Abstract
Peritrichous bacteria exploit bundles of helical flagella for propulsion and chemotaxis. Here, changes in the swimming direction (tumbling) are induced by a change of the rotational frequency of some flagella. Employing coarse-grained modeling and simulations, we investigate the dynamical properties of helical flagella bundles driven by mismatched motor torques. Over a broad range of distances between the flagella anchors and applied torque differences, we find a stable bundled state, which is important for a robust directional motion of a bacterium. With increasing torque difference, a phase lag in the flagellar rotations develops, followed by slippage and ultimately unbundling, which sensitively depends on the anchoring distance of neighboring flagella. In the slippage and drift states, the different rotation frequencies of the flagella generate a tilting torque on the bacterial body, which implies a change of the swimming direction as observed experimentally.
Introduction
Motile bacteria exploit bundles of rotating helical flagellar filaments for propulsion and chemotaxis [1]–[4]. Various arrangements of the flagella on the bacterial cell membrane have developed in the evolutionary process [5]. The flagella of peritrichous bacteria self-organize into bundles by (typically) counterclockwise rotation of the flagella motors leading to nearly straight swimming. To change the swimming direction, this “running” phase is interrupted by short periods of “tumbling” [6]–[8]. A specific flagella design [9] implies a distinct tumbling mechanism of a particular bacterium. In Escherichia coli bacteria, clockwise rotation of one or several flagella leads to a polymorphic transition, [10]–[14] disintegration of the bundle and bacterial tumbling [1], [15], [16]. In contrast, the flagella of Rhizobium meliloti or Rhizobium lupini are only capable of limited polymorphic transitions and their motors are unidirectional [9], [17], [18]. These bacteria modulate the rotation speed of individual motors to induce tumbling [9], [17].
The bundling process of bacterial flagella has been studied experimentally [3], [17], [19]–[21], theoretically [22]–[24], and by computer simulations [5], [8], [16]. An essential aspect of bundling is synchronization of the flagella driven by motors of nonuniform strength. Synchronization of flagellar rotation is, aside from bacterial motion, of fundamental importance for a broad range of phenomena in biology [25]–[29], ranging from fluid transport in the respiratory system [30], to embryonic left-right asymmetry [31], and intercellular communication [32]. Thus, a theoretical understanding of flagella synchronization is of paramount importance. Synchronization in fluid systems can be achieved by hydrodynamic interactions as demonstrated by various studies [16], [27], [28], [33]–[40]. In bacterial bundling, steric interactions between the various flagella may also play an important role due to the opposite rotation of the flagella bundle and the cell body [16].
Much less is known about the tumbling mechanism of bacteria in general, and for bacteria such as R. lupini or R. meliloti with unidirectional motors in particular. In Ref. [17], bundle disintegration has been observed when a flagellar filament slows down or stops. However, tumbling should also be possible without complete disintegration of the bundle, but might be more difficult to observe experimentally.
Here, we study synchronization and unbundling of driven flagella, properties which are of fundamental importance for bacterial tumbling. Of particular interest is the dephasing of flagella rotational frequencies with increasing torque difference between the various flagella. Qualitatively, our results suggest the following classification of the bundle dynamics. At small torque differences, the bundle remains stable with a phase lag between the various flagella. For very large torque differences, the bundle disintegrates and the flagella rotated asynchronous and independently; the phase differences of neighboring helices are drifting. In between, there is an intermittent regime, where phase slippage occurs, i.e., the synchronized rotational motion is interrupted by events, where the flagellum with the larger torque leaves the bundle, rotates faster, and rejoins the bundle. The interval between individual slippages decreases with increasing torque difference and ultimately drifting is obtained. We predict that this slippage or drift leads to a tumbling motion of the bacteria.
Models
A hybrid mesoscopic simulation approach is adopted, combining molecular dynamics simulations for the bacterial flagella with the multiparticle collision dynamics (MPC) approach, a mesoscale hydrodynamics simulation technique, for the fluid [41]–[43]. This hybrid method has been successful applied to study the dynamics of various kinds of macromolecular and cellular systems [35], [40], [44], [45]. The bacterial flagellum is represented by a coarse-grained macromolecular system embedded in a MPC fluid with a helical sequence of 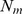 mass points of mass
mass points of mass 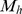 [16] (cf. Fig. 1). These points interact with each other by the following bond, bending, and torsional potentials:
[16] (cf. Fig. 1). These points interact with each other by the following bond, bending, and torsional potentials:
| (1) |
| (2) |
| (3) |
Here, 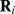 denotes the position of bead
denotes the position of bead  , and
, and 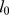 ,
, 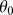 , and φ
0 are the equilibrium bond length, bending angle, and torsional angle, respectively.
, and φ
0 are the equilibrium bond length, bending angle, and torsional angle, respectively. 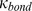 ,
, 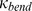 , and
, and 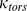 are the bond, bending, and torsional rigidities, respectively. The repulsive and truncated Lennard-Jones potential
are the bond, bending, and torsional rigidities, respectively. The repulsive and truncated Lennard-Jones potential
| (4) |
is applied to account for excluded-volume interactions between the mass points of the helices of distance  , with the size of a point
, with the size of a point  , the energy
, the energy  [16], [46], and
[16], [46], and  the Heaviside step function. In addition, one central bead and four peripheral beads are added in a plane at the base of the helix (cf. Fig. 1). These five beads are trapped in constraining harmonic potentials of the form,
the Heaviside step function. In addition, one central bead and four peripheral beads are added in a plane at the base of the helix (cf. Fig. 1). These five beads are trapped in constraining harmonic potentials of the form,
| (5) |
where 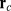 is either the equilibrium position of the central particle or the
is either the equilibrium position of the central particle or the  -coordinate of the peripheral beads; in the latter case, the
-coordinate of the peripheral beads; in the latter case, the  - and
- and 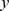 -coordinates are unconstrained. Hence, a flagellum is not allowed to perform any translational motion but rotates around a central bead driven by an external torque applied at two opposite peripheral beads as illustrated in Fig. 1. The dynamics of the mass points is described by Newton's equations of motion, which are integrated by the velocity-Verlet algorithm [46]. Symmetrical forces are applied in counterclockwise direction when watched from the distal end, which generate a torque
-coordinates are unconstrained. Hence, a flagellum is not allowed to perform any translational motion but rotates around a central bead driven by an external torque applied at two opposite peripheral beads as illustrated in Fig. 1. The dynamics of the mass points is described by Newton's equations of motion, which are integrated by the velocity-Verlet algorithm [46]. Symmetrical forces are applied in counterclockwise direction when watched from the distal end, which generate a torque 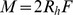 pointing into the positive
pointing into the positive  -direction, where
-direction, where 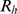 is the helix radius and
is the helix radius and 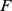 the applied force.
the applied force.
