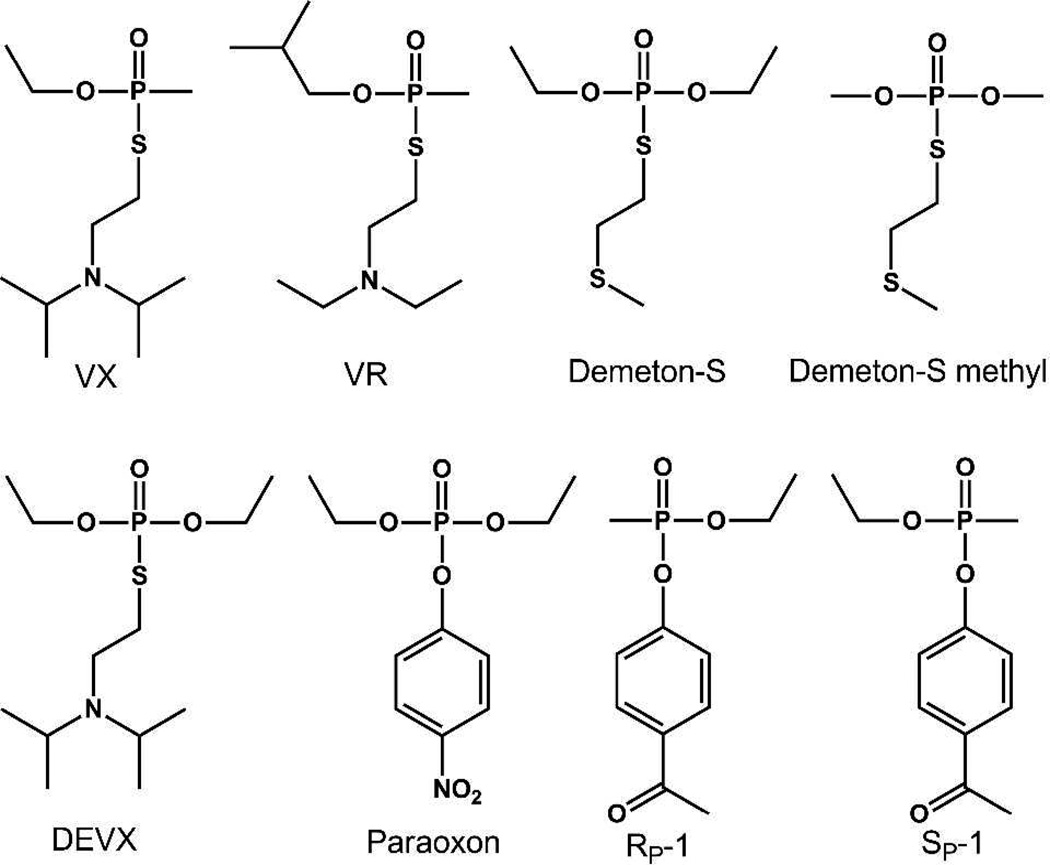
Abstract
The V-type nerve agents (VX and VR) are among the most toxic substances known. The high toxicity and environmental persistence of VX makes the development of novel decontamination methods particularly important. The enzyme phosphotriesterase (PTE) is capable of hydrolyzing VX but with an enzymatic efficiency more than 5-orders of magnitude lower than with its best substrate, paraoxon. PTE has previously proven amenable to directed evolution for the improvement of catalytic activity against selected compounds through the manipulation of active site residues. Here, a series of sequential two-site mutational libraries encompassing twelve active site residues of PTE was created. The libraries were screened for catalytic activity against a new VX analogue (DEVX), which contains the same thiolate leaving group of VX coupled to a di-ethoxy phosphate core rather than the ethoxy, methylphosphonate core of VX. The evolved catalytic activity with DEVX was enhanced 26-fold relative to wildtype PTE. Further improvements were facilitated by targeted error-prone PCR mutagenesis of Loop-7 and additional PTE variants were identified with up to a 78-fold increase in the rate of DEVX hydrolysis. The best mutant hydrolyzed the racemic nerve agent VX with a value of kcat/Km of 7×104 M−1 s−1; a 230-fold improvement relative to the wild-type PTE. The highest turnover number achieved by the mutants created for this investigation was 137 s−1; an enhancement of 152-fold relative to wild-type PTE. The stereoselectivity for the hydrolysis of the two enantiomers of VX was relatively low. These engineered mutants of PTE are the best catalysts ever reported for the hydrolysis of nerve agent VX.
INTRODUCTION
The G-type (sarin, cyclosarin, and soman) and V-type (VX and VR) organophosphonates are among the most toxic compounds known. The toxicity of these compounds is due to their ability to inactivate acetylcholine esterase, an enzyme required for proper nerve function.1 With an estimated lethal dermal exposure of ~6 mg for an average human, contact with VX is ~200-fold more toxic than soman (GD) and 300-fold more toxic than sarin (GB).2 The extreme potential for acute toxicity with VX is due, in part, to the low volatility of this compound, which allows it to persist indefinitely on common surfaces.3 Methods currently utilized for the destruction of organophosphate nerve agents include high temperature incineration and treatment with strong base or concentrated bleach.4,5 Medical treatment of VX toxicity is currently limited to the injection of atropine, which reduces neurological symptoms, and oximes, which can help to reactivate the inactivated acetylcholine esterase.2 Butyrylcholine esterase, which is closely related to acetylcholine esterase, has proven effective in animal models as a stoichiometric scavenger of VX.6 However, the large amount of enzyme required for treatment with a stoichiometric scavenger, and the limited supply of this protein, have prevented butyrylcholine esterase from being an effective antidote for medical use.7,8
Enzymatic hydrolysis of nerve agents provides numerous advantages over harsh physical or chemical methods of decontamination and could provide a catalytic antidote for medical use. Enzymes such as OPAA, DFPase and human PON1 can hydrolytically neutralize the various G-type agents,9–12 but except for PON1, they have no activity against the V-type agents.11 The enzyme phosphotriesterase (PTE) is capable of hydrolyzing a wide variety of organophosphonates including both the G-type and V-type nerve agents.13,14 The best known substrate for PTE is the insecticide paraoxon (Scheme 1) with an enzymatic efficiency that approaches the limits of diffusion (kcat/Km ~108 M−1 s−1).15 For the hydrolysis of the G-type agents by PTE, the values of kcat/Km are between 104 and 105 M−1 s−1.13
Scheme 1.
