Abstract
Synonymous mutations, which do not alter the protein sequence, have been shown to affect protein function [Sauna ZE, Kimchi-Sarfaty C (2011) Nat Rev Genet 12(10):683–691]. However, synonymous mutations are rarely investigated in the cancer genomics field. We used whole-genome and -exome sequencing to identify somatic mutations in 29 melanoma samples. Validation of one synonymous somatic mutation in BCL2L12 in 285 samples identified 12 cases that harbored the recurrent F17F mutation. This mutation led to increased BCL2L12 mRNA and protein levels because of differential targeting of WT and mutant BCL2L12 by hsa-miR-671–5p. Protein made from mutant BCL2L12 transcript bound p53, inhibited UV-induced apoptosis more efficiently than WT BCL2L12, and reduced endogenous p53 target gene transcription. This report shows selection of a recurrent somatic synonymous mutation in cancer. Our data indicate that silent alterations have a role to play in human cancer, emphasizing the importance of their investigation in future cancer genome studies.
Systematic melanoma whole-exome and -genome studies have uncovered numerous recurrent mutations as well as highly mutated genes that show functional consequences on melanoma growth (1–6). These studies focus exclusively on coding mutations and specifically on nonsynonymous mutations, insertion/deletion mutations as well as splice sites. Recently, noncoding mutations in the telomerase reverse transcriptase (TERT) promoter have been shown to generate new E-twenty-six (ETS) transcription factors binding motifs, leading to increased expression of telomerase reverse transcriptase (7, 8). These studies highlight the importance of adjusting our focus beyond the nonsynonymous coding mutations and evaluating all mutations in melanoma.
To gain additional insight into the molecular alterations of melanoma, we report the sequence analysis of 29 melanoma samples and corresponding normal DNA. We performed whole-genome sequencing on 10 matched normal and metastatic tumor DNAs and reanalyzed a previously published melanoma whole-genome study (9, 10). Together with our previous whole-exome analysis of 14 melanoma samples (1) and an additional whole-exome analysis of four matched melanoma and normal samples, this study allows for an unbiased search for unique melanoma genes in a total of 29 samples from treatment naïve patients.
Results
In combined analysis, 13,098 somatic mutations were identified in gene coding regions. Of these mutations, 8,619 caused protein changes, including 7,974 missense, 514 nonsense, 27 small deletion, 11 insertion, and 93 splice site mutations. There were 4,479 silent (synonymous) substitutions (Dataset S1). A nonsynonymous to synonymous ratio of 1.93:1 was calculated, which is not higher than the nonsynonymous to synonymous ratio of 2.5:1 predicted for nonselected mutations (11), suggesting that most are likely passenger mutations. The number of C > T/G > A transitions was significantly greater than other nucleotide substitutions (P < 0.001) (SI Appendix, Fig. S1), which is consistent with a UV radiation (UVR) signature (12).
Recurrent nonsynonymous mutations, including v-raf murine sarcoma viral oncogene homolog B1 (BRAF) V600E and transformation/transcription domain-associated protein (TRRAP) S722F substitutions, were found (1, 13) as well as 16 recurrent synonymous mutations (Table 1). Although synonymous mutations do not alter the protein sequence, they have been shown to affect protein levels and function (14, 15). However, to date, synonymous mutations have not been investigated in numerous published cancer genomes. We sought to determine whether these somatic synonymous mutations have a functional role in melanomagenesis. Additional screening of these 16 synonymous hotspot mutations in an additional 169 melanoma samples identified olfactory receptor family 4 subfamily C, member 3 (OR4C3) and BCL2L12 (SI Appendix, Fig. S2) each to have identical synonymous mutations in three and four additional cases, respectively. The frequency of these recurrent alterations in the validation sample is significantly elevated (P < 1 × 10−7 and P < 1 × 10−11), suggesting that they have either undergone relaxation of purifying selection (16) or been under selection during tumor development. Because BCL2L12 has previously been linked to tumorigenesis (17), we screened the BCL2L12 cytosine to thymine change at position 51 (F17F) in another 87 melanoma samples. This screen identified six additional samples with the same alteration. This mutation, thus, occurred in 10 of 256 melanomas (P < 1 × 10−31) in the combined validation study, strongly suggesting that it has a functional role in melanomagenesis. Consistent with this expectation, this nucleotide position displays evidence of selection (SI Appendix, Fig. S3), suggesting that sequence variation at this site is not well-tolerated.
Table 1.