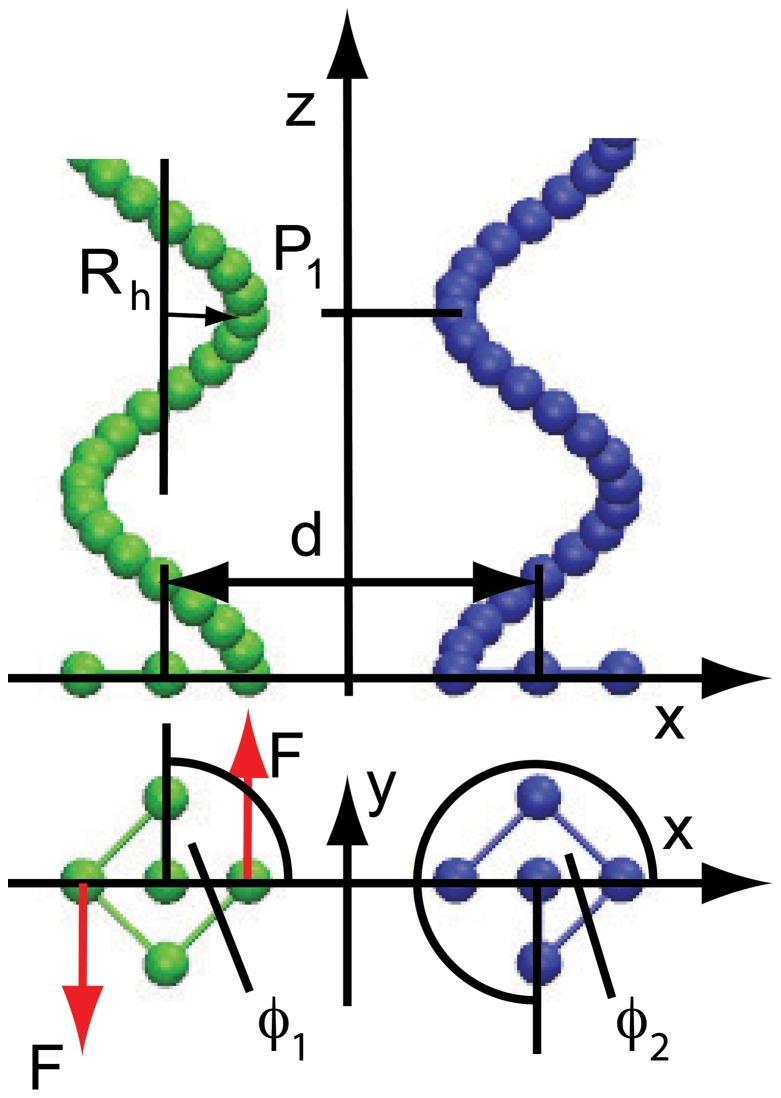
Figure 1. Model of a flagella bundle.
The top part shows a side view of two left-handed flagella, with average distance 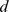 , helix radius
, helix radius 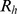 , and pitch
, and pitch 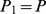 . The bottom part shows a top view with the external forces
. The bottom part shows a top view with the external forces 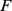 generating a torque and the phase angles
generating a torque and the phase angles 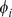 .
.
MPC is a particle-based simulation approach, where the fluid is represented by point particles, and naturally comprises thermal fluctuations. The algorithm consists of alternating streaming and collision steps. In the streaming step, the particles move ballistically; in the collision step, the particles are sorted into cubic cells of length  and their relative velocities, with respect to the center-of-mass velocities of each cell, are rotated around a randomly oriented axis by a fix angle
and their relative velocities, with respect to the center-of-mass velocities of each cell, are rotated around a randomly oriented axis by a fix angle  [16], [43]. A constant temperature is maintained locally by velocity rescaling at every collision cell and every collision step [47]. The MPC parameters are chosen as: average fluid particle number per collision cell
[16], [43]. A constant temperature is maintained locally by velocity rescaling at every collision cell and every collision step [47]. The MPC parameters are chosen as: average fluid particle number per collision cell 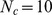 , rotation angle
, rotation angle 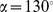 , and collision time step
, and collision time step 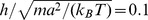 , where
, where 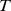 is the temperature,
is the temperature, 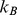 the Boltzmann constant, and
the Boltzmann constant, and  the mass of a fluid particle, which yields the solvent viscosity
the mass of a fluid particle, which yields the solvent viscosity 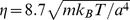 [16], [48]. The parameters for a flagellum are chosen as: mass of a flagellum bead
[16], [48]. The parameters for a flagellum are chosen as: mass of a flagellum bead 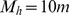 , helix radius
, helix radius 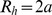 , number of beads for a five-turn helix
, number of beads for a five-turn helix 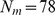 , equilibrium bond length
, equilibrium bond length 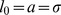 , bending angle
, bending angle 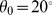 , torsional angle φ
, torsional angle φ
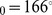 , and
, and 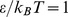 . The bond stretching, bond bending, and torsional rigidities are
. The bond stretching, bond bending, and torsional rigidities are 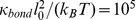 ,
, 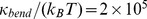 , and
, and 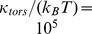 , respectively. The constraining-force constant of the motor part is
, respectively. The constraining-force constant of the motor part is 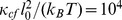 . The distribution of the bond stretching, bending, and torsional energies follows the corresponding Boltzmann distributions.
. The distribution of the bond stretching, bending, and torsional energies follows the corresponding Boltzmann distributions.
The adopted potential parameters and contour length yield a five-turn helix with the pitch angle 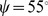 . This closely resembles the shape of a flagellum of R. lupini in the semi-coiled state [17]. The bending rigidity of a Salmonella flagellum has been determined in Ref. [13]. A value of
. This closely resembles the shape of a flagellum of R. lupini in the semi-coiled state [17]. The bending rigidity of a Salmonella flagellum has been determined in Ref. [13]. A value of 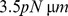 for the bending rigidity was obtained by employing a model with a quadratic Kirchhoff-rod potential for bending and twist, and comparing the theoretical force-extension relation with the respective experimental data. Moreover, the twist rigidity was found to be comparable with the bending rigidity [13]. In order to link the parameters
for the bending rigidity was obtained by employing a model with a quadratic Kirchhoff-rod potential for bending and twist, and comparing the theoretical force-extension relation with the respective experimental data. Moreover, the twist rigidity was found to be comparable with the bending rigidity [13]. In order to link the parameters 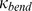 and
and 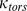 in Eqs. (2) and (3) with the experimental results for the flagellum rigidities, we discretize the continuum bending and torsional energies of the helical wormlike chain model [13], [49] and compare them with the potentials (2) and (3). Using the structural parameters of R. lupini
[17], we find that our
in Eqs. (2) and (3) with the experimental results for the flagellum rigidities, we discretize the continuum bending and torsional energies of the helical wormlike chain model [13], [49] and compare them with the potentials (2) and (3). Using the structural parameters of R. lupini
[17], we find that our 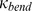 corresponds to an approximately five times larger bending rigidity compared to the experimental value for Salmonella. Since it has been argued that the flagellum of R. lupini is stiffer than that of Salmonella
[50], our chosen value is in reasonable agreement with the biologically relevant scales. The ratio between the bending and the torsional rigidities in our model is approximately four. Hence, also the torsional rigidity is on the order of magnitude of the biological scale.
corresponds to an approximately five times larger bending rigidity compared to the experimental value for Salmonella. Since it has been argued that the flagellum of R. lupini is stiffer than that of Salmonella
[50], our chosen value is in reasonable agreement with the biologically relevant scales. The ratio between the bending and the torsional rigidities in our model is approximately four. Hence, also the torsional rigidity is on the order of magnitude of the biological scale.
The flagella are placed in a cubic simulation box of side length 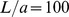 with periodic boundary conditions. When the flagella start to rotate, they set fluid in motion until a stationary (mean) fluid velocity is reached, where the flagella exert no net force on the anchoring plane along its normal. This is equivalent to a free-swimming bacteria with a non-rotating body. In our system, the fluid velocity far away from the flagella corresponds to the swimming velocity of the free swimmer, because the two systems are just related by a Galilean transformation. This point will be discussed in more detail below.
with periodic boundary conditions. When the flagella start to rotate, they set fluid in motion until a stationary (mean) fluid velocity is reached, where the flagella exert no net force on the anchoring plane along its normal. This is equivalent to a free-swimming bacteria with a non-rotating body. In our system, the fluid velocity far away from the flagella corresponds to the swimming velocity of the free swimmer, because the two systems are just related by a Galilean transformation. This point will be discussed in more detail below.
