Abstract
Although a functional role in copper binding has been suggested for the prion protein, evidence for binding at affinities characteristic of authentic metal-binding proteins has been lacking. By presentation of copper(II) ions in the presence of the weak chelator glycine, we have now characterized two high-affinity binding sites for divalent transition metals within the human prion protein. One is in the N-terminal octapeptide-repeat segment and has a Kd for copper(II) of 10−14 M, with other metals (Ni2+, Zn2+, and Mn2+) binding three or more orders of magnitude more weakly. However, NMR and fluorescence data reveal a previously unreported second site around histidines 96 and 111, a region of the molecule known to be crucial for prion propagation. The Kd for copper(II) at this site is 4 × 10−14 M, whereas nickel(II), zinc(II), and manganese(II) bind 6, 7, and 10 orders of magnitude more weakly, respectively, regardless of whether the protein is in its oxidized α-helical (α-PrP) or reduced β-sheet (β-PrP) conformation. A role for prion protein (PrP) in copper metabolism or transport seems likely and disturbance of this function may be involved in prion-related neurotoxicity.
The prion diseases include Creutzfeldt–Jakob disease, Gerstmann–Sträussler–Scheinker disease, and kuru in humans, and scrapie and bovine spongiform encephalopathy in animals (1, 2). Disease states are associated with the accumulation of an abnormal isomer (PrPSc) of host-derived cellular prion protein (PrPC). Neither amino acid sequencing nor systematic study of known covalent posttranslational modifications have shown any consistent differences between PrPC and PrPSc (3). The only consistent differences reported are in chain conformation and state of assembly, as shown by biophysical measurements on secondary structure content of the two isoforms. Both circular dichroism (CD) and Fourier transform infrared (FTIR) spectroscopic methods showing PrPC to be rich in α-helix, whereas PrPSc has a high β-sheet content (4).
Several observations have excited interest in the binding of transition metals to the prion protein and in the structural and functional consequences of such interactions. For instance, it has been demonstrated that different PrPSc types, characteristic of clinically distinct subtypes of sporadic CJD, can be interconverted in vitro by altering the metal ion occupancy (5). In addition, copper chelators can induce spongiform change in experimental animals (6) and it has been claimed that the levels of copper in the brains of PrP-null mice are lower than in wild-type mice, although this has not been replicated by other workers (7, 8). Moreover, it has been reported that recombinant PrP possesses copper-dependent superoxide dismutase, albeit at low levels (9). The prion protein has also been proposed to function as a copper transport protein for internalization of copper(II) ions (10). A number of lines of evidence argue, therefore, that PrP may be a metalloprotein in vivo.
With regard to physical measurements of metal interactions, it has been shown that synthetic peptides corresponding to the octapeptide repeat region of PrP bind copper(II) ions (11, 12). The authors concluded that copper ions bound specifically to a peptide encompassing residues 60–91 with a dissociation constant of 6.7 μM and a 4:1 stoichiometry. A similar binding affinity (Kd = 5.9 μM) was reported for a longer fragment encompassing residues 23–98 (7). In this study, positive cooperativity was observed that indicated the presence of a tight-binding site. Indeed, copper ions were presented in the presence of glycine as a chelator, but no correction was made for this in the determination of the binding affinity. The stoichiometry was reported to be 5.6 copper(II) ions per peptide, which suggests the tight binding was missed because of the dominance of the additional weak sites. Binding of metal ions to full-length recombinant hamster protein (sHaPrP29–231) has also been studied and here the authors concluded that binding was specific for copper(II) ions (13) with an apparent stoichiometry of ≈1.8 copper ions per PrP molecule and an average dissociation constant of 14 μM. Peptides corresponding to varying copy lengths of the octapeptide motif were studied by CD spectroscopy and NMR (14), revealing an apparent binding affinity of 6 μM and a stoichiometry of four copper(II) ions per four copies of the octapeptide repeat. Binding to the octapeptide repeat region was studied further by mass spectrometry with similar findings as regards stoichiometry (15). Here the authors describe a pH dependency to copper binding with only three sites available at pH 6.0 and a total of five copper ions bound at pH 7.4. Binding to a short peptide encompassing residues 23–60 of hamster PrP indicated copper bound in a cooperative manner (15).
