Abstract
Aims
High mobility group box 1 (HMGB1) is an abundant and ubiquitous nuclear DNA-binding protein that has multiple functions dependent on its cellular location. HMGB1 binds to DNA, facilitating numerous nuclear functions including maintenance of genome stability, transcription, and repair. However, little is known about the effects of nuclear HMGB1 on cardiac hypertrophy and heart failure. The aim of this study was to examine whether nuclear HMGB1 plays a role in the development of cardiac hypertrophy induced by pressure overload.
Methods and results
Analysis of human biopsy samples by immunohistochemistry showed decreased nuclear HMGB1 expression in failing hearts compared with normal hearts. Nuclear HMGB1 decreased in response to both endothelin-1 (ET-1) and angiotensin II (Ang II) stimulation in neonatal rat cardiomyocytes, where nuclear HMGB1 was acetylated and translocated to the cytoplasm. Overexpression of nuclear HMGB1 attenuated ET-1 induced cardiomyocyte hypertrophy. Thoracic transverse aortic constriction (TAC) was performed in transgenic mice with cardiac-specific overexpression of HMGB1 (HMGB1-Tg) and wild-type (WT) mice. Cardiac hypertrophy after TAC was attenuated in HMGB1-Tg mice and the survival rate after TAC was higher in HMGB1-Tg mice than in WT mice. Induction of foetal cardiac genes was decreased in HMGB1-Tg mice compared with WT mice. Nuclear HMGB1 expression was preserved in HMGB1-Tg mice compared with WT mice and significantly attenuated DNA damage after TAC was attenuated in HMGB1-TG mice.
Conclusion
These results suggest that the maintenance of stable nuclear HMGB1 levels prevents hypertrophy and heart failure by inhibiting DNA damage.
Keywords: HMGB1, Heart failure, Acetylation, Translocation, Pressure overload
1. Introduction
Cardiac hypertrophy is associated with many forms of heart disease, including ischaemic disease, hypertensive heart disease, and valvular stenosis, and is a major risk factor for the development of heart failure and death.1,2 Despite advances in the treatment of heart failure, it is still one of the leading causes of death in industrialized countries.3 Therefore, elucidation of the mechanisms underlying the progression of cardiac hypertrophy to heart failure is important to develop effective therapeutic strategies for the treatment of heart failure.4
High mobility group box 1 (HMGB1) is a nuclear DNA-binding protein present in various types of cells, which functions in maintaining nucleosome structure, and regulating gene transcription, replication, and DNA repair.5,6 The high degree of conservation among species and organs implies that HMGB1 plays a critical role in the modulation of cellular functions. HMGB1 knockout mice die shortly after birth and show severe hypoglycaemia, indicating that HMGB1 is essential for survival.7 Recent reports investigating the functions of HMGB1 have mainly focused on the extracellular regulation of cells by HMGB1,8 because in addition to its nuclear activity, HMGB1 functions as a cytokine.9 In inflammatory diseases such as sepsis, HMGB1 is translocated from the nucleus to the cytoplasm and is actively secreted into the extracellular environment, where it interacts with several surface molecules, including the receptor for advanced glycation end-products and Toll-like receptors.9,10 Extracellular HMGB1 also regulates both inflammation and regenerating processes.11 In the presence of tissue damage, HMGB1 released from inflammatory cells and necrotic cells stimulates monocytes/macrophages to secrete inflammatory cytokines amplifying the inflammatory response. We have shown that coronary artery ligation in transgenic mice with cardiac-specific overexpression of HMGB1 (HMGB1-TG) resulted in enhanced angiogenesis, improved survival, and restored cardiac function compared with wild-type (WT) mice.12 In contrast, Andrassy et al.13 have shown that serum levels of HMGB-1 are associated with infarct size and the degree of cardiac remodelling in patients with myocardial infarction.
It has been reported in recent years that DNA damage is involved in the progression of heart failure.14 HMGB1 was discovered as a chromatin-binding protein, and is known to adjust transcription and indirectly adjust DNA damage.15 Previous studies have shown that post-translational modification by acetylation of lysine residues is crucial for the translocation of HMGB1 in hepatocytes and immune cells.16,17 Because nuclear HMGB1 plays a critical role in the regulation of DNA repair systems,15 mitochondrial functions,18 gene-specific DNA-binding,19 and morphology in aging and neurodegenerative disorders,20,21 it is suspected that translocation of HMGB1 from the nucleus might modify cellular function. However, the role of nuclear HMGB1 in cardiac hypertrophy and heart failure is not known. In the present study, we examined the role of nuclear HMGB1 in protecting the heart during pressure overload.
2. Methods
The methods/protocols used in the present study are detailed in the Supplementary material online.