Recurrent synonymous mutations in analysis of 29 melanoma tumors
| Gene name | Ref_seq ID | Nucleotide change | Amino acid change | Tumor name |
| FCRL1 | NM_052938.4 | C741T | I247I | 96T |
| 91T | ||||
| OR2T6 | NM_001005471.1 | C339T | F113F | 7T |
| 32T | ||||
| PNLIPRP1 | NM_006229.2 | C600T | F200F | 32T |
| 55T | ||||
| OR4C3 | NM_001004702.1 | C114T | F38F | 17T |
| 108T | ||||
| OR8J3 | NM_001004064.1 | C186T | F62F | 55T |
| 01T | ||||
| CPT1A | NM_001876.3 | C1638T | F546F | 05T |
| 43T | ||||
| DNAH9 | NM_001372.3 | C6333T | F2111F | 24T |
| 01T | ||||
| BCL2L12 | NM_138639.1 | C51T | F17F | 55T |
| 81T | ||||
| PNKP | NM_007254.3 | C75T | P25P | 32T |
| 56T | ||||
| TTN | NM_133378.4 | C10167T | F3389F | 130T |
| 23T | ||||
| POTED | NM_174981.3 | G864A | V288V | 12T |
| 26T | ||||
| GTSE1 | NM_016426.6 | C1782T | S594S | 24T |
| 55T | ||||
| OR5H6 | NM_001005479.1 | C654T | F218F | 24T |
| Colo-829 | ||||
| FILIP1 | NM_015687.2 | G2475A | R825R | 17T |
| 24T | ||||
| PPP1R3A | NM_002711.3 | G2844A | T948T | Colo-829 |
| 51T | ||||
| COL14A1 | NM_021110.1 | G4050A | R1350R | 23T |
| 32T |
Identification of 16 recurrent mutations and their effects on their transcripts. All mutations were validated through Sanger sequencing and subsequently evaluated in additional cohorts of melanoma.
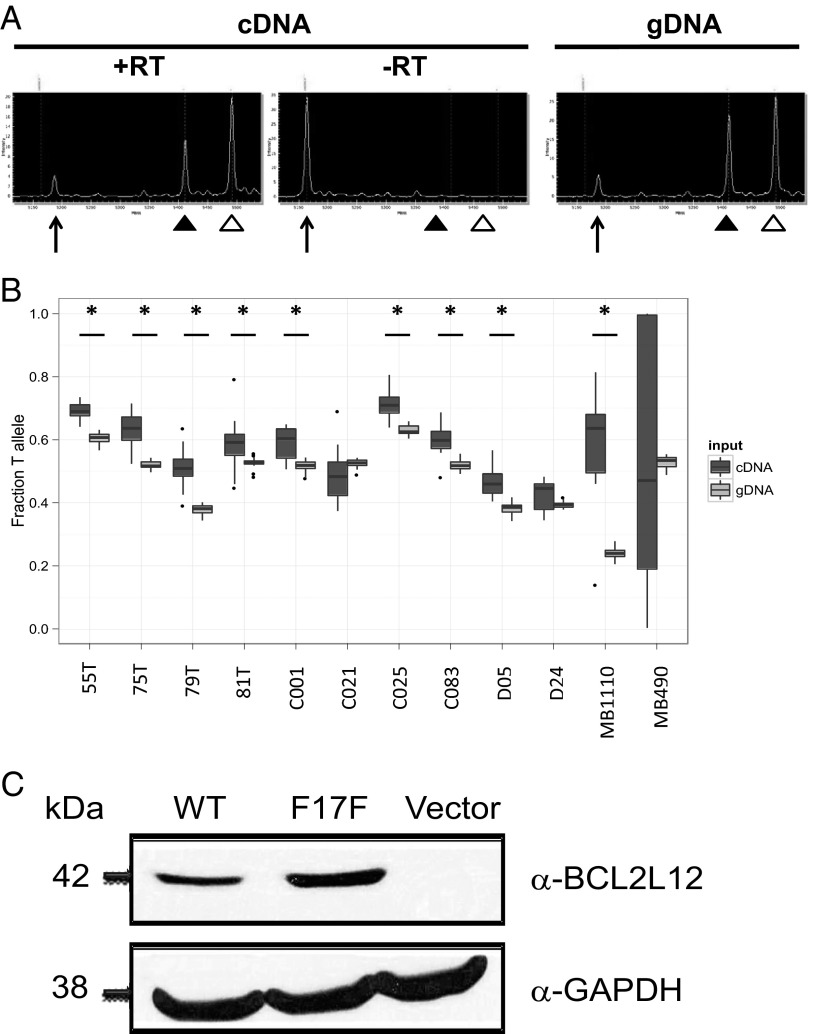
Synonymous mutations have been shown to affect gene function by multiple mechanisms, including but not limited to those mechanisms exerting effects on mRNA splicing, protein translation, and expression (18). Our analyses suggest that the synonymous alteration in BCL2L12 does not affect splicing, because the mutation does not create a guanine thymine (GT) splicing consensus dinucleotide that could compete with the donor splice site of the first exon or encourage the use of seven cryptic GT splice donor sites within its vicinity (SI Appendix, Table S1). Next, we determined whether BCL2L12 allelic expression is affected by the mutation by comparing the levels of mutant and WT BCL2L12 alleles. We used MALDI-TOF (Sequenom) analysis to quantitatively assess relative allelic abundance in paired cDNA and genomic DNA (gDNA) from melanoma samples and found that, for 9 of 12 samples, the mutant BCL2L12 T allele was more abundantly expressed than the WT C allele (P < 0.01, Wilcoxon rank sum test) (Fig. 1 A and B). To test if the protein made from mutant BCL2L12 transcript is expressed more abundantly than WT BCL2L12, we constructed WT and mutated versions of BCL2L12 cDNA and transiently transfected them. We found that the mRNA (SI Appendix, Fig. S4) and protein levels (Fig. 1C) of mutant BCL2L12 were significantly increased relative to WT in multiple independent cotransfection experiments using GFP to control for transfection efficiency.