The puzzling aspect of these studies is the extremely weak binding affinity for copper(II). Binding constants in the micromolar range would lead to the conclusion that such interactions are physiologically irrelevant. Also, given the nature of the octapeptide repeat region alone, with its five histidine side chains, one would expect copper(II) to bind with a far greater affinity. For instance, for the square planar coordination of Cu(II) by four independent imidazole groups, the effective binding affinity is 3 × 10−13 M and even for simple organic oxo acids such as malonate the affinity is 10−8 M (16). To address these issues we have investigated the binding of divalent metals to both the octapeptide-repeat region (residues PrP52–98) and to the recombinantly expressed PrP91–231 protein with a view to determining, rigorously, both the affinity and the location of sites of interaction.
Materials and Methods
Purification of Human PrP.
Recombinant human PrP91–231 in either the oxidized α-form or the reduced β-form was prepared by a modification of the method described (17). To ensure the proteins were free of metal ions before titration they were refolded in the presence of 100 mM EDTA and dialyzed extensively against 5 mM 3-(N-morpholino)propanesulfonic acid (Mops, pH 8.0). Protein concentration was determined by UV absorption using a calculated molar extinction coefficient of 19632 M−1⋅cm−1 at 280 nm.
Preparation of Peptide.
The peptide QG(GGGWGQPH)4GGGWGQGGGTHSQ corresponding to residues 52–98 of the human PrP was synthesized on a MilliGen 9050 automated peptide synthesizer using fluorenylmethoxycarbonyl (Fmoc) chemistry with HBTU (O-benzotriazol-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate) activation on a MilliGen PEG-PS resin using customized protocols. Side-chain protection was as follows: trityl for Gln and His, t-butyl for tyrosine, and protonation for arginine. After completion of the synthesis the resin was washed with methanol and dichloromethane and dried under high vacuum overnight. Peptides were cleaved from the resin by using TFA:ethanediol:anisole:H2O in the ratio 94:2:2:2 for 2 h. The peptides were then recovered by ether precipitation and purified by HPLC. Electrospray mass spectrometry performed on a VG Analytical Quattro spectrometer (Manchester, U.K.) showed this peptide to be greater than 95% pure with a molecular mass of 4479.31 Da. Before use peptide was treated with 100 mM EDTA and dialyzed extensively against 5 mM Mops (pH 8.0).
Determination of Dissociation Constants for Metal Ions.
Samples of either human α-PrP91–231 or β-PrP91–231 were prepared at 1 μM concentration in 5 mM Mops (pH 8.0) with varying concentrations of glycine between 1 mM and 100 μM. For binding studies to the peptide 5 μM peptide was used in similar buffers. Chelation of copper ions was performed by the addition of aliquots of the appropriate concentrations of CuSO4 to the samples with binding monitored by quenching of intrinsic tryptophan fluorescence. Fluorescence was measured in a Jasco FP-750 spectrofluorimeter (Easton, MD) with an excitation wavelength of 285 nm and an emission wavelength of 350 nm. All experiments were performed at 20°C.
Competition for the Metal Binding with Other Divalent Metal Ions.
Competition experiments were performed as above with the addition of 100 μM divalent metal ions before titration with copper. All metals were presented as chloride salts with the exception of copper, which was used as the sulfate.
NMR Spectroscopy.
NMR spectra shown were acquired at 298 K on a Bruker DRX-600 spectrometer (Billerica, MA). Sample conditions were as follows: 500 μM α-PrP91–231 uniformly labeled with 15N in 5 mM Mops (pH 6.0) + 10% D2O (vol/vol) was titrated with 125 μM CuSO4 increments to a final concentration of 1000 μM. Glycine was not used in these experiments as the aim was to identify the copper(II)-binding site. Signal was observed at substoichiometric copper concentrations because only a small proportion of the molecules are complexed with copper. As the molar ratio of copper(II) to protein was increase the proportion of molecules with quenched resonances also increased. 1H,15N heteronuclear single quantum correlation (HSQC) spectra were collected with acquisition times of 328 ms and 168 ms in the direct and indirect dimensions, respectively. NMR data were processed and analyzed on Silicon Graphics Workstations (Mountain View, CA), using FELIX 97 software (kindly donated by Molecular Simulations, San Diego). 1H,15N HSQC peaks were assigned by using the previously determined assignment of human α-PrP91–231 (18). Peak intensities following each addition were determined from the 1H,15N HSQC spectra (19,20) and normalized against the peak intensity of the C-terminal residue (S230). The relatively long electron relaxation time of Cu2+ causes an increase in the line width and a loss of intensity of nuclear resonances in its close proximity, the effect diminishing according to the inverse-sixth power of the interatomic distance.
