Abstract
Infection by pathogenic fungi, such as Candida albicans, begins with adhesion to host cells or implanted medical devices followed by biofilm formation. By high-throughput phenotypic screening of small molecules, we identified compounds that inhibit adhesion of C. albicans to polystyrene. Our lead candidate compound also inhibits binding of C. albicans to cultured human epithelial cells, the yeast-to-hyphal morphological transition, induction of the hyphal-specific HWP1 promoter, biofilm formation on silicone elastomers, and pathogenesis in a nematode infection model as well as alters fungal morphology in a mouse mucosal infection assay. We term this compound filastatin based on its strong inhibition of filamentation, and we use chemical genetic experiments to show that it acts downstream of multiple signaling pathways. These studies show that high-throughput functional assays targeting fungal adhesion can provide chemical probes for study of multiple aspects of fungal pathogenesis.
Keywords: hyphal morphogenesis, small molecule screening, fungal pathogens
In humans, the most widespread fungal pathogen is Candida albicans, which is also one of the most frequent causes of hospital-acquired infections (1, 2). As an opportunistic pathogen, C. albicans is responsible for common clinical conditions, including oral thrush and vaginitis, but it can also lead to life-threatening systemic infections (candidemia) in immunocompromised patients (3), resulting in ∼30–50% mortality rates (2). Furthermore, the estimated annual cost of treating systemic Candida infections exceeds $1 billion per year (1, 2). A contributing factor to these statistics is the ability of C. albicans to develop resistance to antifungal drugs (4). For these reasons, new strategies for combating fungal infections without toxicity to humans are a high medical priority.
Adhesion to surfaces is the first step in establishing a fungal infection. Candida cells with a planktonic yeast morphology initiate adhesion, and a subsequent transition from yeast to hyphal morphology leads to tissue invasion and biofilm formation (5–8). Biofilm formation is an important step in pathogenesis, because after biofilms form on intravascular devices and catheters, they normally do not respond to conventional treatment and subsequently, can give rise to life-threatening systemic infections (9). Therefore, biofilm-associated medical devices often have to be surgically replaced (10).
Here, we report a chemical screen for compounds that prevent adhesion of C. albicans to polystyrene surfaces. One of the identified compounds also inhibited adhesion to human cells. Furthermore, this compound prevented adhesion of other Candida species to polystyrene, diminished the C. albicans yeast-to-hyphal transition, impaired biofilm formation on silicone elastomers, reduced fungal pathogenesis in a nematode infection model, and altered biofilm morphology in a mouse mucosal infection model. Based on its strong inhibition of filamentation, we term this compound filastatin, and we show that it blocks filamentation induced by some but not all external signals. In sum, our chemical screen identified an inhibitor of multiple pathogenesis-related functions that will also be a useful probe of morphological mechanisms as well as a lead for the development of unique antifungal therapeutics.
Results
Identifying Small Molecule Inhibitors of C. albicans Adhesion.
To detect small molecules that inhibit C. albicans adhesion, we modified a previous protocol for measuring adhesion of Saccharomyces cerevisiae to polystyrene (11). We observed that C. albicans cells bind strongly to 96-well polystyrene plates that have been optimized for protein binding (Methods), and we used this robustly bound substrate to set a high threshold for inhibitors of adhesion. Bound cells were stained with crystal violet followed by washing to remove unbound dye and cells. Cells that remained bound after washing were then quantified by measuring dye absorbance. Wells that displayed low A590 values were confirmed visually (Fig. 1A).
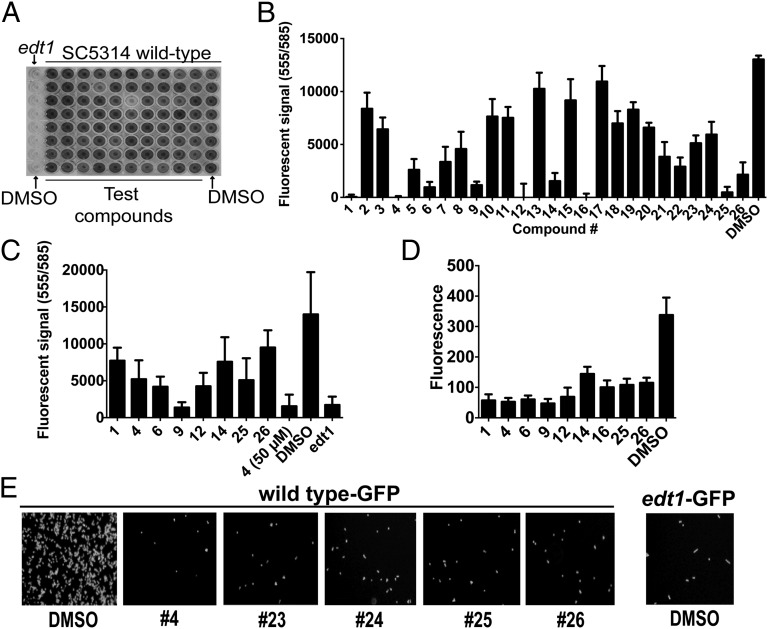
Fig. 1.
Chemical inhibition of C. albicans adhesion to polystyrene. (A) Crystal violet-stained plate from small molecule screen. WT C. albicans strain SC5314 or adhesion-defective edt1−/− cells were plated in the presence of 1% DMSO vehicle or small molecules at 50 μM followed by extensive washing to remove unbound cells. Cells that remained were then stained with crystal violet and quantified by A590. (B) alamarBlue-based polystyrene adhesion assay. Using the vital dye alamarBlue as the detection reagent, 26 primary candidates from the screen were retested at 25 μM alongside a DMSO control. The mean and SD of data from eight wells are shown. (C) The same as in B but with compounds at 7.5 μM. The mean and SD of data from eight wells are shown. (D) GFP-based adhesion assay. C. albicans strains expressing GFP (SC5314-GFP and edt1−/−-GFP) or untagged SC5314 were mixed with compounds at 25 μM or DMSO and bound to polystyrene plates. Unbound cells were removed by washing. GFP fluorescence was measured, and background fluorescence of wells containing untagged SC5314 was subtracted from signals. Mean and SD For eight wells are shown. (E) Wells from the experiment in D were photographed using a 20× objective and FITC filters.
