Abstract
Breast cancer gene 1 (BRCA1) deficient cells not only are hypersensitive to double-strand breaks but also are hypersensitive to UV irradiation and other agents that cause replication blockade; however, the molecular mechanisms behind these latter sensitivities are largely unknown. Here, we report that BRCA1 promotes cell survival by directly regulating the DNA damage tolerance pathway in response to agents that create cross-links in DNA. We show that BRCA1 not only promotes efficient mono- and polyubiquitination of proliferating cell nuclear antigen (PCNA) by regulating the recruitment of replication protein A, Rad18, and helicase-like transcription factor to chromatin but also directly recruits translesion polymerases, such as Polymerase eta and Rev1, to the lesions through protein–protein interactions. Our data suggest that BRCA1 plays a critical role in promoting translesion DNA synthesis as well as DNA template switching.
DNA damage response (DDR) and DNA repair are vital for preserving genomic integrity during a normal cell cycle and after genotoxic stress (1). In humans, many factors involved in DDR and DNA repair, such as BRCA1 and BRCA2, are well-known tumor suppressors. BRCA1 has been linked to both familial and sporadic breast and ovarian cancers, and it operates in multiple biological pathways including DDR and DNA repair (2, 3). BRCA1 is essential for the homologous recombination (HR) repair pathway, and therefore, cells deficient in BRCA1 function are hypersensitive to ionizing radiation and other drugs that directly induce double-strand breaks (DSBs) (4). The fact that BRCA1-deficient cells are also hypersensitive to agents that form adducts on DNA or cross-link DNA, including UV, cis-platin (cis-diamminedichloroplatinum [II]; CDDP), and mitomycin C (MMC), suggests that BRCA1 is important for other responses as well (Fig. 1 A and B) (6–8). Furthermore, Nussenzweig and coworkers recently showed that BRCA1 activity is important for resistance to DNA interstrand cross-links in a manner that is separable from its role in HR (9). One common feature of the lesions created by UV, CDDP, and MMC is that they all have the potential to block the progression of replicative DNA polymerases and activate the DNA damage tolerance (DDT) pathway in mammalian cells (10–12). The DDT pathway is critical for cell viability when replication-blocking lesions are encountered by replicative DNA polymerases by promoting the replicative bypass (i.e., translesion DNA synthesis, or TLS) or template-switching bypass of blocking lesions present in DNA (13, 14). The ubiquitination of PCNA plays an important role in the activation of both TLS and template switching. In budding yeast, monoubiquitination of PCNA is catalyzed by the E2-E3 ubiquitin ligase enzymes Rad6 (E2) and Rad18 (E3), and this modification promotes TLS. Another set of E2-E3 enzymes, UBC13/MMS2 (E2)-Rad5 (E3), can extend this ubiquitin molecule attached to PCNA into a K63-linked polyubiquitin chain, which is thought to activate the error-free template-switching pathway as an alternative mechanism to bypass the lesion and restart a blocked replication fork (15).
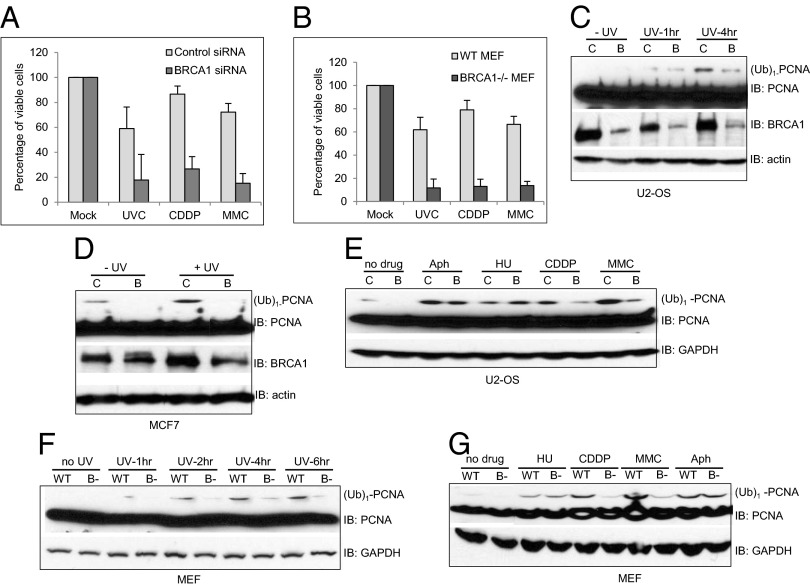
Fig. 1.
BRCA1 deficiency impedes DNA damage-induced monoubiquitination of PCNA. siRNA-transfected U2-OS (A) or WT and BRCA1-deficient (BRCA1−/−) MEFs (B) were treated with 20 J/m2 for U2-OS or 8 J/m2 UVC for MEFs, 800 nM CDDP, or 100 ng/mL MMC, and the percentage of viable cells was assayed as previously described (5). Control siRNA (C) or siRNA against BRCA1-transfected (B) U2-OS cells were either left untreated (−UV) or were treated with 60 J/m2 UVB and then collected at the indicated times. Monoubiquitinated PCNA is labeled as (Ub)1-PCNA. (D) siRNA-transfected MCF7 cells were either left untreated (−UV) or were treated with 60 J/m2 UVB (+UV) and then collected 4 h later. (E) siRNA-transfected U2-OS cells were either left untreated (no drug) or were treated with various genotoxic agents: 1 mM HU, 10 μM CDDP, 30 μM MMC, 3 μM Aph for 24 h. (F) UV-induced monoubiquitination of PCNA is reduced in BRCA1-deficient MEFs. WT or BRCA1-deficient (B−) MEFs were either left untreated (no UV) or were treated with 60 J/m2 UVB and then collected at the indicated times. (G) DNA cross-linking agents CDDP- and MMC-induced monoubiquitination of PCNA is reduced in BRCA1-deficient MEFs. WT or BRCA1-deficient MEFs were either left untreated or were treated with various genotoxic agents, as in E. All error bars are SD and were obtained from three independent experiments.
