The GEF Bcr promotes RhoA-dependent actin remodeling and MAL/SRF signaling in keratinocytes, which in turn promotes differentiation via regulation of desmoglein-1 expression.
Abstract
Although much is known about signaling factors downstream of Rho GTPases that contribute to epidermal differentiation, little is known about which upstream regulatory proteins (guanine nucleotide exchange factors [GEFs] or GTPase-activating proteins [GAPs]) are involved in coordinating Rho signaling in keratinocytes. Here we identify the GEF breakpoint cluster region (Bcr) as a major upstream regulator of RhoA activity, stress fibers, and focal adhesion formation in keratinocytes. Loss of Bcr reduced expression of multiple markers of differentiation (such as desmoglein-1 [Dsg1], keratin-1, and loricrin) and abrogated MAL/SRF signaling in differentiating keratinocytes. We further demonstrated that loss of Bcr or MAL reduced levels of Dsg1 mRNA in keratinocytes, and ectopic expression of Dsg1 rescued defects in differentiation seen upon loss of Bcr or MAL signaling. Taken together, these data identify the GEF Bcr as a regulator of RhoA/MAL signaling in keratinocytes, which in turn promotes differentiation through the desmosomal cadherin Dsg1.
Introduction
The stratification and differentiation of keratinocytes is a crucial process required for the maintenance and regeneration of healthy skin (Delva et al., 2009; Simpson et al., 2011). In keratinocytes destined for terminal differentiation, structural and signaling cues instruct cells to halt proliferation, transit into the superficial layers, and undergo transcriptional reprogramming to produce the structural and chemical products required for creating the epidermal barrier. Many of these cues arise from proteins involved in maintenance of cell–cell junctions. Whereas adherens junction proteins have been identified as key players in epidermal growth, polarity, and barrier formation (Tunggal et al., 2005; Müller et al., 2008; McCaffrey and Macara, 2011), desmosomal proteins have emerged as critical modulators of signaling pathways involved in differentiation (Green and Simpson, 2007; Thomason et al., 2010).
The desmosome is composed of the transmembrane cadherins (desmogleins, desmocollins), armadillo proteins (plakophilins and plakoglobin), and the cytoskeletal linker protein desmoplakin (DP). Desmosomal cadherins physically link cells together in the extracellular space (Dusek et al., 2007). Armadillo proteins serve as the bridge linking the cadherins to DP, but they also have several nonjunctional roles (Hatzfeld, 2007; Bass-Zubek et al., 2009; Wolf and Hatzfeld, 2010). In turn, DP is responsible for tethering the junctional complex to intermediate filaments (Hatsell and Cowin, 2001). Multiple desmosomal proteins such as desmoglein-1 (Dsg1), plakophilin-1 (PKP1), and DP are required for epidermal integrity, as evidenced by the cutaneous diseases caused by mutations in these proteins (McGrath et al., 1997; McGrath, 2005; Kottke et al., 2006; Amagai and Stanley, 2012). We have recently shown that Dsg1 is a crucial mediator of differentiation, as its loss reduces expression of multiple differentiation markers (Getsios et al., 2009).
Other studies have pointed to the importance of Rho GTPases in epidermal differentiation. Rho GTPases (RhoA, Rac1, and Cdc42) are multifunctional proteins that regulate many biological processes such as cell migration and morphogenesis (Etienne-Manneville and Hall, 2002; Burridge and Wennerberg, 2004; Jaffe and Hall, 2005). These proteins cycle between an active GTP-bound form and an inactive GDP-bound form. Guanine nucleotide exchange factors (GEFs) activate GTPases by catalyzing the exchange of GDP for GTP, whereas GTPase activating proteins (GAPs) inactivate GTPases by stimulating the intrinsic ability of GTPases to hydrolyze GTP (Moon and Zheng, 2003; Rossman et al., 2005). The increased focus on studying GEFs and GAPs in recent years has served to considerably increase our understanding on how Rho GTPase signaling is propagated during different biological processes (Rossman et al., 2005).
