Abstract
There is considerable debate over the capacity of the cell wall polymer lignin to incorporate unnatural monomer units. We have identified Tnt1 retrotransposon insertion mutants of barrel medic (Medicago truncatula) that show reduced lignin autofluorescence under UV microscopy and red coloration in interfascicular fibers. The phenotype is caused by insertion of retrotransposons into a gene annotated as encoding cinnamyl alcohol dehydrogenase, here designated M. truncatula CAD1. NMR analysis indicated that the lignin is derived almost exclusively from coniferaldehyde and sinapaldehyde and is therefore strikingly different from classical lignins, which are derived mainly from coniferyl and sinapyl alcohols. Despite such a major alteration in lignin structure, the plants appear normal under standard conditions in the greenhouse or growth chamber. However, the plants are dwarfed when grown at 30 °C. Glycome profiling revealed an increased extractability of some xylan and pectin epitopes from the cell walls of the cad1-1 mutant but decreased extractability of others, suggesting that aldehyde-dominant lignin significantly alters cell wall structure.
Keywords: model legume, monolignol pathway, transposon mutagenesis
Lignin is an abundant plant aromatic heteropolymer in secondary cell walls, where it plays crucial roles in mechanical support, water conductance, and pathogen defense. Generally, lignin is derived from three phenylpropanoid monomers, the monolignols 4-coumaryl, coniferyl, and sinapyl alcohols, that produce the 4-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units in the polymer; H units are typically minor (<2%) in angiosperm lignins. Most genes involved in lignin biosynthesis have now been characterized (1). Because reduction of lignin content or alteration of lignin composition and structure can improve the processing of plant biomass for liquid biofuels (2, 3) or the digestibility of forages (4), there is considerable interest in engineering novel lignins into plants (1, 5, 6). Although there has been some debate over the extent to which lignin polymerization is biochemically controlled (7, 8), an increasing body of evidence supports the theory that lignin polymerization is a strictly chemical process that occurs subject to simple chemical radical (cross-)coupling compatibility (6, 8, 9).
Targeted down-regulation of lignin biosynthetic enzymes can result in a polymer in which the normal proportions of the H, G, and S units are significantly altered. For example, depletion of hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (HCT) or coumaroyl shikimate 3′-hydroxylase (C3H) activities results in lignins with abnormally high proportions of H units (10, 11). Depletion of caffeic acid/5-hydroxyconiferaldehyde 3-O-methyltransferase activity results in a lignin composed of a large proportion of nonconventional 5-hydroxyguaiacyl residues (12). Such malleability in lignin biosynthesis is further illustrated by the recent discovery of naturally occurring lignins composed of caffeyl alcohol- or 5-hydroxyconiferyl alcohol-derived units in seed coats of members of the Orchidaceae and Cactaceae (13, 14). Severe genetic alteration of lignin to elevate incorporation of nontraditional monomers is, however, often accompanied by plant dwarfing or developmental abnormalities such as irregular xylem; the mechanisms underlying such growth defects remain unclear (15).
Cinnamyl alcohol dehydrogenase (CAD) is an enzyme specifically involved in the last step of monolignol biosynthesis, reducing the hydroxycinnamaldehydes to their corresponding hydroxycinnamyl alcohols, before their transport to the wall for lignification. Loss of function or down-regulation of CAD generally leads to reduction of lignin content and red coloration of xylem tissue (16–18). Demonstration that hydroxycinnamaldehydes are integrally incorporated into the lignin, either as end groups or part of the polymer backbone, has been via the release of diagnostic markers during analytical thioacidolysis (18–20), and by NMR (21, 22). However, previous studies have overlooked the extent to which cell wall lignin can be derived from hydroxycinnamaldehydes.
Here, we describe the effect of CAD gene knockout in the model legume Medicago truncatula (23–25). Retrotransposon insertions in the M. truncatula CAD1 (MtCAD1) gene result in stable mutant plants that grow normally under standard growth conditions. However, 2D NMR analysis revealed that lignins in MtCAD1 mutants are, surprisingly, composed almost exclusively (∼95%) of hydroxycinnamaldehyde-derived units.
Results
Isolation of MtCAD1 Mutants.
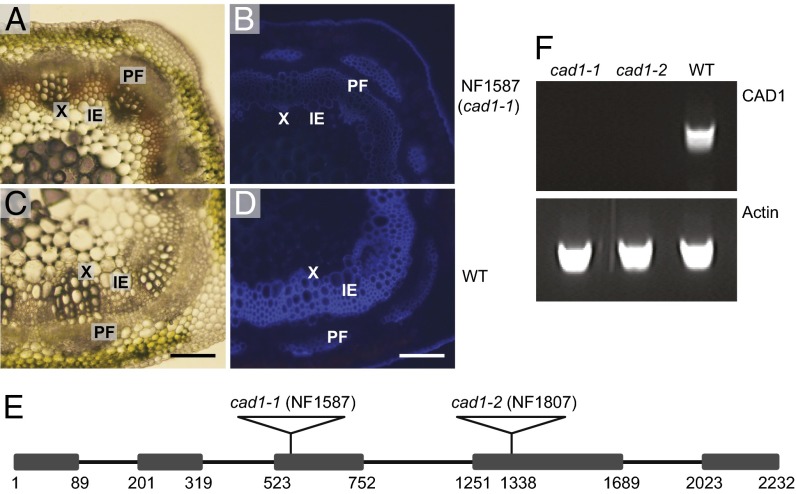
To identify mutants with lignification defects, an R1 population of 3,600 independent R1 lines (around 10,000 plants) of tobacco transposable element of Nicotiana tabacum (Tnt1) retrotransposon insertion-mutagenized M. truncatula was screened by UV microscopy of cross-sections of the sixth stem internodes, which are lignified in wild-type plants. One line, NF1587, showed not only a strong reduction of blue lignin autofluorescence, but a red coloration in interfascicular fibers and vascular bundles was also visible under bright-field microscopy (Fig. 1 A–D).
