Abstract
Endopolyphosphatases (Ppn1) from yeast and animal cells hydrolyze inorganic polyphosphate (poly P) chains of many hundreds of phosphate residues into shorter lengths. The limit digest consists predominantly of chains of 60 (P60) and 3 (P3) Pi residues. Ppn1 of Saccharomyces cerevisiae, a homodimer of 35-kDa subunits (about 352-aa) is of vacuolar origin and requires the protease activation of a 75-kDa (674-aa) precursor polypeptide. The Ppn1 gene (PPN1) now has been cloned, sequenced, overexpressed, and deleted. That PPN1 encodes Ppn1 was verified by a 25-fold increase in Ppn1 when overexpressed under a GAL promoter and also by several peptide sequences that match exactly with sequences in a yeast genome ORF, the mutation of which abolishes Ppn1 activity. Null mutants in Ppn1 accumulate long-chain poly P and are defective in growth in minimal media. A double mutant of PPN1 and PPX1 (the gene encoding a potent exopolyphosphatase) loses viability rapidly in stationary phase. Whether this loss is a result of the excess of long-chain poly P or to the lack of shorter chains (i.e., poly P60 and P3) is unknown. Overexpression of the processed form of Ppn1 should provide a unique and powerful reagent to analyze poly P when the chain termini are unavailable to the actions of polyPase and poly P kinase.
Inorganic polyphosphate (poly P), a linear chain of many hundreds of phosphate (Pi) residues linked by high-energy phosphoanhydride bonds, is ubiquitous in nature, having been found in every organism examined, from bacteria to mammals (1). Poly P kinases (PPKs) in bacteria synthesize poly P from ATP and in reverse, ATP from poly P (2, 3). PPKs, as well as exopolyphosphatases from bacteria and yeast, act at the ends of poly P chains (2–6). PPKs transfer a phosphate residue to ADP, whereas Ppx1s hydrolyze poly P to Pi (2–6). These enzymes have provided crucial reagents needed to estimate poly P and to examine its metabolic roles. The genes encoding PPK and Ppx1 have been cloned, mutated, and overexpressed thereby disclosing insights into the physiologic functions of poly P (7).
In addition to several Ppx1s (5, 6, 8–14), an endopolyphosphatase (Ppn1) that cleaves poly P internally has been discovered in eukaryotic cells and purified to apparent homogeneity from Saccharomyces cerevisiae (15). The enzyme is of vacuolar origin and requires protease cleavage for activity (15). Ppn1 limit-digestion products of long-chain poly P (i.e., poly P of 700 residues, P700) are short chains: tripoly P (P3) and an intermediate size of about 60 residues (P60). We describe here the isolation of the gene encoding Ppn1 and phenotypes of the mutant.
In the interval since the previous study of Ppn1, it has become apparent that Ppx1, when used as a reagent to characterize and measure poly P, fails when the chain ends are masked and unavailable. One example of masked termini is poly P extracted from Escherichia coli grown under certain conditions where it is resistant to degradation by PPK or Ppx1 but becomes susceptible when the poly P is nicked internally by mild acid hydrolysis (N.N.R. and A.K., unpublished data). Thus, Ppn1 should serve as a valuable reagent to degrade and analyze masked poly P.
The objectives of this study were (i) to isolate Ppn1 in sufficient quantity to reexamine the kinetics and products of action, (ii) to identify the gene encoding Ppn1, and (iii) to clone the gene and determine the phenotypic consequences of deleting PPN1. Further studies will be needed to determine the sequence of protease-processed activated gene product, so that the gene corresponding to the processed form could be cloned and overexpressed and thereby provide another reagent for manipulating the levels of poly P in cells and to characterize it in extracts.
Materials and Methods
Reagents.
The ATP, ATP bioluminescence kit, DNase I, RNase IIIa, and creatine kinase were from Roche Molecular Biochemicals. The restriction enzymes were from New England BioLabs; [γ-32P]ATP and carrier-free 32Pi from Amersham Pharmacia Corp.; PPK (2) and rScPpx1 (6) as described previously; other reagents from Sigma.
Strains and Plasmids.