The G- and V-type nerve agents all contain a chiral phosphorus center where the SPenantiomers are significantly more toxic than the corresponding RP-enantiomers.16,17 In general, wild-type PTE preferentially hydrolyzes the RP-enantiomers of these compounds. The overall selectivity depends on the relative size of the substituents attached to the phosphorus center, with larger differences in size resulting in greater stereoselectivity.18 Chiral chromophoric analogues of the G-type agents have been utilized to guide the evolution of PTE for the identification of variants that prefer the more toxic SP-enantiomers of sarin, cyclosarin, and soman.13,16,18 The catalytic activity of PTE for the more toxic SP-enantiomer of cyclosarin (GF) has been increased by more than 4-orders of magnitude.13 The catalytic efficiencies for the hydrolysis of the more toxic SP-enantiomers by the enhanced variants of PTE for the hydrolysis of GB, GD, and GF approach 106 M−1 s−1.13
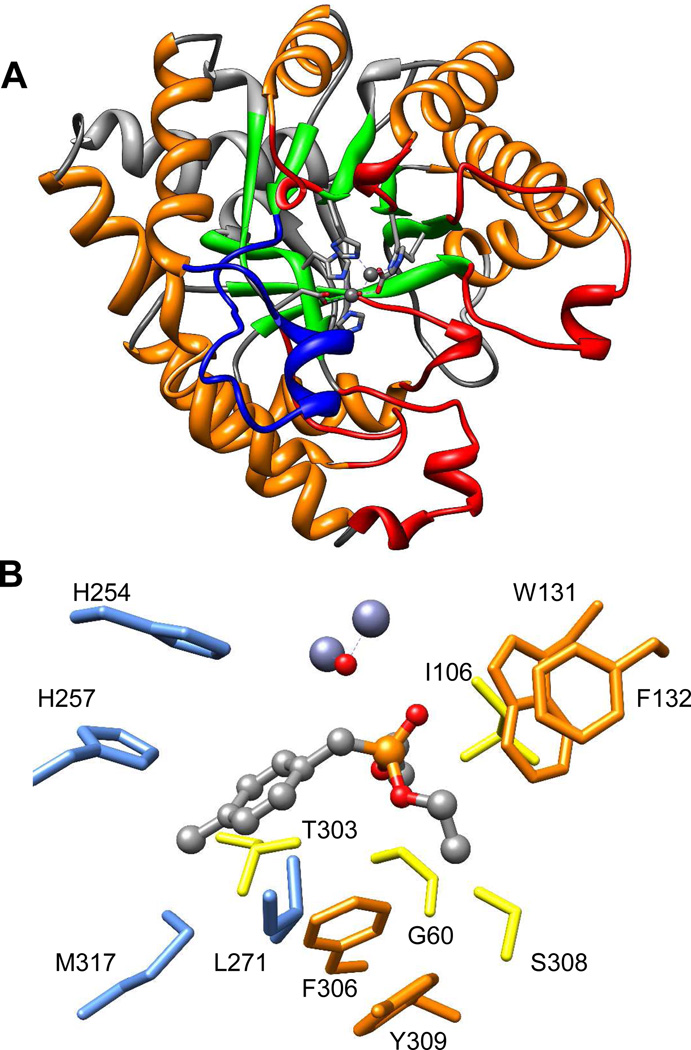
Unfortunately, the activity of PTE against the V-type agents is about 3-orders of magnitude lower than that with the G-type agents (kcat/Km < 103 M−1s−1).14,19 The net rate of VX hydrolysis by PTE is thought to be limited more by the chemistry of the leaving group than by the stereochemistry of the phosphorus center.13,14,20 The X-ray crystal structure of PTE shows that this enzyme folds as a distorted (β/α)8-barrel and that the bulk of the active site is formed from the 8 loops that connect the core β-strands to the subsequent α-helices (Figure 1A).21 The twelve residues which make up the substrate binding site of PTE can be subdivided into three pockets that accommodate the small, large and leaving-group moieties of the substrate (Figure 1B).21 The residues in the active site have been shown to be largely responsible for the observed substrate specificity.22 Loop-7 is the largest of the loops that contribute to the substrate binding site (Figure 1A), and is known to tolerate substantial sequence variation.18,21,23 Previous attempts to evolve PTE for the hydrolysis of VX have utilized the insecticide demeton-S with modest success (Scheme 1).24,25 In this paper we describe the use of a new analogue and mutation strategies that have greatly facilitated the optimization of PTE for the hydrolysis of VX by more than two orders of magnitude.
Figure 1.
The structure of phosphotriesterase. (A) (β/α)8 -barrel structure of PTE. Core β- strands are colored green, core α-helices are colored orange, N-terminal loops are gray, and Cterminal loops are red. Loop-7 is colored in blue and the metals are shown as spheres. (B) The substrate binding site of PTE with a bound substrate analogue. Large group pocket residues are colored blue. Small group pocket residues are colored yellow, and the leaving group pocket residues are colored orange. The 3-dimensional coordinates are taken from PDB id: 1dpm.
MATERIALS and METHODS
Most chemicals were obtained from Sigma Chemical Company. The pfuTurbo DNA polymerase was obtained from Agilent Technologies and the various restriction enzymes were acquired from New England Biolabs. The two enantiomers of compound 1 were synthesized as previously reported.18 The synthesis of DEVX is described in the Supplementary Information. VX samples were Chemical Agent Standard Analytical Reference Material (CASARM). All of the organophosphorus compounds used in this investigation are highly toxic and should be used with the appropriate precautions.
Active Site Library Construction
The gene for PTE lacking the leader peptide (residues 29–365) was inserted into a pET 20b+ vector between the NdeI and EcoRI restriction sites as previously described.18 The 306X/309X, 131X/132X and 254X/257X double substitution libraries were constructed by QuikChange site-directed mutagenesis (Agilent Technologies) using single sets of primers containing NNS (N = any base, S = G or C) codons at the positions of interest. The 271X/308X library was constructed by sequential QuikChange reactions. The 106X/303X and 60X/317X libraries were constructed using a PCR overlap extension technique.26 Plasmids from at least 10 colonies of each library were sequenced to ensure the randomization at the positions of interest. The identities of specific mutations for the variants are given in Table S1.
Construction of Targeted Error-Prone Library
To construct the Loop-7 error-prone library, a set of 30-bp primers corresponding to the DNA sequences upstream and downstream of Loop-7 (amino acid residues 253–276) were used to amplify the PTE gene in three fragments. The DNA coding region of Loop-7 was amplified in an error-prone PCR reaction while the two remaining fragments were amplified using standard PCR techniques. The final gene was constructed using PCR overlap-extension, resulting in a gene library with errors only in the coding region for residues 253–276.
Optimization of Error-Prone Variants
The five residue positions (254, 265, 270, 272, and 276) identified in the best Loop-7 error prone variant and residues 257 and 274 were further optimized by construction of two two-site (254X/257X and 272X/274X) and three single-site libraries (265X, 270X, 276X). Libraries were constructed via QuikChange mutagenesis using degenerate primers to allow all 20 amino acids at the positions of interest. Approximately 200 colonies from each single-site library were screened, and approximately 1200 colonies from each two-site library were screened. The two enhanced variants identified in the Loop-7 error prone library were used as the template for a second round of targeted error-prone PCR of Loop-7.