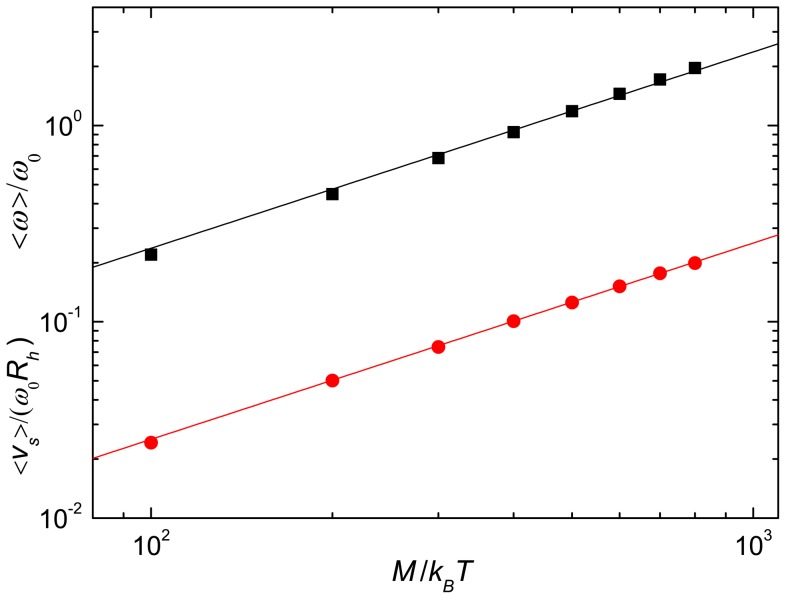
We have performed several test to validate the selected model using a single flagellum. As expected and shown in Fig. 2, we find a linear relation between the applied torque and the rotation frequency of the helix. Since the helix is fixed at the anchoring point, the fluid acquires a constant mean velocity in the stationary state. This velocity also depends linearly on the applied torque, see Fig. 2. The reference frequency 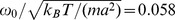 corresponds to the rotation frequency of a bundle of three helices each driven by the torque
corresponds to the rotation frequency of a bundle of three helices each driven by the torque 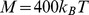 at the distance
at the distance 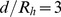 . A single helical flagellum exhibits only a few percent smaller rotation frequency for the same torque.
. A single helical flagellum exhibits only a few percent smaller rotation frequency for the same torque.
Figure 2. Single flagellum rotation frequency and swimming speed.
Mean values of the rotation frequencies (squares) and mean induced fluid velocities (bullets) of a single helix for various applied torques. The lines indicate the linear dependence on the torque.
Results and Discussion
Corresponding to the averaged number of experimentally observed flagella for R. lupini
[17], we study a system of three helices with their anchoring points fixed on an equilateral triangle of side length 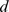 . Figure 1 illustrates the initial parallel alignment for two helices and the phase angles
. Figure 1 illustrates the initial parallel alignment for two helices and the phase angles 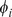 (in general,
(in general, 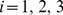 ). The initial angles of the three helices are
). The initial angles of the three helices are 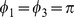 and
and 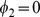 . To study the bundle response to differences in the applied torques
. To study the bundle response to differences in the applied torques 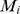 at the various helices, we utilize the fixed torque
at the various helices, we utilize the fixed torque 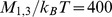 of helices
of helices  and
and  and vary that of helix
and vary that of helix  in the range
in the range 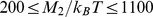 . The experimentally measured torques for flagella of E. coli bacteria are in the range from 1300
. The experimentally measured torques for flagella of E. coli bacteria are in the range from 1300 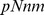 to 4600
to 4600 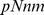 , i.e.,
, i.e., 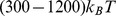 [7], [51]–[53]. Thus, our simulated torques cover approximately the same range at room temperature.
[7], [51]–[53]. Thus, our simulated torques cover approximately the same range at room temperature.
Flagella bundle stability
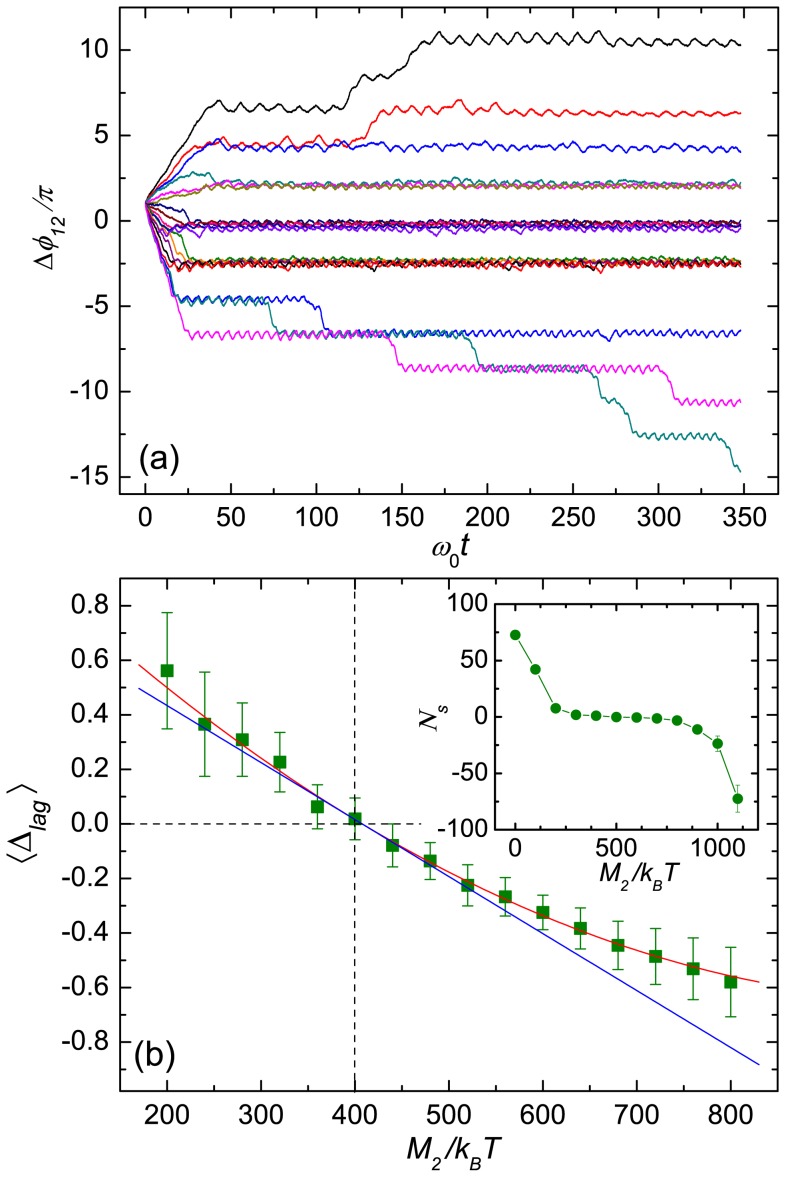
Figure 3(a) displays the phase-angle difference 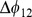 (
(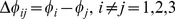 ) as a function of time for various torques
) as a function of time for various torques 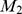 . The time scale is normalized by the reference angular velocity
. The time scale is normalized by the reference angular velocity 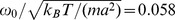 of a bundle of three helices, as already introduced in the previous section. For torques in the range
of a bundle of three helices, as already introduced in the previous section. For torques in the range 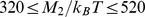 , the helices form bundles and exhibit a phase-locked synchronized rotational dynamics with the same average angular frequency after a short time. As discussed in Ref. [16], synchronization is achieved by hydrodynamic and steric interactions between the various helices. When the torque difference
, the helices form bundles and exhibit a phase-locked synchronized rotational dynamics with the same average angular frequency after a short time. As discussed in Ref. [16], synchronization is achieved by hydrodynamic and steric interactions between the various helices. When the torque difference 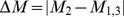 exceeds a critical value, phase slips occur. Here, flagella with the larger torque leave the bundle and perform one additional rotation and then rejoin the bundle; this corresponds to a change in phase difference by
exceeds a critical value, phase slips occur. Here, flagella with the larger torque leave the bundle and perform one additional rotation and then rejoin the bundle; this corresponds to a change in phase difference by 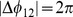 . With increasing
. With increasing 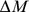 , the frequency of phase slips increases and ultimately the drift state is reached, where
, the frequency of phase slips increases and ultimately the drift state is reached, where 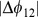 increases linearly in time. The appearance of slips is governed by the interplay of excluded volume interactions and elastic deformations of the flagella as well as thermal fluctuations.