RhoA activates effector proteins such as Rho kinase (ROCK) and Dia, which promote actomyosin contractility and F-actin polymerization, respectively (Bishop and Hall, 2000). In addition, the transcription factor SRF (serum response factor) is activated by Rho GTPases and is responsible for expression of multiple different proteins (Posern and Treisman, 2006; Busche et al., 2008; Olson and Nordheim, 2010). The ability of Rho GTPases to regulate SRF-mediated transcription depends on a family of myocardin-related transcription factors (MRTFs), which include Myocardin, MAL/MRTF-A, and MRTF-B. Rho-mediated F-actin polymerization drives MAL nuclear localization and consequent SRF-dependent transcription (Posern and Treisman, 2006; Olson and Nordheim, 2010). Rho GTPases and SRF have emerged as key signaling players in the process of epidermal differentiation. Blocking the Rho–ROCK pathway inhibits the differentiation of keratinocytes, and expression of active ROCK-II promotes differentiation (Sugai et al., 1992; McMullan et al., 2003).
SRF and MAL have also been shown to positively regulate epidermal differentiation in both animal and in vitro models (Koegel et al., 2009; Connelly et al., 2010; Verdoni et al., 2010; Luxenburg et al., 2011). In contrast to RhoA, Rac1 is required for maintenance of basal proliferating stem cell populations, and loss of Rac1 was shown to promote terminal differentiation (Benitah et al., 2005; Nikolova et al., 2008). Together these data suggest that activities of RhoA and Rac1 need to be dually coordinated for proper differentiation to occur. Unique among the GEF/GAP families are the proteins Bcr and Abr because they contain DH-PH domains capable of activating RhoA, as well as a RhoGAP domain, which selectively inactivates Rac/Cdc42 (Chuang et al., 1995; Vaughan et al., 2011). Although previous work on Bcr suggests that it predominantly functions as a Rac GAP in certain cell types (Cho et al., 2007; Oh et al., 2010), the ability of Bcr to regulate RhoA or Rac1 in keratinocytes has not been investigated.
In this study we demonstrate that Bcr silencing significantly reduces RhoA activity (and stress fibers/focal adhesions) in keratinocytes, with relatively minor effects on global Rac1 or Cdc42 activity. Loss of Bcr decreased the ability of keratinocytes to differentiate, and also decreased MAL nuclear localization and SRF activity. We demonstrate that Dsg1 mRNA levels are reduced upon Bcr and MAL KD, and restoring Dsg1 expression rescues Bcr-induced differentiation defects. These data therefore highlight the importance of Dsg1 in coordinating the process of epidermal differentiation downstream of Bcr-induced RhoA/MAL signaling.
Results
Bcr is required for maintaining RhoA activity in epidermal keratinocytes
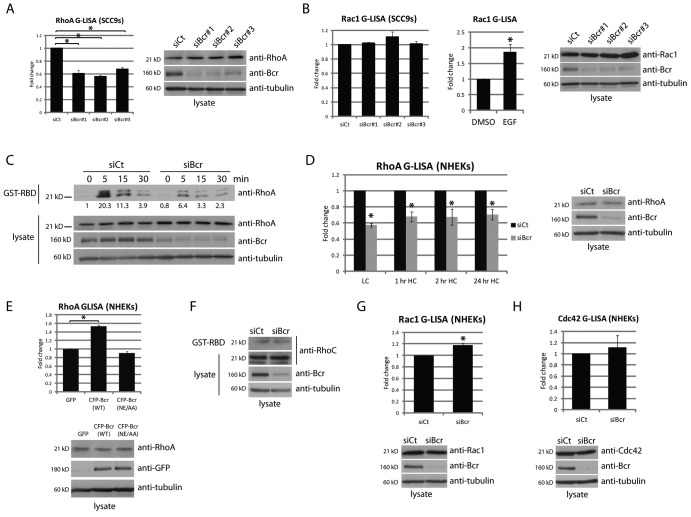
Previous work has suggested that Bcr functions mainly as a Rac GAP in certain cell types, but the net effect of Bcr on RhoA and Rac1 activity has not been determined in keratinocytes. We therefore used three different siRNA oligonucleotides specific for Bcr to knockdown (KD) the protein, each resulting in >80% reduction in protein levels (Fig. 1 A). Bcr KD resulted in a significant decrease in RhoA activity in SCC9 cells (as tested by G-LISA assays), indicating that the DH domain of Bcr has functional exchange activity on RhoA in these cells (Fig. 1 A). All three siRNAs demonstrated a reduction in RhoA activity, indicating that this effect is not due to nonspecific effects of a single siRNA. In contrast to RhoA, there was no significant change in global Rac1 activity upon KD of Bcr (Fig. 1 B). The sensitivity of the G-LISA assay to detect changes in Rac1 activity in SCC9s was tested using EGF, which produced an ∼1.8-fold increase in Rac activity (Fig. 1 B).