2.1. Materials and reagents
Endothelin-1 (ET-1) and angiotensin II (Ang II) were purchased from Sigma-Aldrich Japan (Tokyo, Japan). The antibody against HMGB1 was obtained from Shino-Test Corporation (Sagamihara, Japan). Antibodies against acetyl-lysine and β-tubulin were obtained from Cell Signaling Technology (Danvers, MA, USA). The anti-8-hydroxy-2′-desoxiguanosine (8-OHdG) antibody was obtained from Nikken Seil (Tokyo, Japan), and the anti-histone H3 antibody was obtained from MAB Institute, Inc. (Sapporo, Japan). The anti-actinin was obtained from Sigma-Aldrich Japan (Tokyo, Japan). The anti-platelet endothelial cell adhesion molecule was obtained from Cedarlane Laboratories Limited (Ontario, Canada). Primers for quantitative real-time reverse transcriptase–PCR were designed on the basis of GenBank sequences [mouse atrial natriuretic peptide (ANP), K02781; mouse brain natriuretic peptide (BNP), NM 008726; rat ANP, NM 012612.2; β- myosin heavy chain (β-MHC), AY 056464 and GAPDH, NM001001303]. Luciferase reporter constructs (hANP/luc and BNP/luc) were generated, and were used to examined the effect of nuclear HMGB1 on foetal gene expression.22,23 The pGEX-5X-1 plasmids were kindly provided by Ikuo Maruyama (Kagoshima University Faculty of Medicine, Kagoshima, Japan).
2.2. Assessment of HMGB1 localization in human heart samples
Samples of the ventricle of three patients with heart failure and three control patients who were assessed to rule out cardiomyopathy and had normal cardiac function were used in the study (Table 1). Written informed consent was obtained from all the patients before the study. The protocol was performed in accordance to the Helsinki Declaration and was approved by the human investigations committee of our institution. Biopsy samples were immediately washed in phosphate buffered saline (PBS) before being snap-frozen in liquid nitrogen for immunofluorescent co-staining and biomechanical measurements. Fresh frozen 20 μm tissue sections were treated with a blocking agent prior to the addition of primary antibody at a dilution of 1:100. Sections were then treated with goat anti-rabbit Alexa Fluor 568 (Invitrogen, Carlsbad, CA, USA) at a dilution of 1:200. Samples were counterstained with Phalloidin (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI) (Lonza, Walkersville, MD, USA). For analysis, nuclear positive cells were counted (five random fields to yield ∼300 cardiomyocyte nuclei/section) and expressed as the percentage of the total number of cardiomyocytes.
Table 1.
Characteristics of patients with heart failure and healthy controls
| Age (years) | Gender | Status | Aetiology of heart failure | BNP (pg/mL) | LVEF (%) | LVDd (mm) |
|---|---|---|---|---|---|---|
| 62 | Male | Normal | — | 40 | 74 | 43 |
| 55 | Male | Normal | — | 8 | 77 | 53 |
| 19 | Male | Normal | — | 31 | 79 | 50 |
| 82 | Female | Heart failure | dHCM | 376 | 36 | 48 |
| 71 | Male | Heart failure | HHD | 334 | 29 | 62 |
| 78 | Female | Heart failure | dHCM | 270 | 44 | 55 |
BNP, brain natriuretic peptide; LVEF, left-ventricular ejection fraction; LVDd, left-ventricular end-diastolic dimension; LVMI, LV mass index; dHCM, dilated phase of hypertrophic cardiomyopathy; HHD, hypertensive heart disease.
2.3. Cultured neonatal rat cardiomyocytes
Hearts were collected from 1- to 2-day-old neonatal rat pups, promptly after euthanasia by decapitation, and primary cultures of neonatal rat cardiomyocytes were performed as described previously.24,25 After serum starvation, neonatal rat cardiomyocytes were stimulated with ET-1 or Ang II, and samples were collected to examine the expression levels of HMGB1 by western blot analysis and ANP mRNA by quantitative RT–PCR. HMGB1 siRNA was purchased from Thermo Scientific Dharmacon (Lafayette, CO, USA) and used to transfect cardiomyocytes by using GenomOne-Neo (Ishihara Sangyo Kaisha, Osaka, Japan) according to the manufacturer's instructions. ET-1 inducible ANP and BNP promoter activities were evaluated by luciferase reporter gene assay with hANP/luc and BNP/luc, and the pRL-TK vector (Promega, Madison, WI, USA).26,27 Expression vector transfection was performed using Lipofectamine LTX plus (Invitrogen) according to the manufacturer's instructions. Transcriptional activities were calculated from three separate assays performed in triplicate. Cardiomyocytes transfected with pmax-GFP (Lonza) and an HMGB1 expression vector or control vector for 24 h were incubated with or without ET-1 (100 nM) for an additional 48 h for cell surface area measurements using Image J software (US National Institutes of Health, Bethesda, MD, USA).