Fig. 1.
Abundance of the BCL2L12 transcript and BCL2L12 protein. (A) Image of MALDI-TOF (Sequenom) spectrographs indicating peaks for unextended primer (arrows), C allele (filled arrowheads), and T allele (open arrowheads) for sample 55T cDNA. Unextended primer peak in the −RT control confirms that allelic representation differences in cDNA samples are not because of gDNA contamination. Paired gDNA from each sample was used as a control. (B) Box plots show significantly higher T-allele representation in cDNA (dark gray) compared with gDNA (light gray) in 9 of 12 melanoma samples. Significance was calculated from 12 measurements of each cDNA and gDNA sample using the Wilcoxon rank sum test; asterisks indicate samples with P < 0.01. (C) BCL2L12 protein levels in transiently transfected HEK293T cells (Western blot analysis). Cells were transiently transfected with BCL2L12 WT or mutant cDNA, and Western blotting was done posttransfection.
There could be a number of reasons leading to the elevated BCL2L12 protein expression levels: (i) increased mRNA levels (noticed above), (ii) enhanced mRNA translation, (iii) stabilization of protein against degradation, or (iv) all of the above. We, however, found no change in elongation/translation rates of the mutant BCL2L12 mRNA compared with the WT message (SI Appendix, Fig. S5) and no change in stability of protein (expressed in vivo and in vitro) to limited proteolysis. These results largely rule out the F17F mutation having effects on BCL2L12 translation and protein stability.
The elevated levels of mutant BCL2L12 mRNA could be caused by increased transcription or increased RNA stability. The position corresponding to the mutation in BCL2L12 displays high conservation across the mammalian lineage, suggesting functional constraints other than purely amino acid encoding (SI Appendix, Fig. S6A). However, position weight matrix scanning and ChIP analyses provide no support for a mechanism involving preferential binding of expressed transcription factors to the WT or mutated BCL2L12 alleles (SI Appendix, Fig. S6 B–E). Increased stability of the mutant BCL2L12 mRNA could be caused by differential binding of protein or microRNA (miRNA) to mutant and WT BCL2L12 mRNA. Computational analysis showed that several RNA binding proteins may interact with WT and mutant mRNAs in the region close to the site of mutation. However, gel-shift experiments of top candidate proteins did not reveal any differential binding between the two mRNAs (SI Appendix, Fig. S7).
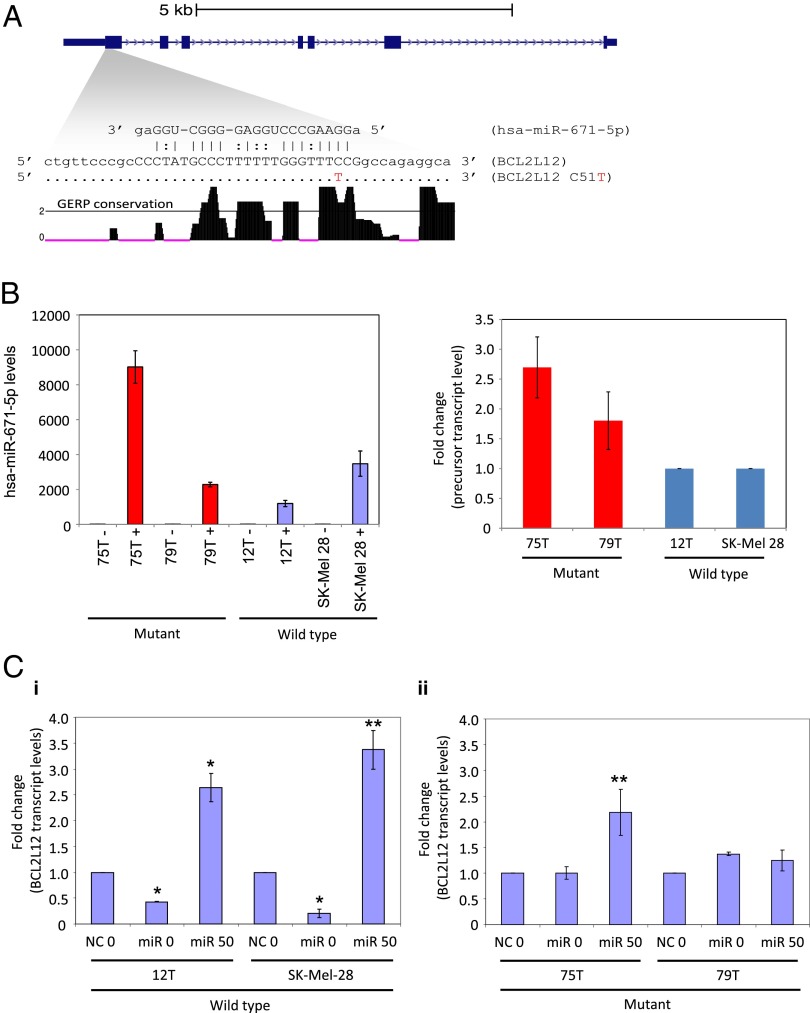
Finally, to evaluate if the mutation affects miRNA binding, we used the miRNA target prediction programs PITA (19) and miRanda (20). A single miRNA (miR) common to both programs, hsa-miR-671–5p, was predicted to bind the WT but not mutant BCL2L12 transcripts. The miRNA target site in its WT form has high complementarity to mature hsa-miR-671–5p. Furthermore, Genome Evolutionary Rate Profiling (GERP) analysis (21), which identifies evolutionarily constrained positions in multiple genome alignments by quantifying substitution deficits across species, indicates that