Fig. 1.
The Medicago cad1-1 mutant shows a lignin deposition defect. (A–D) Cross-sections of stems from sixth internodes of 9-wk-old NF1587 line (cad1-1) and wild-type R108 plants. A and C are light microscopy images. Total lignin in B and D was visualized by UV autofluorescence. IE, interfascicular element; PF, phloem fiber; X, xylem. (Scale bar: 100 µm.) (E) Diagram of the structure of the CAD1 gene and positions of retrotransposon insertions. The numbers indicate nucleotide positions from the site of initiation of translation. The boxes represent exons. The black lines represent introns. (F) RT-PCR analysis of CAD1 transcripts in cad1-1 and cad1-2 mutant and wild-type lines. The actin gene was used as positive control.
Microarray analysis with RNA isolated from stem internodes of the mutant was used to identify the gene responsible for the above phenotype. As Tnt1 retrotransposon insertion-mutagenized M. truncatula plants usually contain 20–50 insertions per plant (25), a progeny line segregating from the same parent plant but showing normal lignin deposition was used as control to minimize the transcript changes resulting directly or indirectly from other insertions. Total RNA samples were subjected to Affymetrix microarray analysis. In total, 108 probe sets were down-regulated, and 190 probe sets up-regulated, in the NF1587 line by at least twofold. The second and third most down-regulated probe sets (Mtr.8589.1.S1_at and Msa.1908.1.S1_at) were annotated as encoding CAD. To check for insertion of Tnt1 in the CAD gene in line NF1587, PCR was performed with a Tnt1 primer and primers designed from the probe set sequences. This failed to amplify a PCR product, indicating that there was no Tnt1 insertion in the CAD locus. However, PCR with primers designed from both ends of the probe set amplified a large product of around 5 kb, suggesting that there is indeed an insertion in the CAD locus. Partial sequencing of the insertion (Fig. S1) indicted a native retrotransposon of M. truncatula instead of the expected Tnt1 retrotransposon.
A cDNA BLAST search was performed against the M. truncatula genome sequence from the Dana-Faber Cancer Institute (DFCI) bioinformatics Web site (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago). The first hit with the lowest e value was TC176769, which contains a 1,656-bp cDNA sequence, including the entire Mtr.8589.1.S1_at and Msa.1908.1.S1_at probe sequences. TC176769 shows high sequence similarity to Arabidopsis CAD4 (At3g19450), one of the two primary lignin-specific CADs in Arabidopsis (16, 26). We named the full-length sequence M. truncatula CAD1 and the NF1587 mutant cad1-1.
The Medicago CAD1 gene contains five exons and four introns and the insertion in NF1587 is in the third exon (Fig. 1E). One additional insertion line (NF1807) of MtCAD1 was subsequently obtained by reverse genetic screening. The Tnt1 insertion was in the fourth exon (Fig. 1E), and the mutant was termed cad1-2. RT-PCR indicated that there is no full-length transcript of MtCAD1 in either of the mutants (Fig. 1F).
Chemical Characterization of MtCAD1 Mutants.