2.4. Pressure overload models
Eight to 10-week-old transgenic mice with cardiac-specific overexpression of HMGB1 (HMGB1-Tg)12 and WT littermates were anaesthetized by intraperitoneal injection with a mixture of ketamine (80 mg/kg/h) and xylazine (8 mg/kg/h), intubated, and artificially ventilated as previously described. Pressure overload was then produced by thoracic transverse aortic constriction (TAC). Standard lead II ECG was recorded throughout the experiment, and the adequacy of anaesthesia was monitored from the disappearance of pedal withdrawal reflex. Cardiac function at 4 weeks after TAC or sham operation was evaluated by transthoracic echocardiography using an FFsonic 8900 (Fukuda Denshi Co., Tokyo, Japan) equipped with a 13 MHz phased-array transducer under anaesthesia with an intraperitoneal administration of pentobarbital sodium (35 mg/kg). Adequacy of anaesthesia was monitored at all times by assessment of skeletal muscle tone, respiration rate and rhythm, and response to tail pinch. Left-ventricular fractional shortening (LVFS) was calculated as [(LVEDD−LVESD)/LVEDD] × 100 (%). These mice and sham-operated ones were sacrificed by intraperitoneal injection of ketamine (1 g/kg) and xylazine (100 mg/kg), and hearts were rapidly excised. Mouse ANP and BNP mRNA levels were determined by quantitative real-time RT–PCR. Myocardial sections from HMGB1-Tg and WT mice were stained with anti-8-OHdG antibodies to evaluate the degree of DNA damage in the heart. The staining was visualized by treatment with a solution of 3,3′-diaminobenzidine (Dako Cytomation Liquid DAB Substrate Chromogen System, Dako Japan, Tokyo, Japan) for 40 s. 8-OHdG positive area was measured (five random fields to yield around 400 cardiomyocyte) using Image J software and expressed as fold increase over HMGB1-TG TAC mice. Results were normalized by arbitrarily setting the area of the 8-OHDG positive cells in HMGB1-TG TAC mice to 1.0. All experimental procedures were performed according to the animal welfare regulations of Yamagata University School of Medicine, and the study protocol was approved by the Animal Subjects Committee of Yamagata University School of Medicine. The investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, 8th Edition, 2011).
2.5. Co-immunoprecipitaton and immunoblotting
After samples were collected, protein extracts were prepared in modified radio-immunoprecipitation assay 1 (RIPA) buffer. Immunoprecipitation was performed with 1 µg of antibodies against acetyl-lysine or HMGB1 in 400 µg whole lysate protein or 150 µg nuclear protein. Normal rabbit or mouse IgG was used as a negative control. Lysates were incubated with anti-acetyl-lysine or HMGB1 overnight, and then incubated for 1 h with protein A/G-agarose beads. Samples were washed four times with buffer and subjected to western blot analysis.
2.6. Statistical analysis
Data are presented as means ± standard error of the mean (SEM). Differences between groups were evaluated using one-way analysis of variance with post hoc Bonferroni test. Survival curves after TAC were generated using the Kaplan–Meier method and compared using the log-rank test. A P-value <0.05 was considered statistically significant. Statistical analysis was performed with a standard statistical program package (JMP version 8; SAS Institute Inc., Cary, NC, USA).
3. Results
3.1. HMGB1 expression and localization in human failing hearts
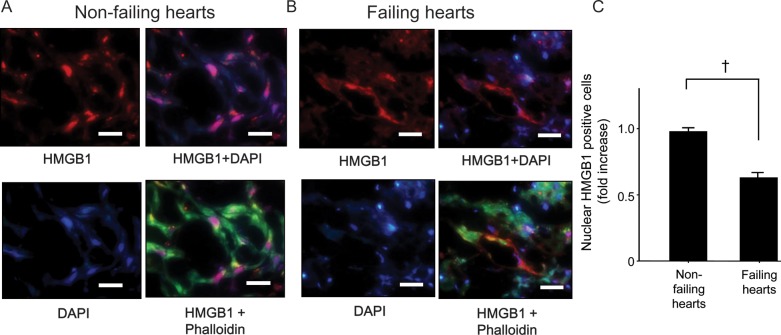
To investigate the expression and localization of HMGB1 in human failing hearts, myocardial samples of patients with heart failure and healthy controls (Table 1) were analysed by immunohistochemistry. HMGB1 was localized in the nuclei of cardiomyocyte in control hearts (Figure 1A). On the contrary, decreased nuclear staining and increased cytosolic staining of HMGB1 were detected in the failing heart (Figure 1B). Comparison of the percentage of HMGB1 positively stained nuclei in failing and non-failing heart samples showed that nuclear HMGB1 expression was clearly lower in failing hearts than in control hearts (Figure 1C) because of the translocation of nuclear HMGB1 to the cytoplasm in failing hearts. These findings suggested that nuclear localization of HMGB1 in cardiomyocytes might be associated with cardiac dysfunction during cardiac remodelling.
Figure 1.
Decreased nuclear HMGB1 in the failing heart. (A) Non-failing (left) and failing (right) human heart tissues were labelled with antibodies against HMGB1 (red) and phalloidin (green). Nuclei were stained with DAPI (blue). Bars, 50 μm. (B) Quantification of nuclear HMGB1 immunofluorescence data. Bars represent means ± SEM.