increases linearly in time. The appearance of slips is governed by the interplay of excluded volume interactions and elastic deformations of the flagella as well as thermal fluctuations.
Figure 3. Phase slip, lag, and drift.
(a) Phase angle difference 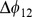 as a function of time for various applied torques on helix
as a function of time for various applied torques on helix  and the distance
and the distance 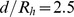 . The torque
. The torque  is changed from 200 (top) to 920 (bottom) with an increment of 40; the constant torque is
is changed from 200 (top) to 920 (bottom) with an increment of 40; the constant torque is 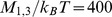 . (b) Average phase lag
. (b) Average phase lag 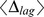 as a function of the torque
as a function of the torque 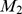 . The red line indicates the fit
. The red line indicates the fit 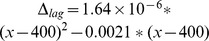 , where
, where 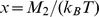 . The blue line is the tangent at
. The blue line is the tangent at 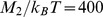 . The inset provides an approximate measure of the number of occurring slips during the time interval
. The inset provides an approximate measure of the number of occurring slips during the time interval 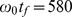 as a function of the applied moment
as a function of the applied moment 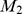 .
.
In the synchronized and bundled state, the helices exhibit a phase lag 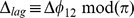 , which depends on the torque difference. The obtained phase lags exhibit considerable fluctuations around the average. The magnitude of the average
, which depends on the torque difference. The obtained phase lags exhibit considerable fluctuations around the average. The magnitude of the average 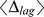 increases with increasing torque difference. As shown in Fig. 3(b) by a quadratic fit function,
increases with increasing torque difference. As shown in Fig. 3(b) by a quadratic fit function, 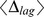 is asymmetric with respect to
is asymmetric with respect to 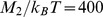 with a slower variation in the region of large
with a slower variation in the region of large 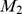 . This “asymmetry” appears due to the increase of the bundle rotational frequency with increasing
. This “asymmetry” appears due to the increase of the bundle rotational frequency with increasing 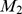 . The larger frequencies imply stronger flows and therefore stronger hydrodynamic interactions and tighter bundles.
. The larger frequencies imply stronger flows and therefore stronger hydrodynamic interactions and tighter bundles.
The inset of Fig. 3 displays the quantity 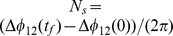 for the fixed time interval
for the fixed time interval 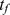 , which is an approximate measure of the number of occurring slips. For momenta
, which is an approximate measure of the number of occurring slips. For momenta 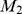 in the vicinity of
in the vicinity of 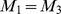 , phase slips are rare. However, we find a strong increase of slippage, when the torque difference
, phase slips are rare. However, we find a strong increase of slippage, when the torque difference 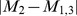 exceeds a threshold.
exceeds a threshold.
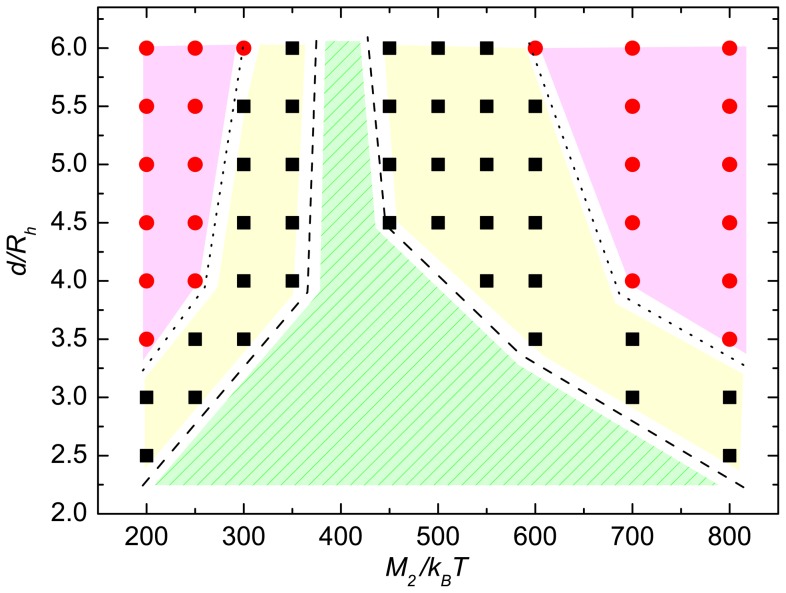
Data for the bundling, slippage, and drifting states for different torques and helix separations can be combined in a dynamic phase diagram, as shown in Fig. 4. Interestingly, there is a broad range of distances and torques, where stable bundles are obtained with no slips over the total considered time interval 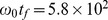 . The bundles are evidently rather robust for
. The bundles are evidently rather robust for 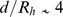 , and are able to sustain considerable torque differences. The asymmetry of the phase diagram with respect to the reference torque
, and are able to sustain considerable torque differences. The asymmetry of the phase diagram with respect to the reference torque 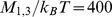 is a consequence of the stronger hydrodynamic interactions at larger torques (cf. Fig. 3(b)). The stable bundle regime is bounded by the intermittent slippage regime, where
is a consequence of the stronger hydrodynamic interactions at larger torques (cf. Fig. 3(b)). The stable bundle regime is bounded by the intermittent slippage regime, where 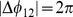 for each individual slippage event. Since this part of the diagram is also broad, the model system yields stable or nearly stable bundles over a wide range of torques for all the considered distances. For even larger distances and torque differences phase-drift appears, where
for each individual slippage event. Since this part of the diagram is also broad, the model system yields stable or nearly stable bundles over a wide range of torques for all the considered distances. For even larger distances and torque differences phase-drift appears, where 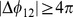 before a loose bundle might be reformed. Naturally, a clear distinction between the various regimes is difficult and the change from one to the other is gradual rather than abrupt.
before a loose bundle might be reformed. Naturally, a clear distinction between the various regimes is difficult and the change from one to the other is gradual rather than abrupt.
Figure 4. Phase diagram of bundle integrity.
Phase diagram indicating stable bundles (shaded), intermittent slippage (squares), and drift states (bullets) for various flagella distances and torques on helix 2. The torques on helices 1 and 3 are 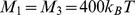 .
.