We screened a library of 30,000 small molecules for compounds that inhibit adhesion of WT clinical isolate C. albicans strain SC5314 (12). All compounds were dissolved in DMSO, and therefore, we used SC5314 cells exposed only to DMSO as a negative control (Fig. 1A, column 12). As a positive control for poor adhesion, we used a C. albicans edt1−/− strain (13) that lacks a cell wall adhesion protein (Fig. 1A, column 1). We ranked the normalized scores for each compound (Methods). Of 30,000 compounds, 40 (1.3%) compounds inhibited adhesion by >75%, and many of these compounds fell into two scaffold families [1-benzoyl-4-phenylpiperazines (termed Scaffold 1) and N-phenylbenzamides (termed Scaffold 2)] (Table S1). Omitting compounds within this list that were very similar, we reordered 26 of the candidate compounds (termed 1–26) (Table S1) for subsequent characterization.
We prioritized the candidate compounds by testing the dose dependence of their effects. Because of the laborious washing required to remove background crystal violet staining, we used an alternative detection reagent, the vital dye alamarBlue. We found that, after two rounds of washing, alamarBlue robustly detected the adhesion differences between the WT SC5314 and edt1−/− mutant cells (Fig. 1C and Fig. S1A). The majority of the reordered compounds were effective at reducing Candida adhesion at 50 μM, which is the concentration originally tested during the screen (Methods). However, on reducing the concentration to 25 μM, some of these compounds were substantially less effective (Fig. 1B). However, many candidates were able to inhibit adhesion by >50%, even at concentrations as low as 7.5 μM (Fig. 1C).
Because alamarBlue measures the metabolic activity of live cells, we confirmed these data with an assay that directly detected cells, regardless of viability or metabolism, by measuring adhesion using a C. albicans strain encoding GFP (13). Both fluorescence measurements and microscopy (Fig. 1 D and E) of the GFP-expressing strain confirmed that all of the compounds effective in the alamarBlue-based assay impair C. albicans adhesion to polystyrene. We conclude that our chemical screen detected compounds that block adhesion and yielded consistent results, regardless of the detection method.
In addition to experiments with inert surfaces, we also tested how the candidate compounds would affect C. albicans adherence to human cells by measuring binding of GFP-encoding C. albicans cells to monolayers of human lung epithelial A549 cells, which are an effective substrate for Candida adhesion (14). We tested each of the reordered compounds and observed that compound 4 was, by far, the strongest inhibitor of adhesion to human epithelial cells, which was shown by both fluorescence quantitation (Fig. 2A) and microscopy (Fig. 2B). We, therefore, prioritized study of this compound (Fig. 2F), which we termed filastatin based on its inhibition of C. albicans filamentation as described below. We showed that filastatin does not affect the viability of human A549 cells, even at concentrations much larger than those concentrations used in the adhesion assay (e.g., 250 μM) (Fig. 2C). We conclude that filastatin is not toxic to this human cell line under our assay concentrations, but it is unique among our candidates in that it can impair fungal adhesion to both inert surfaces and cultured human epithelial cells.
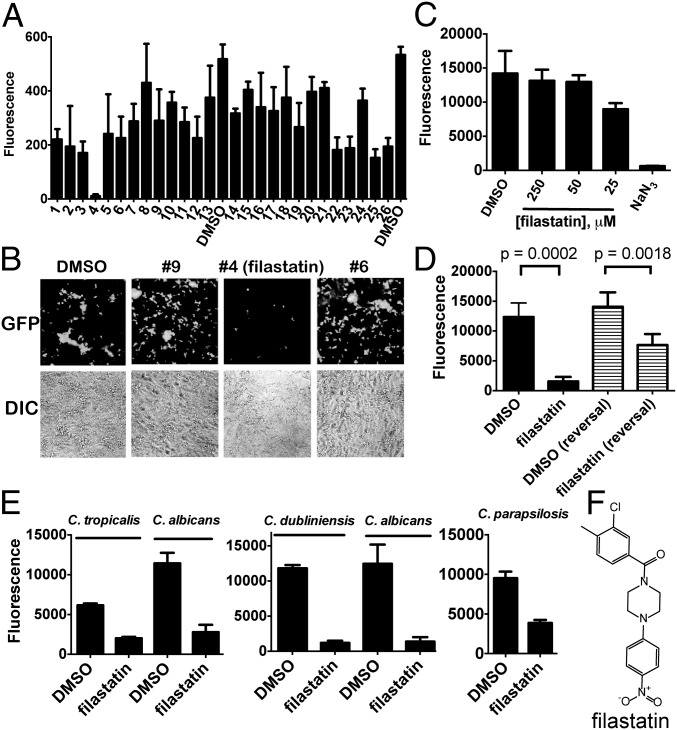
Fig. 2.