Figure 1.
Bcr regulates RhoA activity in keratinocytes, but has a minor effect on Rac. (A) RhoA activity was analyzed in SCC9 cells upon Bcr KD with three different siRNAs (siBcr#1–3). G-LISA experiments indicate a decrease in RhoA activity upon Bcr KD with all three siRNA oligonucleotides. (B) G-LISA experiments indicated no change in global Rac1 activity upon Bcr KD in SCC9 cells (the observed change for siBcr#2 was not significant). Stimulation of SCC9 cells with 0.1 ng/µl EGF for 2 min was used as a positive control for this assay. (C) Control and Bcr KD SCC9 cells were subjected to calcium switch, followed by analysis of RhoA activity using traditional GST-RBD pull-downs. Fold-change values over control quantified by densitometry are noted below blots. (D) G-LISA experiments for RhoA activity were performed upon loss of Bcr in NHEKs at different time points after addition of 1.2 mM Ca2+-containing media. (E) RhoA activity was analyzed in NHEKs expressing GFP, CFP-tagged WT Bcr, or a GEF-dead Bcr mutant (NE/AA), after addition of high calcium media for 1 h. (F) RhoC activity was analyzed in control and Bcr KD NHEKs using a traditional GST-RBD pull-down. (G and H) G-LISA experiments for Rac1 or Cdc42 activity were performed upon Bcr KD in NHEKs. For all graphs, fold-change values from three independent samples are represented with error bars indicating SD. *, P < 0.05.
To further examine a role for Bcr in regulating RhoA activity, we performed a calcium switch assay (Godsel et al., 2010). Control and Bcr KD SCC9s were placed in low calcium media for 12–16 h, followed by reintroduction of normal calcium media to trigger reformation of junctional complexes (Fig. 1 C). RhoA activity was assayed using traditional GST-tagged Rhotekin Rho-binding domain (GST-RBD) pull-downs. As we have previously shown (Godsel et al., 2010), RhoA activity follows a bi-phasic pattern in control SCC9s after a calcium switch, where a transient activation at 5 min is followed by a decrease in activity by 15–30 min. In contrast to control cells, we observed a defect in the ability of Bcr KD cells to activate RhoA in response to calcium switch (Fig. 1 C).
We next addressed whether Bcr functions similarly in normal human epidermal keratinocytes (NHEKs). Successful (>80%) KD of Bcr in NHEKs was obtained by a pool of oligonucleotides as described in Materials and methods. KD of Bcr significantly decreased RhoA activity in these cells in low calcium media, and at 1, 2, or 24 h after addition of high calcium media (Fig. 1 D). Further, overexpression of wild-type (WT) Bcr increased RhoA activity in keratinocytes (Fig. 1 E). Importantly, a previously described GEF-dead mutant of Bcr (NE/AA; Cho et al., 2007) was not able to activate RhoA (Fig. 1 E). RhoB expression was not detected in NHEKs, and activity of RhoC was minimally affected upon KD of Bcr (Fig. 1 F). Although a small but significant increase in Rac1 activity was observed, we did not detect any changes in Cdc42 activity in NHEKs upon loss of Bcr (Fig. 1, G and H). Collectively, these results suggest Bcr plays a major role as a RhoA GEF in human keratinocytes.