Library Screening
Plasmid libraries were transformed into BL21 (DE3) E. coli competent cells and grown on LB plates. For all library transformations, the amount of DNA was kept low (< 10 ng) to avoid the potential complication of double transformants.27 Single colonies were used to inoculate 0.75 mL cultures of Super Broth (32 g tryptone, 20 g yeast extract, 5 g NaCl, and 0.4 g NaOH in 1 L H2O) supplemented with 0.5 mM CoCl2 in a 96-well block format. Cultures were grown at 37 °C for 8 hours. The temperature was reduced to 30 °C and protein expression induced by addition of 1 mM IPTG. Following 16 hours of additional growth, the bacteria were harvested by diluting a portion of the culture in a 1:1 ratio with 1× BugBuster (50 mM HEPES pH 8.0, 100 µM CoCl2, 10% BugBuster 10× (EMD Chemicals)). Cultures were tested for activity against DEVX using a standard 250 µL assay that consisted of 50 mM HEPES, pH 8.0, 100 µM CoCl2, 0.3 mM 5,5'-dithiobis(2-nitro-benzoic acid)(DTNB) and 0.2 – 0.5 mM DEVX. The reactions were initiated by the addition of 10 µL of cell lysate. Reactions proceeded at room temperature until color was clearly visible (1–4 hours). Product formation was determined by the change in absorbance at 412 nm using a Spectra Max 364 plate reader. The variant used as the starting template for each library was included as a control on each plate. To account for differential culture growth, the final change in absorbance was normalized using the OD600 for each culture compared to the average OD600 of controls. The colonies giving the best results were re-grown as 5 mL overnight cultures and the plasmids harvested and sequenced to identify the variants.
Kinetic Measurements
All assays with DEVX, paraoxon, SP−1, RP−1, and demeton-S were 250 µL in total volume and followed for 15 minutes in a 96-well Spectra Max 364 plate reader at 30 °C. Assays with VX were conducted in a volume of 500 µL in 1 mL cuvettes. DEVX, demeton-S, and VX assays monitored the release of the product thiol at 412 nm (Δε412 = 14,150 M−1 cm−1) by the inclusion of DTNB in the reaction mixture (50 mM HEPES, pH 8.0, 100 µM CoCl2, and 0.3 mM DTNB). Assays with paraoxon and compound 1 were conducted in 50 mM CHES, pH 9.0, and 100 µM CoCl2. Assays of compound 1 contained 10% methanol. Paraoxon hydrolysis was followed by the release of p-nitrophenol at 400 nm (Δε400 = 17,000 M−1 s−1) and the hydrolysis of compound 1 was followed at 294 nm (Δε294 = 7,710 M−1cm−1). Reactions were initiated by the addition of enzyme. The data were fit to equation 1 to obtain values of Km, kcat, and kcat/Km. A representative data set is provided in Figure S1.
| (1) |
Stereoselelctive Hydrolysis of Racemic VX
Low initial concentrations (19 to 160 µM) of racemic VX were hydrolyzed by variants of PTE in a solution containing 0.1 mM CoCl2, 0.3 mM DTNB, and 50 mM Hepes, pH 8.0. The reactions were followed to completion and the fraction of VX hydrolyzed plotted as a function of time. The time courses were fit to equations 2 and 3 where F is the fraction of substrate hydrolyzed, a and b are the magnitudes of the exponential phases, t is time, and k1 and k2 are the rate constants for each phase
| (2) |
| (3) |
To identify which one of the two enantiomers of VX was preferentially hydrolyzed by the PTE variants, one gram of racemic VX was hydrolyzed in a 400 mL reaction mixture containing 50 mM bis-tris-propane (pH 8.0), 100 µM CoCl2, and 36 nM of the QF mutant (H254Q/H257F) at 33 °C. The reaction was monitored by determining the concentration of the thiol product with DTNB. When the reaction was approximately 50% complete, the remaining VX was extracted with 200 mL of ethyl acetate. The volume of the extract was reduced to approximately 2 mL by rotary evaporation at 41 °C. The unreacted VX was analyzed with an Anton Paar MCP 500 polarimeter and observed to rotate plane polarized light in a positive direction (+0.055° to +0.075°) which corresponds to an enantiomeric preference for hydrolysis of the SP-enantiomer of VX by the QF mutant.17
RESULTS
Construction and Screening of Active Site Libraries
The variant QF (H254Q/H257F) was previously identified as being improved against the chiral centers in VX and VR.18 Testing the catalytic activity of this mutant with the VX analogue, DEVX, revealed that this variant has an enhanced activity for the hydrolysis of the phosphorothiolate bond, relative to wild-type PTE. The variant QF then served as the starting point for the construction of the F306X/Y309X and W131X/F132X double-substitution protein libraries. Screening 920 colonies from the F306X/Y309X library with DEVX failed to identify any variant that was improved relative to the QF parent. From the W131X/F132X library, a total of 1100 colonies were screened with DEVX and the two best mutants were identified as LQF (QF + F132L) and VQF (QF + F132V). The LQF variant served as the starting template for the 254X/257X library. Approximately 1650 colonies were screened from this library with DEVX, but none proved to be better for the hydrolysis of DEVX.
The 271X/308X library was created using sequential QuikChange procedures; first at position 271 then at position 308 using the LQF template. Approximately 2200 colonies from this library were screened and the best variant was LQFL (LQF + S308L). Incorporation of the new mutation (S308L) into the previously identified VQF variant further enhanced the catalytic activity. The variant VQFL (VQF + S308L) was utilized as the parent for the 106X/303X library. Approximately 1100 colonies were screened with DEVX and the best variant identified was CVQFL (VQFL + I106C). The variant CVQFL was carried forward in the construction of the 60X/317X library. Nearly 1500 colonies from this library were screened with DEVX, but improved variants were not detected.
A number of mutations are known to improve protein expression levels for PTE, including A80V, K185R, and I274N.28,29 These mutations do not typically result in significant changes in the kinetic constants for a given substrate, but they dramatically improve the amount of enzyme produced per liter of cell culture. Adding these expression-enhancing mutations to VQFL resulted in an additional variant, VRN-VQFL (A80V/K185R/I274N + VQFL) with a 26-fold improvement in the value of kcat/Km, relative to the wild-type PTE. The inclusion of two additional expression-enhancing mutations (D208G/R319S) resulted in a decrease in catalytic activity.29 Kinetic constants for the PTE variants with DEVX as the target substrate are presented in Table 1. Kinetic constants for additional variants are provided in Table S2.
Table 1.