the target region exhibits high sequence conservation (Fig. 2A) (22, 23). We hypothesized that loss of this target site in mutant BCL2L12 may lead to increased BCL2L12 transcript levels. hsa-miR-671–5p has been shown previously to be expressed in melanoma (24). Before targeting endogenous BCL2L12 with hsa-miR-671–5p, we used quantitative RT-PCR (qRT-PCR) analysis to detect the presence of transiently transfected miR mimic in melanoma cell lines (Fig. 2B). Furthermore, to show specificity of the miR mimic to target WT BCL2L12, we cotransfected WT or mutant BCL2L12 melanoma cell lines with negative control miR or hsa-miR-671–5p mimic in the presence of a specific miR inhibitor (anti–hsa-miR-671–5p). qRT-PCR analysis shows that anti-miR inhibited and reversed the effect on WT BCL2L12 message by hsa-miR-671–5p. In mutant cell lines, little to no effect was observed (Fig. 2C). The suppression of mature hsa-miR-671–5p mimic by cotransfection with anti–hsa-miR-671–5p was determined by qRT-PCR analysis (SI Appendix, Fig. S8). Our results indicate that WT BCL2L12 mRNA is a target for hsa-miR-671–5p regulation, which leads to its steady state reduction. However, the recurrent BCL2L12 mutation reduces the affinity of hsa-miR-671–5p binding, thus allowing mutant BCL2L12 mRNA and protein accumulation.
Fig. 2.
hsa-miR-671–5p represses WT BCL2L12 expression. (A) Schematic representation of the BCL2L12 locus at hg18 coordinates chr19:54860211–54868985. Based on miRanda and PITA target scanning predictions, hsa-miR-671–5p binds at the first coding exon and has high affinity to the WT version but not the C51T version. GERP single nucleotide resolution evolutionary conservation scores show that this region is highly conserved. The horizontal line at GERP score = 2 indicates the general threshold that defines evolutionarily constrained bases. For this plot, we only show GERP scores ≥0. (B) qRT-PCR analysis of precursor hsa-miR-671 in melanoma cells. Graphs show experimental replicates of qRT-PCR analysis of precursor hsa-miR-671 in mutant BCL2L12 (75T and 79T) compared with WT BCL2L12 (12T and SK-Mel-28) cells. Results are representative of two independent experiments. Error bars are SD. (C) Anti–miR-671–5p rescues hsa-miR-671–5p–mediated knockdown of WT BCL2L12 in melanoma cells. Graphs show experimental replicates of qRT-PCR of endogenous BCL2L12 levels in (i) wild type BCL2L12 (12T and Sk-Mel-28) and (ii) mutant BCL2L12 (75T and 79T) cell lines in the presence of negative control miR (NC) or hsa-miR-671–5p (miR) plus 0 or 50 nM anti–miR-671–5p. Results are representative of two independent experiments. Error bars are SD. Comparison of NC0 with miR0 (*P < 0.01) or comparison of miR0 with miR50 (**P < 0.04; Student t test).
BCL2L12 was previously shown to be amplified in glioblastoma, to bind p53, and to inhibit apoptosis (17). Together with our identification of a BCL2L12 hotspot mutation that increases BCL2L12 expression levels, this finding suggests BCL2L12 to be a candidate unique melanoma oncogene. We, therefore, investigated whether the identified BCL2L12 C51T mutation may affect apoptosis.