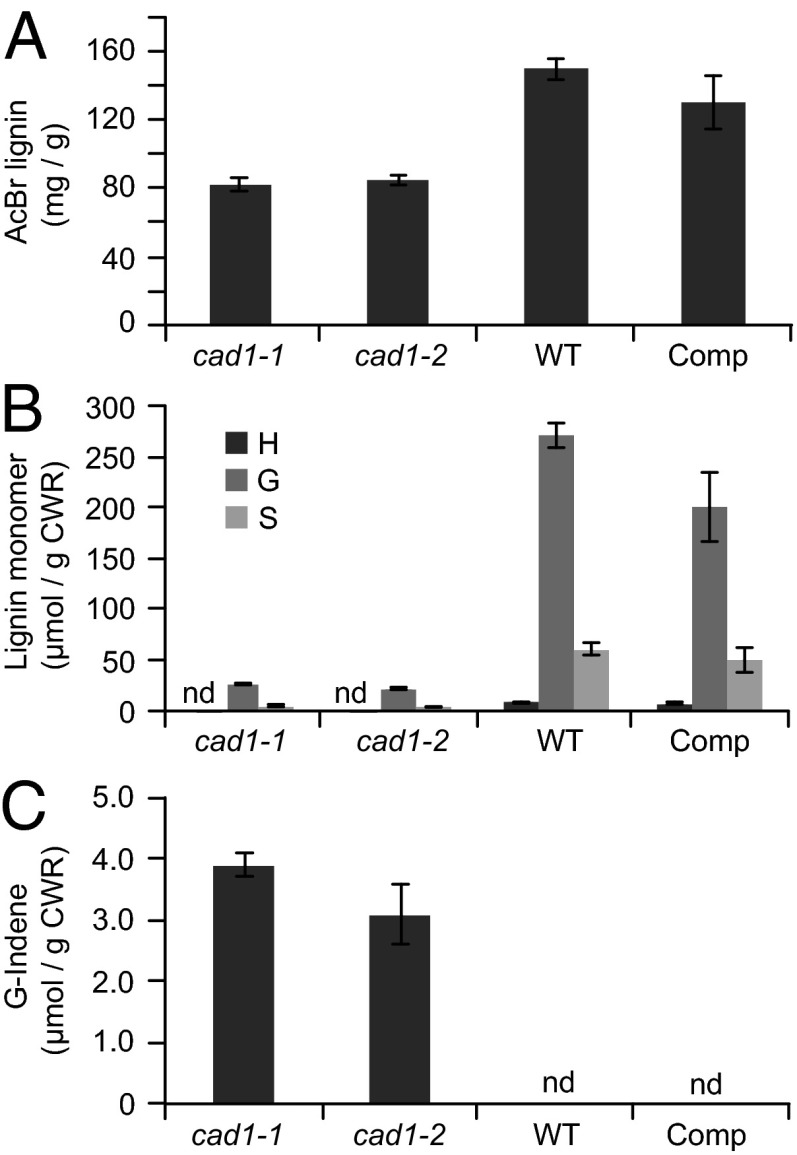
Homozygous lines of both mutants showed a strong reduction in total lignin measured by the acetyl bromide method (Fig. 2A); it should be noted, however, that any lignin assay has an unknown accuracy when the lignin structure is drastically changed (see below). Analytical thioacidolysis revealed that the levels of traditional G and S monomers released by cleaving β–O–4-linked units were severely reduced (Fig. 2B). In addition, thioacidolysis of the mutants released coniferaldehyde-derived indene derivatives, markers that are diagnostic for the incorporation of hydroxycinnamaldehyde monomers into lignins via 8–O–4 coupling (19, 20), whereas the indenes were undetectable in the wild-type plants (Fig. 2C).
Fig. 2.
Lignin levels in the Medicago cad1-1 and cad1-2 mutants and a complemented cad1-1 line. (A) Acetyl bromide lignin content of stems (fifth to eighth internodes) of 4-mo-old M. truncatula wild type (WT), cad1-1 and cad1-2 mutants, and cad1-1 line complemented with the wild-type Medicago CAD1 sequence (Comp). (B) Thioacidolysis yields of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin monomers from the above lines. (C) Quantification of wall-bound coniferaldehyde-derived indene compounds. Bars show means and SDs; n = 6. nd, not detected. CWR, cell wall residue.
Complementation of MtCAD1 Mutations.
The above microarray analysis revealed that a suite of genes involved in secondary cell wall biosynthesis had reduced expression by at least twofold in the cad1-1 mutant line (Table S1). These include a putative homolog of AtMYB46, a master switch of secondary wall biosynthesis (27), 4-coumaroyl-CoA ligase, laccase17, phenylalanine ammonia-lyase 1 (PAL1) and PAL2 involved in the monolignol pathway, and cellulose synthase7 and GAUT12 involved in biosynthesis of cellulose and hemicelluloses/pectins, respectively (28, 29). To confirm that the lignification phenotype was indeed the result of insertional mutagenesis of CAD1, rather than from reduction in expression of another cell wall biosynthetic enzyme, the coding sequence of CAD1 driven by the 35S promoter was used for complementation of the cad1-1 mutant. The lignin level in the mutant was significantly restored, and the indene signature disappeared, in six independent transformation events (Fig. 2). MtCAD1 could also rescue the phenotype of the Arabidopsis cad4/cad5 double knockout mutant; the red coloration in the fibers of the double mutant was no longer visible in the complemented line (Fig. S2A), and acetyl bromide lignin levels were also partially rescued (Fig. S2B).
Biochemical Properties of MtCAD1.
The ORF of Medicago CAD1 was expressed in Escherichia coli. The histidine-tagged recombinant protein was purified to homogeneity by nickel affinity chromatography and, after removal of the His-tag, was assayed for activity against a range of potential substrates. MtCAD1 catalyzed the reduction of 4-coumaraldehyde, coniferaldehyde, and sinapaldehyde, with similar kinetic constants, high affinities, and a slight catalytic preference for 4-coumaraldehyde (Table S2).
NMR Analysis of Cell Wall Structure in MtCAD1 Mutants.
For detailed characterization of cell wall structures in MtCAD1 mutants, we used 2D NMR analysis (30, 31). For comparison, we analyzed, in parallel, synthetic lignins [dehydrogenation polymers (DHPs)] prepared by in vitro peroxidase-catalyzed polymerization of hydroxy-cinnamaldehydes, and cell walls from alfalfa (Medicago sativa) plants in which CAD had been less severely down-regulated by an antisense strategy (32).