Activity of PTE Variants against VX analogues DEVX and compound 1a
| DEVX | RP−1 | SP−1 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variant |
kcat (s−1) |
Km (mM) |
kcat/Km (M−1s-1) |
kcat (s−1) |
Km (µM) |
kcat/Km (M−1s−1) |
kcat (s−1) |
Km (µM) |
kcat/Km (M−1s−1) |
| WT | 1.1 | 0.87 | 1.2 × 103 | 100 | 3700 | 2.7 × 104 | 92 | 320 | 2.9 × 105 |
| QF | 6.1 | 1.4 | 4.2 × 103 | 120 | 230 | 5.3 × 105 | 34 | 18 | 1.8 × 106 |
| LQF | 15 | 1.7 | 9.0 × 103 | 112 | 910 | 1.2 × 105 | 27 | 13 | 2.1× 106 |
| VQF | 18 | 1.0 | 1.9 × 104 | 82 | 340 | 2.4 × 105 | 25 | 11 | 2.2 × 106 |
| LQFL | 10 | 0.76 | 1.4 × 104 | 76 | 160 | 4.7 × 105 | 45 | 7.4 | 6.1 × 106 |
| VQFL | 14 | 0.65 | 2.2 × 104 | 69 | 129 | 5.3 × 105 | 32 | 7.4 | 4.3 × 106 |
| CVQFL | 16 | 0.76 | 2.1 × 104 | 54 | 170 | 3.2 × 105 | 39 | 24 | 1.6 ×106 |
| VRN-VQFL | 22 | 0.73 | 3.1 × 104 | 124 | 160 | 7.8 × 105 | 65 | 26 | 2.5 × 106 |
| VRNGS-VQFL | 11 | 0.99 | 1.1 × 104 | 204 | 350 | 5.8 × 105 | 93 | 22 | 4.3 × 106 |
| L7ep-1 | 16 | 0.60 | 3.2 × 104 | 590 | 8600 | 6.8 × 104 | 670 | 2800 | 2.3 × 105 |
| L7ep-2 | 48 | 0.63 | 7.6 × 104 | 240 | 730 | 3.3 × 105 | 90 | 400 | 2.3 × 105 |
| L7ep-3 | 29 | 0.69 | 4.3 × 104 | 143 | 560 | 2.5 × 105 | 50 | 22 | 2.3 × 106 |
| L7ep-2a | 135 | 1.4 | 9.4 × 104 | 235 | 950 | 2.5 × 105 | 180 | 1090 | 1.7 × 105 |
| L7ep-2b | 76 | 1.0 | 7.4 × 104 | 136 | 610 | 2.2 × 105 | 95 | 1090 | 8.6 × 104 |
| L7ep-3a | 51 | 0.6 | 8.5 × 104 | 290 | 1500 | 1.9 × 105 | 90 | 36 | 2.5 × 106 |
| L7ep-3b | 64 | 1.0 | 6.2 × 104 | 202 | 1800 | 1.1 × 105 | 48 | 34 | 1.4 × 106 |
Standard errors from fits of the data to equation 1 are less than 20% of the stated values.
Construction of Targeted Error-Prone Library
The CVQFL variant was used as the parent for the construction of an error-prone library with an average of six mutations per gene, targeted exclusively to Loop-7 of PTE (residues 253–276). Approximately 4000 colonies from this library were screened with DEVX and a total of 12 variants were identified as being more active than the parent, CVQFL. The values of kcat/Km for the best variants, L7ep-1, L7ep-2 and L7ep-3, were improved 27-, 63-, and 36-fold, respectively, for the hydrolysis of DEVX.
The variant L7ep-2 (CVQFL + H254R/N265D/A270D/L272M/S276T) has 5 amino acid changes to the sequence of Loop-7, relative to the parent. These five sites, and residue positions 257 and 274, were subjected to further optimization. Two, two-site libraries (R254X/F257X, and M272X/I274X) and three, single-site libraries (D265X, D270X, and T276X) were constructed to ensure that the optimum amino acid residue is represented at each position. Screening the libraries with DEVX revealed no improvements at residue positions 254, 257, 265, or 270, but numerous improved combinations were identified in the 272X/274X library. One of these variants, L7ep-2a (L7ep-2 + I274T), has a kcat of 135 s−1 and a kcat/Km 78- fold improved over wild type enzyme. To further optimize the L7ep-3 variant (CVQFL + H257Y/A270V/L272M), Loop-7 was subjected to a second round of targeted error-prone PCR. Screening with DEVX identified two variants, L7ep-3a (L7ep-3 + I274N) and L7ep-3b (L7ep-3 + A270D) that were substantially improved over the parent enzyme. Similar experiments were attempted using L7ep-2 as the starting template, but no improved variants were identified. The kinetic constants for the hydrolysis of DEVX by these mutants are presented in Table 1 and Table S2.
Stereoselectivity of PTE Variants for Chiral VX Analogues
In this investigation there was no attempt to include a stereochemical preference in the screening of mutant libraries. Wild-type PTE is known to have a slight preference for the SP-enantiomer of the VX chiral center.18 To determine that there were no perturbations in stereoselectivity, the variants with improved catalytic activity against DEVX were analyzed using the chromophoric analogues RP-1 and SP-1, and the results are presented in Table 1. With the exception of L7ep-2, L7ep-2a, and L7ep-2b, the evolved variants have values of kcat/Km that are greater for the Sp-enantiomer than for the Rp-enantiomer.
Enzymatic Specificity
To assess changes in substrate specificity, the enzyme variants were tested with paraoxon and demeton-S as alternative substrates. The results are provided in Table 2. The variants from the active site evolution experiments maintain a high enzymatic efficiency for paraoxon, although it is reduced nearly an order of magnitude from wild-type enzyme. The catalytic activity using demeton-S did not show any significant improvement for most of the variants tested. The exceptions are L7ep-2, L7ep-2a, and L7ep-2b, which have increased kcat and decreased Km values for demeton-S.
Table 2.
Kinetic parameters for PTE variants with paraoxon and demeton-Sa.
| Paroxon | Demeton-S | |||||
|---|---|---|---|---|---|---|
| Variant |
k
cat (s−1) |
Km (µM) |
kcat/Km (M−1s−1) |
kcat (s−1) |
Km (mM) |
kcat/Km (M−1s−1) |
| WT | 6700 | 100 | 6.7 × 107 | 1.4 | 1.2 | 1.1 × 103 |
| QF | 41 | 5.3 | 7.7 × 106 | 6.3 | 2.6 | 2.7 × 103 |
| LQF | 72 | 11.1 | 6.5 × 106 | 4.4 | 3.3 | 1.3 × 103 |
| VQF | 108 | 10.5 | 1.0 × 107 | 10 | 6.1 | 1.7 × 103 |
| LQFL | 90 | 11 | 8.2 × 106 | 5.2 | 3.1 | 1.7 × 103 |
| VQFL | 66 | 5.4 | 1.2 × 107 | 4.2 | 3.9 | 1.1 × 103 |
| CVQFL | 38 | 5.6 | 6.8 × 106 | 6.1 | 2.5 | 2.4 × 103 |
| VRN-VQFL | 116 | 8 | 1.5 × 107 | 7.1 | 2.8 | 2.5 × 103 |
| VRNGS-VQFL | 227 | 8.5 | 2.7 × 107 | 6.8 | 3.1 | 2.2 × 103 |
| L7ep-1 | 1590 | 116 | 1.4 × 107 | 0.12 | 1.0 | 1.2 × 102 |
| L7ep-2 | 245 | 93 | 2.6 × 106 | 73 | 0.84 | 8.7 × 104 |
| L7ep-3 | 146 | 11.2 | 1.3 × 107 | 3.4 | 2.4 | 1.4 × 103 |
| L7ep-2a | 243 | 46 | 5.3 × 106 | 32 | 0.58 | 5.4 × 104 |
| L7ep-2b | 280 | 13.5 | 2.1 × 107 | 53 | 0.57 | 9.2 × 104 |
| L7ep-3a | 85 | 7.2 | 1.2 × 107 | 1.8 | 2.5 | 7.4 × 102 |
Standard errors from fits of the data fit to equation 1 are less than 10% of the stated values.