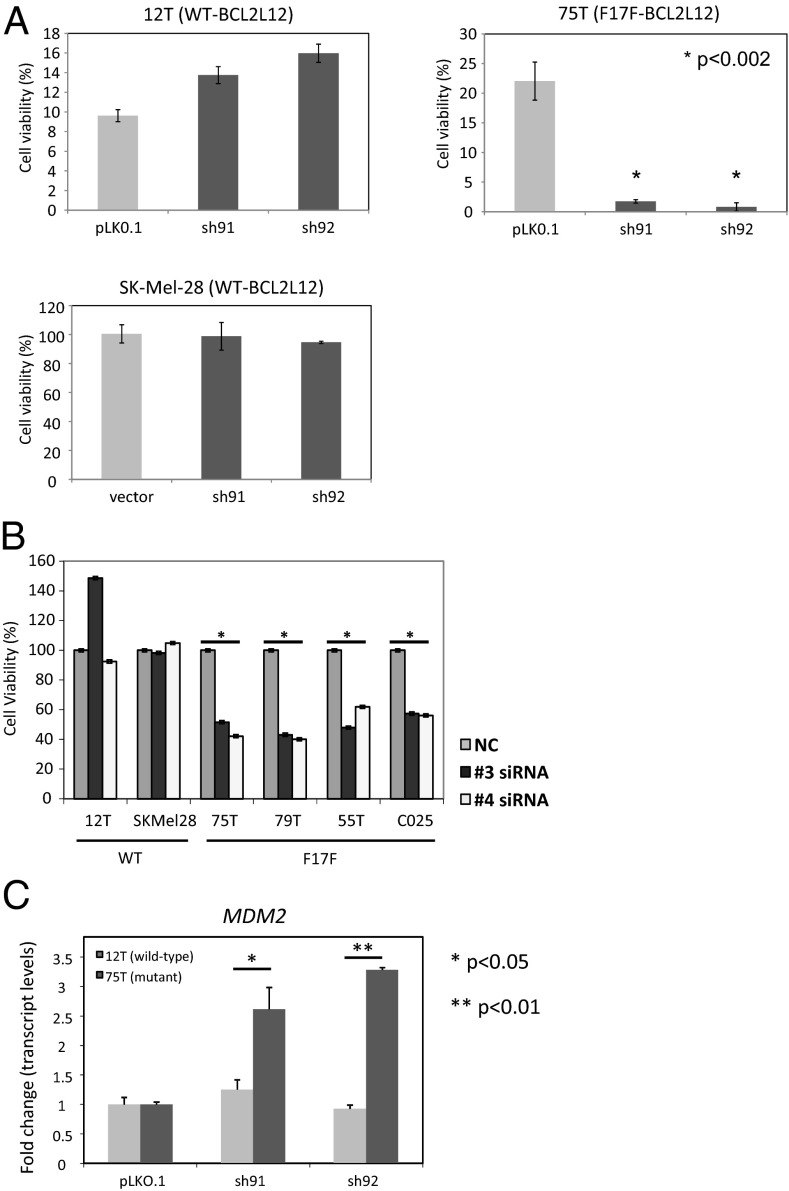
As a first step in assessing this possibility, we confirmed that the mutation does not interfere with p53 binding. Efficient complex formation between endogenous p53 and overexpressed protein transcribed from either WT or mutant BCL2L12 transcript was seen in HEK293 cells (SI Appendix, Fig. S9). The observed interaction with p53, together with the enhanced expression of protein made from mutant BCL2L12 transcript, may repress p53 activity. This repressed activity may lead to an increased ability of melanoma cells to resist p53-dependent induced apoptosis. To directly test this possibility, we assessed the BCL2L12 antiapoptotic activity after genotoxic stress in melanoma cells that harbor either WT or mutant BCL2L12. We used shRNA or siRNA to knockdown BCL2L12 expression in BCL2L12 WT (12T and SK-Mel-28) or mutant [75T, 79T (specifically siRNA for 55T), and C025] cells (SI Appendix, Figs. S10 and S11). In each case, the knockdown had little to no effect on cells harboring WT BCL2L12 but significantly reduced the viability of cells harboring mutant BCL2L12 post-UVR exposure (Fig. 3 A and B). Because BCL2L12 has previously been shown to affect the expression of p53-dependent target genes (17), we tested if depletion of mutant BCL2L12 in melanoma cells results in differential activation of p53-dependent transcription on UVR treatment compared with BCL2L12 depletion in WT-expressing cells. Indeed, we observed significant increases in p53-dependent transcription of MDM2 in mutant cells stably depleted of BCL2L12 compared with stably depleted WT cells after UVR exposure (Fig. 3C). Our results show that synonymous somatic mutations have important roles to play in cancer and also suggest the potential for the prosurvival nature of BCL2L12 in melanoma.
Fig. 3.
Effects of the BCL2L12 (C51T; F17F) recurrent mutation on BCL2L12 function. (A) Graphical representation showing the antiapoptotic effect of BCL2L12 mutant cells compared with WT BCL2L12 cells post-UV treatment. The relative cell numbers after the cells were treated for 48 h with UV as estimated by CellTiter-Glo and plotted as percent survival. Error bars are SD. (B) Melanoma cells (WT, 12T or SK-Mel-28; mutant, 75T, 79T, 55T, or C025) transiently transfected with BCL2L12-specific siRNA were tested for sensitivity to UV-induced cell death. Shown are representative graphs from all cell lines exposed to 50 kμJ UV light. Results were analyzed using Microsoft Excel and GraphPad Prism v5, and graphs are representative of experimental replicates. Error bars are SD (*P < 0.04 comparing siRNA with NC). (C) qRT-PCR analysis shows that depletion of mutant forms of BCL2L12 using specific shRNA increases p53-dependent target gene expression compared with depletion of WT BCL2L12. Graphs show qRT-PCR analysis of WT BCL2L12 (12T) and mutant BCL2L12 (75T) pooled clone mRNA expression levels for Mdm2. Results shown are experimental replicates analyzed using Student unpaired t test. Error bars are SD. * P < 0.05; **P < 0.01.
Discussion
We identified a recurrent synonymous somatic mutation in BCL2L12. This mutation appears in 12 of 285 samples, suggesting that this mutation is being selected for during tumor development. Analysis of the publicly available TCGA melanoma dataset supports this conclusion, as it also contains this mutation in 8 of 255 samples. Our study shows that the BCL2L12 synonymous mutation has no effect on normal protein function but instead, causes an accumulation of BCL2L12 mRNA and protein.