Bcr regulates stress fibers and focal adhesion formation in epidermal keratinocytes
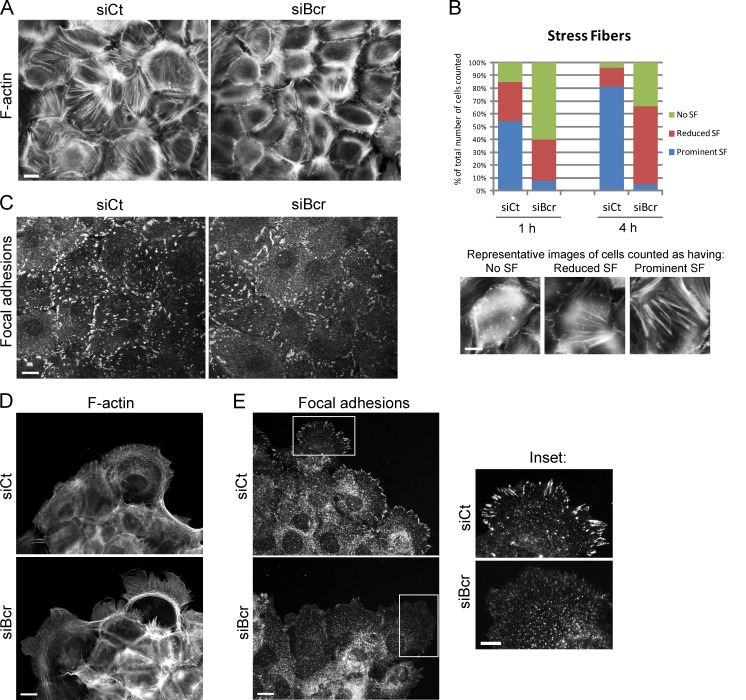
Considering the effect of Bcr on RhoA activity, we next investigated whether Bcr can regulate the actin cytoskeleton in keratinocytes. In control NHEKs, calcium-induced junction formation stimulated the formation of stress fibers, an effect that was markedly reduced in Bcr KD NHEKs (Fig. 2, A and B). Focal adhesion formation was also impaired in Bcr KD NHEKs, which demonstrated smaller and fewer focal adhesions compared with control NHEKs (Fig. 2 C). Actin cytoskeletal organization and focal adhesion formation was also visualized in SCC9 cells (Fig. 2, D and E). Compared with control cells, Bcr KD SCC9s demonstrated larger membrane protrusions (Fig. 2 D), and a significant decrease in the size and number of peripheral focal adhesions (Fig. 2 E, inset). The increase in membrane protrusion upon loss of Bcr may indicate localized changes in Rac1 activity, which was not detected in the G-LISA for global Rac1 activity. Loss of stress fibers is a common phenotype observed upon loss of RhoA activity (Chrzanowska-Wodnicka and Burridge, 1996). We confirmed that RhoA KD reduces stress fiber formation in human keratinocytes (Fig. S1 A). These data demonstrating a reduction in stress fiber and focal adhesion formation upon Bcr KD is therefore consistent with the effect of Bcr on RhoA activity.
Figure 2.
Bcr regulates actin stress fibers and focal adhesions in keratinocytes. (A–C) Control and Bcr KD NHEKs on coverslips were subjected to high calcium for 4 h, fixed, and processed to visualize F-actin using Alexa Fluor 488–conjugated phalloidin (A) or focal adhesions using an anti-vinculin antibody (C). Bar, 20 µm. (B) Representative images of cells with prominent, reduced, or no stress fibers are shown and quantified as described in Materials and methods. (D and E) Control and Bcr KD SCC9 cells were fixed and processed for immunofluorescence. F-actin was visualized using Alexa Fluor 488–conjugated phalloidin (D) and focal adhesions were visualized using an anti-phosphotyrosine antibody (PY99; E, inset). Bar, 20 µm. All images shown are representative of three independent experiments. KD of Bcr in SCC9 cells demonstrated a decrease in focal adhesions along the periphery of cells.