Hydrolysis of Racemic VX
Wild-type PTE and selected variants were characterized using racemic VX as a substrate and the results are presented in Table 3. Wild-type PTE has a low kcat (0.9 s−1) and relatively high Km, resulting in a diminished kcat/Km for the hydrolysis of VX. The variant QF dramatically improves both kcat and Km values resulting in a 100-fold increase in kcat/Km. The VRN-VQFL variant had the highest value of kcat/Km that was increased more than 230-fold over wild-type enzyme. The L7ep-3a had the highest kcat and was increased more than 150-fold, relative to wild-type PTE. The QF mutant was shown to preferentially hydrolyze the SP-enantiomer of VX by polarimetry.
Table 3.
Activity of PTE variants with racemic VXa.
| Variant | kcat (s−1) | Km (mM) | kcat/Km (M−1s−1) | Stereochemical Preferenceb |
|---|---|---|---|---|
| WT | 0.9 ± 0.1 | 2.9 ± 0.9 | 3 ± 1 × 102 | 1:1 |
| QF | 16 ± 1 | 0.5 ± 0.1 | 3.0 ± 0.6 × 104 | 12:1 (SP) |
| CVQFL | 45 ± 6 | 2.1 ± 0.7 | 2.2 ± 0.8 × 104 | 1:1 |
| VRN-VQFL | 44 ± 1 | 0.59 ± 0.09 | 7 ± 1 × 104 | 3:1 |
| L7ep-1 | 11 ± 1 | 0.8 ± 0.2 | 1.4 ± 0.3 × 104 | ND |
| L7ep-2 | 25 ± 2 | 1.3 ± 0.3 | 2.2 ± 0.3 × 104 | 5:1 |
| L7ep-3 | 31 ± 2 | 0.5 ± 0.2 | 6 ± 2 × 104 | 4:1 |
| L7ep-2a | 56 ± 4 | 3.4 ± 0.7 | 1.6 ± 0.4 × 104 | 4:1 |
| L7ep-2b | 56 ± 14 | 8 ± 3 | 7 ± 4 × 103 | 12:1 (RP) |
| L7ep-3a | 137 ± 22 | 5 ± 2 | 3 ± 1 × 104 | 4:1 |
Standard errors from fits of the data to equation 1.
Identity of preferred enantiomer was not determined for variants with less than a 10-fold preference.
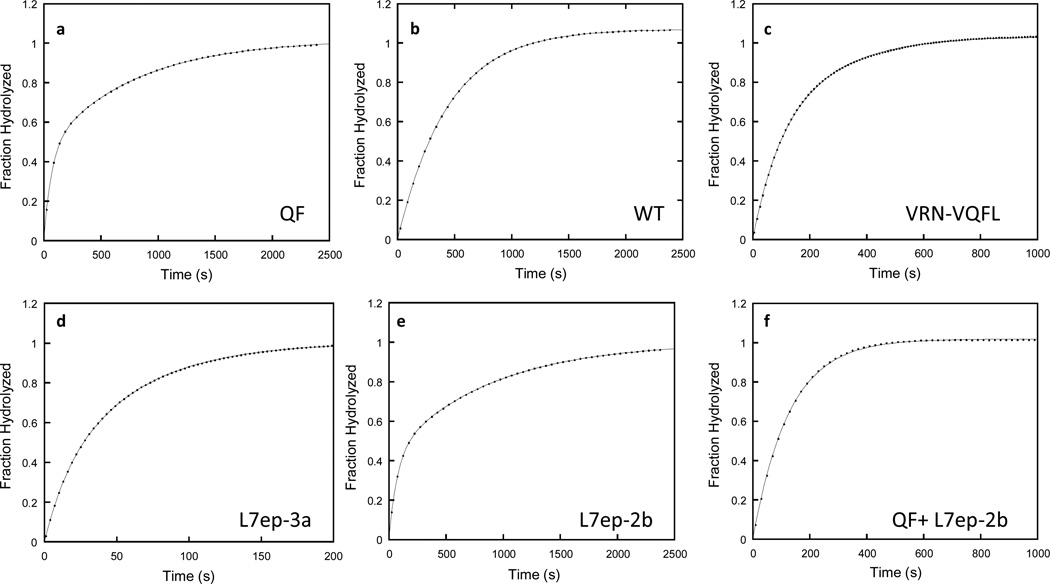
Stereoselective hydrolysis of racemic VX by the PTE mutants was evaluated by analyzing the time courses for the complete hydrolysis of VX at concentrations below the Michaelis constant. The presence of stereoselectivity in these time courses is manifested as the appearance of two exponential phases as observed with the QF variant where the ratio of rate constants is 12:1 (Figure 2a). In contrast, the wild-type enzyme exhibited no selectivity (Figure 2b), whereas the variants VRN-VQFL (3:1) and L7ep-3a (4:1) displayed relatively low selectivity (Figures 2c and 2d). For the L7ep-2b mutant, the observed stereoselectivity was 12:1 (Figure 2e). The enantiomeric specificity of the L7ep-2b variant was determined by its ability to complement the hydrolysis of the slower RP-enantiomer of VX after the addition of the variant QF. Plots of the fractional hydrolysis of VX as a function of time give two well-defined phases for each of these two variants (Figure 2a and Figure 2e). Mixing the two variants together resulted in a single well-defined monophasic curve demonstrating that the two variants prefer the opposite enantiomers (Figure 2f). Therefore, the L7ep-2b variant preferentially hydrolyzes the Rp-enantiomer of VX. The ratios of rate constants for the L7ep-2, L7ep-3, and L7ep-2a mutants were 5:1, 4:1, and 4:1, respectively (Table 3). In the absence of enantiomerically pure VX, the modest selectivities for these variants prevented the definitive assignment of the preferred enantiomer.
Figure 2.