Functional analysis of the mutated form of BCL2L12 suggests that the mutant’s stability leads to overexpression, increasing the antiapoptotic signaling in melanoma cells and promoting cell survival, which may lead to increased resistance to p53-dependent apoptosis. BCL2L12 knockdown experiments support this finding. The reduced viability of mutant lines after BCL2L12 knockdown and UVR exposure suggests that these lines are dependent on BCL2L12 expression for survival, a common occurrence known as oncogene addiction (25). In fact, for several of the mutant lines, stable knockdown of BCL2L12 led to cell death without any exposure to UVR, further supporting the role of BCL2L12 in tumor survival.
Interplay between altered miRNA binding and synonymous mutations has been shown previously. A synonymous mutation in the immunity-related GTPase family M gene altered miR-196 binding and deregulated immunity-related GTPase family M-dependent xenophagy in Crohn disease, implicating a synonymous mutation as a likely causal variant for this disease (26). However, a direct link between a synonymous mutation and the origin of cancer has not been shown. This study, thus, shows that synonymous mutations may be selected in cancer and play a role in tumorigenesis. Importantly, the selection mechanism may possibly be through the relaxation of purifying selection and/or the plasticity–relaxation–mutation mechanism as well as some other alternatives (16, 27–31) rather than positive selection. The data presented here cannot unambiguously select one specific mechanism. However, the presented genetic and functional data support our view that synonymous mutations should receive increasing attention, not only in their detection but also in their functional assessment and elucidation of their role in cancer.
Materials and Methods
Tumor Tissues.
All DNA samples used in this study were derived from metastases. Samples used for whole-exome capture and prevalence screen were extracted from cell lines established directly from patients’ tumors as described previously (32). DNA subjected to whole-genome sequencing was extracted from optimum cutting temperature compound-embedded specimens as described previously (32). The clinical information associated with the melanoma tumors used in this study is provided in SI Appendix, Table S2.
DNA Extraction.
DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instructions. DNA was eluted in 35 μL elution buffer. DNA measurements were made using an ND-1000 UV-Vis spectrophotometer from NanoDrop Technologies.
Whole-Genome Single Nucleotide Variants.
For variant calling, only reads with mapping quality of Q30 or greater and bases with quality of Q20 or greater were considered. We used two related algorithms to make single position genotype calls in the normal and melanoma genomes. For all genomes, we used a Bayesian genotype caller named Most Probable Genotype (MPG) that has been described previously (33).
To identify variant positions, we developed an algorithm similar to MPG called Most Probable Variant (MPV). An important distinction between MPG and MPV is that MPV identifies variant positions relative to a reference genotype, whereas MPG identifies genotypes without an a priori reference assumption.
Statistical Calculation of Significance.
To evaluate whether the frequency of a synonymous mutation is significantly higher than would be expected if the mutation were neutral, we performed a statistical test. We only considered the validation samples to avoid biases. The null hypothesis is that the probability of a mutation at a specific base is the neutral rate of 11.4 mutations/Mb (i.e., P = 11.4e-6). We computed a one-sided P value using the pbinom function in the R statistical software.
Allelic Expression Analyses of Melanoma Samples.
Allelic mRNA/cDNA representation analyses shown in Fig. 1A were determined using iPlex Gold SBE (Sequenom) using the mel_1 amplification and the mel_X extension primer sets. For each melanoma sample, 12.5 ng cDNA or 20 ng gDNA were aliquotted in 384-well format. Allelic cDNA representation was compared with paired gDNA representation for each sample to identify statistically significant differences. Because cDNA variance did not appear to exhibit a normal distribution, the more conservative Wilcoxon rank sum statistical test was used to determine statistical significance.
Immunoprecipitation and Western Blotting.
BCL2L12 subcellular fractionation was performed as previously described (34). Briefly, the BCL2L12 overexpressed HEK293T cells were harvested, washed with ice-cold PBS, and lysed in a hypotonic lysis buffer [10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM EDTA, 1 mM EGTA, a mixture protease inhibitors (78415; Thermo Scientific)]. The cells were resuspended in 200 µL lysis buffer, incubated on ice for 10 min, titurated through a p2 tip 15–20 times, and sonicated 2–3 times; the total fraction was centrifuged for 15 min at 375 × g at 4 °C, resulting in a pellet that is the nuclear fraction and a supernatant that is the postnuclear fraction. Postnuclear fractions were loaded on 10% (vol/vol) Bris⋅Tris gel and further analyzed using mouse monoclonal BCL2L12 (1:1,000 dilution; Abcam) fragment antibody. The same membrane also processed later with GAPDH antibody.