S. cerevisiae strains (CRY, CRX, CBO23, and CBX) are derived from W 303-1A (ref. 6; Tables 1 and 2). Strains CRN and CNX were prepared in this study. pUC19 harboring the Candida glabrata TRP1 fragment (pCgTRP1; courtesy of N. Ogawa, Stanford University) was used for deletion of PPN1 and pYES2. The 1/V5-HIS-TOPO cloning kit (Invitrogen) was used for overexpression of Ppn1.
Table 1.
S. cerevisiae strains
| Strains* | Genotype |
|---|---|
| CRY | MATa ade2 his3 leu2 trp1 ura3 |
| CRN | MATa ade2 his3 ura3 ppn1Δ∷CgTRP1 |
| CRX | MATa ade2 his3 trp1 ura3 ppx1Δ∷LEU2 |
| CNX | MATa ade2 his3 ura3 ppn1Δ∷CgTRP1 ppx1Δ∷LEU2 |
| CBO23 | MATa ade2 his3 trp1 leu2 ura3 pep4Δ∷HIS3 prb1Δ∷hisG prc1Δ∷hisG |
| CBX | MATa ade2 his3 trp1 leu2 ura3 pep4Δ∷HIS3 prb1Δ∷hisG prc1Δ∷hisG ppx1Δ∷LEU2 |
Strains CRN and CNX were prepared in this study. The other strains were from Wurst et al. (6).
Table 2.
Protease and PolyPases in S. cerevisiae strains
| Strains | Activity
|
||
|---|---|---|---|
| Protease | Ppx1 | Ppn1 | |
| CRY | + | + | + |
| CRN | + | + | − |
| CRX | + | − | + |
| CNX | + | − | − |
| CBO23 | − | + | − |
| CBX | − | − | − |
Media and Growth Conditions.
Yeast strains were grown aerobically at 30°C either in YPD (1% yeast extract/2% tryptone/2% glucose) or in SDC [2% glucose, 0.5% (NH4)2SO4, 7.35 mM phosphate, trace elements, salts, and vitamins; ref. 16] medium. For radiolabeling cultures, 32Pi was added at 10 μCi/ml of SDC medium. Growth was monitored at A600. For Ppn1 assays, 1-ml cultures, and for poly P analyses, 10-ml cultures, were collected, centrifuged at 14,000 × g for 10 min at 4°C in a microfuge, and the pellets were stored at −80°C.
Overexpression of Ppn1.
For overexpression of native and recombinant Ppn1, pYES2.1/V5-HIS-TOPO cloning kit (Invitrogen) was used. For native Ppn1, the primers used to amplify the PPN1 gene were forward primer 5′-GCCATGGTTGTAGTGGGTAAG-3′ (ATG start codon in bold followed by a 14-mer stretch of DNA downstream of the stop codon) and reverse primer 5′ GCGCTCGAGCTGTAATTGAAGAATGATATGC-3′ (GCG clamp in italics, XhoI in bold followed by a 22-mer stretch of DNA downstream of the stop codon). This reverse primer was designed to cover the native stop codon of PPN1. For truncation of Ppn1 at amino acid residue 372, the reverse primer 5′-GCG- GAATTCTTAATCCGTTGGGATGAAGTG-3′ (GCG clamp in italics, EcoRI in bold, the stop codon underlined, followed by a 18-mer stretch of DNA upstream of the truncated residue) was used with the above forward primer. For overexpression of N terminus His-tagged Ppn1, the primers were forward primer 5′-GCCATGGCGCATCATCACCATCACCATGTTGTAGTGGGTAAG-3′ (ATG start codon in bold, the 6 × HIS sequences underlined, followed by a 15-mer stretch of DNA downstream of the start codon) and reverse primer 5′-GCGCTCGAGCTGTAATTGAAGAATGATATGC-3′ (GCG clamp in italics, XhoI in bold, and followed by a 22-mer stretch of DNA upstream of the stop codon). For overexpression of C terminus His-tagged Ppn1, the primers were forward primer 5′-GCGGGATCCATGGTTGTAGTGGGTAAG-3′ (GCG clamp in italics, BamHI in bold, the ATG start codon underlined. followed by a 15-mer stretch of DNA seen downstream of the start codon) and reverse primer 5′-CGCATCATCTTTATATCCAGAAGA-3′ (CGC clamp in italics followed by a 21-mer DNA sequence upstream of the stop codon; the stop codon was intentionally