3.2. Translocation and acetylation of HMGB1 induced by hypertrophic stimulation
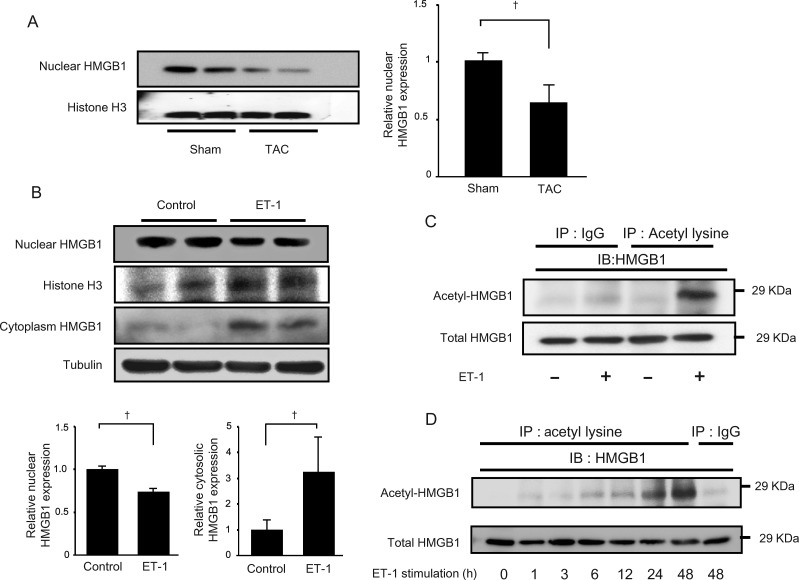
To confirm whether translocation of HMGB1 was observed during LV remodelling, we evaluated the localization of HMGB1 in heart samples of mice in which pressure overload was generated by TAC. Similar to the observations in human failing heart samples, nuclear HMGB1 was decreased after TAC (Figure 2A). HMGB1 is mainly expressed in the nuclei of cardiomyocytes in sham mice. On the other hand, decreased nuclear staining and increased cytosolic staining of HMGB1 were observed in the cardiomyocytes after pressure overload (Supplementary material online, Figure S1A and B). However, HMGB1 expressions in endothelial cells were similar between sham- and TAC-operated mice (Supplementary material online, Figure S1B). Nuclear HMGB1 expression was decreased in neonatal rat cardiomyocytes after ET-1 (Figure 2B) or Ang II (Supplementary material online, Figure S2A) stimulation, whereas cytosolic HMGB1 was increased compared with without stimulation. To determine whether HMGB1 acetylation is associated with the localization of HMGB1 in cardiomyocytes, neonatal rat cardiomyocytes were stimulated with ET-1 or Ang II, and HMGB1 acetylation status was evaluated. ET-1 stimulation increased the acetylation of HMGB1, and in a time-dependent manner with a maximum at 48 h (Figure 2C and D). Ang II also induced HMGB1 acetylation as shown in Supplementary material online, Figure S2B. These findings indicated that nuclear HMGB1 translocated from the nucleus to the cytoplasm during cardiac remodelling.
Figure 2.
Translocation of HMGB1 induced by pressure overload and hypertrophic stimulation. (A) Decrease in nuclear localization of HMGB1 after pressure overload. Data are expressed as means ± SEM. †P < 0.05 vs WT TAC mice. (B) HMGB1 expression in the nucleus and cytoplasm of neonatal rat cardiomyocytes after ET-1 (100 nM) stimulation. Data are expressed as means means ± SEM. †P < 0.05 vs control. (C) Lysates of neonatal rat cardiomyocytes treated with or without ET-1 were immunoprecipitated with an anti-acetyl-lysine antibody and immunoblotted against an anti-HMGB1 antibody. (D) Lysates of neonatal rat cardiomyocytes following stimulus with ET-1 (100 nM) were immunoprecipitated with an anti-acetyl-lysine antibody and immunoblotted against HMGB1.
3.3. The impact of nuclear HMGB1 on the cardiac foetal gene expression
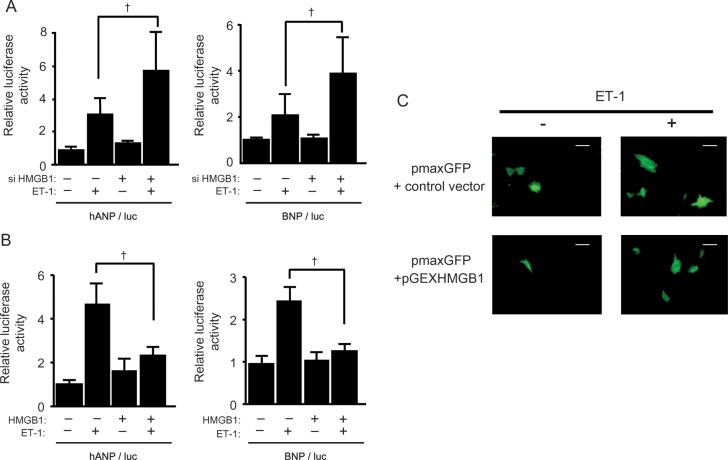
To determine the impact of nuclear HMGB1 on cardiac hypertrophy, foetal cardiac gene expression in cardiomyocytes was assessed using HMGB1 siRNA. Specific siRNA against HMGB1 blocked its expression by >70% (Supplementary material online, Figure S3A). ANP and BNP promoter activities were increased by ET-1 stimulation in cardiomyocytes. Moreover, co-transfection with HMGB1 siRNA enhanced ANP and BNP promoter activities compared with control siRNA transfection (Figure 3A), and increased ANP RNA expression (Supplementary material online, Figure S3B), suggesting that loss of nuclear HMGB1 might be associated with cardiomyocyte hypertrophy during ET-1 stimulation.
Figure 3.
Protective effect of nuclear HMGB1 on ET-1 induced hypertrophy. (A) Increase in ANP and BNP promoter activity by HMGB1 SiRNA transfection after ET-1 stimulation. Data represent means ± SEM from three independent experiments; †P < 0.05. (B) Suppressed ANP and BNP promoter activity after ET-1 stimulation in HMGB1 vector transfected cardiomyocytes. Data represent means ± SEM; †P < 0.05. (C) Effect of HMGB1 overexpression on the cell surface area of neonatal rat cardiomyocytes after ET-1 stimulation. Cardiomyocytes were transfected with pmax-GFP with either an HMGB1 expression vector or a control vector, and incubated for 48 h with or without ET-1. Scale bar = 20 μm.