Filastatin is a nontoxic inhibitor of adhesion to human cells, and it affects multiple Candida species. (A) Adhesion to A549 cells. Human A549 cells were grown to confluence on 48-well plates. SC5314-GFP, edt1−/−-GFP, or untagged SC5314 were added to triplicate wells and incubated at 37 °C for 90 min with 1% DMSO or 25 μM indicated compounds in 1% DMSO. Wells were washed extensively to remove unbound fungi, and GFP fluorescence was measured on a plate reader. Mean and SD for three wells are shown. (B) Representative differential interference contrast (DIC) and corresponding GFP fluorescence images from the experiment in A. (C) Human cell toxicity. A549 cells were coincubated with 1% DMSO, 250, 100, or 50 μM filastatin, or 1% NaN3 for 24 h, and cell viability was then measured using alamarBlue. Mean and SD for three wells are shown. (D) Reversal of adhesion. WT C. albicans SC5314 cells were seeded onto 96-well plates. One-half the samples were incubated continuously in the presence of 1% DMSO or 50 μM filastatin as indicated for 8 h at 37 °C (left black bar). One-half of the samples were allowed to adhere for 4 h in the absence of compounds, and then, 1% DMSO or 50 μM filastatin was added followed by additional incubation for 4 h at 37 °C (reversal; left striped bar). After incubations, plates were washed two times, and bound cells were detected with alamarBlue. Means and SDs (n = 5 independent experiments, 24 wells measured for each) with P value comparisons are shown (unpaired two-tailed t test). (E) Other Candida species. Adhesion assays are the same as in Fig. 1B, with 25 μM filastatin tested for C. dubliniensis and C. tropicalis and 50 μM filastatin tested for C. parapsilosis. Mean and SD for 16 wells are shown. (F) Structure of filastatin.
We next tested whether filastatin could affect adhesion after cells had already bound polystyrene, because this activity could prove useful in a clinical setting. We compared cells continuously incubated with filastatin for 8 h with cells adhered for 4 h (as in our previous assays) (Fig. 1B) followed by 4 h in the presence of filastatin. We observed that filastatin did, indeed, reduce the amount of bound cells when added after adhesion, although not as efficiently as when the compound and cells are coincubated from the start of the experiment (Fig. 2D). We conclude that filastatin does display some activity against adhered cells, consistent with potential use against pathogen biofilms.
To test the applicability of filastatin to other fungal pathogens, we examined adhesion by additional pathogenic Candida species (15). Specifically, we observed that filastatin inhibited polystyrene adhesion by C. dubliniensis and C. tropicalis and to a lesser extent, C. parapsilosis (Fig. 2E). Filastatin, therefore, inhibits adhesion by multiple pathogenic Candida species. Additionally, filastatin (molecular weight = 360 g/mol) has an IC50 ∼ 3 μM in the GFP-based adhesion assay (Fig. S1B), equivalent to a concentration of ∼1.1 μg/ml. This value is similar to or less than the effective concentrations of existing antifungal compounds (16).
Additionally, data in Figs. S2, S3, and S4 compare the activities of filastatin with other compounds from this study and prior studies (17). Of all these studies, only filastatin inhibited C. albicans adhesion to human cell monolayers, and therefore, we focused additional studies on this molecule.
Filastatin Inhibits C. albicans Biofilm Formation.
Fungal biofilm formation on implanted medical devices is a serious medical problem, because these biofilms can lead to life-threatening systemic infections (18). An established in vitro assay to study device colonization measures biofilm formation on surgical silicone elastomers (19). In the presence of DMSO vehicle, we confirmed that WT C. albicans cells efficiently formed biofilms on silicone elastomers; in contrast, edt1−/− mutant cells did not, resulting in the majority of the cells dispersing throughout the media rather than adhering to the elastomer surface (Fig. 3A). Visual inspection showed that filastatin, but not compounds 6 or 9, effectively kept the cells dispersed in the media rather than on the elastomer surface (Fig. 3A). Measurements of the media turbidity and biofilm dry weight (Fig. S5) confirmed these assessments. Additionally, we observed that compounds 27, Q1, and Q2 also inhibited biofilm formation in this assay (Fig. S5). Therefore, filastatin has multiple activities, with inhibition of human cell adhesion unique among our candidate compounds (Tables S1 and S2).
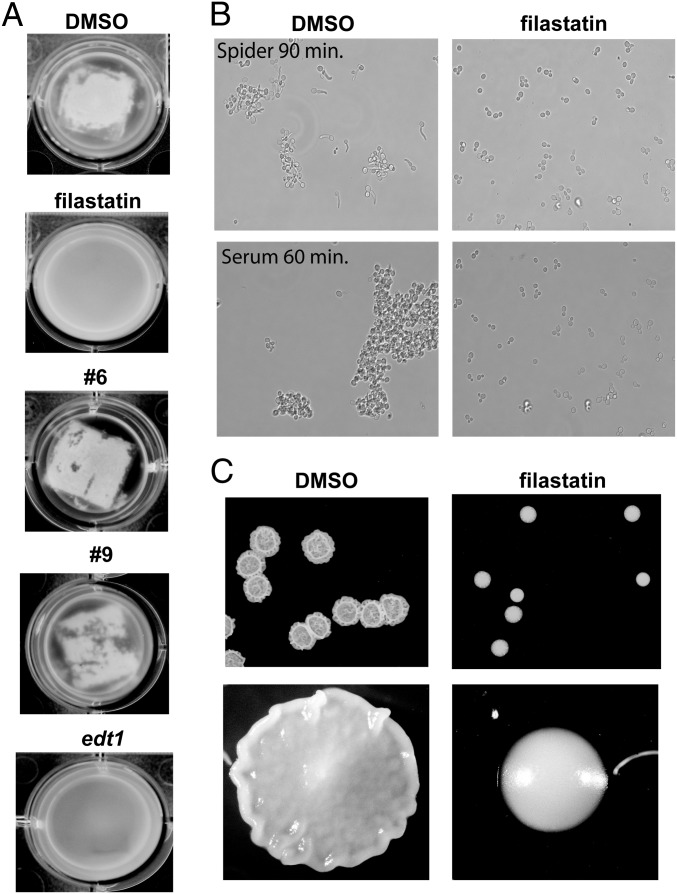
Fig. 3.