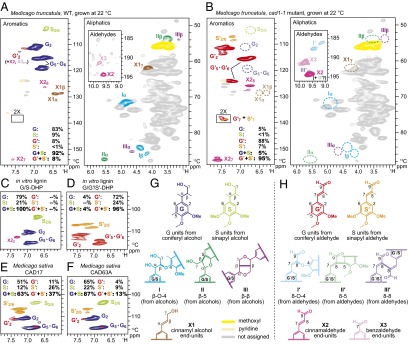
In the aromatic regions of the heteronuclear single-quantum coherence (HSQC) NMR spectra of cell walls from the cad1-1 mutant (Fig. 3B), the “normal” guaiacyl (G) and syringyl (S) units derived from monolignols were present at extremely low levels. Instead, dominant signals appeared from unusual guaiacyl (G′) and syringyl (S′) units derived from polymerization of hydroxycinnamaldehydes (21, 22). In terms of the HSQC contour intensity ratio, the aldehyde-derived units (G′ + S′) accounted for ∼95% of the total lignin aromatics detected (Fig. 3B). In addition, correlations from unsaturated cinnamaldehyde end-units, X27 and X28, and hydroxycinnamaldehyde side-chain signals, G′7 and S′7, were also clearly observed in the spectra from the cad1-1 mutant. As expected, the cell wall lignins in the wild-type plants are typical G-rich G/S-lignins, and the natural presence of aldehyde units was minimal (Fig. 3A); particularly in the wild-type plants, G′2 signals might be overestimated by the presence of oxidized β-aryl ether units with α-carbonyl carbons (30). The S/G ratios in the wild-type samples were nearly identical to the S′/G′ ratios in the cad1-1 mutants. The signal patterns in the cad1-1 mutant spectra were well matched with those observed in the spectra of DHPs prepared from coniferaldehyde and sinapaldehyde (Fig. 3 C and D). The NMR spectra from two independent CAD down-regulated alfalfa lines (32) revealed significant G′ and S′ signals (37% and 13% of the total lignin aromatic signal), but classical G and S units remained predominant (Fig. 3 E and F).
Fig. 3.
Partial HSQC spectra of ball-milled whole cell walls from M. truncatula and M. sativa, and synthetic lignins (DHPs). (A) M. truncatula wild-type plant grown at 22 °C. (B) M. truncatula cad1-1 mutant grown at 22 °C; contours for G′7 and S′7 are artificially enlarged (by a factor of 2) to enhance visibility. (C) G/S-DHP, DHP prepared from coniferyl and sinapyl alcohols (4:1, mol/mol); (D) G′/S′-DHP, DHP prepared from coniferaldehyde and sinapaldehyde in combination with coniferyl alcohol (7:2:1, mol/mol/mol). (E and F) Two independent alfalfa lines with CAD activity reduced by antisense expression (partial spectra) generated in a previous study (32). (G and H) Conventional and new lignin subunits derived from polymerization of coniferyl and sinapyl alcohols (G) and coniferaldehye and sinapaldehyde (H).
In the aliphatic regions of the HSQC spectra of whole cell walls from wild-type plants, typical lignin side-chain signals from β-aryl ether (I), phenylcoumaran (II), and resinol (III) structures, as well as cinnamyl alcohol end groups (X1), were readily visible (Fig. 3A). Those signals were, however, practically absent in spectra from the cad1-1 mutants (Fig. 3B). Instead, in the aldehyde region (Fig. 3 A and B, Insets), aldehyde signals uniquely derived from hydroxycinnamaldehyde polymerization (21, 22) were clearly observed (Fig. 3B). The 8–O–4- (I′ and I″) and 8–8- (III′) units with unsaturated side chains as well as augmented cinnamaldehyde end units (X2) were resolved; similar signal patterns were also observed in the spectra of DHPs prepared from coniferaldehyde and sinapaldehyde (Fig. S3A), and less apparently but also visible in the spectra of the two CAD down-regulated alfalfa lines (Fig. S3A). These aldehyde signals, except for the naturally occurring cinnamaldehyde (X2) and benzaldehyde (X3) end units, were not visible in the spectra from the wild-type plants (Fig. 3A) nor from a conventional DHP from coniferyl and sinapyl alcohols (Fig. S3A).
We also analyzed cell wall polysaccharide unit profiles by NMR (Fig. S3B). The HSQC signal patterns observed in polysaccharide anomeric regions of the whole-cell wall NMR spectra (30, 33) appeared to be similar for wild-type and cad1-1 M. truncatula grown in the growth chamber at 22 °C (Fig. S3B).
Phenolic Metabolites in MtCAD1 Mutants.