left out to take advantage of the His-tag sequence located downstream of the plasmid cloning site). PCR amplification of PPN1 (30 cycles at 94°C, 45 s; 56°C, 45 s; and 72°C, 300 s) was performed by using cloned pfu DNA polymerase (Stratagene) with S. cerevisiae genomic DNA (Promega) as the template. For cloning the amplified PCR fragments into pYES2.1 TOPO vector, Invitrogen's procedures were followed. Recombinant plasmids were amplified in high competency E. coli (Invitrogen) and verified for orientation of insert PPN1 by restriction analysis: PvuII and XhoI (full-length native Ppn1 and N-His-tag Ppn1); PvuII and EcoRI (truncated Ppn1); and BamHI and XbaI (C terminus His-tagged Ppn1). Insert DNA was verified also by using the sequencing primers, GAL1 forward and V5 C terminus reverse primers, supplied by Invitrogen. Plasmid constructs with the insert PPN1 in the correct orientation were used to transform CNX (protease-positive, Ppn1- and Ppx1-deficient), and the transformants were selected on SDC agar plates deficient in uracil. CNX transformants containing the expression-control plasmid (pYES2.1/V5-His/lacZ) were used as negative controls to check Ppn1 expression. Transformation was performed as above. For induction of Ppn1, Invitrogen's protocols were followed. Cell extracts were prepared and tested for Ppn1 activity at levels of 1 and 10 ng of protein.
Other Methods.
Protein assays and SDS/PAGE were performed as described (15).
Results
Purification of Ppn1.
Ppn1 was purified from the CRX strain, a mutant in the PPX1 (Ppx1) gene (Table 1). Breaking the cells with glass beads yielded extracts 20-fold higher in specific activity compared with sonication (15). Subsequent steps were essentially the same as before (15), leading to a 2,100-fold enrichment.
Mass spectrophotometric analysis of the purified enzyme identified the ORF YDR452w as the gene for Ppn1. The gene is 2,025-bp long and encodes a hypothetical polypeptide of 674 amino acids (about 75 kDa; Fig. 1). Tryptic peptides of Ppn1 were sequenced; overlap of these was found only in the N terminus half (amino acids 19–352), indicative of truncation by protease action. The absence of Ppn1 activity in protease-deficient strains (see below) is further evidence that the polypeptide is modified posttranslationally. Microarrays of cDNAs also have identified ORF YDR452w as one of the genes involved in poly P metabolism (17). A mutant of this gene (PHM5) accumulates long-chain poly P, suggestive of a gene product that degrades long-chain poly P (17). Further studies of disruption and overexpression of the gene (see below) confirmed that YDR452w is the structural gene for Ppn1. The gene sequence has been registered with GenBank (accession no. AF322107). Structural features of Ppn1 (Fig. 2A) indicate a putative transmembrane region (about 20-aa long) near the N terminus, proteolytic cleavage sites near residue 352, and two glycosylation sites at Asn-11 and Asn-505. Also present is a region of homology with the gene for human sphingomyelin phosphodiesterase.
Figure 1.
Correspondence of tryptic digest peptides of Ppn1 to ORF YDR452w; 674 amino acids (GenBank accession no. AF322107). Two sequenced peptides (184–198 and 199–212) are shown in bold, and those underlined were identified by mass spectrophotometry. The identity is 100%.
Figure 2.