Filastatin inhibits C. albicans biofilm formation and hyphal morphogenesis. (A) Biofilm formation on silicone elastomers. Biofilms formed by SC5314-GFP (or edt1−/−-GFP where indicated) cells were photographed after 60 h. Indicated compounds were added at 50 μM. Turbid media indicate planktonic cells unattached to silicone elastomers when biofilm formation is inhibited. Clear media is observed when robust biofilm formed. Representatives of triplicate wells are shown. Additional biofilm data are in Fig. S5. (B) Hyphal morphogenesis. SC5314 cells were diluted into Spider media or Supplemented YNB + 10% bovine serum with DMSO or 50 μM filastatin as indicated, grown at 37 °C for the indicated times, and photographed. Additional hyphal development experiments are in Fig. S6. (C) Colony morphology. SC5314 cells were spotted onto solid Spider media agar plates overlaid with either 1% DMSO or 50 μM filastatin and incubated at 37 °C for 2 d before photography.
Filastatin Inhibits Hyphal Morphogenesis and Colony Morphology.
Visual inspection of cells remaining at the end of adhesion assays suggested that some of the candidate compounds inhibit generation of hyphae. Because the ability to interconvert between yeast and hyphal morphologies is usually correlated with pathogenicity (5, 7, 8), we explored this finding in more detail, examining induction of hyphae on carbon starvation (Spider media) (20). We used a strain that contains the Red Fluorescent Protein ORF driven by the hyphal-specific HWP1 promoter (21), which provides a molecular reporter for hyphae formation in addition to cell morphology. We observed that filastatin as well as a subset of the other compounds (Fig. S6A and Table S1) blocked formation of hyphae. Together, our data indicate that the related compound pairs (filastatin/27 and Q1/Q2) were effective inhibitors of both hyphal morphogenesis and biofilm formation on silicone elastomers (Fig. 3A and Figs. S5 and S6). Therefore, the shared backbone of these compounds correlates with inhibition of multiple activities (summarized in Table S2).
On titration, we observed that filastatin is effective at inhibiting hyphae formation at concentrations > 2.5 μM (Fig. S6B), similar to the results observed in the polystyrene adhesion assay (Fig. S1). Using this protein reporter assay, the effect of filastatin could be detected within 2 h of Spider media addition (Fig. S6C). Also, because filastatin blocked filamentation, even after 16 h of incubation (Fig. S6A), it seemed unlikely that this inhibitory function relied on a reduced growth rate. Although filastatin did modestly reduce growth rates of yeast-form cells in rich media (Fig. S7A), this effect was less pronounced during hyphal induction, where we assessed growth by both OD and metabolic activity (Fig. S7 B and C). Furthermore, filastatin maintained the yeast morphology of cells throughout an 8-h time course (Fig. S7D). We conclude that filastatin is a long-lasting inhibitor of C. albicans filamentation.
Hyphal morphogenesis is induced by multiple stimuli that act through multiple signal transduction pathways (5, 22, 23) (Fig. 4E) and mediated by a complex transcriptional network (8, 24). In addition to nutrient-poor Spider media, mammalian serum also induces hyphae (25, 26), and we observed that filastatin inhibited the response to both these stimuli (Fig. 3B). We detected morphological difference between control and filastatin-treated cultures as early as 60 min in 10% (vol/vol) serum and 90 min in Spider media. Additionally, a wrinkled colony morphology is displayed by C. albicans when grown on solid Spider media, reflecting transitions between hyphal and yeast forms (5, 27). Notably, this wrinkled phenotype was also abolished by filastatin (Fig. 3B). Together, our data indicate that filastatin inhibits hyphal morphogenesis and colony morphology phenotypes that depend on the hyphal transition.
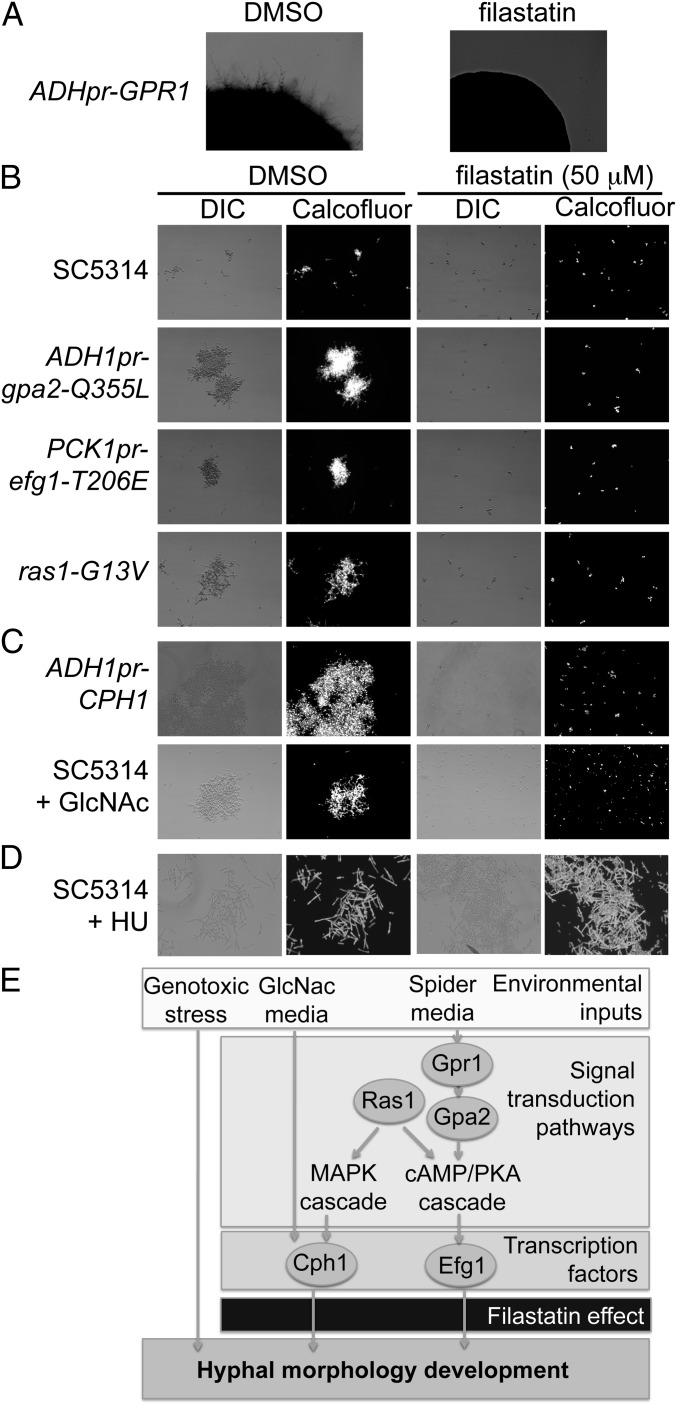
Fig. 4.