HEK293 cells transiently transfected with BCL2L12-FLAG (WT, mutant, or empty vector) were gently washed two times in PBS and then lysed using 1.0 mL 1% Nonidet P-40 lysis buffer [1% Nonidet P-40, 50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, Complete Protease Inhibitor tablet, EDTA-free (Roche), 1 μM sodium orthovanadate, 1 mM sodium fluoride, 0.1% β-mercaptoethanol] per T-75 flask for 20 min on ice. Lysed cells were scraped and transferred into a 1.5-mL microcentrifuge tube. Extracts were centrifuged for 10 min at 20,000 × g at 4 °C; 800 μL supernatant were immunoprecipitated overnight using 30 μL anti-FLAG (M2) beads (Sigma Aldrich). The immunoprecipitates were washed and subjected to SDS/PAGE and Western blotting as previously described (32).
Lentiviral shRNA.
Constructs for stable depletion of BCL2L12 (RHS45330-NM_138639) were obtained from Open Biosystems and confirmed to efficiently knockdown BCL2L12 at the protein level. Lentiviral stocks were prepared as previously described (35). Melanoma cell lines were infected with shRNA lentiviruses for each condition (vector and two different BCL2L12-specific shRNAs). Selection of stable pooled clones was done in the presence of 3 µg/mL puromycin containing normal medium for 3–5 d before determining knockdown efficiency.
siRNA Depletion of Endogenous BCL2L12 in Melanoma Cells.
Specific siRNA was purchased from Dharmacon (Thermo Fisher Scientific) designed using their siRNA design program for human BCL2L12. Four independent siRNA molecules were used to transiently deplete BCL2L12 in malignant melanoma cells. Using DharmaEffect transfection reagent #1 specific for siRNA, melanoma cells were tranfected with 50 nM siRNA molecules (#3 and #4) in the presence of OptiMEM-I medium after cells were seeded into 96-well plates at a density of 2,000 cells/well 24 h before transfection. Cells were incubated for 24 h posttransfection before application of any genotoxic stressors.
Cell Viability Assays.
Stably depleted pooled clones were seeded into 96-well clear bottom opaque plates at 1,000 cells per well. Cells were incubated 24 h before exposure to UV light (50 kμJ) using a UV Stratalinker 2400 (Stratagene). Plates were then incubated for an additional 48 h before testing for cell viability using Cell-Titer-Glo (G7571). Plates were analyzed on a Thermo Electron Luminoskan reader. Data were then analyzed using Microsoft Excel to generate graphs and statistics.
miRNA Target Site Prediction.
We used the two miRNA target site prediction platforms PITA (19) and miRanda (20) to search for miRNA target site predictions that overlap the C51T mutated position in BCL2L12. We executed them with default prediction parameters and found that both platforms predict hsa-miR-671–5p to target the WT BCL2L12 mRNA overlapping position 51 but not the mutant mRNA.
miRNA Depletion of Endogenous BCL2L12 in Melanoma Cells A.
Specific miRNA mimetic (hsa-miR-671–5p) was purchased from Sigma Aldrich (HMI0901) that was determined to potentially target human BCL2L12. A negative control scrambled miR (NC) was purchased from Dharmacon (CN-001000-01; Thermo Fisher Scientific). Using DharmaFECT transfection reagent #1 (T-2001) specific for siRNA or miRNA, melanoma cells were tranfected with 20 nM has-miR-671–5p or NC molecules in the presence of OptiMEM-I medium after cells were seeded into six-well plates at a density of 200,000 cells/well 24 h before transfection. Cells were incubated for 24 h posttransfection before extraction of miRNA and mRNA and qRT-PCR analysis.
Anti–miR-671–5p Rescue Assay.
A specific anti-miRNA mimic (anti–hsa-miR-671–5p) was purchased from Qiagen (MIN0003880), which was determined to inhibit the hsa-miR-671–5p mimic. An NC was purchased from Dharmacon (Thermo Fisher Scientific). Using DharmaEffect transfection reagent #1 specific for siRNA or miRNA, melanoma cells were cotranfected with 20 nM hsa-miR-671–5p or NC molecules plus either 0 or 50 nM anti–miR-671–5p in the presence of OptiMEM-I medium after cells were seeded into six-well plates at a density of 200,000 cells/well 24 h before transfection. Cells were incubated for 24 h posttransfection before extraction of miRNA and mRNA and qRT-PCR analysis.
Real-Time qPCR of miRNA Targeted Cell Lines.
miRNA and mRNA were extracted from transiently transfected melanoma to assess for knockdown of endogenous BCL2L12 following the manufacturer’s protocol for the miRNeasy Mini Kit (217004; QIAGEN); 1 μg total RNA was used for cDNA synthesis using an miScript II Reverse Transcription Kit (218193; QIAGEN). cDNA was amplified with the 5× HiFlex buffer to quantitate in parallel the miRNA and mRNA. To test for loss of BCL2L12 message, we followed the manufacturer’s protocol and mixed primers and cDNA with QuantiTect SYBR Green PCR master mix at a final volume of 10 μL in triplicate (QIAGEN). qRT-PCR analysis was done using the ABI 7900HT Fast Real-Time PCR System. Results were analyzed using Microsoft Excel and GraphPad Prism v5.0.
miRNA Rescue Experiment.