Representative time courses for the complete hydrolysis of 160 µM racemic VX by selected PTE variants. (a) QF (850 nM); (b) Wild-type (27 µM); (c) VRN-VQFL (110 nM); (d) L7ep-3a (82 nM); (e) L7ep-2b (603 nM); and (f) L7ep-3b (301 nM) and QF (440 nM). The time courses for panels b and f were fit to equation 2, while the data for panels a, c, d, and e were fit to equation 3.
DISCUSSION
Library Design, Analogue Selection and Variant Identification
The P-S bond in VX is chemically more stable than the P-F bond found in the G-agents.20 Previous attempts to evolve PTE for the hydrolysis of VX have utilized the commercially available insecticides, demeton-S and demeton-S methyl and have yielded only modest improvements in activity.24,25 Demeton- S, while it contains the requisite P-S bond, does not contain the tertiary amine of VX, which is likely to be protonated at the relevant pH values. The new analogue, DEVX, contains the authentic leaving group of VX and was expected to yield results more directly applicable to the hydrolysis of VX itself.
The QF variant of PTE was previously shown to be improved against the SP-enantiomer of a chiral VX analogue.18 This mutant was found to be significantly better for the hydrolysis of the P-S bond in DEVX and demeton-S than wild-type PTE. The synergistic mutations in this variant suggested that further improvements in catalytic activity could be facilitated by simultaneously mutating pairs of residues in the active site. The initial mutant libraries targeted pairs of residues in the active site that modulated the size and shape of the three substrate binding pockets. Sequential optimization of the active site residues resulted in an 18-fold improvement in catalytic activity against DEVX. Combining the best variant (VQFL) with expression enhancing mutations resulted in the variant VRN-VQFL, with a 26-fold improvement for the hydrolysis of DEVX.
To further enhance the activity of PTE against the phosphorothiolate bond more advanced strategies were needed. Error-prone PCR is a useful technique for enzyme evolution, but the mutation frequencies are typically restricted to 1–3 base pair changes per gene because of the significant chance of introducing deleterious mutations. Unfortunately, substantial improvements in enzyme activity may require numerous amino acid changes, which are not typically achievable by error-prone PCR. Targeting error-prone PCR to only Loop-7 (residues 253–276) resulted in a mutation library with an average of 6 mutations per gene but still retained > 20% active colonies. The hydrolysis of DEVX for one of the variants (L7ep-3) was improved to 36-fold over wild-type PTE due to 3 additional amino acid changes. The best variant identified (L7ep-2) was improved 63-fold for the hydrolysis of DEVX by 5 additional amino acid changes. Further optimization of L7ep-2 and L7ep-3 resulted in additional mutations that improved the activity to 78-fold (L7ep-2a) and 71-fold (L7ep-3a) over wild-type PTE, and achieved turnover numbers for the hydrolysis of the phosphorothiolate bond in excess of 100 s−1.
Hydrolysis of VX
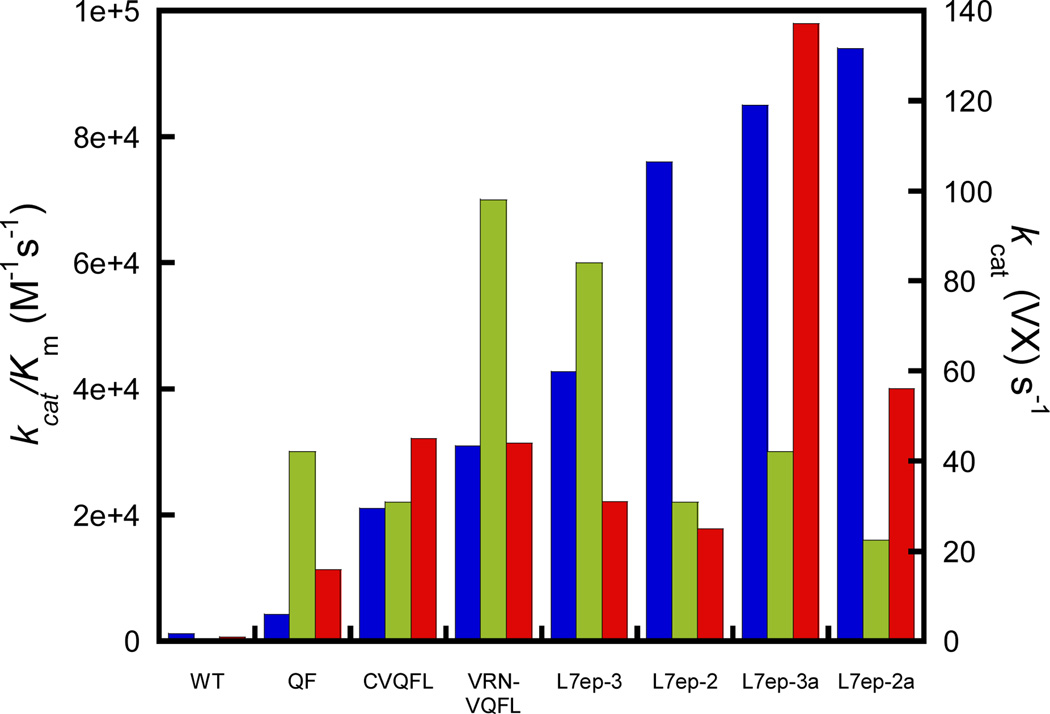
A full kinetic characterization of wild-type and improved variants of PTE using racemic VX was conducted. The wild-type enzyme exhibited low activity against VX, but there was a dramatic improvement with the QF mutant. The mutations in the large group pocket resulted in substantial improvements to kcat. The best variant identified (VRN-VQFL) against VX combines active site mutations in all three pockets and has a kcat/Km value that is increased 235-fold over wild-type PTE (Figure 3). The Loop-7 optimized variants show good activity against VX, but did not demonstrate improved activity relative to the VRN-VQFL variant. The changes to Loop-7 resulted in substantial improvements in kcat but little change in the catalytic efficiency.
Figure 3.
Enhancement in the catalytic properties for the hydrolysis of VX and DEVX by variants of PTE. The values of kcat/Km for evolved variants of PTE are presented for DEVX (blue) and VX (green). The kcat values for the hydrolysis of VX are shown in red.