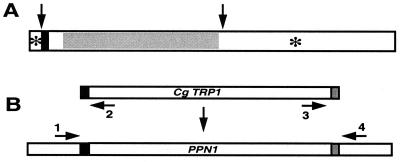
Structural features of Ppn1 and disruption of the gene. (A) Structural features. Arrows indicate putative proteolytic processing sites (near residues 20 and 352). ■, transmembrane region (residues 21–40); *, putative glycosylation sites (residues 11 and 505). Gray region has homologies to human sphingomyelin phosphodiesterase. (B) Disruption of PPN1. PCR-mediated gene disruption was performed as described (18). pCgTRP1 harboring the C. glabrata TRP1 fragment on pUC19 was used as the template to generate the PCR fragment for PPN1 disruption. The forward primer, 5′-CTATTGTTTTCATTGAGTAGGGGTAGAGCTAGTTAGCTGCTTTTCGCACAGGAAACAGCTATGACC-3′ (46-mer PPN1 region upstream of the ATG start codon in italics and 20-mer TRP1 fragment homolog in bold), and the reverse primer, 5′-GAAACTGTAATTGAAGAATGATATGCATTTCTATGTGTATATTAACGTTGTAAAACGACGGCCAGT-3′ (46-mer PPN1 region downstream of the stop codon in italics and 20-mer TRP1 homolog in bold), were used to amplify the TRP1 fragment of pCgTRP1. The initial 2 cycles of PCR conditions were at 94°C, 60 s; 56°C, 30 s; and 72°C, 90 s, followed by 30 cycles of amplifications at 94°C, 30 s; 60°C, 30 s; and 72°C, 90 s. The PCR-amplified fragment was used to transform CRY and CRX strains to produce the PPN1 knockout strains CRN (ppn1Δ) and CNX (ppn1Δ ppx1Δ), respectively. After transformation of yeast strains with the CLONTECH kit, transformants were selected on YPD agar plates deficient in tryptophan. Disruption on the genome of the transformants was verified by colony-PCR (30 cycles at 94°C, 30 s; 56°C, 30 s; and 72°C, 60 s; ref. 19), using two pairs of primers: upstream forward (primer 1) (5′-TTTTGGTTGTTGGGTTGGAG-3′) and reverse (primer 2) (5′-GGTCATAGCTGTTTCCTGTG-3′) primers, and downstream forward (primer 3) (5′-ACTGGCCGTCGTTTTACAAC-3′) and reverse (primer 4) (5′-TCTTCGCACGATAACGCTAG-3′) primers. Both pairs of primers were designed to produce about 150-bp PCR fragments from the estimated upstream and downstream recombination regions on the genome of the successful transformants.
Disruption of PPN1 and the Growth Defects of the Mutant.
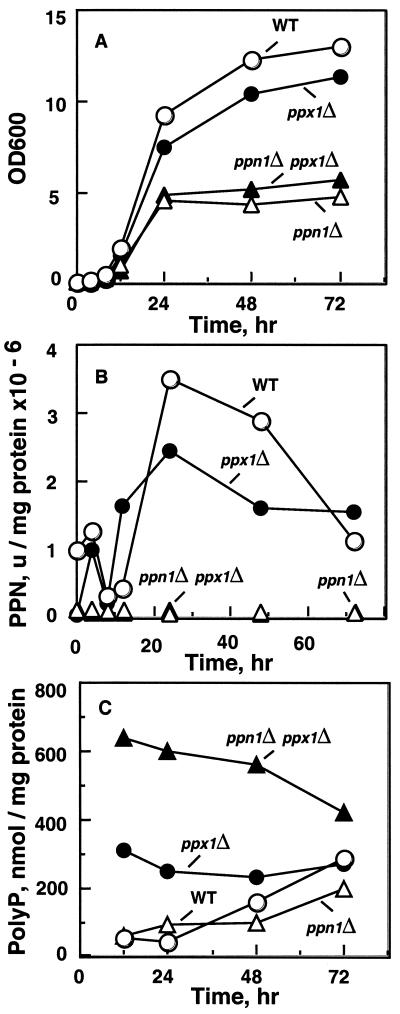
The protocol followed in creating a null mutant of PPN1 is described in Fig. 2B. With inactivation of PPN1, there was a significantly altered growth pattern in rich medium (Fig. 3A). Both the single mutant (CRN) and the double mutant (CNX) exhibited poor growth, only half that of the wild type (WT) after 24 h and even less after 72 h (Fig. 3A). In addition, the Ppn1-deficient strains formed much smaller colonies when plated on YPD agar plates. The growth phenotype was specific for the PPN1 deletion and not for the PPX1 deletion. The Ppn1-deficient strains were also lacking in Ppn1 activity (Fig. 3B). Like the PPN1-deletion strains, those lacking genes for three vacuolar proteases (CBO23), and those lacking both the vacuolar proteases and Ppx1 (CBX derived from CBO23), showed no Ppn1 activity (YPD medium, 24 h; Fig. 4). The absence of Ppn1 activity in vacuolar protease-deficient strains, CBO23 and CBX, confirms a need for proteolytic activation of Ppn1. The significant amount of polyphosphatase activity in the CRX strain (Fig. 3B) was misinterpreted earlier to be a vacuolar protease-dependent Ppx1 activity and was named ScPpx2 (6). Protease and polyPase activities of the yeast strains are summarized in Table 2. Poly Ps extracted from samples in Fig. 3A were higher in the double mutant and were at least double that in the other strains (Fig. 3C).