To confirm whether the maintenance of HMGB1 levels in the nucleus could suppress the development of cardiomyocyte hypertrophy, the HMGB1 construct was co-transfected with ANP or BNP promoter luciferase constructs. Overexpression of HMGB1 significantly attenuated ANP and BNP promoter activity after ET-1 stimulation (Figure 3B). Similarly, the increase in ANP RNA expression after ET-1 stimulation was also suppressed by HMGB1 overexpression (Supplementary material online, Figure S4A). In addition, ET-1 induced hypertrophic changes in cardiomyocytes were significantly attenuated by HMGB1 overexpression (Figure 3C, Supplementary material online, Figure S4B). These findings implied that preservation of the expression of nuclear HMGB1 may suppress cardiomyocyte hypertrophy.
3.4. HMGB1 in the hypertrophic and heart failure model
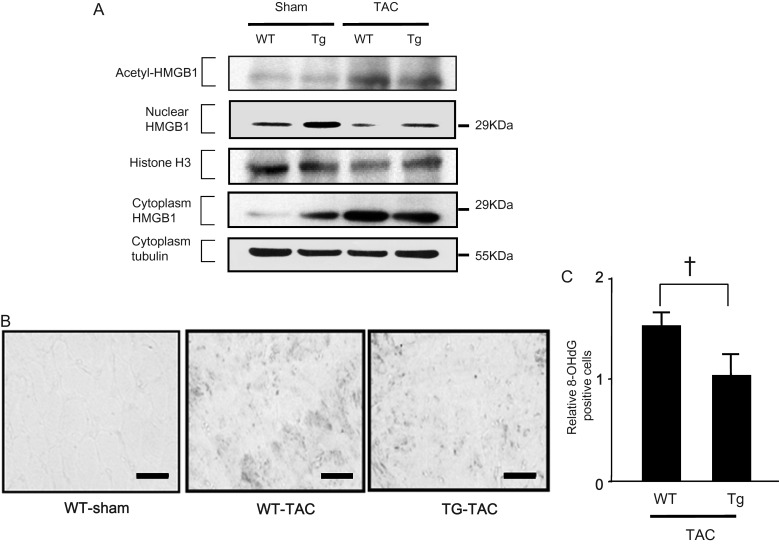
To examine the role of nuclear HMGB1 in the development of cardiac hypertrophy and heart failure in vivo, cardiac-specific HMGB1 overexpressing (HMGB1-Tg) mice and WT mice were subjected to TAC or sham surgery. The levels of acetylated and cytosolic HMGB1 were similarly increased and cardiac nuclear HMGB1 was decreased after TAC in WT and HMGB1-Tg mice compared with those undergoing sham surgery (Figure 4A). Notably, nuclear HMGB1 levels after TAC were higher in HMGB1-Tg mice than in WT mice, indicating that nuclear HMGB1 levels were preserved in HMGB1-Tg mice (Figure 4A). To investigate the role of nuclear HMGB1 in protecting cardiomyocytes from DNA damage in cardiac hypertrophy, we performed immunohistochemical staining of TAC-operated hearts using an anti- 8-OHdG antibody (Figure 4B). Sham-operated mice did not show 8-OHdG positive cardiomyocytes. On the other hand, in mice undergoing TAC, 8-OHdG expression was significantly increased in both HMTG1-TG and WT mice, whereas the induction of 8-OHdG was suppressed in HMGB1-Tg mice. Comparison of the relative 8-OHDG positively stained cells in WT and Tg mice after TAC showed that 8-OHDG expression was clearly lower in Tg mice than in WT mice (Figure 4C).
Figure 4.
Preserved amounts of nuclear HMGB1 prevented DNA damage induced by pressure overload. (A) Acetylation and translocation of cardiac HMGB1 from the nucleus to the cytoplasm after pressure overload. (B) Immunohistochemical staining of heart tissues with an anti-8-OHdG antibody 4 weeks after TAC and quantification of histological data (n = 5). Scale bar, 50 mm. (C) Results were normalized by arbitrarily setting the area of the 8-OHDG positive cells in HMGB1-TG TAC mice to 1.0. Bars represent means ± SEM. (n = 5). †P < 0.05 vs. WT TAC mice.