Filastatin inhibits hyphal development induced by some but not all external signals. (A) Filastatin inhibits the hyperfilamentous growth of a strain that constitutively overexpresses Gpr1. Colonies were grown on solid Spider media agar overlaid with either 1% DMSO or 50 μM filastatin and incubated at 37 °C for 3 d. (B) Effects of filastatin on strains hyperactivating the cAMP-PKA pathway. Cells of the indicated genotypes were grown in the presence of 1% DMSO or 50 μM filastatin in Spider medium at 37 °C for 4 h, stained with Calcofluor, and photographed using DIC and fluorescence microscopy. (C) Filastatin blocks filamentation induced by GlcNAc or constitutive overexpression of the GlcNac-activated transcription factor Cph1. (D) Filastatin does not block genotoxic stress-induced filamentation. SC5314 cells were grown in yeast extract-peptone-dextrose (YPD)-rich media + 50 μM HU at 30 °C. (E) Schematic diagram of some of the signaling pathways that govern hyphal morphogenesis (22, 26). For simplicity, only those genes investigated are shown. Filastatin is shown blocking hyphal development induced by Spider media and GlcNac but not genotoxic stress.
Filastatin Inhibits Induction of Hyphal Morphogenesis by Multiple but Not All Signaling Pathways.
To explore the mechanism of filastatin action, we tested whether it would alter hyphal induction in hyperfilamentous mutants. For example, hyphal induction by Spider media requires the cAMP-PKA pathway (23, 26) (Fig. 4E). Cells constitutively overexpressing the G protein-coupled receptor Gpr1 become hyperfilamentous on solid Spider media by PKA stimulation (26, 28), and we observed that filastatin blocked hyphal morphogenesis in these cells (Fig. 4A). Stimulation of the PKA pathway also drives phosphorylation of transcription factor Efg1, activating Efg1 to increase expression of genes required for hyphal morphogenesis. We confirmed that cells with a hyperactive Ras1 signaling protein (ras1-G13V) (29), an overexpressed, constitutively active G-α subunit that acts upstream of Efg1 (gpa2-Q355L) (28), or a constitutively expressed Efg1 transcription factor with a phosphomimetic mutation that simulates constitutive PKA signaling (PCKpr-efg1-T206E) (30), indeed, were all hyperfilamentous compared with WT cells in Spider media (Fig. 4B). However, in the presence of filastatin, cells from all these mutant strains retained a planktonic, budded morphology (Fig. 4A). Together, these data suggest that filastatin acts downstream of transcription factor Efg1 (Fig. 4E).
However, other experiments suggested that filastatin affects more than one signaling pathway. For example, the modified sugar GlcNac also stimulates hyphal morphogenesis but does so independently of the cAMP-PKA pathway; instead, it activates the transcription factor Cph1 (26). On testing morphogenesis driven by GlcNac-containing media or constitutive overexpression of Cph1, we found that filastatin also inhibits formation of hyphae in these cases (Fig. 4C). These data indicated that filastatin may affect multiple signaling pathways or could act by destroying the ability of the cell to form elongated structures, regardless of the inducing signal.
In addition to nutrient-mediated signals, genotoxic stress promotes hyphal morphogenesis in C. albicans, and the DNA damage signaling kinase Rad53 is required for the hyphal transition (31). In contrast to the above studies, cells treated with the DNA replication inhibitor hydroxyurea (HU) form hyphae, regardless of the presence of filastatin (Fig. 4D). Together, these data suggest that filastatin is not incompatible with hyphal morphogenesis per se but blocks multiple signaling mechanisms that promote it (Fig. 4E).
Filastatin Exhibits Antifungal Activity in a Nematode Model of Infection.