The phase diagram of Fig. 4 highlights a strong phase-locking for bundles of flexible helical flagella. Even for distances as large as 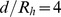 , phase locking occurs for torque differences as large as
, phase locking occurs for torque differences as large as 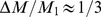 . This can be compared with the synchronization of rigid, three-arm colloidal micro-rotators discussed in Ref. [38], which can tolerate only torque differences of the order of
. This can be compared with the synchronization of rigid, three-arm colloidal micro-rotators discussed in Ref. [38], which can tolerate only torque differences of the order of 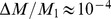 at much smaller distances of
at much smaller distances of 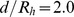 to
to 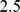 . This largely enhanced stability is due to the flexibility of the bacterial flagella, which allows them to wrap around each other. A high stability of the bundled state is very important for the directional motion of a bacterium because of the substantial noise inherent in biological systems, either originating from internal sources like variations in motor activities, or from external sources such as other swimming bacteria.
. This largely enhanced stability is due to the flexibility of the bacterial flagella, which allows them to wrap around each other. A high stability of the bundled state is very important for the directional motion of a bacterium because of the substantial noise inherent in biological systems, either originating from internal sources like variations in motor activities, or from external sources such as other swimming bacteria.
Intra-bundle distances
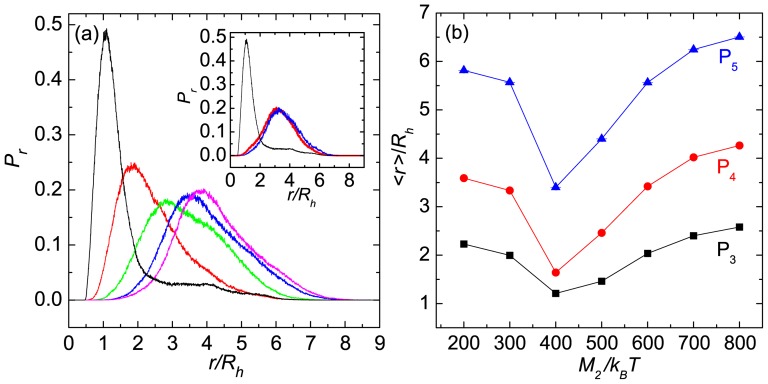
Slippage or drift of individual helices leads to partial or full flagella unbundling, which is reflected in the distribution 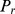 of distances between the equivalent beads of the various helices. Figure 5(a) provides an example for the distributions at
of distances between the equivalent beads of the various helices. Figure 5(a) provides an example for the distributions at 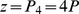 , where
, where 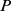 is the pitch, and
is the pitch, and 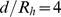 . For equal torques, we obtain a tight bundle with a narrow distribution and a maximum at the single monomer diameter. As the torque difference increases, the peak maximum shifts to large distances and the width broadens. The inset of Fig. 5 shows distance distributions for
. For equal torques, we obtain a tight bundle with a narrow distribution and a maximum at the single monomer diameter. As the torque difference increases, the peak maximum shifts to large distances and the width broadens. The inset of Fig. 5 shows distance distributions for 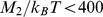 . Here, the maximum of
. Here, the maximum of 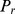 also moves to large distances with increasing torque difference, but saturation seems to be reached already at rather small
also moves to large distances with increasing torque difference, but saturation seems to be reached already at rather small 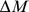 , much smaller than for
, much smaller than for 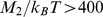 . The mean values of the distances between helical beads are shown in Fig. 5(b) as a function of torque
. The mean values of the distances between helical beads are shown in Fig. 5(b) as a function of torque 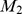 at the points
at the points 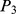 ,
, 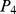 , and
, and 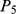 along the bundle. Evidently, the helices form a tight bundle for equal torques (
along the bundle. Evidently, the helices form a tight bundle for equal torques (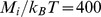 ). As the torque difference
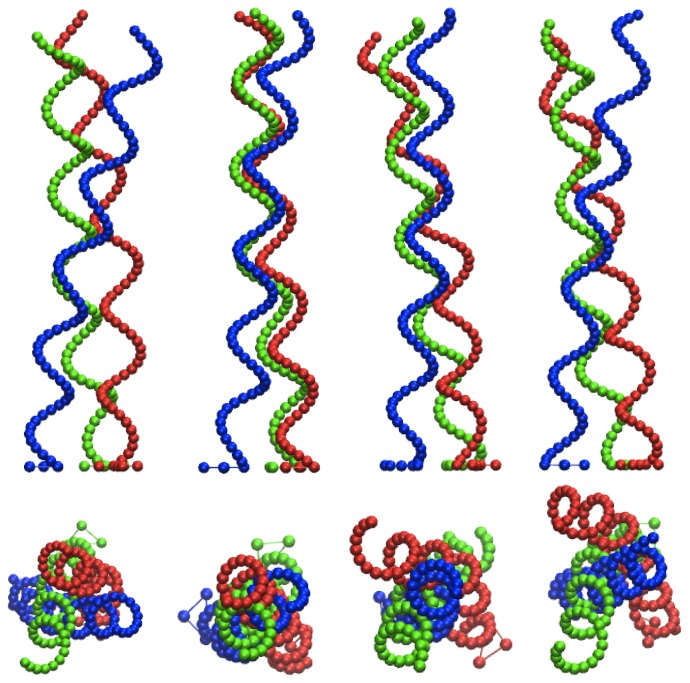
). As the torque difference 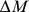 increases, the bundle disintegrates and the helices move apart. The snapshots of Fig. 6 illustrate the bundle geometry for various
increases, the bundle disintegrates and the helices move apart. The snapshots of Fig. 6 illustrate the bundle geometry for various 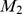 (see also videos S1 and S2). Interestingly, the bundle does not completely disintegrate. The hydrodynamic interactions continuously force the helices back into a bundle. Nevertheless, generation of a large torque difference is an effective strategy to impose helix unbundling.
(see also videos S1 and S2). Interestingly, the bundle does not completely disintegrate. The hydrodynamic interactions continuously force the helices back into a bundle. Nevertheless, generation of a large torque difference is an effective strategy to impose helix unbundling.
Figure 5. Bead distance distributions and mean distances.
(a) Normalized bead-distance distribution functions 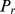 between helices at
between helices at 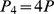 ,
, 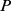 is the helical pitch (cf. Fig. 1), for the torque
is the helical pitch (cf. Fig. 1), for the torque 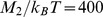 (black), 500 (red), 600 (green), 700 (blue), and 800 (magenta) with
(black), 500 (red), 600 (green), 700 (blue), and 800 (magenta) with 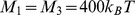 . The distance between anchored helix ends is fixed at
. The distance between anchored helix ends is fixed at 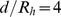 . The inset shows the distribution functions for
. The inset shows the distribution functions for 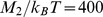 (black), 300 (red), and 200 (blue). (b) Average bead distances between the helices at
(black), 300 (red), and 200 (blue). (b) Average bead distances between the helices at 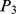 (black),
(black), 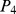 (red), and
(red), and 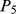 (blue) (
(blue) (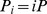 ) as a function of the torque
) as a function of the torque 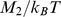 .
.
Figure 6. Snapshots of helices for various torque differences.
Snapshots of side (top) and top (bottom) views for the torque  , 400, 600, and 800 (from left to right) at
, 400, 600, and 800 (from left to right) at 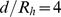 . See also videos S1 8 and S2 8 for
. See also videos S1 8 and S2 8 for 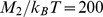 and 800, respectively.
and 800, respectively.