Loss of Bcr impairs differentiation in submerged cultures of epidermal keratinocytes
Previous work has demonstrated that inhibition of all Rho isoforms with C3 causes a decrease in differentiation (McMullan et al., 2003). To extend this analysis, we specifically knocked down RhoA in NHEKs and placed the cells in high calcium–containing media (1.2 mM CaCl2) to induce differentiation (Hennings et al., 1980). An ∼50% knockdown of RhoA resulted in reduced expression of multiple differentiation markers, such as the desmosomal cadherins Dsg1 and desmocollin-1 (Dsc1), and the cornified envelope protein loricrin (Fig. S1 C). Importantly, 50% knockdown of RhoA did not perturb endogenous levels of other GTPases, as has been shown to occur in other cell types in response to competition for RhoGDI1 binding (Boulter et al., 2010). Nevertheless, when greater than 90% knockdown of RhoA was achieved, expression of both RhoC and Rac1 were indeed increased, as previous evidence would predict (Fig. S1 D). Under these conditions, the specific effect of RhoA on differentiation is partially lost, likely a result of compensatory changes in expression of these other GTPases.
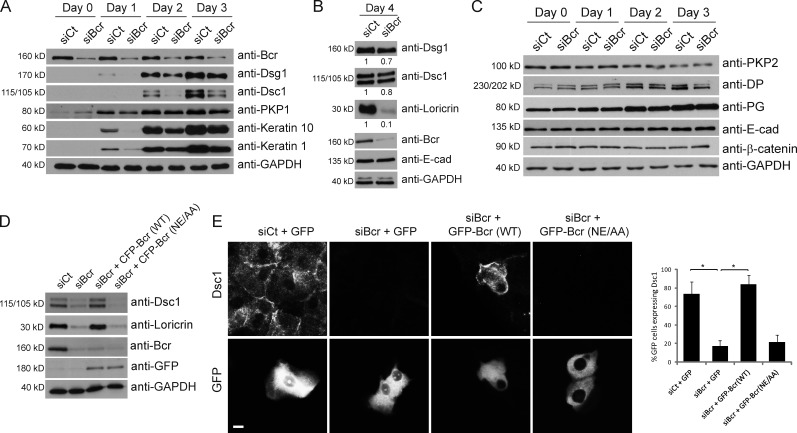
Considering the effect of Bcr on RhoA activity, we wanted to determine if loss of Bcr would also affect the ability of keratinocytes to differentiate. Control or Bcr KD NHEKs were grown to confluence, placed in high calcium media, and samples were analyzed for differentiation markers at different days (day 0 indicates low calcium). Expression of Dsg1, Dsc1, and PKP1 was induced in control cells within 1–2 d of calcium induction, along with the suprabasal keratins 1 and 10. In contrast, Bcr KD cells exhibited reduced expression of all these differentiation markers (Fig. 3 A). By day 4, expression of Dsg1 and Dsc1 had partially recovered, but expression of the cornified envelope protein loricrin was dramatically reduced in Bcr KD cells. These data therefore demonstrate a defect in the ability of NHEKs to differentiate upon Bcr KD (Fig. 3 B).
Figure 3.
Loss of Bcr causes a delay in the onset of differentiation markers in submerged cultures of epidermal keratinocytes. (A–C) Control and Bcr KD NHEKs were grown to confluence and switched to high calcium media for 1–4 d to induce differentiation. (A) Control and Bcr KD samples were blotted for Bcr, GAPDH, and a panel of differentiation markers including Dsg1, Dsc1, PKP1, keratin 10, and keratin 1. (B) Samples from lysates taken at 4 d after induction of differentiation were also lysed for Dsg1, Dsc1, loricrin, Bcr, E-cad, and GAPDH. Fold-change values over control quantified by densitometry are noted below blots. These data indicate that Bcr KD causes a decrease in expression of all differentiation markers tested. (C) Samples were also blotted for a range of proteins in the desmosome and adherens junction families, such as PKP2, DP, PG, E-cad, and β-catenin, indicating that expression of cell–cell adhesion proteins is not compromised upon loss of Bcr. All Western blots shown are representative of three independent experiments. (D) Control or Bcr KD NHEKs expressing WT CFP-tagged Bcr or a GEF-dead Bcr mutant (NE/AA) were grown to confluence, induced to differentiate, and samples blotted with Bcr, GFP, Dsc1, loricrin, and GAPDH antibodies (note: anti-Bcr [N-20] antibody does not recognize exogenous expression of CFP-Bcr likely due to interference of the N-terminal CFP tag with the antibody epitope which is at the N terminus of Bcr). (E) Similar samples using GFP-tagged Bcr constructs were induced to differentiate on coverslips and stained for Dsc1. Re-expression of WT Bcr (but not the GEF-dead mutant) can rescue the loss of loricrin and Dsc1 expression seen upon Bcr KD.