To determine whether hydroxycinnamaldehydes spill over into other metabolites in the cad1 mutant, liquid chromatography (LC)-MS coupled with photodiode array detection was used for metabolite profile analysis of organic extracts from stem samples of 4-mo-old mature cad1-1 and wild-type plants. The main differences between the soluble phenolic profiles of mutant and wild-type plants were the overaccumulation of the predicted substrate of CAD1, coniferaldehyde, and feruloyl glucose, in the mutant (Fig. S4 and Table S3). Feruloyl glucose could be formed from coniferaldehyde via the action of an aldehyde dehydrogenase followed by glucosylation (34, 35); neither coniferaldehyde nor feruloyl glucose was detectable in extracts from wild-type tissue (Fig. S4A). Levels of flavone (apigenin) glucuronoside, a major phenolic constituent of Medicago species (36), were essentially unchanged in the cad1-1 mutant (Table S3).
Levels of wall-bound phenolic compounds were determined by HPLC analysis of alkaline hydrolysates of isolated cell walls. The cad1-1 cell wall samples released significantly higher amounts of vanillin, ferulic acid, and p-coumaric acid than the wild-type controls (Fig. S4B and Tables S2 and S3B). In addition, the cad1-1 mutant accumulated wall-associated coniferaldehyde and syringaldehyde, neither of which was detected in cell walls from wild-type plants (Fig. S4B and Tables S3 A and B).
Altered Extractability of Cell Wall Polysaccharides in MtCAD1 Mutants.
We used glycome profiling to determine whether the abnormal lignin structure of the cad1-1 mutant impacts polysaccharides within the cell walls. Cell wall residues from the cad1-1 mutant and control wild-type plants were fractionated by increasingly harsh sequential extractions, and each fraction subjected to ELISA using a panel of 155 plant cell wall glycan-directed monoclonal antibodies (37) (Table S4). Changes observed in the cad1-1 mutant cell walls are highlighted by dotted blocks in Fig. S5. The alterations of lignin structure in the cad1-1 mutant were associated with significantly enhanced abundance of xylan epitopes in the carbonate and 4 M KOH extracts compared with the control samples. Pectin-directed epitopes (homogalacturonan-1 and rhamnogalacturonan-I) also showed stronger signals in the oxalate and 4 M KOH fractions of the cad1-1 mutant cell wall than the controls, and the abundance of pectic-arabinogalactan epitopes was also significantly enhanced in the 4 M KOH extracts of the mutant cell wall sample. These results suggest that subpopulations of several classes of polysaccharides are altered in their cross-linking into the cell wall matrix. The reduced overall level of polysaccharide extractability per gram of cell wall residue in the cad1 mutant (Fig. S5, upper bar graph panel) suggests an overall greater extent of wall cross-linking in the mutant, although the structural basis for this observation is not clear at present.
MtCAD1 Mutants Exhibit a Temperature-Sensitive Growth Phenotype.
MtCAD1 mutant lines appeared to grow normally in the greenhouse at 22 °C (Fig. 4A). However, when grown at higher temperature (30 °C), the cad1 mutant lines were strongly dwarfed compared with wild-type plants (Fig. 4B). Both insertion lines showed the same growth defect, which could be rescued by complementation with the wild-type copy of MtCAD1. The overall growth of wild-type plants at 30 °C was very similar to their growth at 22 °C (Fig. 4 A and B).
Fig. 4.
The Medicago cad1-1 and cad1-2 mutants have a conditional growth defect at elevated temperature. Each pot has two 3-mo-old plants. Comp. is the complemented cad1-1 mutant. (A) Plants grown at 22 °C. (B) Plants grown at 30 °C.
Based on 2D NMR analysis, growth at high temperature had only small effects on lignin composition and structure, with slightly increased S′/G′ ratio in the cad1-1 mutants, but not S/G ratio in the wild-type plants (Fig. S3 C and D, and Fig. 3). Growth at high temperature induced reductions of unknown cause in galacturonate (GalpA) signals (30, 33) in both the wild type and cad1-1 mutant (Fig. S3B).
Microarray analysis was performed with RNA isolated from mutant and wild-type plants grown at 22 and 30 °C. Genes of interest were those expressed differentially between mutant and wild type only at high temperature. Within this class were many pathogen response-related genes, and a set of heat shock-related genes that were not up-regulated in the wild type at elevated temperature (Table S5).
Discussion
CAD genes usually exist as a multigene family in angiosperms, and the corresponding enzymes have significant affinity for both coniferaldehyde and sinapaldehyde. Of the nine Arabidopsis CADs (38), CAD5 is the most catalytically active and can use all potential monolignol pathway hydroxycinnaldehydes effectively. However, loss of function of CAD5 has only a small effect on lignin content and composition (26), and simultaneous disruption of CAD4 and CAD5 is required to generate a significant lignin biosynthesis defect (16). In contrast, disruption of only CAD1 in M. truncatula results in a large reduction in lignin levels and a striking alteration in lignin structure. Similar to the apparent lack of redundancy in CAD function in M. truncatula, knockout of the single Medicago NST1 gene phenocopies the Arabidopsis NST1/NST3 and NST1/NST2 double-knockout mutants, showing defects in both secondary cell wall biosynthesis and anther dehiscence (39).