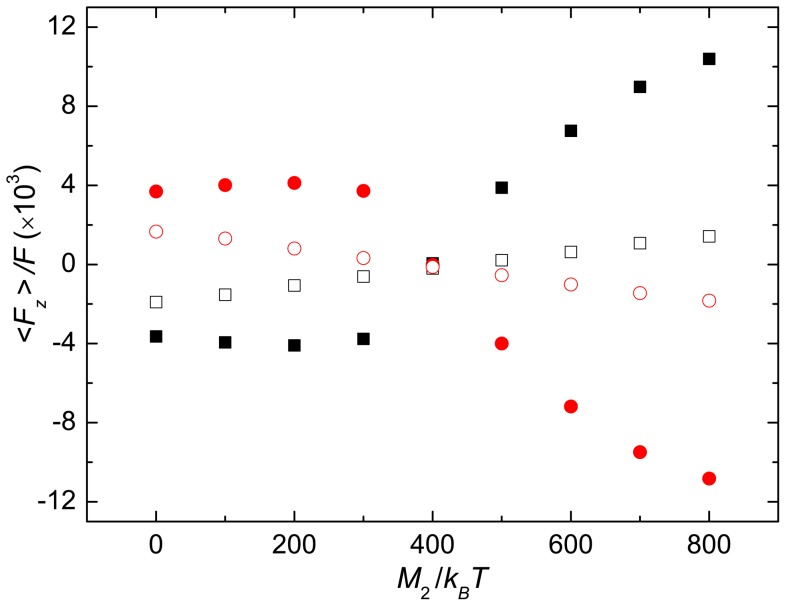
Forces on the bacterial body by flagella rotation
In the stationary state, the flagella exert no net force on the anchoring plane parallel to its normal for equal torques. Fluid is pumped by the flagella until an stationary fluid velocity is reached and the system is force free. For equal torques this applies also to the individual flagella. However, the situation is changed for non-zero torque differences, where the various helices exert different forces on the anchoring plane. Figure 7 shows average forces for various torque difference. For torques 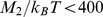 , helix
, helix  experiences a positive force
experiences a positive force 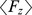 , i.e., pointing along the positive
, i.e., pointing along the positive  -direction, and the other two experience a negative force, i.e., pointing in negative
-direction, and the other two experience a negative force, i.e., pointing in negative  -direction. The force orientation is reverse for
-direction. The force orientation is reverse for 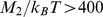 . However, the sum of the corresponding forces in the
. However, the sum of the corresponding forces in the  -direction still vanishes, as expected, i.e., the total system is force free.
-direction still vanishes, as expected, i.e., the total system is force free.
Figure 7. Average forces on bacteria body.
Average forces per monomer on the anchoring plane of three helices as a function of the torque 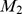 for
for 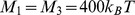 at
at 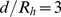 . The bullets indicated the forces by helix two and the solid squares those by helix one and three. The circles and open squares are the contributions by the corresponding hydrodynamics forces for unbundled helices.
. The bullets indicated the forces by helix two and the solid squares those by helix one and three. The circles and open squares are the contributions by the corresponding hydrodynamics forces for unbundled helices.
These forces are of various origin, depending on the bundling state. When the helices are bundled and show a phase lag, particularly for 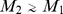 , they mainly appear due to excluded-volume interactions between the beads. The calculation of the total Lennard-Jones force between the various helices approximately agrees with the total force
, they mainly appear due to excluded-volume interactions between the beads. The calculation of the total Lennard-Jones force between the various helices approximately agrees with the total force 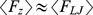 . In contrast, for unbundled helices, i.e., large torque differences, hydrodynamic interactions yield a significant contribution to the forces. In case of the slower rotating helix, the fast rotating helices create a fluid flow which drags the other helix along, since the mean fluid velocity originating from their rotation is faster than the fluid velocity due to the rotation of the slow helix. The situation is reverted for a fast rotating helix. Here, the correspondingly fast moving fluid is slowed down by the other helices, which adds an additional drag to the fast moving helix.
. In contrast, for unbundled helices, i.e., large torque differences, hydrodynamic interactions yield a significant contribution to the forces. In case of the slower rotating helix, the fast rotating helices create a fluid flow which drags the other helix along, since the mean fluid velocity originating from their rotation is faster than the fluid velocity due to the rotation of the slow helix. The situation is reverted for a fast rotating helix. Here, the correspondingly fast moving fluid is slowed down by the other helices, which adds an additional drag to the fast moving helix.
Since it is difficult to estimate the contribution of hydrodynamics on the total force quantitatively in the time-dependent slippage state, we confine the individual beads of the three helices in harmonic potentials, such that the helices remain parallel aligned along the  -axis. Explicitly, the potentials are
-axis. Explicitly, the potentials are
| (6) |
where 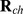 has the same
has the same  - and
- and 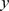 -component as the central bead
-component as the central bead 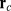 and the same
and the same  -component as
-component as 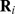 initially [16]. Thus, in this special case, the helices rotate freely and do not form a bundle. Now, the force exerted on the anchoring plane is of hydrodynamic origin only. As shown in Fig. 7, this hydrodynamic force is comparable to that by excluded-volume interactions and the sum is very close to the total force obtained for free helix bundles for
initially [16]. Thus, in this special case, the helices rotate freely and do not form a bundle. Now, the force exerted on the anchoring plane is of hydrodynamic origin only. As shown in Fig. 7, this hydrodynamic force is comparable to that by excluded-volume interactions and the sum is very close to the total force obtained for free helix bundles for 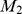 significantly smaller than
significantly smaller than 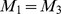 . We expect even more significant hydrodynamic contributions for bundles in the slippage and drift states.
. We expect even more significant hydrodynamic contributions for bundles in the slippage and drift states.
For flagellated bacteria, different torques on the various flagella should be the rule rather than the exception. Based in our simulation results, we expect that the forces created by torque differences in the bundled state lead to a wobbling motion, because both, the body and the helices, rotate. In the slipping state, the larger forces should give rise to a pronounced helical swimming trajectory of a bacterium. In the case of unbundling, either by reverse rotation or stopping of a motor, the rotational motion of the body is strongly reduced, and a large change in body orientation should be generated. Hence, the different dynamical states of flagella (cf. Fig. 4)) imply different swimming modes. A sampling of a larger spatial area is achieved by helical paths in the bundled and slippage state, whereas major directional changes are achieved in the unbundled state.