To date, most studies in mammalian cells on DDT have focused on the regulation of TLS because of the lack of robust assays for template switching and the difficulty in detecting the polyubiquitinated form of PCNA. TLS can be either error-free or error-prone, depending on the specific lesion being bypassed and the TLS polymerase involved in inserting nucleotides opposite the lesion. For example, TLS polymerase eta (Polη) can bypass a thymidine dimer induced by UV with high fidelity (16, 17). In addition to the enzymes involved in ubiquitinating PCNA, the single-stranded DNA (ssDNA) binding protein, replication protein A (RPA), is also important for the ubiquitination of PCNA and functions to recruit the E3 ligase, Rad18 (12, 18). Both branches of the DDT pathways appear to be more complex in mammals. For example, two Saccharomyces cerevisiae Rad5 homologs have been identified in mammals, HLTF and SHPRH (19–22). Both can bind the E2 heterodimer, UBC13 and MMS2, and promote the polyubiquitination of PCNA in vitro and in vivo. Many additional regulatory factors have also been identified in mammalian cells that influence the DDT pathway (13, 14, 23).
Even though the core reaction and enzymes involved in DDT have been identified, the detailed mechanism of how this process is precisely regulated in mammalian cells remains unclear. Here, we show that BRCA1 affects cell survival after exposure to UV, CDDP, and MMC by directly promoting the DDT pathway. BRCA1 promotes efficient mono- and polyubiquitination of PCNA by regulating the recruitment of RPA, Rad18, and HLTF to stalled replication forks. Our data suggest that BRCA1 likely plays an important role in both TLS and template switching.
Results
BRCA1 Promotes the Monoubiquitination of PCNA in Response to UV, CDDP, and MMC.
In addition to being hypersensitive to DSBs, BRCA1-deficient mammalian cells are also hypersensitive to agents that block DNA replication, including the DNA lesions created by UV, CDDP, and MMC, all of which can activate the DDT pathways (Fig. 1 A and B) (6–8).
To investigate the potential role of BRCA1 in the DDT pathway and whether defective DDT contributes to the UV, CDDP, and MMC hypersensitivities exhibited by BRCA1-deficient cells, we first examined the effects of BRCA1 deficiency on the monoubiquitination of PCNA in human cells. As shown in Fig. 1 C–E, depletion of endogenous BRCA1 in two human cancer cell lines, U2-OS and MCF7, with small interfering RNA (siRNA) severely impaired the induction of monoubiquitinated PCNA [labeled as (Ub)1-PCNA in all figures] in response to UV, CDDP, and MMC. Intriguingly, the requirement of BRCA1 for efficient monoubiquitination of PCNA appears to be lesion-specific. BRCA1 deficiency only caused very mild effects, if any effects at all, with respect to the monoubiquitination of PCNA in response to direct inhibition of DNA polymerase activity by either the DNA polymerase inhibitor, Aphidicolin (Aph), or through deoxynucleotide deprivation caused by the ribonucleotide reductase inhibitor, hydroxyurea (HU) (Fig. 1E). We observed similar defects in BRCA1 knockout mouse embryonic fibroblasts (MEFs) (24) compared with wild-type (WT) MEFs, suggesting these phenotypes are not limited to human cancer cell lines (Fig. 1 F and G and Fig. S1). The inefficient monoubiquitination of PCNA observed in both human cell lines and MEFs deficient of BRCA1 is not caused by pronounced alterations in the cell cycle (Figs. S2 and S3) or off-target effects of siRNA, as we observed similar defects with four different siRNAs that target BRCA1 mRNA (Fig. S4). Together, these results strongly suggest that BRCA1 activates the DDT pathway by promoting the monoubiquitination of PCNA specifically in response to replication blocking base damage, and that it plays a much lesser role in promoting the monoubiquitination of PCNA after direct DNA polymerase inhibition. Moreover, these results imply that there may be distinct signal transduction pathways that sense and activate DDT pathways in response to different replication stresses.
BRCA1 Associates with and Promotes the Chromatin Binding of DDT-Related Factors, Including RPA and Rad18.
To further examine the role of BRCA1 in regulating DDT, we first tested whether BRCA1 can interact with some of the key players of DDT (Fig. 2A). The anti-BRCA1 antibody used for immunoprecipitation is a polyclonal antibody very specific to BRCA1 (Fig. S5). Consistent with previous reports (25), we observed strong interactions on chromatin between BRCA1 and DDR proteins, including ATR, TopBP1, and Chk1. Several associations are strongly stimulated by DNA damage, such as BRCA1-TopBP1 and BRCA1-Chk1, whereas other interactions do not appear to change much, such as BRCA1-ATR. Interestingly, we were able to immunoprecipitate RPA70, the largest subunit of the RPA heterotrimer, as well as Rad18 with the anti-BRCA1 antibody. Importantly, the interactions between BRCA1-Rad18 and BRCA1-RPA70 are strongly enhanced when cells were treated with UV, suggesting there may be active functional interplays among them. Furthermore, we also detected pronounced binding between BRCA1 and RPA70 as well as BRCA1 and Rad18, using tagged proteins (Fig. S6). In response to a variety of DNA damage, BRCA1, RPA70, and Rad18 can all be phosphorylated by the ATM or ATR kinases.