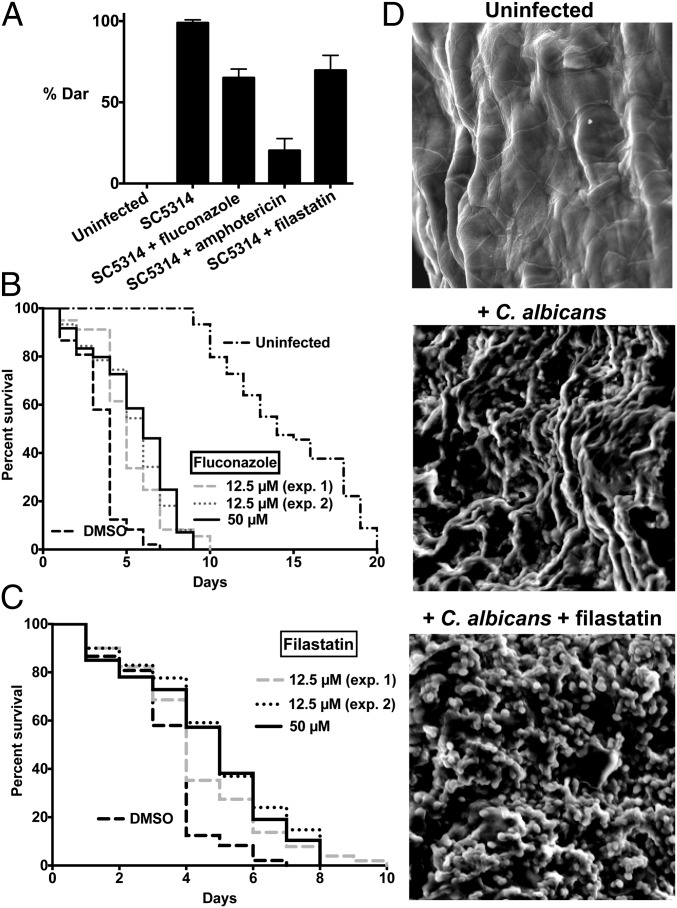
In addition to our in vitro assays, we tested whether filastatin affects fungal infections in vivo. We used the previously described deformed anal region (Dar) tail-swelling phenotype (16, 32) as a biomarker to monitor fungal infections in nematode hosts. A small quantity of C. albicans was introduced with the nematode’s standard diet of Escherichia coli OP50 cells, and E. coli growth was attenuated with the antibiotic streptomycin to avoid interactions with C. albicans (33). Here, we used a dose of C. albicans that caused all untreated worms to display Dar disease (Fig. 5A). In contrast, ∼30% of worms treated with either 12.5 μM filastatin or fluconazole during this experiment maintained a healthy phenotype and did not display Dar disease. Worms treated with the same concentration of amphotericin B displayed an even larger decrease in Dar disease, perhaps due to amphotericin B being a fungicidal drug, while fluconazole is fungistatic (34). In any case, these data indicate that filastatin can ameliorate a fungal pathogenesis phenotype in vivo.
Fig. 5.
Effects on metazoan hosts. (A) Nematodes were either cultured without C. albicans (Uninfected) or with WT C. albicans SC5314 cells with or without 12.5 μM indicated compounds. Dar disease rates were compared for worms treated with filastatin and untreated worms (P = 2.9 × 10−3). Infected worms were treated with antifungal agents fluconazole (P = 2.6 × 10−4) and amphotericin B (P = 2.9 × 10−3) as positive controls. Experiments were done in triplicate, with 14–28 animals in each biological replicate sample, with 63–81 total animals for each experimental condition. Student t test (one tail, equal variance) was used to calculate P values. (B) Lifespan of C. elegans exposed to C. albicans treated with DMSO, 12.5 µM fluconazole (two experiments, plotted separately), or 50 µM fluconazole; n = 60 individuals for each experiment. All fluconazole-treated populations were significantly different from the DMSO control, with P values for each <0.0001 by the Mantel–Cox test. Lifespan of uninfected C. elegans is plotted on the same graph. (C) As in B, except worms were treated with filastatin at 12.5 (two experiments, plotted separately) or 50 µM. All filastatin-treated populations were significantly different from the DMSO control, with P < 0.012 for 12.5 µM experiment 1, P < 0.0001 for 12.5 µM experiment 2, and P < 0.0002 for the 50 µM experiment. (D) Microscopic analysis of ex vivo vaginal mucosal biofilm formation. Tissues were processed for scanning EM. (Magnification: 1,000×.) These representative images show areas of biofilm growth from two independent repeats (n = 6 vaginal explants per experiment).
We next tested whether filastatin protects Caenorhabditis elegans against killing by C. albicans. C. elegans was exposed to C. albicans in the presence of DMSO, fluconazole, or filastatin. The C. albicans infections greatly shortened the lifespan of the C. elegans hosts compared with uninfected controls (Fig. 5B). We observed that both fluconazole and filastatin increased the lifespan of the infected worms (Fig. 5 B and C), indicating that both these compounds are protective during infection of a live animal. Increased lifespan in the presence of filastatin was statistically significant as confirmed by Kaplan–Meier statistical analysis (P value < 0.012 or less for all three experiments). Therefore, consistent with its in vitro activities, filastatin functions as an antifungal agent in this model infection system.
Filastatin Alters Biofilm Morphology in a Mouse Model of Vulvovaginal Candidiasis.
As an initial test of filastatin in a mammalian infection model, we examined a mouse model of vulvovaginal candidiasis (VVC). This system is ideal for evaluating our compounds, because VVC infections depend on fungal morphogenesis and biofilm formation (35). This assay is medically important, because VVC affects 75% of all women at least once in their lifetimes, and because C. albicans is responsible for 85--95% of these infections (36).
In this assay, excised murine vaginal mucosal tissue is tested as a substrate for infection by Candida cells. This protocol provides a rapid and simple method for optimizing treatment conditions before in vivo infection assays, although it does not measure adhesion to the tissue, because the tissue is not washed before processing for microscopy. One readout in this assay is a measure of the number of viable fungal cells expressed as cfu. We did not observe significant differences between the cfu recovered from untreated and filastatin-treated samples, although because unadhered cells were not washed away, direct comparison with the in vitro adhesion assays cannot be made. Also, in the presence of filastatin, the Candida cells still formed a carbohydrate-rich ECM, which was detected by Con A staining and confocal microscopy (Fig. S8). However, filastatin modulated the morphology of the C. albicans biofilm during ex vivo VVC, which was revealed by scanning EM (Fig. 5D), with a predominance of individual yeast-form cells appearing. Therefore, filastatin restrained formation of hyphae in this infection model, consistent with its hyphal inhibition activity in vitro (Figs. 3 and 4). Future experiments will be required to determine how filastatin affects ultrastructural aspects of biofilm formation and how it may affect adhesion during biofilm formation on mammalian cells.