Considering the effect of Bcr on expression of Dsg1 and Dsc1, we analyzed whether expression of other junctional proteins are affected by Bcr KD. Compared with control, KD of Bcr did not alter the expression patterns of PKP2, DP, PG, E-cad, or β-catenin (Fig. 3 C), and we have observed no change in the junctional localization of these proteins upon Bcr KD (Fig. S2). In particular, assembly of E-cad at cell–cell borders is not altered in Bcr KD cells (Fig. S2).
Interestingly, knockdown of the closely related protein Abr did not cause any changes in the expression of differentiation markers (Fig. S3). To confirm that the effects of Bcr KD are specific, we rescued Bcr expression using constructs containing siRNA-refractory silent mutations. Re-expression of WT Bcr was able to restore the loss of Dsc1 and loricrin expression seen upon Bcr KD, but the GEF-dead mutant of Bcr could not (Fig. 3, D and E). These data indicate that the GEF activity of Bcr is crucial for its effect on differentiation.
Bcr deficiency impairs cellular organization and differentiation in an organotypic raft model
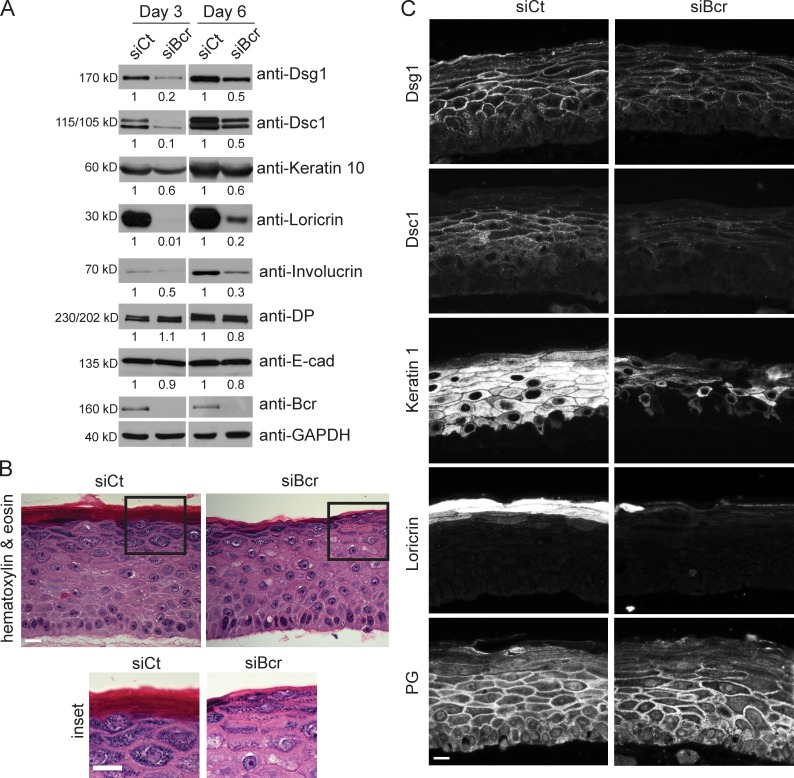
To study differentiation in a more physiologically relevant setting, an organotypic model of human epidermis was used. Control and Bcr KD NHEKs were induced to undergo stratification in 3D by exposing them to an air–medium interface (Asselineau and Prunieras, 1984; Meyers and Laimins, 1994; Getsios et al., 2009). Consistent with data obtained from 2D cultures, loss of Bcr in rafts resulted in a decrease in expression of differentiation-specific proteins (Dsg1, Dsc1, keratin 10, loricrin, and involucrin), without any effect on other cell–cell adhesion proteins (E-cad and DP; Fig. 4 A). Analysis of the morphology of rafts by hematoxylin and eosin staining indicated that Bcr KD resulted in reduced granulation in the superficial layers and impaired stratum corneum development (Fig. 4 B). In addition, although localization of Dsg1 to cell–cell borders was slightly affected, border localization of Dsc1 was dramatically reduced (Fig. 4 C). Junctional localization of PG was marginally perturbed in the suprabasal layers, likely an effect of decreased Dsg1, PG’s primary binding partner in this region of the epidermis. The staining intensity of keratin 1 and loricrin is also reduced upon loss of Bcr (Fig. 4 C). The effects of Bcr on keratinocyte differentiation are not related to alterations in cell proliferation, as we did not observe significant changes in the number of Ki67-positive cells upon loss of Bcr (Fig. S4). Taken together, these data demonstrate that Bcr promotes the differentiation of keratinocytes in organotypic raft cultures.