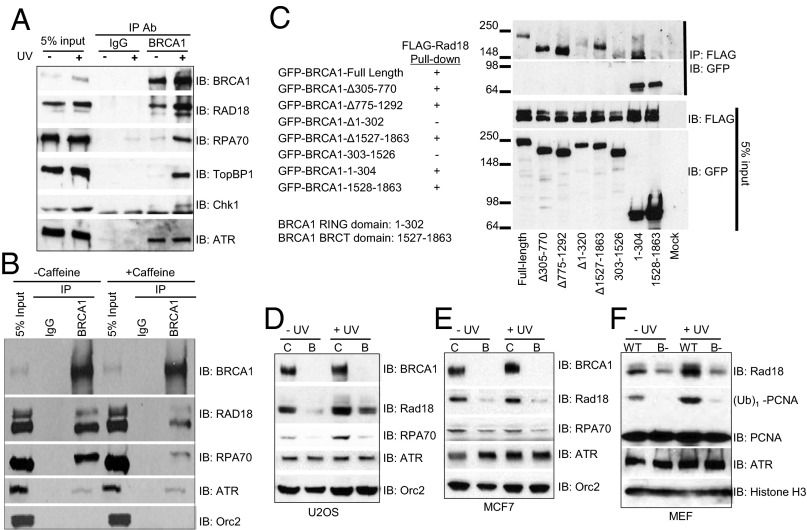
Fig. 2.
Chromatin association of key factors involved in the monoubiquitination of PCNA is compromised in BRCA1-deficient cells. (A) Association of BRCA1 with factors involved in the monoubiqutination of PCNA on chromatin. 293T cells were either left untreated (−UV) or were treated with 60 J/m2 UVB (+UV) and collected 4 h later. Chromatin fractions were prepared, and equal amounts of the chromatin fraction were used for immunoprecipitation (IP). Normal rabbit IgG was used as the negative control for IP. Precipitates were then blotted for the indicated proteins. (B) Association of BRCA1 with Rad18 and RPA70 is dependent on ATR. Experiments were done the same as in A except 293T cells were treated with 5 mM caffeine overnight before being treated with 60 J/m2 UVB. (C) BRCA1 associates with Rad18 through its RING domain and BRCT domain. FLAG-tagged Rad18 was cotransfected with either GFP-tagged, full-length BRCA1 or different truncation variants in 293T cells. Cells were lysed in lysis buffer and used for IP with anti-FLAG antibody. Precipitates and WCLs were then blotted by the indicated antibodies. (D–F) Chromatin association of factors involved in the monoubiquitination of PCNA is compromised in BRCA1-depleted U2-OS cells (D), MCF7 cells (E), and MEFs (F). Control siRNA (C) or siRNA against BRCA1-transfected (B) cells, or WT or BRCA1-deficient (B−) MEFs were either left untreated (−UV) or treated with 60 J/m2 UVB (+UV) and then used for chromatin fractionation, as previously described (26). Chromatin fractions were blotted with the indicated antibodies. Orc2 and Histone H3 were used as loading controls for the chromatin fractions of human cells and MEFs, respectively.
Next we tested whether the interactions between BRCA1 and RPA70, or BRCA1 and Rad18, are dependent on these phosphorylation events. Caffeine is a commonly used ATM/ATR inhibitor. As shown in Fig. 2B, treatment with caffeine dramatically impeded the UV-induced interactions between BRCA1 and RPA70, and between BRCA1 and Rad18, but had no effects on the interaction between BRCA1 and ATR. These data suggest that in response to UV damage, ATM/ATR phosphorylates BRCA1, RPA70, and Rad18 and promotes the interactions among them.
To map the Rad18-interacting region in BRCA1, we coexpressed FLAG-tagged Rad18 with either GFP-tagged full-length BRCA1 or different deletion mutants of BRCA1 (27) in 293T cells and immunoprecipitated Rad18 using anti-FLAG antibody. BRCA1 has two prominent domains: the really interesting new gene (RING) domain, located in 1–302 aa, and the tandem BRCA1 C terminus (BRCT) domains, located in 1527–1683 aa. As shown in Fig. 2C, Rad18 preferably binds to the RING domain of BRCA1. Deletion of the RING domain alone (Δ1–302) and in combination with BRCT domain (303–1526) severely weakened the interaction between BRCA1 and Rad18. Furthermore, Rad18 also robustly pulled down the isolated RING domain (1–302) of BRCA1. Rad18 may also loosely associate with the BRCT domain (1528–1863), as when the BRCT domain is overexpressed, its interaction with Rad18 can be easily detected. These data demonstrate that Rad18 likely associates with both the RING and BRCT domains of BRCA1.
Next, we performed chromatin fractionation studies using BRCA1-deficient cells to test whether the binding of RPA70 and Rad18 to chromatin are dependent on BRCA1. Consistent with poor monoubiquitination after UV irradiation, UV-induced chromatin binding of Rad18 and RPA70 is severely impaired in BRCA1-deficient cells (Fig. 2 D–F and Figs. S5B and S7). In contrast, the chromatin binding of other BRCA1-associated proteins, such as ATR, was not affected by the absence of BRCA1. These results suggest that BRCA1 interacts with Rad18 primarily through its RING domain and promotes the monoubiquitination of PCNA by actively recruiting both RPA70 and Rad18 to the vicinity of a replication-blocking DNA lesion.