Analytical Methods.
To determine the apparent dissociation constant (Kd(App) ), in conditions where the concentration of metal-binding sites was smaller than Kd(App), quenching data were fitted to the equation
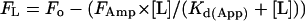 |
where FL is the fluorescence at a given concentration of ligand, [L], Fo is the starting fluorescence, and FAmp is the fluorescence amplitude. Stoichiometries for metal⋅protein complexes were determined in conditions where the concentration of metal-binding sites (Mo) was greater than Kd(App). In these circumstances binding curves were fitted to the quadratically derived function
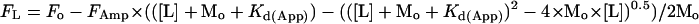 |
Apparent dissociation constants were converted to actual dissociation constants (Kd(Real) ) by using the equation
 |
where [G] is the concentration of glycine used in the experiment and Kd1 and Kd2 are the serial dissociation constants for the binding of divalent metal ions to glycine (16). For competitive binding, the apparent dissociation constant for the competing metal ion (Kd(App) ) is given by
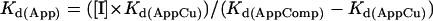 |
where [I] is the molar concentration of the competing metal ion, Kd(AppCu) is the apparent dissociation constant for copper binding in the presence of glycine (but in the absence of competitor), and Kd(AppComp) is the apparent dissociation constant for copper in the presence of glycine and competitor. The apparent dissociation constant for the inhibiting metal ion (Kd(App) ) is then converted to Kd(Real) by correcting for glycine concentration as described above.
Molecular Modeling.
Models were generated by calculating the energetic stabilities of all possible combinations of exactly repeating structure in the octapeptide repeat. To enable exhaustive structural evaluation within the repeating unit, a simplified energetic and structural model was applied. Those conformations that satisfied the requirement for coordination of four histidine side chains around the central transition metal were then assessed for energetic stability by using a united atom model and physicochemical potentials describing hydrophobic, hydrophilic, and backbone hydrogen bonding energies. Conformational space was reduced by restricting residues to adopt one of six alternative backbone φ–ψ torsion angles. The system is described in detail elsewhere (21).
Results
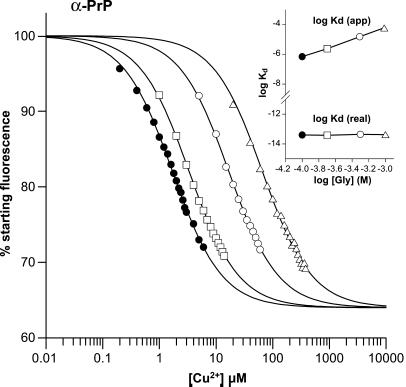
Fig. 1 shows binding curves for Cu(II) when introduced to the PrP (91) protein in its oxidized, native α-helical conformation. The signal recorded was the fluorescence arising from the single tryptophan residue at position 99. The experiment was performed at a series of glycine concentrations and the effective dissociation constant measured (see Inset in Fig. 1). When the real binding constant was back-calculated from these data (see Materials and Methods) it was the same for each determination, demonstrating the robustness of the method. In the presence of glycine, which abolishes weak binding, the stoichiometry of the interaction was 1:1 and the dissociation constant ≈4 × 10−14 M. By the same method the binding affinity for Ni(II) was determined to be ≈2 × 10−8 M and this interaction is competitive with Cu(II), showing that the two metals bind at the same site. The binding affinity for Ni(II) calculated from competition data (see Materials and Methods)—i.e., by its ability to increase the apparent Kd for Cu(II)—was identical to that determined by direct titration.
Figure 1.
Metal binding to recombinant human α-PrP91–231. The main figure shows the data for the binding of copper(II) to human PrP91–231 in the presence of increasing concentrations of glycine. The quenching of intrinsic fluorescence from tryptophan-99 is shown as a percentage of the starting fluorescence and is plotted versus the concentration of copper(II) ions. The data in the presence of 100 μM glycine are shown by solid circles, 200 μM by open squares; 500 μM by open circles, and 1 mM by open triangles. The lines superimposed on the data represent an optimal fit of the data to the equation described above. (Inset) A plot of the apparent dissociation constants versus glycine concentration, which clearly vary. Overlaid are the real dissociation constants derived as described above, which are constant with respect to glycine concentration.