Figure 3.
Yeast strains in YPAD medium. (A) Growth at 30°C. (B) Ppn1-specific activity (106 units/mg of protein) in samples from A. The Ppn1 activity assay was measured as described (15). Crude extracts were prepared from cell pellets suspended in 5 volumes of lysis buffer (0.25 M sucrose/10 mM Tris⋅HCl, pH 7.0/1 mM EDTA) with 250 mg of glass beads. Lysis was obtained by vortexing the suspension 10 times for 30 s each with intermittent cooling on ice. The suspension, after removal of glass beads, was centrifuged at 10,000 × g for 10 min, and the supernatant was used for enzyme assays. A unit of Ppn1 activity was defined as 1 pmol of poly P (as Pi residues) used per minute; units/mg of protein give the specific activity (15). [32P]poly P used as substrate in Ppn1 assays was obtained by synthesis from [γ-32P]ATP using purified E. coli PPK (20); the purified poly P (20) had a chain length of ≈750 Pi residues (P750). (C) Poly P levels in samples from A. Frozen cells were thawed and suspended in equal volume of extraction buffer (50 mM Tris⋅HCl, pH 7.4/100 mM KCl/1 mM EDTA). Cells were lysed as above. Poly P was extracted from the lysate with a buffer of phenol/chloroform saturated with 10 mM Tris⋅HCl, pH 7.5, and 1 mM EDTA, followed by chloroform and ether extractions. Poly P extracted from unlabeled sources was estimated with E. coli PPK (20); poly P extracted from radiolabeled sources was estimated as described (21).
Figure 4.
Activation of Ppn1 in protease-deficient strains. Strains were grown in YPAD medium for 24 h at 30°C. Ppn1 was determined by PAGE analysis after incubation of [32P]PolyP700 at 37°C (see Materials and Methods). The specific activity of CRY (WT) was about 2.5 × 106 units/mg of protein as was CRX (ppx1Δ). Ppn1 was not detected in CRN (ppn1Δ) and CNX (ppn1Δ ppx1Δ) or the protease-deficient strains CBO23 and CBX (ppx1Δ)
Loss of Viability in the PPN1 Mutant.
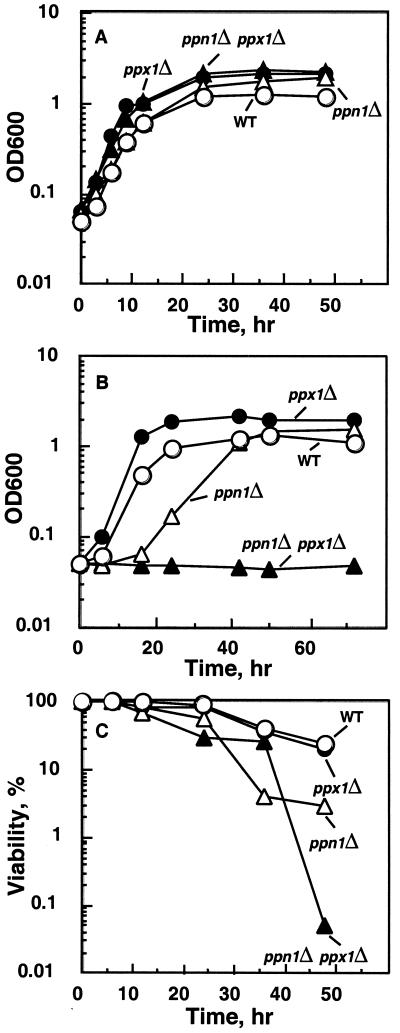
The growth phenotype exhibited by the PPN1 deletion strains was more marked when cultured in a synthetic (SDC) medium. Virtually no difference in growth was observed when the medium was inoculated directly with colonies picked from YPD agar plates (Fig. 5A). However, when the 48-h samples shown in Fig. 5A were used as inoculum and grown in the SDC medium, the CRN strain showed a long lag (24–36 h) but finally recovered, whereas the double mutant (CNX) failed to grow even after 72 h (Fig. 5B). The vacuolar protease-deficient strains (CBO23 and CBX) also exhibited a growth phenotype similar to the corresponding CRN strains when cultured in the synthetic medium (data not shown). When checked for viability, a sharp decline was observed with the double mutant (CNX) about 40 h after inoculation (Fig. 5C); the loss of viability was less prominent with the CRN strain (Fig. 5C).