BRCA1 Deficiency Affects the UV-Induced Foci Formation of DDT-Related Factors, Including RPA, Rad18, and TLS Polymerases Polη and REV1.
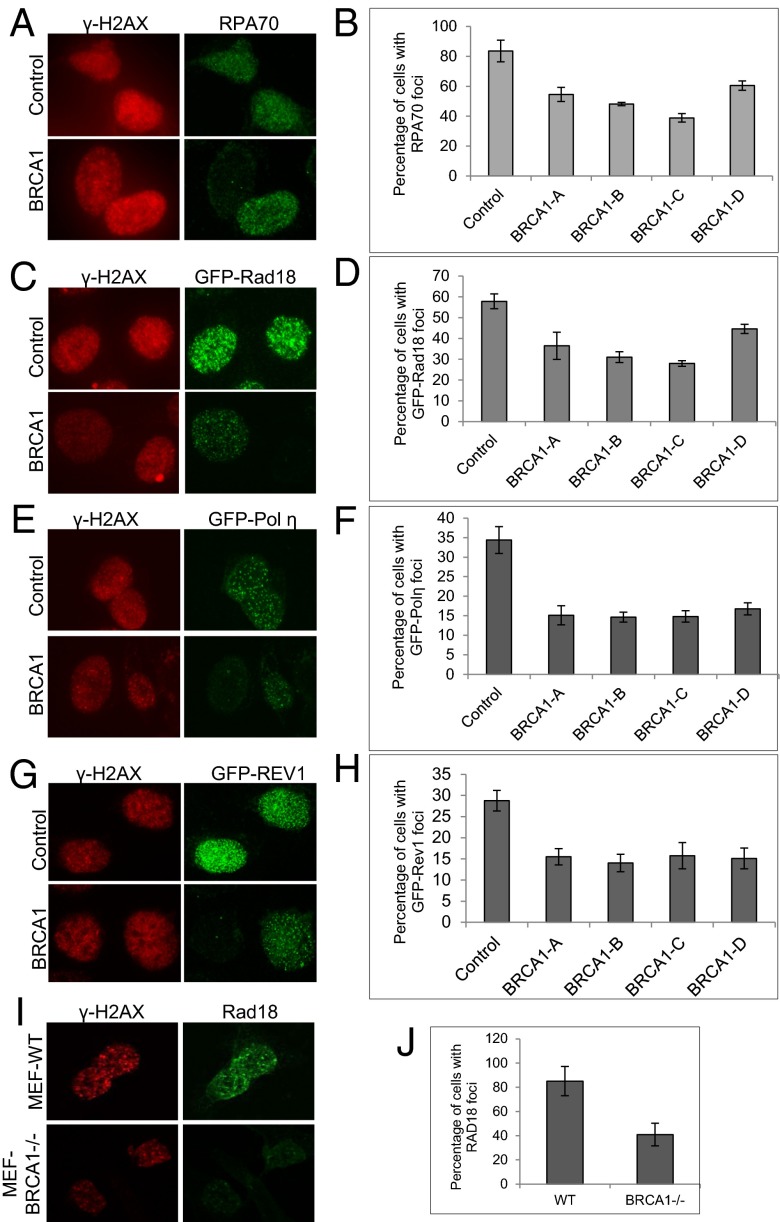
To gain further evidence supporting the role of BRCA1 in the activation of DDT, we performed UV-induced foci (UVIF) formation assays that directly monitor the recruitment of proteins to sites of DNA damage. Many DDR and DNA repair factors are recruited to the vicinity of stalled replication forks and form foci that can be detected by immunofluorescent microscopy. Interestingly, BRCA1, RPA70, Rad18, and Polη, which is the major TLS polymerase that bypasses UV induced lesions, can all form UVIF (Fig. S8). More important, the UVIF formed by RPA70, Rad18, and Polη colocalize nicely with those of BRCA1. Next we tested whether the formation of UVIF consisting of RPA70, Rad18, or Polη is dependent on BRCA1. We depleted endogenous BRCA1 using four individual siRNAs against BRCA1 (BRCA1-A to BRCA1-D) in U2-OS cells. As demonstrated in Fig. 3, BRCA1 deficiency severely impaired the formation of UVIF containing either RPA70 (Fig. 3 A and B and Fig. S9) or Rad18 (Fig. 3 C and D for U2-OS cells and Fig. 3 I and J for MEFs). Consistent with defective RPA70 and Rad18 UVIF formation and poor PCNA monoubiquitination, BRCA1 deficiency also impaired the recruitment of TLS polymerases Polη and REV1 into UVIF (Fig. 3 E and F for Polη and Fig. 3 G and H for REV1). Together with the chromatin fractionation data shown in Fig. 2, these results strongly indicate that BRCA1 functions upstream of RPA and Rad18 to regulate the monoubiquitination of PCNA and the subsequent recruitment of TLS polymerases.
Fig. 3.