Neither Zn(II) nor Mn(II) induce a significant fluorescence change when added to PrP (91), however the affinities of each can be determined by their effect on the Kd for Cu(II) as described above. Zn(II) binds five times more weakly than Ni(II) and the affinity for Mn(II) is extremely low (Kd = 2 × 10−4 M). Hence, the single binding site for transition metals on PrP (91) is selective and follows the order Cu(II) >> Ni(II) > Zn(II) >> Mn(II). When the protein is reduced and acidified to produce the β-PrP conformation the binding affinities for some of the transition metals are altered (see Table 1). There is a marginal increase in the affinity for Ni(II) and a near abolition of Zn(II) and Mn(II) binding.
Table 1.
Binding affinities of transition metals to α- and β-PrP91-231 and PrP52-98
| Metal (measurement) | α-PrP91-231 | β-PrP91-231 | PrP52-98 |
|---|---|---|---|
| Cu2+ (direct) | 41 fM | 33 fM | 8 fM |
| Ni2+ (direct) | 16 nM | 0.8 nM | 14 nM |
| Ni2+ (competition) | 17 nM | 1.1 nM | 16 nM |
| Zn2+ (competition) | 98 nM | >5 mM | 410 nM |
| Mn2+ (competition) | 202 μM | >5 mM | 2.95 mM |
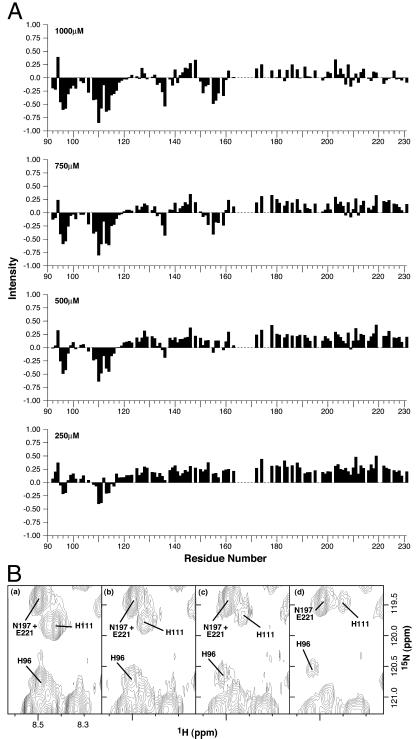
Having established the binding properties of this site, we then wished to determine its location. The HSQC spectrum of human α-PrP91–231 has been assigned (18), thus allowing us to look at site-specific perturbations on binding of metals; these data are summarized in Fig. 2. In this protein, residues 91–123 are unstructured and the NMR analysis shows that during titration to stoichiometry the major shifts occur in this region, chiefly centered around histidine residues 96 and 111. However, within the structured region (124) there are perturbations occurring remote from the binding site (data not shown), implying cooperative changes in conformation upon metal binding. An additional weak site can be seen to titrate above stoichiometric binding centered around residues 135 and 155.
Figure 2.
NMR signal intensity versus residue number. (A) HSQC peak intensities shown sequentially at 250 μM, 500 μM, 750 μM, and 1000 μM CuSO4 relative to 0 μM CuSO4; showing the effects of addition of Cu2+ to the human PrP91–231. Residues 94–98, 108–114, 135–136, and 153–159 exhibit significant differential alteration of resonances, indicating copper binding. (B) 1H-15N HSQC spectra with 0 μM (a), 375 μM (b), 625 μM (c), and 1000 μM CuSO4 (d), showing the region surrounding peaks arising from H96 and H111. As Cu2+ is titrated into the NMR sample, a general broadening of NMR resonances is observed, due to the presence of Cu2+ in solution. In addition, the peaks arising, for example, from the backbone amide resonances of H96 and H111 exhibit significant further broadening because of their proximity to the binding site of the paramagnetic ion. The peaks arising from the backbone amides of residues N197 and E221 are shown for comparison.