The L7ep-3a variant has a kcat of 137 s−1 for the hydrolysis of VX. While there is relatively little data in the literature for the enzymatic hydrolysis of VX, this value is the highest ever reported.11,14,24,25 Single concentration experiments with the H254R/H257L mutant of PTE showed an improvement of 10-fold against racemic VX.24 Another variant exhibited a 26-fold improvement over wild-type PTE at 0.5 mM VX .25 By contrast, the VRN-VQFL and L7ep-3 variants are improved by more than 200-fold in the value of kcat/Km. Human PON1 has been evolved in the laboratory for the hydrolysis of VX, but the reported value of kcat/Km for the best variant is 2.5×103 M−1 s−1, whereas the best PTE variant (VRN-VQFL) identified in this investigation has a value of kcat/Km of 7×104 M−1 s−1.11
Stereochemical Preferences of Active Site Mutants
The toxicity of the organophosphate nerve agents depends on the stereochemistry of the phosphorus center.17 With VX, it is estimated that the SP-enantiomer is about 100-fold more toxic than the RPenantiomer. We have demonstrated that the QF mutant prefers to hydrolyze the more toxic SP-enantiomer of VX by a factor of 12, relative to the Rp-enantiomer, whereas the L7ep-2b mutant prefers to hydrolyze the RP-enantiomer by a factor of 12. The stereochemical preferences for the hydrolysis of VX are fully consistent with the stereoselective properties of these two mutants for the hydrolysis of Sp-1 and Rp-1, suggesting that the variants VRN-VQFL L7ep-3, and L7ep-3a also prefer the SP-enantiomer of VX. While the modest selectivity prevented definitive assignment of the preferred enantiomer, complete neutralization of VX by the VRN-VQFL mutant via the hydrolysis of both enantiomers was demonstrated by 31P-NMR spectroscopy (Figure S2).
Conclusions
The reconstruction of PTE for the hydrolysis of VX has resulted in dramatic improvements in the values of kcat and kcat/Km, relative to the wild type enzyme. We propose that the increase in the catalytic constants has been achieved by an increase in the rate constant for cleavage of the P-S bond (k3) rather than changes in the formation of the ternary complex (k1, k2) or the rate constant for product release (k5) as illustrated in a minimal kinetic mechanism (Scheme 2). The thiol leaving group of VX has a higher pKa than the fluoride leaving group of the G-agents, and the p-nitrophenol group of paraoxon. It has been previously demonstrated with the wild-type PTE that the chemical step (k3) is rate limiting for substrates with leaving groups having pKa values higher than 8.20 Disruption of the hydrogen bonded network from D301-H254-D233 reduced the rate of hydrolysis of substrates with leaving groups having low pKa values but increased the rate of hydrolysis of substrates with leaving groups of higher pKa values.30 Introduction of a glutamine at residue position 254 (as in the initial QF mutant), which apparently cannot support the transport of a proton away from the active site, may now facilitate the protonation of the thiol group by Asp-301 as the phosphorothiolate bond is cleaved.
Scheme 2.
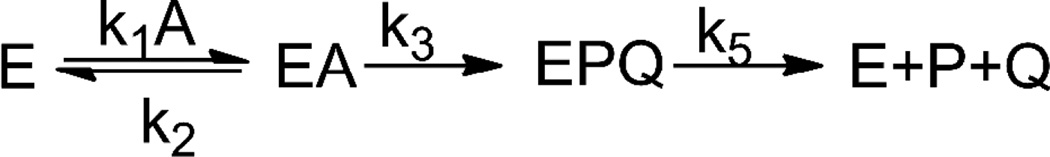
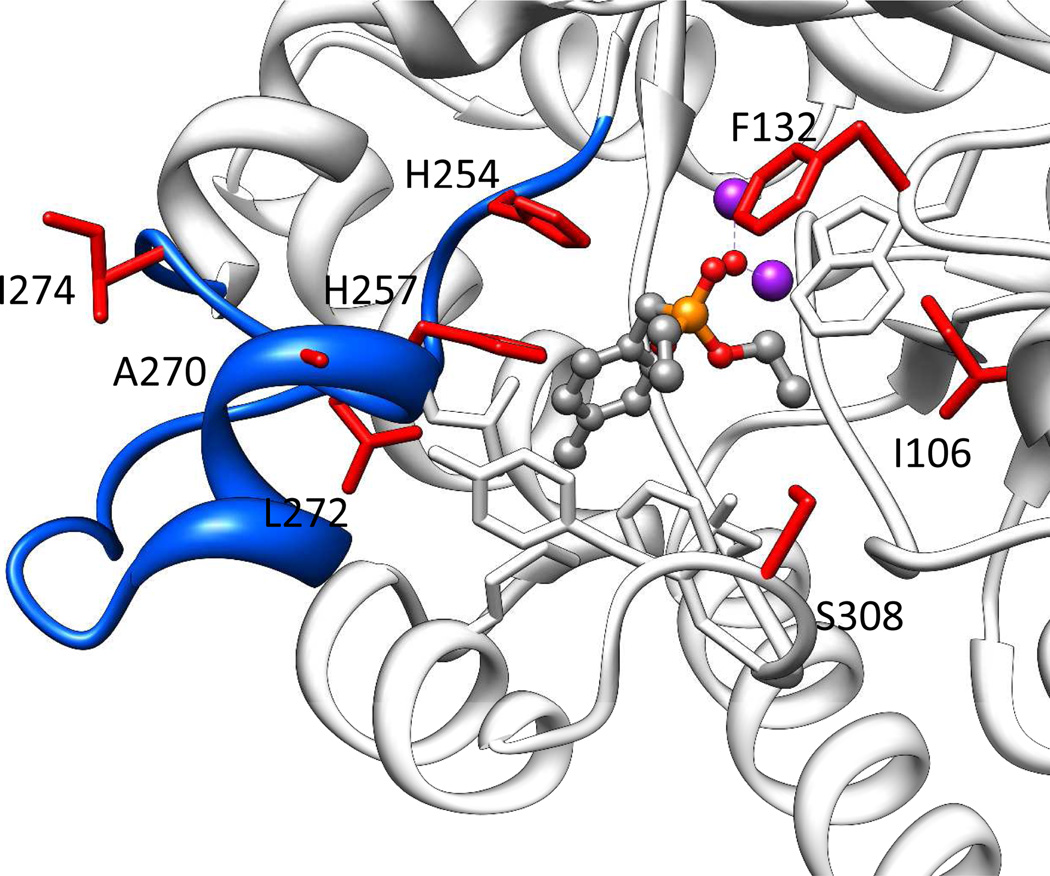
The turnover numbers for some slow substrates of PTE are thought to be reflective of the ability of the enzyme to align the substrate with the nucleophilic hydroxyl group attached to the binuclear metal center.13 There is a strong likelihood that for some of the variants, subtle changes in the conformation of the active site will facilitate a better alignment between the substrate and attacking hydroxide, thereby achieving higher enzymatic rates of hydrolysis. In particular, the Loop-7 variants have been modified at residues that are somewhat distant from the active site, but are expected to bring about changes in the positioning of the Loop-7 α-helix (Figure 4).13,28 This alignment effect would, of course, differ between the di-ethoxy phosphorus center of DEVX and the methylphosphonate core of VX, which may explain the differences in the kcat values for DEVX and VX with the variants L7ep-2a and L7ep-3a. While the exact molecular mechanism for the improvement in the catalytic properties of these mutants remains undetermined, these changes have resulted in variants with high enzymatic efficiency and exceptional kinetic constant for the hydrolysis of VX.
Figure 4.