Discussion
In recent years, the rate at which new drugs are brought to market has slowed (37). Traditional drug discovery methods have often relied on rational drug design, where agents that inhibit a particular protein are selected. However, this approach is time-consuming and can miss important but unanticipated targets. Here, we describe an alternate function-based approach that relies on phenotypic screening without bias to a particular target. To our knowledge, there has not, to date, been a published screen that directly targets fungal adhesion. By targeting adhesion, our screen required removal of unbound cells. Although laborious in a high-throughput setting, this extra effort allowed us to detect a class of compounds that has unique properties. Importantly, because our assays involve intact cells, we avoided the complication of compounds unable to cross the cell wall and/or membranes.
Here, we report discovery of filastatin, which displays multiple activities related to fungal pathogenesis. Filastatin inhibits fungal adhesion to polystyrene and human cells (Figs. 1 and 2), hyphal morphogenesis and colony morphology (Figs. 3 and 4), biofilm formation (Fig. 3), pathogenesis in a worm infection model (Fig. 5C), and biofilm morphology on mouse tissue explants (Fig. 5D). Notably, inhibition of hyphal morphogenesis occurs in liquid culture in the absence of adhesion, suggesting that filastatin affects multiple cellular processes. The effects on morphology were not selected in our primary screen, showing the need for multiple assays to characterize our candidate compounds fully.
We find that small changes in the compounds can lead to different combinations of activities. This observation was most pronounced in the comparison of filastatin with compound 27, which differ by the substitution of a methyl group for a chlorine substitution on one aryl ring (Fig. S3 and Table S1). Filastatin, but not compound 27, effectively inhibited human cell binding, but these two compounds shared the ability to inhibit polystyrene binding, hyphae formation, and biofilm formation (Table S1). As another example, the quinoline ring-containing compounds Q1 and Q2 do not inhibit polystyrene binding or human cell binding, but they do inhibit hyphae formation and biofilm formation. Conversely, filastatin is not synergistically toxic to fluconazole-resistant cells like Q1 and Q2 (Fig. S4). More systematic structure–activity relationship studies are underway to better understand the complex pharmacophore of filastatin and explore its molecular target(s). We note that the multiple activities of filastatin leave open the possibility that multiple targets exist.
Several previous experiments suggest a strong link between hyphal morphogenesis and fungal pathogenesis, including tissue-associated biofilm formation. For example, experiments with a doxycycline-inducible transcription factor (NRG1) that governs hyphae formation have shown that the ability to form hyphae is continually required for the lethality associated with systemic candidaisis (6). Likewise, many mutants defective in hyphae formation are nonpathogenic (7) and poor at biofilm formation, although this correlation is not absolute (38).
Our data show that filastatin blocks the transcriptional induction of the HWP1 promoter (Fig. S6), which is an early and essential event in the process of hyphal development (23). Notably, filastatin blocks hyphal formation induced by serum, Spider media, and GlcNac but not the genotoxic stress agent HU (Fig. 4). These data indicate that the structural machinery required for cell elongation and hyphae formation is able to function in the presence of the compound. Therefore, our screen has provided a powerful chemical probe of the signaling mechanisms that govern hyphal formation. Biochemical and molecular genetics experiments are underway to better understand how filastatin affects hyphal development and fungal adhesion.
Methods
During the screen for small molecule adhesion inhibitors, one 5-mL culture of SC5314 cells and two 5-mL cultures of edt1−/− cells were inoculated with single colonies in yeast nitrogen broth without amino acids supplemented with 30 µg/mL l-leucine, 20 µg/mL l-histidine, 20 µg/mL l-tryptophan, and 25 µg/mL uridine (Supplemented YNB) + 0.15% glucose media and grown overnight with shaking at 200 rpm at 30 °C. The next day, these starter cultures were used to inoculate 100-mL cultures for each strain grown under the same conditions. One day later, OD at 600 nm was measured for both cultures. Cells were pelleted at 3,000 × g and resuspended in fresh Supplemented YNB + 0.15% glucose media to a final concentration of 0.5 OD/mL; 200 µL/well 0.5 OD/mL edt1−/− cells were added to the first column of each 96-well Immulon 2HB flat-bottom microtiter plate (Part No. 3455; Thermo Scientific), and 200 µL/well 0.5 OD/mL SC5314 cells were added to the remaining columns. Cell plating was followed by robotic addition of 2 µL DMSO vehicle to columns 1 and 12; compounds from the University of Massachusetts Medical School Small Molecule Facility DIVERset Library (Chembridge) at a stock concentration of 5 mM were added to columns 2–11 (Fig. 1A). The wells were mixed by robotic pipetting three times, yielding a final compound concentration of 50 µM. The plates were covered with foil and incubated at 37 °C for 4 h. The contents of the wells were then decanted, and 50 µL 0.5% crystal violet (Sigma) in water were added to each well. The plates were covered again and incubated at room temperature for 45 min. The dye was removed by decanting, and the plates were gently rinsed by 10 rounds of submersion in an ice bucket filled with distilled water followed by decanting the water. The water in the bucket was changed after the fifth wash. The plates were then gently inverted onto a paper towel to remove excess water; 200 µL 75% methanol were then added to each well. The plates were incubated for 30 min at room temperature, and then, absorbance at 590 nm was measured.