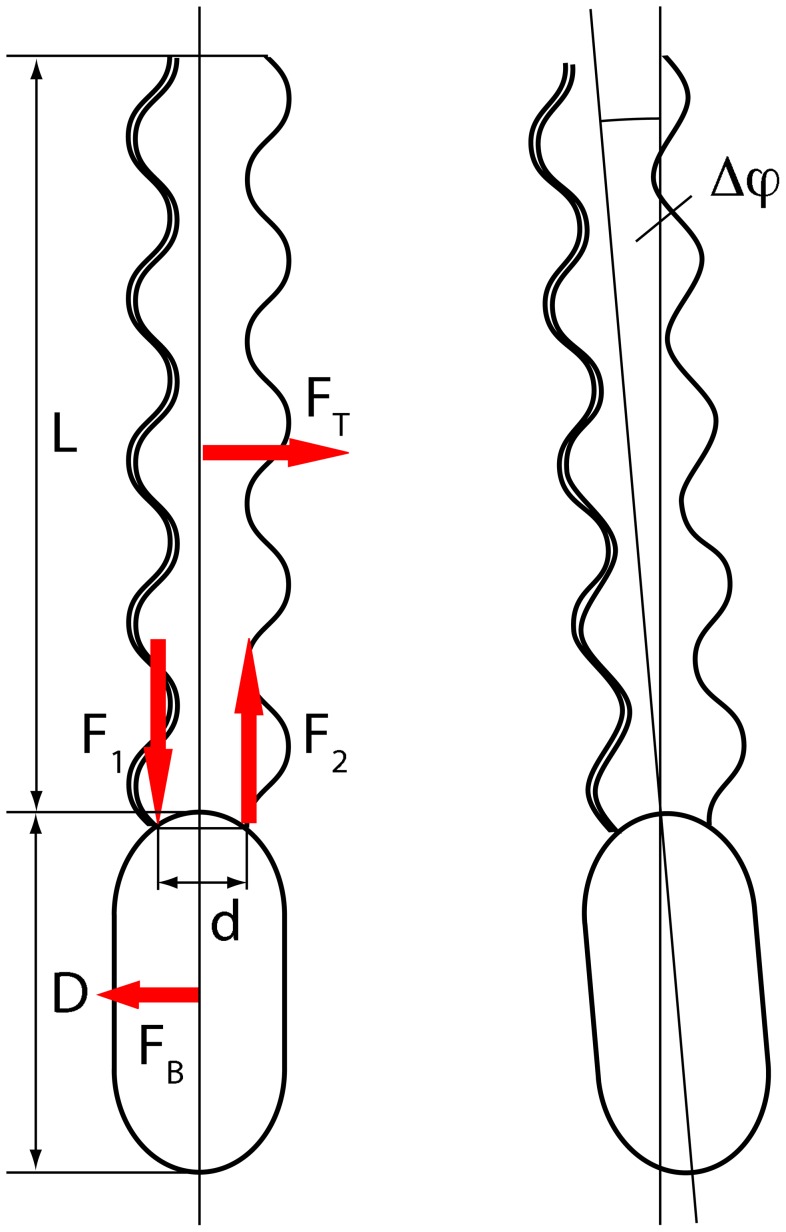
An estimation of the change in the bacterial orientation 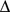 φ during the time interval
φ during the time interval 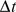 by the forces
by the forces 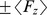 can be obtained from the following simple arguments. The torque due to friction with the fluid of the bundle and cell body is
can be obtained from the following simple arguments. The torque due to friction with the fluid of the bundle and cell body is 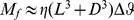 φ
φ
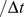 [54], where
[54], where 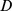 is the length of the cell body and
is the length of the cell body and 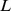 the length of the bundle (cf. Fig. 8). This torque is balanced by the torque
the length of the bundle (cf. Fig. 8). This torque is balanced by the torque 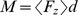 . Hence, for one period of the bundle rotation, i.e.,
. Hence, for one period of the bundle rotation, i.e., 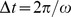 , we find
, we find 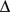 φ
φ
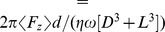 , with the bundle rotation frequency
, with the bundle rotation frequency 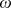 . For values obtained from the simulations,
. For values obtained from the simulations, 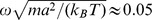 ,
, 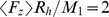 , and the estimated parameters of the bacterial geometry of R. lupini
[17] of
, and the estimated parameters of the bacterial geometry of R. lupini
[17] of 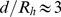 ,
, 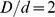 , and
, and 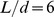 , we find
, we find 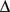 φ
φ
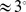 for a single flagellar rotation. This adds up to approximately
for a single flagellar rotation. This adds up to approximately 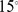 after about five rotations, a value in reasonable agreement with the rotation angle for R. lupini extractable from the images in Fig. 3 of Ref. [17].
after about five rotations, a value in reasonable agreement with the rotation angle for R. lupini extractable from the images in Fig. 3 of Ref. [17].
Figure 8. Bacterial reorientation.
Illustration of the forces and torques on a bacterium and the estimated change in orientation 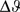 .
. 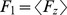 and
and 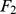 are the excess forces after the mean driving force has been subtracted. The induced torque rotates the whole structure, which gives rise to the drag forces
are the excess forces after the mean driving force has been subtracted. The induced torque rotates the whole structure, which gives rise to the drag forces 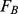 on the cell body and
on the cell body and 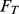 on the tail.
on the tail.
Summary
We have been investigating the properties of rotating bacteria flagella, where individual flagella are driven by motor torques of different strengths. In particular, we have focused on the stability of the flagella bundle, an aspect which is important for both, the swimming of an bacterium and its tumbling by partial degradation of the bundle. Our coarse-grained mesoscale simulations demonstrate that hydrodynamic interactions between flagella, short-range volume exclusion which prevents the intersection of flagella on contact, and the flagellar flexibility which allows for partial wrapping of flagella around each other, are all essential ingredients in the bacterial tumbling process. The interplay of these physical mechanisms not only cause synchronization and bundling of rotating flagella, but also guarantee the robustness of bundle formation in the presence of small differences in motor torques, which is essential for directed bacterial locomotion. For larger torque differences, the competition of hydrodynamic interactions with volume exclusion and elastic deformation of the flagellum leads to slippage and drift, which implies a reorientation of the swimming direction. It would be interesting to search for flagellar slippage in motile bacteria experimentally.
Supporting Information
Flagella bundle of three helices with the motor torques
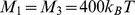 (green, blue) and
(green, blue) and
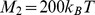 (red).
(red).
(MPG)
Flagella bundle of three helices with the motor torques
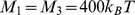 (green, blue) and
(green, blue) and
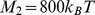 (red).
(red).
(MPG)
Funding Statement
Financial support by the VW Foundation (VolkswagenStiftung) within the program Computer Simulation of Molecular and Cellular Bio-Systems, as well as Complex Soft Matter, is gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berg HC, Brown DA (1972) Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239: 500–504. [DOI] [PubMed] [Google Scholar]
- 2. Berg HC, Anderson RA (1973) Bacteria swim by rotating their flagella filaments. Nature 245: 380–382. [DOI] [PubMed] [Google Scholar]
- 3.Berg HC (2004) E. Coli in Motion. New York: Springer.