Active site of wild-type PTE with diethyl 4-methylbenzylphosphonate bound in active site (PDB id: 1dpm). Loop-7 is colored in blue and the active site metals are shown as purple spheres. The residues mutated in the L7ep-3a variant (I106C/ F132V/ H254Q/ H257Y/ A270V/ L272M/ I274N/ S308L) are shown as red sticks. Substrate binding site residues not mutated in the L7ep-3a variant are shown as white sticks.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by the NIH (GM68550). We gratefully acknowledge the Defense Threat Reduction Agency for funding the work conducted at the Edgewood Chemical Biological Center.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Additional details for mutant library construction and Table S1 and S2. These tables provide a complete list of characterized mutants and the kinetic constants for the hydrolysis of DEVX. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
References
- 1.Maxwell DM, Brecht KM, Koplovitz I, Sweeney RE. Mol. Toxicol. 2006;80:756. doi: 10.1007/s00204-006-0120-2. [DOI] [PubMed] [Google Scholar]
- 2.Leikin JB, Thomas RG, Walter FG, Klein R, Meislin HW. Crit. Care. Med. 2002;30:2346. doi: 10.1097/00003246-200210000-00026. [DOI] [PubMed] [Google Scholar]
- 3.Columbus I, Waysbort D, Marcovitch I, Yehezkel L, Mizrahi DM. Environ. Sci. Technol. 2012;46:3921. doi: 10.1021/es300404y. [DOI] [PubMed] [Google Scholar]
- 4.Yang YC. Chem. Ind. 1995;9:334. [Google Scholar]
- 5.Yang YC. Accounts Chem. Res. 1999;32:109. [Google Scholar]
- 6.Saxina A, Sun W, Fedorko JM, Koplovitz I, Doctor BP. Biochem. Pharmacol. 2011;81:164. doi: 10.1016/j.bcp.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Ashani Y, Pistinner S. Toxicol. Sci. 2004;77:358. doi: 10.1093/toxsci/kfh012. [DOI] [PubMed] [Google Scholar]
- 8.Saxena A, Tipparaju P, Luo C, Doctor BP. Process Biochem. 2010;45:1313. [Google Scholar]
- 9.Cheng TC, Liu L, Wang B, Wu J, DeFrank JJ, Anderson DM, Rastogi VK, Hamilton AB. J. Ind. Microbiol. Biotechnol. 1997;18:49. doi: 10.1038/sj.jim.2900358. [DOI] [PubMed] [Google Scholar]
- 10.Melzer M, Chen JCH, Heidenreich A, Gab J, Koller M, Kehe K, Blum MM. J. Am. Chem. Soc. 2009;131:17226. doi: 10.1021/ja905444g. [DOI] [PubMed] [Google Scholar]
- 11.Kirby SD, Norris JR, Smith JR, Bahnson BJ, Cerasoli DM. Chem. Biol. Interact. 2012;203:181. doi: 10.1016/j.cbi.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Gupta RD, Goldsmith M, Ashani Y, Simo Y, Mullokandov G, Bar H, Ben-David M, Leader H, Margalit R, Silman I, Sussman JL, Tawfik DS. Nat. Chem. Biol. 2011;7:120. doi: 10.1038/nchembio.510. [DOI] [PubMed] [Google Scholar]
- 13.Tsai PC, Fox N, Bigley AN, Harvey SP, Barondeau DP, Raushel FM. Biochemistry. 2012;51:6463. doi: 10.1021/bi300811t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai K, Grimsley JK, Kuhlmann BD, Scapozza L, Harvey SP, DeFrank JJ, Kolakowski JE, Wild JR. Chimia. 1996;50:430. [Google Scholar]
- 15.Caldwell SR, Newcomb JR, Schlecht KA, Raushel FM. Biochemistry. 1991;30:7438. doi: 10.1021/bi00244a010. [DOI] [PubMed] [Google Scholar]
- 16.Benschop HP, De Jong LPA. Accounts Chem. Res. 1988;21:368. [Google Scholar]
- 17.Ordentlich A, Barak D, Sod-Moriah G, Kaplan D, Mizrahi D, Segall Y, Kronman C, Karton Y, Lazar A, Marcus D, Velan B, Shafferman A. Biochemistry. 2004;43:11255. doi: 10.1021/bi0490946. [DOI] [PubMed] [Google Scholar]
- 18.Tsai PC, Bigley AN, Li Y, Ghanem E, Cadieux CL, Kasten SA, Reeves TE, Cerasoli DM, Raushel FM. Biochemistry. 2010;49:7978. doi: 10.1021/bi101056m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rastogi VK, DeFrank JJ, Cheng T, Wild JR. Biochem. Bioph. Res. Co. 1997;241:294. doi: 10.1006/bbrc.1997.7569. [DOI] [PubMed] [Google Scholar]
- 20.Hong SB, Raushel FM. Biochemistry. 1996;35:10904. doi: 10.1021/bi960663m. [DOI] [PubMed] [Google Scholar]
- 21.Vanhooke JL, Benning MM, Raushel FM, Holden HM. Biochemistry. 1996;35:6020. doi: 10.1021/bi960325l. [DOI] [PubMed] [Google Scholar]
- 22.Chen-Goodspeed M, Sogorb MA, Wu F, Hong SB, Raushel FM. Biochemistry. 2001;40:1325. doi: 10.1021/bi001548l. [DOI] [PubMed] [Google Scholar]
- 23.Afriat-Jurnou L, Jackson CJ, Tawfik DS. Biochemistry. 2012;51:6047. doi: 10.1021/bi300694t. [DOI] [PubMed] [Google Scholar]
- 24.Reeves TE, Wales ME, Grimsley JK, Li P, Cerasoli DM, Wild JR. Protein Eng. Des. Sel. 2008;21:405. doi: 10.1093/protein/gzn019. [DOI] [PubMed] [Google Scholar]
- 25.Schofield DA, DiNovo AA. J. Appl. Microbiol. 2010;109:548. doi: 10.1111/j.1365-2672.2010.04672.x. [DOI] [PubMed] [Google Scholar]
- 26.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Gene. 1989;77:61. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 27.Goldsmith M, Kiss C, Bradbury ARM, Tawfik DS. Protein Eng. Del. Sel. 2007;20:315. doi: 10.1093/protein/gzm026. [DOI] [PubMed] [Google Scholar]
- 28.Cho CM, Mulchandani A, Chen W. Protein Eng. Del. Sel. 2006;19:99. doi: 10.1093/protein/gzj007. [DOI] [PubMed] [Google Scholar]
- 29.Roodveldt C, Tawfik DS. Protein Eng. Des. Sel. 2005;18:51. doi: 10.1093/protein/gzi005. [DOI] [PubMed] [Google Scholar]
- 30.Aubert SD, Li Y, Raushel FM. Biochemistry. 2004;43:5707. doi: 10.1021/bi0497805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.