Figure 4.
Bcr deficiency impairs differentiation in an organotypic raft model. (A) 3D organotypic raft cultures were grown from control and Bcr KD NHEKs for 3 or 6 d. Samples were lysed, subjected to SDS-PAGE, and blotted for Dsg1, Dsc1, keratin 10, loricrin, involucrin, DP, E-cad, Bcr, and GAPDH. Fold-change values over control quantified by densitometry are noted below blots. All Western blots shown are representative of three independent experiments. These data obtained in organotypic raft cultures corroborate the data obtained in submerged cultures, indicating a loss of differentiation upon Bcr KD. (B, inset) Control and Bcr KD organotypic raft cultures were grown for 6 d, embedded in paraffin, and sections were subjected to hematoxylin and eosin staining. (C) Control and Bcr KD organotypic raft cultures were grown for 6 d, embedded in paraffin, and sections stained for Dsg1, Dsc1, keratin 1, loricrin, and PG. Bar, 40 µm. All images shown are representative of three independent experiments. Bcr KD rafts demonstrate a decrease in Dsc1, keratin 1, and loricrin.
Loss of Bcr abrogates nuclear localization of MAL and SRF signaling in keratinocytes
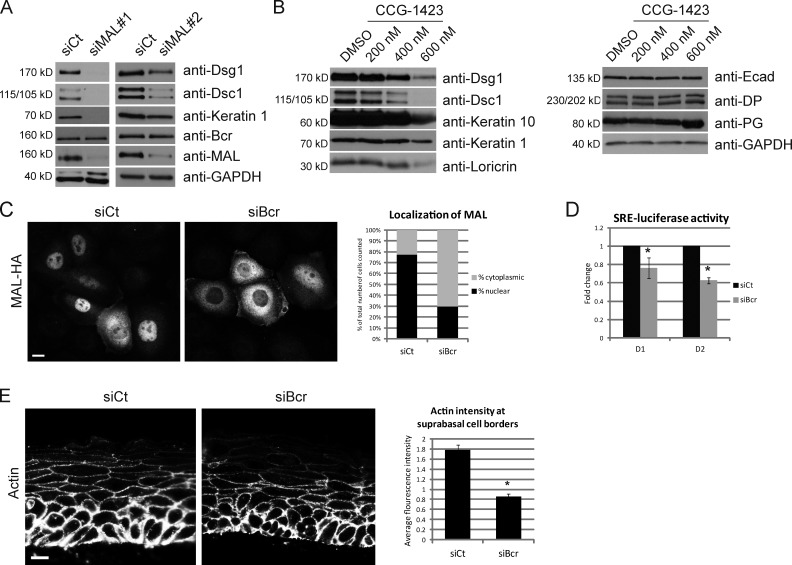
Previous work has demonstrated the importance of MAL and SRF for differentiation (Connelly et al., 2010; Luxenburg et al., 2011). We therefore hypothesized that Bcr may regulate differentiation via the transcriptional activities of MAL/SRF. We first determined whether MAL KD generates a similar phenotype to Bcr KD. MAL KD by two different siRNAs abrogated expression of Dsg1, Dsc1, and keratin 1 without affecting levels of Bcr (Fig. 5 A). As expected, we also observed a similar effect on differentiation upon KD of SRF (Fig. S5 A). Analysis of MAL/SRF function was also performed using the drug CCG-1423, which is an inhibitor of MAL/SRF-dependent transcriptional signaling (Evelyn et al., 2007). A dose–response analysis revealed that higher doses of CCG-1423 demonstrated a selective inhibition of differentiation-specific proteins Dsg1, Dsc1, keratin 1, keratin 10, and loricrin, but not other adhesion molecules such as E-cad, DP, or PG (Fig. 5 B).