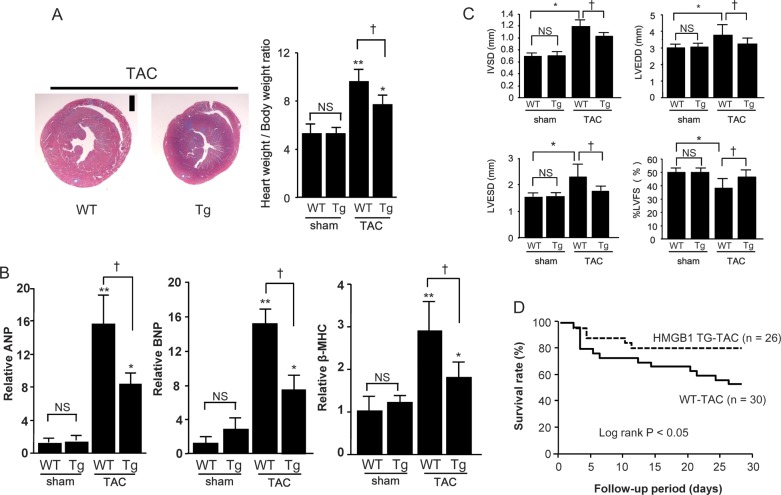
At 4 weeks after the TAC operation, the increase in the weight of the heart was significantly lower in HMGB1-Tg mice than in WT-mice (Figure 5A). We next examined the mRNA expression of foetal cardiac genes after TAC. The expressions of ANP, BNP, and β-MHC were significantly up-regulated in the TAC group compared with the sham-surgery group, and this up-regulation was significantly attenuated in HMGB1-Tg mice (Figure 5B). Moreover, systolic dysfunction and left-ventricular dilatation after TAC were attenuated in HMGB1-Tg mice compared with WT-mice (Figure 5C, Supplementary material online, Table). Furthermore, the survival rate after TAC was significantly higher in HMGB1-Tg mice than in WT mice (Figure 5D).
Figure 5.
Suppression of DNA damage induced by pressure overload in HMGB-1 Tg mice. (A) Left-ventricular transverse sections in WT and HMGB1-Tg mice at 4 weeks after TAC, and heart weight: body ratios in TAC or sham-operated mice. Data are mean ± SEM from eight mice for each group. *P < 0.05 and **P < 0.01 vs. sham-operated mice of the same strain; †P < 0.05 vs. TAC-operated Tg mice. (B) Quantitative analyses of ANP, BNP, and β-MHC gene expression in WT and HMGB1-Tg mice at 4 weeks after TAC. Data are mean ± SEM from eight mice for each group. *P < 0.05 and **P < 0.01 vs. sham-operated mice of the same strain; †P < 0.05 vs. TAC-operated Tg mice. (C) Data showing echocardiographic measurements in WT and HMGB1-Tg mice at 4 weeks after TAC or sham surgery. IVS, interventricular wall thickness; LVEDD, left-ventricular end-diastolic dimension; LVESD, left-ventricular end-systolic dimension; %LVFS, left-ventricular fractional shortening. Data are mean ± SEM from eight mice for each group. *P < 0.05 vs. sham-operated WT mice; †P < 0.05 vs. TAC-operated WT mice. (D) Survival curves in WT and HMGB1-Tg mice after TAC.
4. Discussion
In the present study, we demonstrated the critical role of intracellular HMGB1 in the development of cardiac hypertrophy and heart failure. We found that cardiac HMGB1 was exported from the nucleus to the cytoplasm in patients with heart failure. Our in vitro study showed that nuclear HMGB1 was acetylated and translocated to the cytoplasm in neonatal rat cardiomyocytes in response to ET-1 and Ang II stimulation. We also demonstrated that loss of nuclear HMGB1 aggravated foetal gene expressions induced by ET-1 stimulation. In contrast, this up-regulation of gene expression was suppressed by HMGB1 overexpression. We further showed that preserved HMGB1 expression in the nucleus attenuated DNA damage during pressure overload, and abolished ventricular remodelling and cardiac dysfunction.
HMGB1, which is ubiquitously expressed in all vertebrate nuclei with a uniquely conserved sequence among species, was identified as a chromosomal protein with important structural functions in chromatin organization.5,6 HMGB1 binds to double-stranded DNA and interacts with other DNA-binding proteins, which facilitate chromatin bending.28,29 On the other hand, HMGB1 released or secreted into the circulation has attracted attention for its cytokine-like function and involvement in the pathology of cardiovascular disease.8,12,30,31 Scaffidi et al.8 reported that HMGB1 secreted from inflammatory cells and passively released from necrotic cells promoted inflammation. A role for extracellular HMGB1 in the regulation of inflammation was proposed based on its association with Toll-like receptor family members, the interleukin-1 receptor, and the receptor for advanced glycation end-products.10,16,32,33 However, the localization of HMGB1 in cardiomyocytes and its potential role in the pathogenesis of human heart failure have not been studied in detail. In the present study, HMGB1 localized to the nucleus in the non-failing heart, as expected, whereas nuclear HMGB1 levels decreased and cytosolic HMGB1 increased in the failing human heart. These findings suggest that nuclear HMGB1 may play an important role in the regulation of cardiomyocyte phenotype in relation to the pathogenesis of human heart failure.
In addition to the mechanisms mediating the secretion of HMGB1 from necrotic cells, other pathways involved in the export of HMGB1 from the nucleus have been reported.17 Prior studies have reported that post-translational modification of HMGB1 including acetylation in cultured cells modifies the binding of HMGB1 to DNA and its extranuclear localization.17,34,35 Hyperacetylation of HMGB1 induced by lipopolysaccharide and hydrogen peroxide triggers its translocation into the cytoplasm.17,36 In the present study, stimulation of cardiomyocytes with ET-1 or Ang II and pressure overload in mice induced the acetylation and translocation of nuclear HMGB1. To the best of our knowledge, this is the first report of HMGB1 acetylation and translocation from the nucleus in cardiomyocytes.