UV-induced RPA70, Rad18, Polη, and REV1 containing foci are reduced in BRCA1-deficient cells. (A and B) siRNA-transfected U2-OS cells were treated with 60 J/m2 UVB and, 4 h later, fixed with methanol and stained with antibodies against γ-H2AX and RPA70. More than 100 cells with γ-H2AX-positive foci were counted. The percentage of cells with RAP70-positive foci were also counted and then normalized against cells containing γ-H2AX-positive foci. The results were then plotted. (C–H) siRNA-transfected U2-OS cells were infected with GFP-Rad18–expressing lenti-virus (C and D), GFP-Polη–expressing lenti-virus (E and F), or GFP-REV1–expressing lenti-virus (G and H). Forty-eight hours after infection, cells were treated with 60 J/m2 UVB and stained with antibody against γ-H2AX and GFP. Around 500 cells were counted, and the percentage of cells with GFP-positive foci were plotted and normalized against control siRNA-treated cells. (I and J) WT or BRCA1-deficient (BRCA1−/−) MEFs were treated with 60 J/m2 UVB and, 4 h later, stained with antibodies against γ-H2AX and mouse Rad18. More than 100 cells with γ-H2AX-positive foci were counted. The percentages of cells with Rad18-positive foci were plotted. All error bars are SD and were obtained from at least three independent experiments.
BRCA1 Actively Promotes the Recruitment of Rad18 and the TLS Polymerase, Polη, to UVIF in Addition to RPA.
The binding of RPA to ssDNA is thought to be an earlier event during the activation of DDT (12, 18). Likely through a direct protein–protein interaction, RPA70 recruits Rad18 to ssDNA associated with replication fork stalling, where Rad18 then initiates the ubiquitination of PCNA and the DDT pathway. TLS polymerases such as Polη and REV1 are recruited to these sites by binding to the monoubiquitinated form of PCNA through their ubiquitin-binding domains (28). In addition, some TLS polymerases, such as Polη, can interact with PCNA through an ubiquitination-independent fashion through their PCNA-interacting peptide (29), suggesting that TLS polymerases can be recruited to the lesions through multiple ways. Because BRCA1 colocalizes with Rad18 and Polη (Fig. S8) and affects their recruitment to DNA lesions (Fig. 3), we tested how much of a role RPA and BRCA1 play in terms of the recruitment of Rad18 and Polη. Meiotic recombination 11 homolog A (Mre11) is one of the upstream nucleases that are capable of generating ssDNA, thus promoting the binding of RPA. Indeed, a chemical inhibitor of Mre11 (30), Mirin, dramatically reduced the formation of RPA70 UVIF, suggesting that Mre11 is one of the key nucleases promoting the ssDNA formation in response to UV (Fig. S10A). Depletion of BRCA1 with siRNA in Mirin-treated cells further reduces the RPA70 UVIF, suggesting that BRCA1 either helps recruit additional nucleases to UV lesions or actively recruits RPA to the ssDNA itself. Much to our surprise, inhibition of Mre11 alone affects neither the UVIF of Rad18 (Fig. S10B) nor the UVIF of Polη (Fig. S10C). Moreover, simultaneous inactivation of both Mre11 and BRCA1 has the same effect on the UVIF formation of both Rad18 and Polη, as does depletion of BRCA1 alone (Fig. S10 B and C), suggesting that BRCA1 may use additional mechanisms to recruit Rad18 and Polη to the lesions, for example, through protein–protein interactions. In Fig. 2, we have demonstrated that BRCA1 is capable of interacting with Rad18. We therefore reasoned that BRCA1 may also directly recruit the TLS polymerases through protein–protein interactions. Indeed, we detected strong binding between BRCA1 and Polη, as well as BRCA1 and REV1 (Fig. S10 D and E). Taken together (Fig. 2 and Fig. S10), our data suggest that BRCA1 may play multiple roles during DDT. BRCA1 actively recruits the E3 ligase Rad18 to the vicinity of lesions by affecting the recruitment of RPA and promoting the monoubiquitination of PCNA. In addition, BRCA1 actively recruits TLS polymerases to the UV-induced lesions through protein–protein interactions.
BRCA1 Shows Strong Overlapping Interactions with Factors Involved in DDT in Response to UV, CDDP, and MMC.
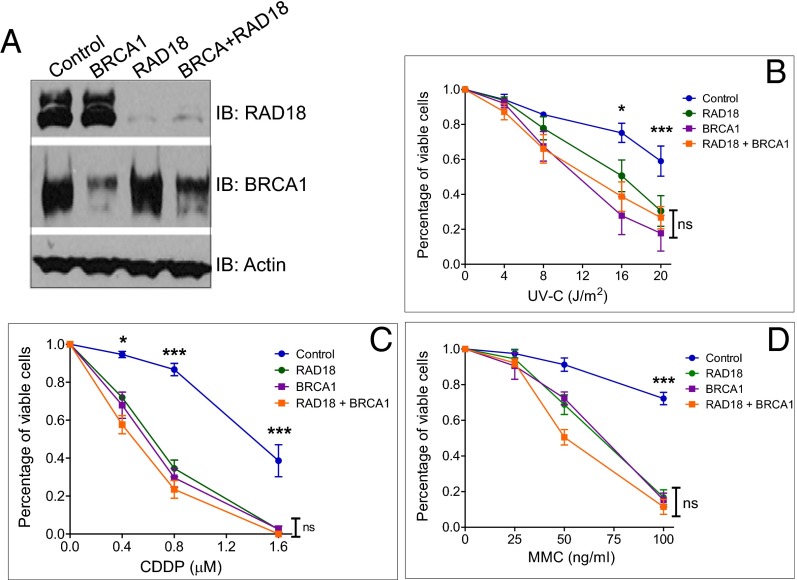
To further prove that BRCA1 acts upstream of Rad18 during DDT, we performed the cell viability assay as described in Fig. 1A. We first transfected U2-OS cells with siRNA against BRCA1, Rad18 alone, or in combination; exposed cells to UV, CDDP, or MMC; and then quantitated relative cell survival after 7–10 d. Consistent with BRCA1 being important for regulating RAD18 activity, we observed strong overlapping interactions between BRCA1 and Rad18 when cells were treated with UV, CDDP, or MMC, as demonstrated by the lack of further sensitization to DNA damage when both proteins were knocked down simultaneously (Fig. 4).