Figure 5.
Growth and viability of strains in synthetic (SDC) medium. (A) Growth at 30°C. SDC medium was inoculated with colonies picked from YPAD agar plates. (B) Growth in SDC medium inoculated with (48-h culture) samples from A. (C) Viability of samples from A. Survival was assayed by growing cells in an SDC medium directly inoculated with colonies picked from YPD plates. Viable cell counts were determined in samples by plating onto YPD agar plates. Cell number was measured with a hemocytometer, and viability was expressed as a percentage of viable cells over the total number. The experiment was repeated twice with virtually identical results.
Poly P Levels and Chain Lengths in Mutant Strains.
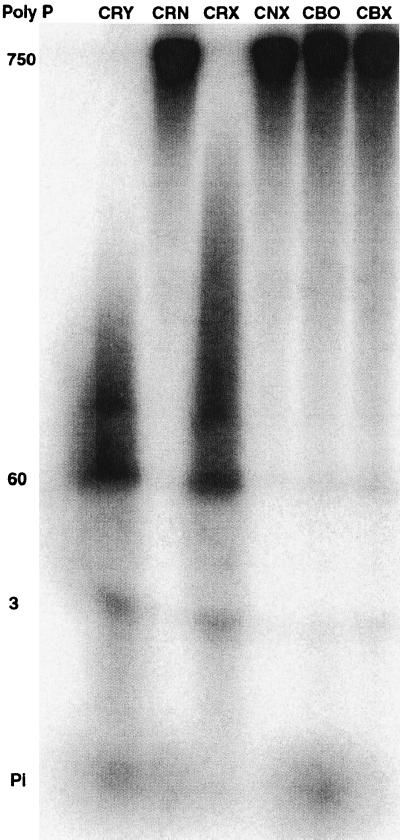
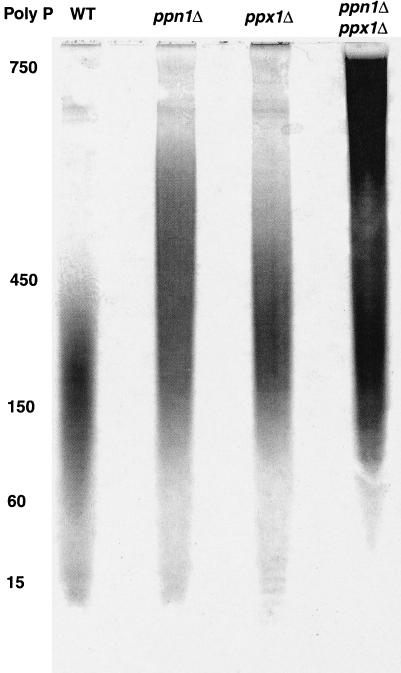
Poly P extracted from cells grown for 48 h in an unlabeled synthetic medium (Fig. 5A) and analyzed by PAGE (20%) exhibited distinctive chain lengths for the WT and mutant strains (Fig. 6). The exopolyPase (Ppx1) and endopolyPase (Ppn1) mutants contained largely medium chain lengths compared with the predominance of the short chains lengths in the WT strain. Mutants in both PPX1 and PPN1 accumulated large amounts of the long chains. The patterns of chain length in mutants deficient in vacuolar proteases were observed to be similar to those of the PPN1 mutants (6). The poly P levels, determined in labeled and unlabeled cultures (Fig. 5A), reached values in the double mutant of PPX1 PPN1 (CNX) of 5.5 μmol/mg of protein, corresponding to about 20% of the cell dry weight. Those in the WT (CRY), ppn1Δ (CRN), and ppx1Δ (CRX) were 3.3, 2.2, and 3.9 μmol/mg of protein, respectively.
Figure 6.
Poly P chain lengths in mutant strains. Cells were grown in an SDC medium containing 7.35 mM Pi. PAGE analysis was as described (22) with poly P5, poly P15, and [γ-32P]ATP as size markers. Electrophoresis was at 300 V until the dye was 7–8 cm from the origin; the lane with [γ-32P]ATP was cut out and analyzed with a PhosphorImager; the nonradioactive portion was soaked in 10% acetic acid, 10% methanol for 15 min, stained with a dye solution [0.5% (wt/vol) toluidine blue (Sigma)/25% (vol/vol) methanol/5% (vol/vol) glycerol/5% (vol/vol) acetic acid] for 15 min, and destained in the same solution without toluidine blue. The migration distance by [γ-32P]ATP corresponds to poly P with a chain length of about P8 (17). See Table 1 for strain descriptions.