- 4. Taylor GI (1951) Analysis of the swimming of microscopic organisms. Proc Roy Soc A 209: 447–461. [Google Scholar]
- 5. Janssen PJA, Graham MD (2011) Coexistence of tight and loose bundled states in a model of bacterial flagella dynamics. Phys Rev E 84: 011910. [DOI] [PubMed] [Google Scholar]
- 6. Turner L, Ryu WS, Berg HC (2000) Real-time imaging of fluorescent flagellar filaments. J Bacteriol 182: 2793–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darnton NC, Turner L, Rojevsky S, Berg HC (2007) On torque and tumbling in swimming Escherichia coli . J Bacteriol 189: 1756–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watari N, Larson RG (2010) The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella. Biophys J 98: 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Platzer J, Sterr W, Hausmann M, Schmitt R (1997) Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti . J Bacteriol 179: 6391–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calladine CR (1975) Construction of bacterial flagella. Nature 225: 121–124. [DOI] [PubMed] [Google Scholar]
- 11. Calladine CR (1978) Change of waveform in bacterial flagella: the role of mechanics at the molecular level. J Mol Biol 118: 457–479. [Google Scholar]
- 12. Macnab RM, Ornston MK (1977) Normal-to-curly flagella transitions and their role in bacterial tumbling. stabilization of an alternative quaternary structure by mechanical force. J Mol Biol 112: 1–30. [DOI] [PubMed] [Google Scholar]
- 13. Darnton NC, Berg HC (2007) Force-extension measurements on bacterial flagella: triggering polymorphic transformations. Biophys J 92: 2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wada H, Netz RR (2008) Discrete elastic model for stretching-induced flagellar polymorphs. Europhys Lett 82: 28001. [Google Scholar]
- 15. Xie L, Altindal T, Chattopadhyay S, Wu XL (2011) Bacterial flagellum as a propeller an as a rudder for efficient chemotaxis. Proc Natl Acad Sci USA 108: 2246–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reigh SY, Winkler RG, Gompper G (2012) Synchronization and bundling of anchored bacterial flagella. Soft Matter 8: 4363–4372. [Google Scholar]
- 17. Scharf B (2002) Real-time imaging of fluorescent flagellar filaments of Rhizobium lupini h13-3: flagellar rotation and ph-induced polymorphic transitions. J Bacteriol 184: 5979–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmitt R (2002) Sinorhizobial chemotaxis: a departure from the enterobacterial paradigm. Microbiology 148: 627–631. [DOI] [PubMed] [Google Scholar]
- 19. Macnab RM (1977) Bacterial flagella rotating in bundles: a study in helical geometry. Proc Natl Acad Sci USA 74: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim M, Bird JC, Parys AJV, Breuer KS, Powers TR (2003) A macroscopic scale model of bacterial flagellar bundling. Proc Natl Acad Sci USA 100: 15481–15485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qian B, Jiang H, Gagnon DA, Breuer KS, Powers TR (2009) Minimal model for synchronization induced by hydrodynamic interactions. Phys Rev E 80: 061919. [DOI] [PubMed] [Google Scholar]
- 22. Reichert M, Stark H (2005) Synchronization of rotating helices by hydrodynamic interactions. Eur Phys J E 17: 493–500. [DOI] [PubMed] [Google Scholar]
- 23. Flores H, Lobaton E, Mendez-Diez S, Tlupova S, Cortez R (2005) A study of bacterial flagellar bundling. Bull Math Biol 67: 137–168. [DOI] [PubMed] [Google Scholar]
- 24. Elfring GJ, Lauga E (2009) Hydrodynamic phase locking of swimming microorganisms. Phys Rev Lett 103: 088101. [DOI] [PubMed] [Google Scholar]
- 25. Polin M, Tuval I, Drescher K, Gollub JP, Goldstein RE (2009) Chlamydomonas swims with two “gears” in a eukaryotic version of run-and-tumble locomotion. Science 325: 487–490. [DOI] [PubMed] [Google Scholar]
- 26. Stocker R, Durham WM (2009) Tumbling for stealth? Science 325: 400–402. [DOI] [PubMed] [Google Scholar]
- 27. Goldstein RE, Polin M, Tuval I (2009) Noise and synchronization in pairs of beating eukaryotic flagella. Phys Rev Lett 103: 168103. [DOI] [PubMed] [Google Scholar]
- 28. Golestanian R, Yeomans JM, Uchida N (2011) Hydrodynamic synchronization at low reynolds number. Soft Matter 7: 30743082. [Google Scholar]
- 29. Friedrich BM, Jülicher F (2012) Flagellar synchronization independent of hydrodynamic interactions. Phys Rev Lett 109: 138102. [DOI] [PubMed] [Google Scholar]
- 30. Afzelius BA (1976) A human syndrome caused by immotile cilia. Science 193: 317–319. [DOI] [PubMed] [Google Scholar]
- 31. Cartwright JHE, Piro O, Tuval I (2004) Fluid-dynamical basis of the embryonic development of left-right asymmetry in vertebrates. Proc Natl Acad Sci USA 101: 7234–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Q, Pan J, Snell WJ (2006) Intraflagellar transport particles participate directly in ciliumgenerated signaling in Chlamydomonas . Cell 125: 549–562. [DOI] [PubMed] [Google Scholar]
- 33. Kim M, Powers TR (2004) Hydrodynamic interactions between rotating helices. Phys Rev E 69: 061910. [DOI] [PubMed] [Google Scholar]
- 34. Kim YW, Netz RR (2006) Pumping fluids with periodically beating grafted elastic filaments. Phys Rev Lett 21: 158101. [DOI] [PubMed] [Google Scholar]
- 35. Yang Y, Elgeti J, Gompper G (2008) Cooperation of sperm in two dimensions: synchronization, attraction, and aggregation through hydrodynamic interactions. Phys Rev E 78: 061903. [DOI] [PubMed] [Google Scholar]
- 36. Uchida N, Golestanian R (2010) Synchronization and collective dynamics in a carpet of microfluidic rotors. Phys Rev Lett 104: 178103. [DOI] [PubMed] [Google Scholar]
- 37. Kotar J, Leoni M, Bassetti B, Lagomarsino MC, Cicuta P (2010) Hydrodynamic synchronization of colloidal oscillators. Proc Natl Acad Sci USA 107: 7669–7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Di Leonardo R, Búzás A, Kelemen L, Vizsnyiczai G, Oroszi L, et al. (2012) Hydrodynamic synchronization of light driven microrotors. Phys Rev Lett 109: 034104. [DOI] [PubMed] [Google Scholar]
- 39. Uchida N, Golestanian R (2011) Generic conditions for hydrodynamic synchronization. Phys Rev Lett 106: 058104. [DOI] [PubMed] [Google Scholar]
- 40. Elgeti J, Gompper G (2013) Emergence of metachronal waves in cilia arrays. Proc Natl Acad Sci USA 110: 4470–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malevanets A, Kapral R (1999) Mesoscopic model for solvent dynamics. J Chem Phys 110: 8605–8613. [Google Scholar]
- 42. Kapral R (2008) Multiparticle collision dynamics: simulation of complex systems on mesoscales. Adv Chem Phys 140: 89–146. [Google Scholar]
- 43. Gompper G, Ihle T, Kroll DM, Winkler RG (2009) Multi-particle collision dynamics: a particlebased mesoscale simulation approach to the hydrodynamics of complex fluids. Adv Polym Sci 221: 1–87. [Google Scholar]
- 44. Mussawisade K, Ripoll M, Winkler RG, Gompper G (2005) Dynamics of polymers in a particlebased mesoscopic solvent. J Chem Phys 123: 144905. [DOI] [PubMed] [Google Scholar]
- 45. Huang CC, Winkler RG, Sutmann G, Gompper G (2010) Semidilute polymer solutions at equilibrium and under shear flow. Macromolecules 43: 10107. [Google Scholar]
- 46.Allen MP (1987) Computer Simulation in Liquids. Clarendon: Oxford.
- 47. Huang CC, Chatterji A, Sutmann G, Gompper G, Winkler RG (2010) Cell-level canonical sampling by velocity scaling for multiparticle collision dynamics simulations. J Comput Phys 229: 168–177. [Google Scholar]
- 48. Ripoll M, Mussawisade K, Winkler RG, Gompper G (2005) Dynamic regimes of fluids simulated by multiparticle-collision dynamics. Phys Rev E 72: 016701. [DOI] [PubMed] [Google Scholar]
- 49. Liu Y, Pérez T, Li W, Gunton JD, Green A (2011) Statistical mechanics of the helical wormlike chain model. J Chem Phys 134: 065107. [DOI] [PubMed] [Google Scholar]
- 50. Götz R, Schmitt R (1987) Rhizobium meliloti swims by unidirectional, intermittent rotation of right-handed flagellar helices. J Bacteriol 169: 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54. [DOI] [PubMed] [Google Scholar]
- 52. Berry RM, Berg HC (1997) Absence of a barrier to backwards rotation of the bacterial flagellar motor demonstrated with optical tweezers. Proc Natl Acad Sci USA 94: 14433–14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reid SW, Leake MC, Chandler JH, Lo C, Armitage JP, et al. (2007) The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA 103: 8066–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doi M, Edwards SF (1986) The Theory of Polymer Dynamics. Oxford: Clarendon Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flagella bundle of three helices with the motor torques
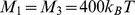 (green, blue) and
(green, blue) and
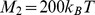 (red).
(red).
(MPG)
Flagella bundle of three helices with the motor torques
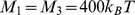 (green, blue) and
(green, blue) and
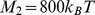 (red).
(red).
(MPG)