Our NMR data reveal that the cell wall lignins synthesized in the Medicago CAD mutants are massively (by ∼95%) composed of polymers derived from hydroxycinnamaldehydes. Several previous studies have reported the impacts of CAD down-regulation on lignin content and composition (16, 19, 40–43). However, none has shown the presence of a lignin with the remarkable structure described here. The prior failure to observe lignin derived predominantly from hydroxycinnamaldehydes is likely the result of two factors: (i) CAD gene redundancy, such that genetically targeted knockdown or knockout fails to reduce CAD activity to a level that blocks formation of hydroxycinnamyl alcohols, or (ii) simple failure to detect the presence of highly aldehyde-rich lignins; this may be due to the previously limited interpretability of spectra, or to the application of analytical techniques having insufficient resolution or that are not specifically targeted to detection or discrimination of incorporated hydroxycinnamaldehydes.
The highly unusual lignin composition in the Medicago cad1 mutants is associated with increased extractability of subpopulations of polymers containing xylan and pectic (rhamnogalacturonan) residues, and reduced extractability of pectic arabinogalactan residues. Although likely, it is not yet proven that this reflects altered lignin–polysaccharide linkages in the walls of the mutant. Reduced lignin polysaccharide cross-linking would, however, be expected for hydroxycinnamaldehyde-derived polymers. This is because the quinone methide intermediate resulting from β–O–4 cross-coupling of a monolignol with the phenolic end of a growing lignin oligomer is capable of being nucleophilically trapped by polysaccharide hydroxyls or pectin uronic acids resulting in lignin-benzyl-polysaccharide ethers or lignin-benzyl-uronate esters, whereas the quinone intermediate resulting from analogous 8–O–4 cross-coupling of a hydroxycinnamaldehyde has a more facile pathway for rearomatization—elimination of the acidic 8-proton (next to the aldehyde carbonyl) is faster than external nucleophile addition (22, 44).
Genetic modification of the lignin biosynthetic pathway in plants can cause moderate-to-severe growth defects depending on the gene targeted. In general, plants deficient in C3′H, hydroxycinnamoyl-CoA: shikimate HCT, cinnamate 4-hydroxylase, or cinnamoyl-CoA reductase exhibit dwarfed phenotypes (10, 45–47), whereas the Arabidopsis CAD4/CAD5 double mutant does not exhibit a significantly reduced growth phenotype even though the lignin content is reduced based on determination with acetyl bromide (16). Whether alteration of lignin structures is primarily responsible for such developmental defects is currently unclear. To date, to the best of our knowledge, no monolignol biosynthetic mutant in any plant species has been reported to have a similar conditional growth phenotype to the Medicago cad1-1 and cad1-2 mutants. The NMR data suggest that growth at elevated temperature does not significantly affect lignin composition.
The phenotype of dwarfing associated with defense gene expression at the restrictive temperature in the M. truncatula cad1 mutants is similar to that observed in HCT-deficient alfalfa (48). Some of the genes activated at the restrictive temperature in the cad1-1 mutant (e.g., two PR genes and a set of DNAJ-type heat shock genes) are up-regulated in HCT down-regulated alfalfa grown at the permissive temperature for the cad1 mutants (48). This suggests that the mechanisms underlying the dwarf phenotypes and defense-gene expression patterns of both HCT-deficient alfalfa and the Medicago cad1-1 mutant may be similar.
Two hypotheses are currently under consideration to explain the dwarf phenotypes of lignin down-regulated plants. One postulates nonspecific stress due to impairment of vascular function (10), which can reduce transpiration capacity, leading to overheating and induction of heat shock proteins (49). The other proposes involvement of chemical signals released from incorrectly assembled cell walls (48). CAD down-regulation may impact cell wall integrity less severely than HCT down-regulation, such that elicitor-active cell wall fragments are not released at the restrictive temperature.
The almost exclusive derivation of lignin polymers in the cad1 mutants from nontraditional lignin monomers extends the scope of modifications that we may yet consider for lignin modification toward improved biomass processing. The present results suggest, however, that transgenic plants with low lignin should be carefully tested under a range of different environmental conditions for possible biomass reduction traits.
Materials and Methods
Plant Materials and Growth Conditions.
A tobacco (Nicotiana tabacum) Tnt1 retrotransposon-tagged mutant collection of M. truncatula (25) was screened for defects in secondary cell wall formation. Plants were grown in MetroMix 350 soil mix at 24/20 °C (day/night), with a 16-h day/8-h night photoperiod, 70–80% relative humidity, and 150 μmol⋅m−2⋅s−1 light intensity. The sixth internodes were harvested when plants had reached around eight internodes, and were stored at −80 °C.
Identification and Molecular Cloning of MtCAD1.
Total RNA samples from the fifth to the eighth internodes were subjected to Affymetrix microarray analysis. Segregating progeny without the reduced lignification phenotype from the same parent plant were used as controls. PCR of down-regulated probe sets was performed using gene-specific primers to confirm that the insertion was linked to the phenotype.
To clone the full-length CAD1 gene, BLAST analysis of the M. truncatula genome from DFCI (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago) was performed using Mtr.8589.1.S1_at as the query probe sequence; this led to a complete cDNA sequence (TC176769). The CAD1 genomic sequence was PCR amplified and sequenced from M. truncatula ecotype R108.
Medicago Transformation.
Cloning of MtCAD1 for complementation was performed as described in SI Materials and Methods, and Medicago transformation performed as described previously (50).