Overexpression of Ppn1.
Proteolytic activation of Ppn1 required a strain (CNX) that is protease-positive and double mutant in PPN1 and PPX1 to reduce the background of polyPase activities. For overexpression of native Ppn1, the full-length gene was cloned into pYES2.1/V-5-HIS-TOPO, and the orientation of the cloned fragment was verified. Lysates from the transformants grown in a rich medium for 24 h contained 25 times higher specific activity than those obtained from WT cells. In contrast to the full-length PPN1 construct, extracts from transformants with the gene truncated at residue 372, presumably including the proteolytic cleavage site, showed no expression of an active Ppn1. Transformants with the full-length gene carrying a His-tag at either the N terminus or the C terminus yielded active lysates but no His-tagged activated Ppn1, either by nickel-affinity column purification or by a His-tag antibody Western blot. Presumably, both termini are removed in the proteolytic processing.
Discussion
Ppn1 that cleaves long-chain poly P of 700 residues (P700) internally to create chains of about 60 residues (P60) and 3 residues (P3) was identified in mammalian tissues and purified from yeast (15). The encoding gene has been identified, cloned, overexpressed, and disrupted. A mutant lacking Ppn1 shows limited growth and fails to survive in a minimal medium in the stationary phase.
The loss of viability of the yeast PPN1 mutant may be the result of an accumulation of large amounts of poly P (up to 20% of the cell dry weight) as long chains. The powerful chelating capacity of this poly P might sequester divalent metal cations (e.g., Ca2+, Mg2+, and Mn2+) that would damage essential cellular functions (23, 24). In addition, we have observed that when poly P is overproduced in E. coli by the overexpression of PPK, cells die within hours, a result that can be prevented by the overproduction of Ppx1, the potent exopolyPase (7). Another possibility is that the production of the shorter poly P chains of P60 and P3 is needed for some important function as observed in E. coli. P60 forms a membrane complex with Ca2+ and polyhydroxybutyrate to mediate the cellular entry of plasmid DNA (25, 26). Poly P of about this size also serves in the plasma membrane calcium pump in human erythrocytes (27). Inasmuch as Ppn1 has been detected only in eukaryotes, production of P60 by Ppn1 may play a role in membrane functions of organelles such as vacuoles, lysosomes, and nuclei.
Our improved enzymatic methods for the analysis of poly P (28) do not distinguish chains of 10–20 residues from those of many hundreds. Yet short- and intermediate-chain lengths of poly P are extracted from E. coli (N.N.R. and A.K., unpublished data) and yeast (22). The chain length of poly P synthesized by E. coli PPK in vitro is uniformly long, about 700 residues. The actions of exopolyPases and Ppn1 could account for the production of shorter chains. Methods are needed for the facile and sensitive determination of these shorter chains to explore their origin and functional significance.
The activated form of Ppn1 is produced by proteolytic truncation of the carboxyl half of the molecule and, likely, the removal of some N termini residues. In its action, Ppn1 produces intermediate chain lengths of poly P in a distributive fashion before reaching a digestion limit of P60; the general accumulation of P3 may be the result of an internal attack by the end of the chain on a subterminal bond, analogous to the generation of trimetaphosphate from poly P at alkaline pH and elevated temperatures (29).
An extension of the present studies is needed to determine the sequence of the catalytically active Ppn1 to clone the properly truncated gene and to overexpress it in bacteria. The use of overexpressed exopolyPases as a means of depleting cellular poly P is limited to poly P chain ends that are unblocked and available to the enzyme. Ppn1 will be useful in the in vitro analysis of the modifications to the chain ends of poly P.
Acknowledgments
We are grateful to D. Winant, Protein and Nucleic Acids Faculty, Stanford University, for peptide sequencing and mass spectrophotometric analysis of purified Ppn1. We thank N. Ogawa for gifts of plasmids and primers. We also thank I. R. Lehman, L. Bertsch, and C. Fraley for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health.
