Research) and Dr. Jianxin Ma (Purdue University) for location information of pericentromeric regions of the soybean genome. This work was funded by the Missouri Soybean Merchandising Council Grants 308F and 320F (to H.T.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in Soykb, http://soykb.org/public_data.php.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222368110/-/DCSupplemental.
References
- 1.Walters SA, Baker KR. Current distribution of five major Meloidogyne species in the United States. Plant Dis. 1994;78:772–774. [Google Scholar]
- 2.Tamulonis J, et al. RFLP mapping of resistance to southern root-knot nematode in soybean. Crop Sci. 1997;37(6):1903–1909. [Google Scholar]
- 3.Li Z, et al. SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. TAG. 2001;103(8):1167–1173. [Google Scholar]
- 4.Song QJ, et al. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE. 2013;8(1):e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird NA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3(10):e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andolfatto P, et al. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 2011;21(4):610–617. doi: 10.1101/gr.115402.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6(5):e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19(6):1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie W, et al. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci USA. 2010;107(23):10578–10583. doi: 10.1073/pnas.1005931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 11.Hyten DL, et al. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA. 2006;103(45):16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5(3):e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broman KW, Sen ´S. A guide to QTL mapping with R/qtl. Berlin: Springer; 2009. [Google Scholar]
- 16.van Ooijen JW. MapQTL 5, software for the mapping of quantitative trait loci in experimental populations. Wageningen, The Netherlands: Kyazma B.V.; 2004. [Google Scholar]
- 17.Hewezi T, et al. Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: Cooperative cell wall modification during parasitism. Plant Cell. 2008;20(11):3080–3093. doi: 10.1105/tpc.108.063065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyten DL, et al. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics. 2010;11:38. doi: 10.1186/1471-2164-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganal MW, Altmann T, Röder MS. SNP identification in crop plants. Curr Opin Plant Biol. 2009;12(2):211–217. doi: 10.1016/j.pbi.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Hyten DL, et al. A High Density Integrated Genetic Linkage Map of Soybean and the Development of a 1536 Universal Soy Linkage Panel for Quantitative Trait Locus Mapping. Crop Sci. 2010;50(3):960–968. [Google Scholar]
- 21.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 22.Luzzi BM, Boerma HR, Hussey RS. Inheritance of resistance to the southern root-knot nematode in soybean. Crop Sci. 1994;34(5):1240–1243. [Google Scholar]
- 23.Milligan SB, et al. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell. 1998;10(8):1307–1319. doi: 10.1105/tpc.10.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R, et al. CaMi, a root-knot nematode resistance gene from hot pepper (Capsium annuum L.) confers nematode resistance in tomato. Plant Cell Rep. 2007;26(7):895–905. doi: 10.1007/s00299-007-0304-0. [DOI] [PubMed] [Google Scholar]
- 25.Brummell DA, Harpster MH. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol Biol. 2001;47(1-2):311–340. [PubMed] [Google Scholar]
- 26.Fries M, Ihrig J, Brocklehurst K, Shevchik VE, Pickersgill RW. Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J. 2007;26(17):3879–3887. doi: 10.1038/sj.emboj.7601816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen MH, Sheng J, Hind G, Handa AK, Citovsky V. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 2000;19(5):913–920. doi: 10.1093/emboj/19.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lionetti V, et al. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007;143(4):1871–1880. doi: 10.1104/pp.106.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuong T, et al. Confirmation of quantitative trait loci for resistance to multiple-HG types of soybean cyst nematode (Heterodera glycines Ichinohe) Euphytica. 2011;181:101–113. [Google Scholar]
- 30.Hussey RS, Boerma R. A greenhouse screening procedure for root-knot nematode resistance in soybean. Crop Sci. 1981;21:794–796. [Google Scholar]
- 31.Wan J, et al. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012;160(1):396–406. doi: 10.1104/pp.112.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.