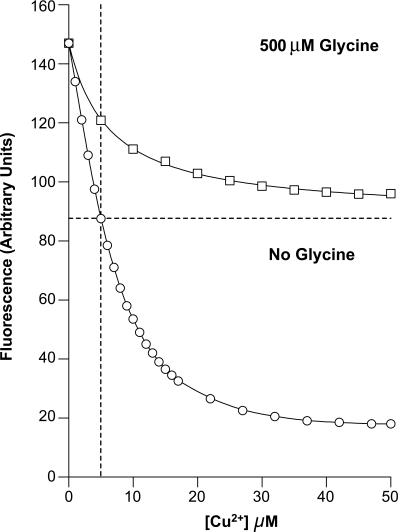
Fig. 3 shows a copper(II) titration of the octapeptide-repeat segment corresponding to residues 52–98. This region contains four exact repeats of a sequence ‘GGGWGQPH’ plus a pseudo repeat (see Materials and Methods). It is the binding of metals to this region that has attracted most attention. In the absence of a glycine as a competitive buffer there is a ≈90% quench of the indole fluorescence and although the binding curve appears to represent relatively weak binding, with half-saturation at ≈10 μM, this is deceptive because the curve is complex, being composed of contributions from both a weak and a tight site. Without glycine the binding of copper(II) ions to the first site is too tight to assign a Kd as the concentration of sites is far in excess of the Kd. The only measurable signal observed in these conditions represents binding to the weak site. Obtaining the affinity for the tight site requires glycine competition to weaken the apparent binding sufficiently to measure. In these conditions weak binding is abolished, which simplifies interpretation. The inclusion of 500 μM glycine into the system reveals a single site that, when occupied, elicits a ≈40% quench of fluorescence. The apparent dissociation constant is 3.2 μM in these conditions, giving a real Kd of 10−14 M, when the competitive buffering effect of glycine is taken into account. Hence, copper(II) binding in the absence of glycine is biphasic; there is an initial phase representing the occupation of a single tight site, giving the ≈40% quench, followed by a weak binding phase with a Kd of 15 μM, which accounts for the remaining 50% loss of indole fluorescence.
Figure 3.
Binding of copper to the octapeptide repeat region PrP52–98. The data for the binding of copper(II) to PrP52–98 in both the presence of 500 μM glycine (open squares) and the absence of glycine (open circles) are shown overlaid. The horizontal dotted line is drawn at a point representing stoichiometry of copper(II) to PrP52–98 (5 μM) to illustrate the loss of the weak-binding site in the presence of glycine. The lines superimposed on the data represent an optimal fit of the data to the equation described above.
Nickel(II) binds with a dissociation constant of 2 × 10−8 M, some six orders of magnitude weaker than copper(II). As in the case of the site on PrP91–231, this selectivity implies nitrogen coordination rather than oxygen and, further, it is known from small molecule crystallography that copper(II) forms a stable, square planar complex with imidazole. Taken with the stoichiometry of one tight site per polypeptide, these factors lead to the conclusion that the only plausible coordination ligands for the tight site are four of the five histidine side chains in the octapeptide-repeat. This would leave the fifth to contribute to the weak site.
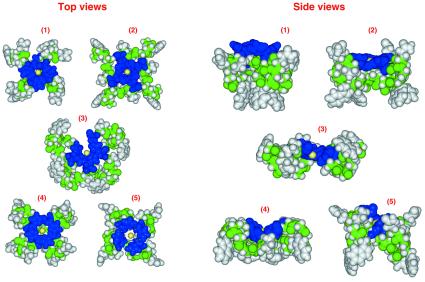
Given these constraints, it is possible to address the question of whether the binding of copper(II) or nickel(II) would induce a single, distinct conformation within the octapeptide-repeat region. To approach this we took a simplified (united atom) representation of the octapeptide-repeat and exhaustively modeled the 1.7 million conformations available to this chain given six φ–ψ angles per residue and repetition between each of the four repeats (i.e., 68 structures) as described in the legend to Fig. 4. Those that satisfied the requirement for coordination of four histidine side chains around the central transition metal were then assessed for energy by using a physicochemical force field, which accounted for steric effects, hydrophobicity, and backbone hydrogen bonding. The five energetically most favored conformations are shown in Fig. 4, and it can be seen that they are very diverse. This prompts the conclusion that although metal binding to the repeat region would render this region more compact than in the apoprotein, it would not lock it into any single rigidly defined conformation.