Fig. 4.
BRCA1 and Rad18 show strong overlapping interactions in response to UVC, CDDP, and MMC treatment. (A) U2-OS cells were transfected with either control siRNA or siRNA targeting BRCA1, Rad18, or both. WCL was used for immunoblotting. (B–D) U2-OS cells were transfected with either control siRNA or siRNA targeting BRCA1, Rad18, or both and then treated with different doses of UVC (B), CDDP (continuous) (C), or MMC (2 h) (D). The percentage of viable cells was determined as in Fig. 1. Data represent the mean ± SEM from at least three independent experiments. ns, not significant. *P < 0.05; ***P < 0.001; one-way ANOVA.
Both the RING Domain and the BRCT Domain Are Crucial for the DDT Function of BRCA1.
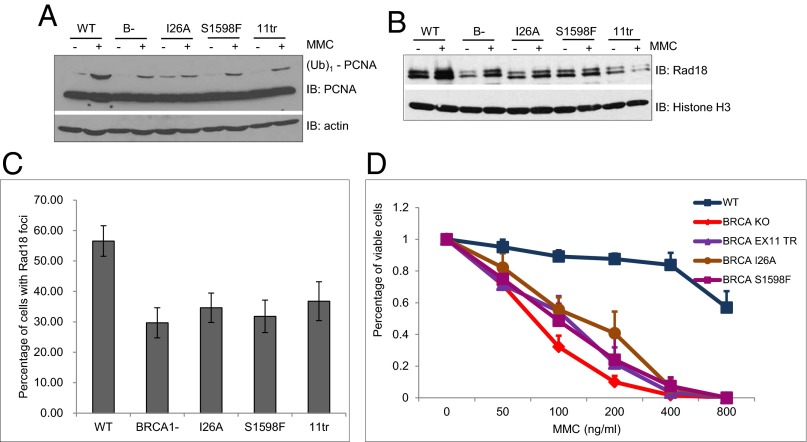
BRCA1 contains two prominent domains: an N-terminal RING domain and two tandem C-terminal BRCT domains. The RING domain is often found in E3 ubiquitin ligases and functions to recruit an E2, ubiquitin-conjugating enzyme to facilitate the ubiquitination reaction. BRCA1 is capable of ubiquitinating itself and several other potential substrates (31). The BRCT domain is a phospho-peptide-binding domain and mediates protein–protein interactions (3). A commonly used BRCA1 RING domain mutant, I26A, impairs its binding to E2 but is still able to bind its partner, BARD1 (32). A mouse BRCA1 BRCT domain mutant, S1598F (corresponding to the S1655 of human BRCA1), disrupts its phospho-peptide binding activity (33). Previously, Ludwig and coworkers generated knock-in mice expressing I26A or S1598F mutant BRCA1 (34). In addition, they also created mice expressing a truncated form of BRCA1 after amino acid 924 (11tr), which lacks both BRCT domains. We first examined whether the RING domain or BRCT domains are important for the monoubiquitination of PCNA when cells were treated with MMC. Interestingly, I26A and S1598F, as well as the 11tr BRCA1 mutant cell lines, all exhibited reduced monoubiquitination of PCNA in response to MMC treatment, almost to the same extent as cells lacking BRCA1 (Fig. 5A). Similar to the BRCA1 knockout line, MEFs expressing I26A or S1598F BRCA1 protein also showed impaired Rad18 chromatin binding, as well as reduced foci formation of Rad18 after MMC treatment (Fig. 5 B and C). Consistent with the defective DDT, I26A, S1598F, and 11tr MEFs are also sensitive to MMC (Fig. 5D), although somewhat less than the BRCA1 knockout MEFs. Together, these data indicate that both the RING domain and the BRCT domain of BRCA1 are critical for its function in DDT.
Fig. 5.
Both the RING domain and the BRCT domain of BRCA1 are important for the DNA damage tolerance pathway. (A) MMC-induced monoubiquitination of PCNA is defective in both RING domain and BRCT domain mutants. WT, BRCA1-deficient (B-), BRCA1 RING mutant (I26A), BRCA1 BRCT mutant (S1598F), or BRCA1 exon-11 truncation mutant (11tr) MEFs were either left untreated (−) or were treated with MMC overnight and then collected for immunoblotting. (B) Chromatin association of Rad18 is defective in BRCA1 mutants. MEF cells either were left untreated (−) or were treated with MMC overnight and then collected for chromatin fractionation. (C) Rad18 foci formation is defective in BRCA1 mutants. MEF cells were either left untreated (−) or were treated with MMC overnight, then stained with antibody against γ-H2AX and Rad18. More than 100 cells with γ-H2AX-positive foci were counted. The percentage of cells with Rad18-positive foci was plotted. All error bars are SD and were obtained from at least three independent experiments. (D) All four BRCA1 mutants are sensitive to MMC. WT or BRCA1 mutant MEFs were treated with MMC for 2 h, and the percentage of viable cells was determined as in Fig. 1. Data represent the mean ± SEM from at least three independent experiments.