Abbreviations
- poly P
inorganic polyphosphate
- PPK
polyphosphate kinase
- Ppn1
endopolyphosphatase
- Ppx1
exopolyphosphatase
- WT
wild type
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF322107).
References
- 1.Kulaev I S. The Biochemistry of Inorganic Polyphosphates. New York: Wiley; 1979. [DOI] [PubMed] [Google Scholar]
- 2.Ahn K, Kornberg A. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 3.Akiyama M, Crooke E, Kornberg A. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 4.Akiyama M, Crooke E, Kornberg A. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 5.Wurst H, Kornberg A. J Biol Chem. 1994;269:10996–11001. [PubMed] [Google Scholar]
- 6.Wurst H, Shiba T, Kornberg A. J Bacteriol. 1995;177:898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornberg A, Rao N N, Ault-Riché D. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 8.Lorenz B, Muller W E G, Kulaev I S, Schröder H C. J Biol Chem. 1994;269:22198–22204. [PubMed] [Google Scholar]
- 9.Kulaev I, Andreeva N A, Lichko L P, Kulakovskaya T V. Microbiol Res. 1997;152:221–226. doi: 10.1016/S0944-5013(97)80031-6. [DOI] [PubMed] [Google Scholar]
- 10.Lichko L, Kulakovskaya T, Kulaev I S. Biochemistry. 1997;62:1146–1151. [PubMed] [Google Scholar]
- 11.Lichko L, Kulakovskaya T, Kulaev I. Biochim Biophys Acta. 1998;1372:153–162. doi: 10.1016/s0005-2736(98)00013-3. [DOI] [PubMed] [Google Scholar]
- 12.Lichko L, Kulakovskaya T V, Dmitriev V V, Kulaev I S. Biochemistry. 1995;60:1465–1468. [Google Scholar]
- 13.Lichko L, Kulakovskaya T, Kulaev I S. Biochemistry. 1996;61:488–495. [PubMed] [Google Scholar]
- 14.Andreeva N A, Kulakovskaya T V, Kulaev I. FEBS Lett. 1998;429:194–196. doi: 10.1016/s0014-5793(98)00591-2. [DOI] [PubMed] [Google Scholar]
- 15.Kumble K D, Kornberg A. J Biol Chem. 1996;271:27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- 16.Rose M D, Winston F, Hieter P. Methods in yeast genetics—A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 17.Ogawa N, DeRisi J, Brown P O. Mol Biol Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakumoto N, Mukai Y, Uchida K, Kouchi T, Kuwajima J, Nakagawa Y, Sugioka S, Yamamoto E, Furuyama T, Mizubuchi H, et al. Yeast. 1999;15:1669–1679. doi: 10.1002/(SICI)1097-0061(199911)15:15<1669::AID-YEA480>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Adams A, Gottschling D E, Kaiser C A, Stearn T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 20.Ault-Riché D, Fraley C D, Tzeng C-M, Kornberg A. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao N N, Liu S, Kornberg A. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumble K D, Kornberg A. J Biol Chem. 1995;270:5818–5822. doi: 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- 23.Dunn T, Gable K, Beeler T. J Biol Chem. 1994;269:7273–7278. [PubMed] [Google Scholar]
- 24.Lee R M, Hartmann P A, Stahr H M, Olson D G, Williams F D. J Food Protect. 1994;57:289–294. doi: 10.4315/0362-028X-57.4.289. [DOI] [PubMed] [Google Scholar]
- 25.Reusch R N, Sadoff H L. Proc Natl Acad Sci USA. 1988;85:4176–4180. doi: 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castuma C E, Huang R, Kornberg A, Reusch R N. J Biol Chem. 1995;270:12980–12983. doi: 10.1074/jbc.270.22.12980. [DOI] [PubMed] [Google Scholar]
- 27.Kulaev I, Kulakovskaya T. Annu Rev Microbiol. 2000;54:709–734. doi: 10.1146/annurev.micro.54.1.709. [DOI] [PubMed] [Google Scholar]
- 28.Ault-Riché D, Kornberg A. In: Progress in Molecular and Subcellular Biology. Schröder H C, Müller W E G, editors. Vol. 23. Berlin: Springer; 1999. pp. 241–252. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg S R. J Biol Chem. 1956;218:23–31. [PubMed] [Google Scholar]