Additional methods are in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by a University of Massachusetts—Worcester Polytechnic Institute Collaborative Pilot Project Initiative Grant (University of Massachusetts Clinical and Translational Science Pilot Project Grants UL1RR031982 and UL1TR000161; to R.P.R. and P.D.K.) and the University of Massachusetts Center for AIDS Research as part of Grant P30 AI042845 (to P.D.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305982110/-/DCSupplemental.
References
- 1.Miller LG, Hajjeh RA, Edwards JE., Jr Estimating the cost of nosocomial candidemia in the united states. Clin Infect Dis. 2001;32(7):1110. doi: 10.1086/319613. [DOI] [PubMed] [Google Scholar]
- 2.Pappas PG, et al. A prospective observational study of candidemia: Epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis. 2003;37(5):634–643. doi: 10.1086/376906. [DOI] [PubMed] [Google Scholar]
- 3.Fidel PL, Jr, Sobel JD. Immunopathogenesis of recurrent vulvovaginal candidiasis. Clin Microbiol Rev. 1996;9(3):335–348. doi: 10.1128/cmr.9.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowen LE, Anderson JB, Kohn LM. Evolution of drug resistance in Candida albicans. Annu Rev Microbiol. 2002;56:139–165. doi: 10.1146/annurev.micro.56.012302.160907. [DOI] [PubMed] [Google Scholar]
- 5.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12(7):317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2(5):1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo HJ, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 8.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9(2):109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 2006;8(9):1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 10.Bauters TG, Moerman M, Vermeersch H, Nelis HJ. Colonization of voice prostheses by albicans and non-albicans Candida species. Laryngoscope. 2002;112(4):708–712. doi: 10.1097/00005537-200204000-00021. [DOI] [PubMed] [Google Scholar]
- 11.Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291(5505):878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 12.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198(1):179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 13.Wheeler RT, Kombe D, Agarwala SD, Fink GR. Dynamic, morphotype-specific Candida albicans β-glucan exposure during infection and drug treatment. PLoS Pathog. 2008;4(12):e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura A, et al. Effect of beta-1,6-glucan inhibitors on the invasion process of Candida albicans: Potential mechanism of their in vivo efficacy. Antimicrob Agents Chemother. 2009;53(9):3963–3971. doi: 10.1128/AAC.00435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Junqueira JC, et al. Oral Candida albicans isolates from HIV-positive individuals have similar in vitro biofilm-forming ability and pathogenicity as invasive Candida isolates. BMC Microbiol. 2011;11:247. doi: 10.1186/1471-2180-11-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okoli I, et al. Identification of antifungal compounds active against Candida albicans using an improved high-throughput Caenorhabditis elegans assay. PLoS One. 2009;4(9):e7025. doi: 10.1371/journal.pone.0007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youngsaye W, et al. Piperazinyl quinolines as chemosensitizers to increase fluconazole susceptibility of Candida albicans clinical isolates. Bioorg Med Chem Lett. 2011;21(18):5502–5505. doi: 10.1016/j.bmcl.2011.06.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguly S, Mitchell AP. Mucosal biofilms of Candida albicans. Curr Opin Microbiol. 2011;14(4):380–385. doi: 10.1016/j.mib.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobile CJ, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2(7):e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan N, Kruppa MD. Standard growth media and common techniques for use with Candida albicans. Methods Mol Biol. 2009;499:197–201. doi: 10.1007/978-1-60327-151-6_18. [DOI] [PubMed] [Google Scholar]
- 21.Ganguly S, et al. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell. 2011;10(11):1448–1454. doi: 10.1128/EC.05196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shareck J, Belhumeur P. Modulation of morphogenesis in Candida albicans by various small molecules. Eukaryot Cell. 2011;10(8):1004–1012. doi: 10.1128/EC.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011;9(7):e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobile CJ, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148(1-2):126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: A genome-wide analysis. Mol Biol Cell. 2005;16(6):2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Midkiff J, Borochoff-Porte N, White D, Johnson DI. Small molecule inhibitors of the Candida albicans budded-to-hyphal transition act through multiple signaling pathways. PLoS One. 2011;6(9):e25395. doi: 10.1371/journal.pone.0025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5(12):e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miwa T, et al. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot Cell. 2004;3(4):919–931. doi: 10.1128/EC.3.4.919-931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181(20):6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bockmühl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157(4):1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell. 2007;18(3):815–826. doi: 10.1091/mbc.E06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain C, Pastor K, Gonzalez AY, Lorenz MC, Rao RP. The role of Candida albicans AP-1 protein against host derived ROS in in vivo models of infection. Virulence. 2013;4(1):67–76. doi: 10.4161/viru.22700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain C, Yun M, Politz SM, Rao RP. A pathogenesis assay using Saccharomyces cerevisiae and Caenorhabditis elegans reveals novel roles for yeast AP-1, Yap1, and host dual oxidase BLI-3 in fungal pathogenesis. Eukaryot Cell. 2009;8(8):1218–1227. doi: 10.1128/EC.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawser S, Islam K. Comparisons of the effects of fungicidal and fungistatic antifungal agents on the morphogenetic transformation of Candida albicans. J Antimicrob Chemother. 1999;43(3):411–413. doi: 10.1093/jac/43.3.411. [DOI] [PubMed] [Google Scholar]
- 35.Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Jr, Noverr MC. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 2010;156(Pt 12):3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobel JD, et al. Vulvovaginal candidiasis: Epidemiologic, diagnostic, and therapeutic considerations. Am J Obstet Gynecol. 1998;178(2):203–211. doi: 10.1016/s0002-9378(98)80001-x. [DOI] [PubMed] [Google Scholar]
- 37.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 38.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42(7):590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.