Microarray Analysis.
Microarray analysis was performed as described in SI Materials and Methods.
Determination of Lignin Content and Composition.
Lignin content and composition were determined by acetyl bromide and thioacidolysis assays as described in SI Materials and Methods.
Expression of MtCAD1 in E. coli and Assay of Enzyme Activity.
Cloning of the CAD1 ORF, transformation into E. coli, protein expression, purification, and assay of enzyme activity and kinetics were performed as described in SI Materials and Methods.
Determination of Soluble and Wall-Bound Phenolics.
Soluble phenolic compounds were extracted as described in SI Materials and Methods. Phenolics were identified by LC–electrospray ionization-MS/MS as described (51). Authentic feruloyl glucose was prepared as described (52). Coniferaldehyde, sinapaldehyde, vanillin, syringaldehyde, p-coumaric acid, and ferulic acid were obtained from Sigma-Aldrich.
DHPs.
DHPs were generated via peroxidase-catalyzed polymerization as described previously (13) and further elaborated in SI Materials and Methods.
NMR Analysis.
NMR spectra of plant cell walls and DHPs were acquired on a Bruker Biospin AVANCE 700-MHz spectrometer fitted with a cryogenically cooled 5-mm TXI gradient probe with inverse geometry (proton coils closest to the sample). The detailed NMR methods used were largely as described previously (30, 31), and as further described in SI Materials and Methods.
Glycome Profiling.
Sequential extraction and glycome profiling of alcohol insoluble cell wall residues were performed as described previously (37).
Supplementary Material
Acknowledgments
We thank Drs. Wolfgang Schieble and Elison Blancaflor for critical reading of the manuscript. The M. truncatula plants used in this work were created through research funded, in part, by National Science Foundation Grant 703285. This work was supported in part by Grant DE-FG02-06ER64303 from the Department of Energy (DOE) Feedstock Genomics program (to R.A.D.), with additional support from Forage Genetics International, The Samuel Roberts Noble Foundation, and the DOE’s Bioenergy Sciences and Great Lakes Bioenergy Research Centers, supported by the Office of Biological and Environmental Research in the DOE Office of Science (BER DE-AC05-00OR22725 and DE-FC02-07ER64494, respectively).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper can be found in the Dana-Farber Cancer Institute Medicago Gene Index (accession no. TC 176769).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312234110/-/DCSupplemental.
References
- 1.Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu Rev Genet. 2010;44:337–363. doi: 10.1146/annurev-genet-102209-163508. [DOI] [PubMed] [Google Scholar]
- 2.Fu C, et al. Genetic manipulation of lignin biosynthesis in switchgrass significantly reduces recalcitrance and improves biomass ethanol production. Proc Natl Acad Sci USA. 2011;108(9):3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25(7):759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 4.Reddy MSS, et al. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.) Proc Natl Acad Sci USA. 2005;102(46):16573–16578. doi: 10.1073/pnas.0505749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudet AM, Grima-Pettenati J. Lignin genetic engineering. Mol Breed. 1996;2(1):25–39. [Google Scholar]
- 6.Vanholme R, et al. Metabolic engineering of novel lignin in biomass crops. New Phytol. 2012;196(4):978–1000. doi: 10.1111/j.1469-8137.2012.04337.x. [DOI] [PubMed] [Google Scholar]
- 7.Davin LB, Lewis NG. Lignin primary structures and dirigent sites. Curr Opin Biotechnol. 2005;16(4):407–415. doi: 10.1016/j.copbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Ralph J, Brunow G, Harris P, Dixon RA, Boerjan W. Lignification: Are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? Rec Adv Polyphenols Res. 2008;1:36–66. [Google Scholar]
- 9.Ralph J. Hydroxycinnamates in lignification. Phytochem Rev. 2010;9(1):65–83. [Google Scholar]
- 10.Franke R, et al. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 2002;30(1):47–59. doi: 10.1046/j.1365-313x.2002.01267.x. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Bonawitz ND, Weng J-K, Chapple C. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell. 2010;22(5):1620–1632. doi: 10.1105/tpc.110.074161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marita JM, et al. Structural and compositional modifications in lignin of transgenic alfalfa down-regulated in caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase. Phytochemistry. 2003;62(1):53–65. doi: 10.1016/s0031-9422(02)00434-x. [DOI] [PubMed] [Google Scholar]
- 13.Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J. A polymer of caffeyl alcohol in plant seeds. Proc Natl Acad Sci USA. 2012;109(5):1772–1777. doi: 10.1073/pnas.1120992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, et al. Novel seed coat lignins in the Cactaceae: Structure, distribution and implications for the evolution of lignin diversity. Plant J. 2012;73(2):201–211. doi: 10.1111/tpj.12012. [DOI] [PubMed] [Google Scholar]
- 15.Bonawitz ND, Chapple C. Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol. 2013;24(2):336–343. doi: 10.1016/j.copbio.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Sibout R, et al. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell. 2005;17(7):2059–2076. doi: 10.1105/tpc.105.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKay JJ, Liu W, Whetten R, Sederoff RR, O’Malley DM. Genetic analysis of cinnamyl alcohol dehydrogenase in loblolly pine: Single gene inheritance, molecular characterization and evolution. Mol Gen Genet. 1995;247(5):537–545. doi: 10.1007/BF00290344. [DOI] [PubMed] [Google Scholar]