Whether extracellular HMGB1 acts as a cardioprotective factor is the subject of controversy, and the effect of nuclear HMGB1 under hypertrophic stimulation is also poorly understood. We showed that the expression of foetal genes induced by ET-1 was inhibited by HMGB1 overexpression. In addition, we showed that pressure overload-induced foetal gene expression is attenuated in HMGB1-Tg mice compared with WT mice. Oxidative stress induced by pressure overload contributes to cardiac DNA damage and DNA repair/synthesis in failing hearts with systolic dysfunction.37 Therefore, DNA damage is thought to be a key pathogenic factor in ventricular dysfunction.14 A recent study showed the involvement of intracellular HMGB1 in protecting against DNA damage in the brain and in neurons.20,21 Nuclear HMGB1 was recruited to sites of cellular oxidative DNA base damage, and cell-based experiments and biochemical data suggested that HMGB1 plays a role in base excision repair.15,20,21 In the present study, 8-OHdG induction after TAC was significantly attenuated in HMGB1-Tg mice compared with WT mice. Taken together, these findings indicate that nuclear HMGB1 might prevent DNA damage during pressure overload, as seen in the decrease in cardiac dysfunction in HMBG1-Tg mice. HMGB1 translocation induced by hypertrophic stimulation might cause DNA damage, increasing the severity of cardiac hypertrophy, affecting foetal gene expression, and causing cardiac dysfunction.
There are several limitations in this study. In the present study, we showed that maintenance of HMGB1-TG in cardiomyocyte and the heart prevent cardiac dysfunction after pressure overload, however, we did not show the direct link between reduced HMGB1 in the heart and cardiac dysfunction in vivo study. To confirm this point, we might need to evaluate cardiac function in cardiac-specific HMGB1 knockout mice in the future study. However, we demonstrated that intranuclear HMGB1 was reduced in failing heart, and HMGB1 silencing promoted foetal gene expressions. Therefore, the findings of this study suggest a novel approach to the investigation of the pathogenesis of heart failure, and nuclear HMGB1 may be a potential novel therapeutic target for the prevention of heart failure.
5. Conclusions
We demonstrated that nuclear HMGB1 was decreased in association with human heart failure and preserved amounts of nuclear HMGB1 could prevent cardiac hypertrophy and improve survival in a pressure overload heart failure model.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported in part by a grant-in-aid for Scientific Research (Nos. 21590923 and 24659380 to I.K., 25860580 to A.F., and 23790830 to T.S.) from the Ministry of Education, Science, Sports, and Culture, Japan, a grant-in-aid from the 21st Global Century Center of Excellence (COE) program of the Japan Society for the Promotion of Science to I.K. T.S. was supported by the Japan Heart Foundation Research Grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
Acknowledgements
We thank Ms Emiko Nishidate and Ms Miyuki Tsuda for their excellent technical assistance.
Conflict of interest: none declared.
References
- 1.Jessup M, Brozena S. Heart failure. N Engl J Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 2.McMurray JJ, Pfeffer MA. Heart failure. Lancet. 2005;365:1877–1889. doi: 10.1016/S0140-6736(05)66621-4. [DOI] [PubMed] [Google Scholar]
- 3.Roger VL, Weston SA, Redfield MM, Hellermann-Homan JP, Killian J, Yawn BP, et al. Trends in heart failure incidence and survival in a community-based population. JAMA. 2004;292:344–350. doi: 10.1001/jama.292.3.344. [DOI] [PubMed] [Google Scholar]
- 4.Rame JE, Ramilo M, Spencer N, Blewett C, Mehta SK, Dries DL, et al. Development of a depressed left ventricular ejection fraction in patients with left ventricular hypertrophy and a normal ejection fraction. Am J Cardiol. 2004;93:234–237. doi: 10.1016/j.amjcard.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 5.Nightingale K, Dimitrov S, Reeves R, Wolffe AP. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 6.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci USA. 2008;105:10320–10325. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 8.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Hreggvidsdottir HS, Palmblad K, Wang H, Ochani M, Li J, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci USA. 2010;107:11942–11947. doi: 10.1073/pnas.1003893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 11.Germani A, Limana F, Capogrossi MC. Pivotal advances: high-mobility group box 1 protein–a cytokine with a role in cardiac repair. J Leukoc Biol. 2007;81:41–45. doi: 10.1189/jlb.0306165. [DOI] [PubMed] [Google Scholar]
- 12.Kitahara T, Takeishi Y, Harada M, Niizeki T, Suzuki S, Sasaki T, et al. High-mobility group box 1 restores cardiac function after myocardial infarction in transgenic mice. Cardiovasc Res. 2008;80:40–46. doi: 10.1093/cvr/cvn163. [DOI] [PubMed] [Google Scholar]
- 13.Andrassy M, Volz HC, Riedle N, Gitsioudis G, Seidel C, Laohachewin D, et al. HMGB1 as a predictor of infarct transmurality and functional recovery in patients with myocardial infarction. J Intern Med. 2011;270:245–253. doi: 10.1111/j.1365-2796.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki S, Shishido T, Ishino M, Katoh S, Sasaki T, Nishiyama S, et al. 8-Hydroxy-2′-deoxyguanosine is a prognostic mediator for cardiac event. Eur J Clin Invest. 2011;41:759–766. doi: 10.1111/j.1365-2362.2010.02465.x. [DOI] [PubMed] [Google Scholar]