BRCA1 Affects Template Switching by Promoting the Chromatin Binding of the E3 Ligase for the Polyubiquitination of PCNA, HLTF in Response to UV.
BRCA1 is best known to suppress genome instability by promoting the repair of DSBs through the error-free HR pathway (2, 3). We tested whether BRCA1 also promotes the error-free DDT pathway through template switching by influencing the polyubiquitination of PCNA. Genetic studies in budding yeast have established essential roles for UBC13, MMS2, and Rad5 during the template switching process (35). Their corresponding mammalian homologs are hUBC13, hMMS2, and two potential Rad5 orthologs, HLTF and SHPRH. Cimprich and coworkers recently demonstrated that HLTF mainly functions in response to UV, whereas SHPRH functions in response to methyl methanesulfate, indicating that the players involved in this pathway can be recruited in a lesion-specific manner (36).
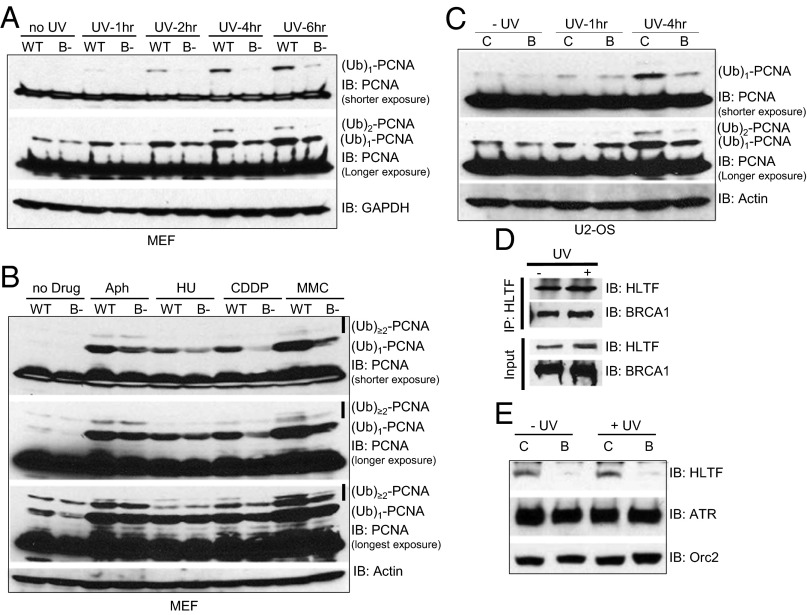
Using the BRCA1-deficient cellular systems described earlier, we observed defective diubiquitinated [labeled as (Ub)2-PCNA] and polyubiquitinated [labeled as (Ub)>2-PCNA] PCNA formation in response to UV, CDDP, and MMC. We observed inefficient polyubiquitination in BRCA1 knockout MEFs (Fig. 6 A and B) and BRCA1 siRNA-treated U2-OS cells (Fig. 6C) in response to UV, CDDP, or MMC. We also detected strong binding between BRCA1 and endogenous HLTF, the E3 ligases responsible for UV-induced K63-linked polyubiquitination of PCNA (Fig. 6D). Most important, the chromatin association of HLTF is largely dependent on BRCA1 in U2-OS cells (Fig. 6E and Fig. S7). These data suggest that in addition to promoting the chromatin recruitment of Rad18, which catalyzes the monoubiquitination of PCNA, BRCA1 may also be involved in the polyubiquitination of PCNA. Consistent with its critical roles in the maintenance of genome stability, our data indicate that BRCA1 also promotes the activation of the error-free branch of DDT by facilitating the efficient recruitment of the HLTF E3 ligase to stalled replication forks, where it polyubiquitinates PCNA and activates template switching.
Fig. 6.
BRCA1 deficiency reduces genotoxic agents induced polyubiquitination of PCNA. (A–C) Lysates from Fig. 1 were run on a 4–12% SDS/PAGE and blotted for PCNA. GAPDH or Actin is used as the loading control. Blots were exposed longer to visualize the polyubiquitinated forms of PCNA, labeled as (Ub)2-PCNA and (Ub)>2-PCNA. (D) UV enhances the interaction between BRCA1 and HLTF. 293T cells were either left untreated (−) or were treated with 60 J/m2 UVB (+). Four hours later, cells were collected and lysed. Equal amounts of WCL were used for IP with antibody against HLTF. Normal rabbit IgG was used as a negative control. (E) U2-OS cells were first transfected with either control siRNA (C) or siRNA against BRCA1 (B). The cells were then either left untreated (−UV) or treated with 60 J/m2 UVB (+UV). Chromatin fractionations were done as in Fig. 2, and chromatin fractions were blotted with antibodies (Right).
Discussion
Here we have demonstrated a function of BRCA1 in DDT. We showed that BRCA1 is necessary for both efficient mono- and polyubiquitination of PCNA. BRCA1 exerts these functions by actively recruiting or stabilizing RPA interactions at stalled replication forks and subsequent recruitment of the E3 ligases necessary for both the mono- and polyubiquitination of PCNA. In addition, BRCA1 physically interacts with multiple DDT factors that mediate PCNA ubiquitination (Rad18 and HLTF) and perform lesion bypass (Polη and REV1), suggesting that the function of BRCA1 during DDT is most likely direct. Furthermore, our data, shown in Fig. 1, suggest that BRCA1 is necessary for proper activation of the DDT response to bona fide DNA lesions, such as those created by UV, CDDP, and MMC, but is dispensable for a proper DDT response to direct DNA polymerase inhibition (e.g., Aph) and deoxynucleotide imbalances (HU). This unexpected finding suggests that physical barriers to DNA replication, such as intra-and interstrand DNA cross-links, are likely sensed, bypassed, and repaired differently (36).