- 18.Ralph J, et al. Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency) Plant J. 2008;53(2):368–379. doi: 10.1111/j.1365-313X.2007.03345.x. [DOI] [PubMed] [Google Scholar]
- 19.Lapierre C, et al. Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry. 2004;65(3):313–321. doi: 10.1016/j.phytochem.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Kim H, et al. Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J Biol Chem. 2002;277(49):47412–47419. doi: 10.1074/jbc.M208860200. [DOI] [PubMed] [Google Scholar]
- 21.Kim H, Ralph J, Yahiaoui N, Pean M, Boudet A-M. Cross-coupling of hydroxycinnamyl aldehydes into lignins. Org Lett. 2000;2(15):2197–2200. doi: 10.1021/ol005906o. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, et al. NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org Biomol Chem. 2003;1(2):268–281. doi: 10.1039/b209686b. [DOI] [PubMed] [Google Scholar]
- 23.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedito VA, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55(3):504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 25.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54(2):335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim S-J, et al. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA. 2004;101(6):1455–1460. doi: 10.1073/pnas.0307987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20(10):2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12(12):2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persson S, et al. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell. 2007;19(1):237–255. doi: 10.1105/tpc.106.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H, Ralph J. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org Biomol Chem. 2010;8(3):576–591. doi: 10.1039/b916070a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansfield SD, Kim H, Lu F, Ralph J. Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc. 2012;7(9):1579–1589. doi: 10.1038/nprot.2012.064. [DOI] [PubMed] [Google Scholar]
- 32.Jackson LA, et al. Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. BioEnergy Res. 2008;1(3):180–192. [Google Scholar]
- 33.Petersen BO, Meier S, Duus JO, Clausen MH. Structural characterization of homogalacturonan by NMR spectroscopy-assignment of reference compounds. Carbohydr Res. 2008;343(16):2830–2833. doi: 10.1016/j.carres.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 34.Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C. The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell. 2004;16(2):544–554. doi: 10.1105/tpc.017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner D, Albert A, Böttcher C, Strack D, Milkowski C. The role of UDP-glucose:hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana. Planta. 2008;228(4):663–674. doi: 10.1007/s00425-008-0768-3. [DOI] [PubMed] [Google Scholar]
- 36.Deavours BE, Dixon RA. Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiol. 2005;138(4):2245–2259. doi: 10.1104/pp.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pattathil S, Avci U, Miller JS, Hahn MG. Immunological approaches to plant cell wall and biomass characterization: Glycome profiling. Methods Mol Biol. 2012;908:61–72. doi: 10.1007/978-1-61779-956-3_6. [DOI] [PubMed] [Google Scholar]
- 38.Sibout R, et al. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 2003;132(2):848–860. doi: 10.1104/pp.103.021048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Q, et al. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 2010;63(1):100–114. doi: 10.1111/j.1365-313X.2010.04223.x. [DOI] [PubMed] [Google Scholar]
- 40.Sattler SE, et al. A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol. 2009;150(2):584–595. doi: 10.1104/pp.109.136408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouvier d’Yvoire M, et al. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 2013;73(3):496–508. doi: 10.1111/tpj.12053. [DOI] [PubMed] [Google Scholar]
- 42.Fu C, et al. Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. BioEnergy Res. 2011;4(3):153–164. [Google Scholar]
- 43.Zhang K, et al. GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 2006;140(3):972–983. doi: 10.1104/pp.105.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ralph J. What makes a good monolignol substitute? In: Hayashi T, editor. The Science and Lore of the Plant Cell Wall. Biosynthesis, Structure and Function. Boca Raton, FL: BrownWalker; 2006. pp. 285–293. [Google Scholar]
- 45.Hoffmann L, et al. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell. 2004;16(6):1446–1465. doi: 10.1105/tpc.020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schilmiller AL, et al. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009;60(5):771–782. doi: 10.1111/j.1365-313X.2009.03996.x. [DOI] [PubMed] [Google Scholar]
- 47.Mir Derikvand M, et al. Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta. 2008;227(5):943–956. doi: 10.1007/s00425-007-0669-x. [DOI] [PubMed] [Google Scholar]
- 48.Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.) New Phytol. 2011;190(3):627–639. doi: 10.1111/j.1469-8137.2010.03621.x. [DOI] [PubMed] [Google Scholar]
- 49.Kaplan F, et al. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136(4):4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosson V, Durand P, d’Erfurth I, Kondorosi A, Ratet P. Medicago truncatula transformation using leaf explants. Methods Mol Biol. 2006;343:115–127. doi: 10.1385/1-59745-130-4:115. [DOI] [PubMed] [Google Scholar]
- 51.Zhao J, et al. MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell. 2011;23(4):1536–1555. doi: 10.1105/tpc.110.080804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y, Ralph J. Stereoselective synthesis of 1-O-β-feruloyl and 1-O-β-sinapoyl glucopyranoses. Tetrahedron Lett. 2011;52(21):3729–3731. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.