- 15.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, et al. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell. 2007;27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evankovich J, Cho SW, Zhang R, Cardinal J, Dhupar R, Zhang L, et al. High mobility group box 1 release from hepatocytes during ischemia and reperfusion injury is mediated by decreased histone deacetylase activity. J Biol Chem. 2010;285:39888–39897. doi: 10.1074/jbc.M110.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enokido Y, Yoshitake A, Ito H, Okazawa H. Age-dependent change of HMGB1 and DNA double-strand break accumulation in mouse brain. Biochem Biophys Res Commun. 2008;376:128–133. doi: 10.1016/j.bbrc.2008.08.108. [DOI] [PubMed] [Google Scholar]
- 21.Qi ML, Tagawa K, Enokido Y, Yoshimura N, Wada Y, Watase K, et al. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol. 2007;9:402–414. doi: 10.1038/ncb1553. [DOI] [PubMed] [Google Scholar]
- 22.Kuwahara K, Saito Y, Ogawa E, Takahashi N, Nakagawa Y, Naruse Y, et al. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol Cell Biol. 2001;21:2085–2097. doi: 10.1128/MCB.21.6.2085-2097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shishido T, Woo CH, Ding B, McClain C, Molina CA, Yan C, et al. Effects of MEK5/ERK5 association on small ubiquitin-related modification of ERK5: implications for diabetic ventricular dysfunction after myocardial infarction. Circ Res. 2008;102:1416–1425. doi: 10.1161/CIRCRESAHA.107.168138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le NT, Takei Y, Shishido T, Woo CH, Chang E, Heo KS, et al. p90RSK targets the ERK5-CHIP ubiquitin E3 ligase activity in diabetic hearts and promotes cardiac apoptosis and dysfunction. Circ Res. 2012;110:536–550. doi: 10.1161/CIRCRESAHA.111.254730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo CH, Massett MP, Shishido T, Itoh S, Ding B, McClain C, et al. ERK5 activation inhibits inflammatory responses via peroxisome proliferator-activated receptor delta (PPARdelta) stimulation. J Biol Chem. 2006;281:32164–32174. doi: 10.1074/jbc.M602369200. [DOI] [PubMed] [Google Scholar]
- 27.Woo CH, Le NT, Shishido T, Chang E, Lee H, Heo KS, et al. Novel role of C terminus of Hsc70-interacting protein (CHIP) ubiquitin ligase on inhibiting cardiac apoptosis and dysfunction via regulating ERK5-mediated degradation of inducible cAMP early repressor. FASEB J. 2010;24:4917–4928. doi: 10.1096/fj.10-162636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallier C, Scaffidi P, Chopineau-Proust S, Agresti A, Nordmann P, Bianchi ME, et al. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol Biol Cell. 2003;14:3414–3426. doi: 10.1091/mbc.E02-09-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai K, Naruse Y, Shaul Y, Agata Y, Mori N. Direct interaction of NRSF with TBP: chromatin reorganization and core promoter repression for neuron-specific gene transcription. Nucleic Acids Res. 2004;32:3180–3189. doi: 10.1093/nar/gkh550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrassy M, Volz HC, Igwe JC, Funke B, Eichberger SN, Kaya Z, et al. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation. 2008;117:3216–3226. doi: 10.1161/CIRCULATIONAHA.108.769331. [DOI] [PubMed] [Google Scholar]
- 31.Limana F, Germani A, Zacheo A, Kajstura J, Di Carlo A, Borsellino G, et al. Exogenous high-mobility group box 1 protein induces myocardial regeneration after infarction via enhanced cardiac C-kit+ cell proliferation and differentiation. Circ Res. 2005;97:e73–83. doi: 10.1161/01.RES.0000186276.06104.04. [DOI] [PubMed] [Google Scholar]
- 32.El Gazzar M. HMGB1 modulates inflammatory responses in LPS-activated macrophages. Inflamm Res. 2007;56:162–167. doi: 10.1007/s00011-006-6112-0. [DOI] [PubMed] [Google Scholar]
- 33.Treutiger CJ, Mullins GE, Johansson AS, Rouhiainen A, Rauvala HM, Erlandsson-Harris H, et al. High mobility group 1 B-box mediates activation of human endothelium. J Intern Med. 2003;254:375–385. doi: 10.1046/j.1365-2796.2003.01204.x. [DOI] [PubMed] [Google Scholar]
- 34.Assenberg R, Webb M, Connolly E, Stott K, Watson M, Hobbs J, et al. A critical role in structure-specific DNA binding for the acetylatable lysine residues in HMGB1. Biochem J. 2008;411:553–561. doi: 10.1042/BJ20071613. [DOI] [PubMed] [Google Scholar]
- 35.Pasheva E, Sarov M, Bidjekov K, Ugrinova I, Sarg B, Lindner H, et al. In vitro acetylation of HMGB-1 and -2 proteins by CBP: the role of the acidic tail. Biochemistry. 2004;43:2935–2940. doi: 10.1021/bi035615y. [DOI] [PubMed] [Google Scholar]
- 36.Ugrinova I, Pashev IG, Pasheva EA. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry. 2009;48:6502–6507. doi: 10.1021/bi9004304. [DOI] [PubMed] [Google Scholar]
- 37.Siggens L, Figg N, Bennett M, Foo R. Nutrient deprivation regulates DNA damage repair in cardiomyocytes via loss of the base-excision repair enzyme OGG1. FASEB J. 2012;26:2117–2124. doi: 10.1096/fj.11-197525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.