Although the basic principles underlining the DNA damage tolerance pathway are evolutionarily conserved, recent studies suggest its regulation is much more complex in mammals. For example, more mammalian factors have been identified as regulating the monoubiquitination of PCNA, including p21, p53, PTIP/Swift, Chk1, Claspin, Timeless, and Tipin (14). Furthermore, Zou and coworkers have shown that Claspin can regulate the chromatin binding of Rad18 while Chk1 indirectly regulates the monoubiquitination of PCNA by stabilizing the protein level of Claspin (37). How Claspin regulates the recruitment of Rad18 is not known; however, an interaction between Claspin and the RPA heterotrimer has been reported (38). An interaction between Claspin and BRCA1 was previously demonstrated by Elledge and coworkers, and this interaction is also enhanced by DNA damage (39). Whether BRCA1 and Claspin act in a linear or parallel pathway in the regulation of ubiquitination of PCNA needs further investigation. Our data and other previous data unambiguously put BRCA1 at the center of DDR, DDT, and DNA repair, underscoring its essential function in the maintenance of genomic stability and the suppression of tumorigenesis.
Materials and Methods
Plasmids, siRNA, and Chemicals.
The following siRNAs used in this article were purchased from DHARMACON. ON-TARGETplus siCONTROL Nontargeting pool (D-001810-10-20) was used as a negative control for all siRNA transfections. BRCA1 in U2-OS and MCF7 cells was depleted with ON-TARGETplus SMARTpool siRNA against BRCA1 (L003461-00-0020) or four single ON-TARGETplus siRNAs (LU-003461-00-0002): BRCA1-A (CAACAUGCCCACAGAUCAA), BRCA1-B (CCAAAGCGAGCAAGAGAAU), BRCA1-C (UGAUAAAGCUCCAGCAGGA), and BRCA1-D (GAAGGAGCUUUCAUCAUUC). The RAD18 siRNA was purchased from Qiagen (ATGGTTGTTGCCCGAGGTTAA) (40). Human cells were transfected with 50 nM siRNA twice, using RNAiMAX (Invitrogen). All chemicals were purchased from Sigma.
Supplementary Material
Acknowledgments
We thank Lee Zou, Karlene Cimprich, and Satoshi Tateishi for reagents and Robin Miskimins for critical reading of the manuscript. This research was supported by the generous starting package from the Sanford School of Medicine (to D.Z.), Sidney Kimmel Foundation and MD Anderson startup fund (to B.W.) and a grant from the National Institutes of Health (CA133046; to C.E.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306534110/-/DCSupplemental.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donovan PJ, Livingston DM. BRCA1 and BRCA2: Breast/ovarian cancer susceptibility gene products and participants in DNA double-strand break repair. Carcinogenesis. 2010;31(6):961–967. doi: 10.1093/carcin/bgq069. [DOI] [PubMed] [Google Scholar]
- 3.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11(2):138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi T, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109(4):459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275(31):23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 7.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61(12):4842–4850. [PubMed] [Google Scholar]
- 8.Quinn JE, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63(19):6221–6228. [PubMed] [Google Scholar]
- 9.Bunting SF, et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46(2):125–135.
- 10.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14(4):491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 11.Albertella MR, Green CM, Lehmann AR, O’Connor MJ. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 2005;65(21):9799–9806. doi: 10.1158/0008-5472.CAN-05-1095. [DOI] [PubMed] [Google Scholar]
- 12.Niimi A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105(42):16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang DJ, Cimprich KA. DNA damage tolerance: When it’s OK to make mistakes. Nat Chem Biol. 2009;5(2):82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8(4):461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol Cells. 2008;26(1):5–11. [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283(5404):1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 17.McCulloch SD, et al. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428(6978):97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 18.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29(5):625–636. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unk I, et al. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA. 2008;105(10):3768–3773. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unk I, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2006;103(48):18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motegi A, et al. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175(5):703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA. 2008;105(34):12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanagihara H, et al. NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol η-dependent translesion DNA synthesis. Mol Cell. 2011;43(5):788–797. doi: 10.1016/j.molcel.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 24.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17(2):201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20(1):34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei L, et al. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol. 2008;28(24):7380–7393. doi: 10.1128/MCB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16(2):198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310(5755):1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 29.Haracska L, et al. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol. 2001;21(21):7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupré A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4(2):119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu W, Koike A, Takeshita T, Ohta T. The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell Div. 2008;3:1. doi: 10.1186/1747-1028-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brzovic PS, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA. 2003;100(10):5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clapperton JA, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11(6):512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 34.Shakya R, et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334(6055):525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unk I, Hajdú I, Blastyák A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9(3):257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol Cell. 2011;42(2):237–249. doi: 10.1016/j.molcel.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22(9):1147–1152. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakaya R, et al. Identification of proteins that may directly interact with human RPA. J Biochem. 2010;148(5):539–547. doi: 10.1093/jb/mvq085. [DOI] [PubMed] [Google Scholar]
- 39.Lin SY, Li K, Stewart GS, Elledge SJ. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc Natl Acad Sci USA. 2004;101(17):6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hicks JK, et al. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30(5):1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.