Figure 4.
Five energetically identical space-filled models of the Cu(II)-binding octapeptide repeat region. Models were generated by calculating the energetic stabilities of all possible combinations of exactly repeating structure in the octapeptide repeat. To enable exhaustive structural evaluation within the repeating unit, a simplified energetic and structural model was applied. Energetic stabilities were calculated by using a united atom model and physicochemical potentials describing hydrophobic, hydrophilic, and backbone hydrogen bonding energies. Conformational space was reduced, by restricting residues to adopt one of six alternative backbone φ–ψ torsion angles. The system is described in detail elsewhere (21). The models presented represent the five most energetically stable conformations of the repeat that also satisfy the proposed coordination of the copper ion (yellow) by the histidine side chains (blue). The remaining side chains are colored gray and backbone atoms are represented by green.
Previously reported structures of the metal-binding site within the octapeptide repeat region have indicated the Cu(II) ion to be coordinated via histidine residues and amide NH groups of the peptide backbone (14). Such structures cannot be stable as the lone pair on the nitrogen atom is delocalized to form a partial double bond in the amide group, thus leaving a partial positive charge on the nitrogen. The only plausible backbone interaction would be with the lone pairs on carbonyl oxygen atoms.
Discussion
With respect to the biological relevance of metal binding it is important to compare the affinities we measure for PrP with those of known metal-binding proteins. For instance, the Cu(II) affinity of superoxide dismutase 1 (22) and cerruloplasmin (23) are both about 10−14 M, which are very similar affinities to those we have measured here for the two tight sites of PrP. Such a tight interaction with copper ions is a prerequisite for a protein to be an authentic cuproprotein. The concentration of free copper within a mammalian cell is unknown. However, studies on yeast that provides the best model system for eukaryotic cells provide an estimation for the concentration of free copper as 10−18 M (22). For comparison a single atom of copper in a typical cell volume of 10−14 liters is a concentration of 10−10 M. Thus, if the concentration of free copper is substantially less than 1 atom per cell, a proteinaceous metal-binding site with micromolar affinity is unlikely to be of any physiological relevance. Although we see evidence for four copper-binding sites in total, we conclude that at neutral pH it is likely that only the two high-affinity sites described would be occupied by Cu(II) ions.
The significant discrepancy between the dissociation constant for the octapeptide repeat region we report here and those previously reported is due to the avidity of the copper site and the presence of additional weak sites. Titrations that others have performed by using copper(II) ions in the micromolar range would have saturated the tight site after the first addition of copper and this effect would then have been masked by the titration of weak sites.
It is only with the addition of a competitive chelator (glycine) with a Kd1 for copper of 8 × 10−9 (16), in effect using glycine as a buffer for copper(II) ions, that the contribution of weak sites can be abolished to reveal the presence of tight-binding sites. In addition, several studies have involved the use of copper(II) chloride, which is known to form insoluble copper hydroxide—indeed, at neutral or basic pH, copper exists almost exclusively as an insoluble copper hydroxyl species (24).
Although we see evidence for four copper-binding sites in total, two in the octapeptide repeat region and two in the C-terminal domain, the two sites with femtomolar affinity are likely to be the only physiologically relevant sites. The two additional sites have dissociation constants in the micromolar range and are unlikely to be occupied in vivo.
The use of glycine as a competitive chelator has revealed an erstwhile unidentified high-affinity site outside of the octapeptide repeat region. Indeed, binding occurs in a region of the molecule known to bind metals in PrPSc and confer prion strain-specific properties (5). NMR techniques have also allowed us to identify the residues involved in forming this site, which maps to a region of the molecule known to be associated with different clinical phenotypes of CJD when occupied or vacant (5). This high-affinity site appears distinct from that which can bind copper to produce low levels of SOD-like activity, because the latter cannot be titrated into the PrP molecule after it has folded into its native conformation (9).