A modified form of the hsa-miR-671–5p was custom made from Sigma Aldrich, with a single site changed to represent the synonymous mutation found in melanoma. Melanoma cells were seeded at ∼300,000 cells per well in six-well plates and incubated overnight before transient transfection. Cells were transfected with hsa-miR-671–5p (miR), mod-hsa-miR-671–5p (mod-miR), or NC in triplicate; total miRNA/RNA was amplified using the miScript miRNA cDNA Kit from Qiagen, and levels of BCL2L12 message were detected using SYBR Green master mix (Qiagen) in triplicate. GAPDH was used as an internal control to normalize between samples and generate graphs using Microsoft Excel. All experiments were repeated two to three times.
Supplementary Material
Acknowledgments
We thank Drs. Chris Schmidt and Peter Parsons for establishment of the majority of melanoma cell lines and V. Maduro, H. Ozel Abaan, and P. Cruz for generating the sequence data analyzed here. We thank Dr. V. G. Prieto for pathologic review of the biospecimens from the Melanoma Informatics, Tissue Resource, and Pathology Core (MelCore) at MD Anderson. We thank Dr. T. Wolfsberg for bioinformatics help, J. Jiang for sequencing help, and J. Fekecs and D. Leja for graphical assistance. We thank Drs. T. Barber and M. Willard for critical comments on the manuscript. S.C.J.P. is supported by an NIGMS Postdoctoral Research Associate (PRAT) Fellowship. This work was supported by the Intramural Research Programs of the National Human Genome Research Institute, by the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics, the estate of Alice Schwarz-Gardos, the estate of John Hunter, the Knell Family, the Peter and Patricia Gruber Award, National Cancer Institute Grant R21CA152432 (to R.K.), the National Institutes of Health, University of Texas MD Anderson Cancer Center Melanoma Specialized Programs of Research Excellence Grant P50 CA093459, Cancer Center Support Grant (CCSG) Core Grant NCI 5P30 CA006516-46 (to G.P.), the Human Frontier Science Program RGP0024 Grant (to A.A.K.), National Health and Medical Research Council of Australia Grants 1026112 and 613686, and a public–private partnership between the Intramural Research Programs of the National Human Genome Research Institute, the National Cancer Institute, and Eli Lilly and Company coordinated by the Foundation for the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequence reported in this paper has been deposited in the dbSNP, ClinVar database (accession no. 1057273).
2A complete list of the NISC Comparative Sequencing Program can be found in SI Text.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304227110/-/DCSupplemental.
Contributor Information
Collaborators: Jesse Becker, Betty Benjamin, Robert Blakesley, Gerry Bouffard, Shelise Brooks, Holly Coleman, Mila Dekhtyar, Michael Gregory, Xiaobin Guan, Jyoti Gupta, Joel Han, April Hargrove, Shi-ling Ho, Taccara Johnson, Richelle Legaspi, Sean Lovett, Quino Maduro, Cathy Masiello, Baishali Maskeri, Jenny McDowell, Casandra Montemayor, James Mullikin, Morgan Park, Nancy Riebow, Karen Schandler, Brian Schmidt, Christina Sison, Mal Stantripop, James Thomas, Pam Thomas, Meg Vemulapalli, and Alice Young
References
- 1.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485(7399):502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44(9):1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikolaev SI, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44(2):133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 6.Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2012;44(2):165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 9.Parker SC, et al. Mutational signatures of de-differentiation in functional non-coding regions of melanoma genomes. PLoS Genet. 2012;8(8):e1002871. doi: 10.1371/journal.pgen.1002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 12.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 14.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 15.Chamary JV, Parmley JL, Hurst LD. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7(2):98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 16.Hughes AL. Evolution of adaptive phenotypic traits without positive Darwinian selection. Heredity (Edinb) 2012;108(4):347–353. doi: 10.1038/hdy.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stegh AH, et al. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24(19):2194–2204. doi: 10.1101/gad.1924710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 19.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 20.John B, et al. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goode DL, et al. Evolutionary constraint facilitates interpretation of genetic variation in resequenced human genomes. Genome Res. 2010;20(3):301–310. doi: 10.1101/gr.102210.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark MS, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5(3):e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3(8):448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 26.Brest P, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43(3):242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 27.Hughes AL. Looking for Darwin in all the wrong places: The misguided quest for positive selection at the nucleotide sequence level. Heredity (Edinb) 2007;99(4):364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- 28.Hughes AL. The origin of adaptive phenotypes. Proc Natl Acad Sci USA. 2008;105(36):13193–13194. doi: 10.1073/pnas.0807440105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M. Selectionism and neutralism in molecular evolution. Mol Biol Evol. 2005;22(12):2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen JD, Thornton KR, Bustamante CD, Aquadro CF. On the utility of linkage disequilibrium as a statistic for identifying targets of positive selection in nonequilibrium populations. Genetics. 2007;176(4):2371–2379. doi: 10.1534/genetics.106.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishi N, Odagaki Y, Koyama T. Pharmacological characterization of metabotropic glutamate receptor-mediated high-affinity GTPase activity in rat cerebral cortical membranes. Br J Pharmacol. 2000;130(7):1664–1670. doi: 10.1038/sj.bjp.0703464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palavalli LH, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat Genet. 2009;41(5):518–520. doi: 10.1038/ng.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teer JK, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20(10):1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fazioli F, et al. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993;12(10):3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prickett TD, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41(10):1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.