Regarding a biological function for the octapeptide-repeat region it is worthy of note that a histidine-coordinated copper site will have a highly pH-dependent binding affinity. For instance, at the cell surface at pH 7.4 the Kd for Cu(II) would be ≈10−14 M, whereas in an acidified cellular vacuole at pH 4.5 and assuming a pK of 6.5 the affinity would drop some eight orders of magnitude. This property might be expected of a protein whose function lay in transporting metals from the extracellular milieu to endosomal and lysosomal vacuoles as has been suggested (10). Perturbation of copper transport by conversion of PrPC to PrPSc during prion propagation could play a central role in prion-induced neuronal damage and death that underlies the pathology of these conditions. The ability of the unstructured N-terminal domain of apo-PrP to adopt a number of conformations on copper binding is also consistent with the observed role of metal occupancy on prion strain determination in the human prion diseases (5).
Acknowledgments
This work was funded by the Medical Research Council and the Wellcome Trust. J.P.W. is a Lister Institute Research Fellow. The Krebs Institute is a BBSRC-funded center.
Abbreviations
- HSQC
heteronuclear single quantum correlation
- Mops
3-(N-morpholino)propanesulfonic acid
- PrP
prion protein
- PrPSc
scrapie isomer of PrP
- PrPC
cellular isomer of PrP
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson G S, Clarke A R. Curr Opin Struct Biol. 2000;10:69–74. doi: 10.1016/s0959-440x(99)00051-2. [DOI] [PubMed] [Google Scholar]
- 3.Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson B W, Burlingame A L, Prusiner S B. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 4.Pan K-M, Baldwin M A, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wadsworth J D F, Hill A F, Joiner S, Jackson G S, Clarke A R, Collinge J. Nat Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 6.Pattison I H, Jebbett J N. Nature (London) 1971;230:115. doi: 10.1038/230115a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown D R, Qin K, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, von Bohlen A, Schulz-Schaeffer W, et al. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 8.Waggoner D J, Drisaldi B, Bartnikas T B, Casareno R L B, Prohaska J R, Gitlin J D, Harris D A. J Biol Chem. 2000;275:7455–7458. doi: 10.1074/jbc.275.11.7455. [DOI] [PubMed] [Google Scholar]
- 9.Brown D R, Wong B S, Hafiz F, Clive C, Haswell S J, Jones I M. Biochem J. 1999;344:1–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Pauly P C, Harris D A. J Biol Chem. 1998;273:33107–33110. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 11.Hornshaw M P, McDermott J R, Candy J M. Biochem Biophys Res Commun. 1995;207:621–629. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 12.Hornshaw M P, McDermott J R, Candy J M, Lakey J H. Biochem Biophys Res Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 13.Stockel J, Safar J, Wallace A C, Cohen F E, Prusiner S B. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 14.Viles J H, Cohen F E, Prusiner S B, Goodin D B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittal R M, Ball H L, Cohen F E, Burlingame A L, Prusiner S B, Baldwin M A. Prot Sci. 2000;9:332–343. doi: 10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson R M C, Elliot D, Elliot W, Jones K M, editors. Data for Biochemical Research. Oxford: Clarendon; 1986. pp. 399–415. [Google Scholar]
- 17.Jackson G S, Hosszu L L P, Power A, Hill A F, Kenney J, Saibil H, Craven C J, Waltho J P, Clarke A R, Collinge J. Science. 1999;283:1935–1937. doi: 10.1126/science.283.5409.1935. [DOI] [PubMed] [Google Scholar]
- 18.Hosszu L L P, Baxter N J, Jackson G S, Power A, Clarke A R, Waltho J P, Craven C J, Collinge J. Nat Struct Biol. 1999;6:740–743. doi: 10.1038/11507. [DOI] [PubMed] [Google Scholar]
- 19.Bax A, Ikura M. J Biomol NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- 20.Davis A L, Keeler J, Laue E D, Moskau D J. J Magn Reson Ser B. 1992;98:207–216. [Google Scholar]
- 21.Gibbs, N., Clarke, A. R. & Sessions, R. B. (2000) Proteins, in press. [DOI] [PubMed]
- 22.Rae T D, Schmidt P J, Pufahl R A, Culotta V C, O'Halloran T V. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 23.Lippard S J, Berg J M. Principles of Bioinorganic Chemistry. Mill Valley, CA.: University Science Books; 1994. [Google Scholar]
- 24.McGrath M E, Haymore B L, Summers N L, Craik C S, Fletterick R J. Biochemistry. 1993;32:1914–1919. doi: 10.1021/bi00059a005. [DOI] [PubMed] [Google Scholar]