Figure 5.
Loss of Bcr abrogates signaling through the MAL/SRF nexus, a process required for the ability of keratinocytes to undergo differentiation. (A) Control NHEKs or those knocked down for MAL (using two different oligonucleotides, siMAL#1 and 2) were subjected to high calcium to induce differentiation, and samples collected at day 2. Lysates were blotted for Dsg1, Dsc1, keratin 1, Bcr, MAL, and GAPDH. KD of MAL with either siRNA decreased expression of Dsg1, Dsc1, and keratin 1, but not Bcr. (B) NHEKs were grown to confluence and subjected to high calcium to induce differentiation for 2 d, either in the presence of DMSO (vehicle) or different concentrations (200–600 nM) of an inhibitor of MAL/SRF-induced transcription (CCG-1423). Higher doses of CCG-1423 inhibited expression of all differentiation-associated proteins tested (Dsg1, Dsc1, keratin 10, and loricrin), but not other adhesion markers (E-cad, DP, and PG). All Western blots shown are representative of three independent experiments. (C) Control and Bcr KD cells were retrovirally transduced with HA-tagged MAL as described in Materials and methods. 48 h after infection, cells were subjected to calcium switch (see Materials and methods) for 1 h, fixed, and processed for immunofluorescence with an anti-HA antibody. Cells from randomly imaged fields were scored for nuclear staining of HA-tagged MAL, and represented as percentage of total cells counted (n = 122). The data shown are representative of three independent experiments. Loss of Bcr resulted in a decrease in the percentage of cells with nuclear MAL. Bar, 20 µm. (D) Control and Bcr KD NHEKs coexpressing an SRE-luciferase construct were induced to differentiate for 1–2 d, and luciferase activity measured using a standard reporter assay. (E) Control and Bcr KD raft sections were stained for actin. Bar, 40 µm. Average fluorescence intensity of junctional actin was measured by measuring pixel intensity per area in a given region of interest formed by tracing cell–cell junctions in randomly selected fields. *, P < 0.05. KD of Bcr results in a significant decrease of junctional actin staining in cells in the suprabasal layers. All images shown are representative of three independent experiments.
We next investigated whether loss of Bcr alters MAL localization in keratinocytes, as nuclear translocation of MAL occurs in response to Rho-mediated F-actin polymerization (Posern and Treisman, 2006; Olson and Nordheim, 2010). Compared with control, loss of Bcr caused a major defect in the ability of MAL to translocate to the nucleus upon a calcium switch (Fig. 5 C). To determine whether Bcr affects SRF activity in keratinocytes, we performed an SRE-luciferase assay, which measures the transcriptional ability of SRF to drive expression of the luciferase gene under control of a serum response element (SRE). KD of Bcr caused a significant reduction in SRF activity in differentiating keratinocytes at day 1 and 2 (Fig. 5 D). Like Bcr, RhoA KD also caused a decrease in SRF activity in keratinocytes (Fig. S1 B). Previous studies have established that loss of SRF in mouse epidermis results in decreased levels of junctional actin (Koegel et al., 2009; Luxenburg et al., 2011). Analysis of junctional actin staining in rafts indicated that Bcr KD also causes a significant decrease in the amount of junctional actin in suprabasal cells (Fig. 5 E). Taken together, these data demonstrate that loss of Bcr abrogates normal MAL/SRF signaling in keratinocytes, a process crucial for epidermal differentiation.
Loss of Bcr or MAL signaling decreases mRNA transcript levels of the differentiation modulator Dsg1
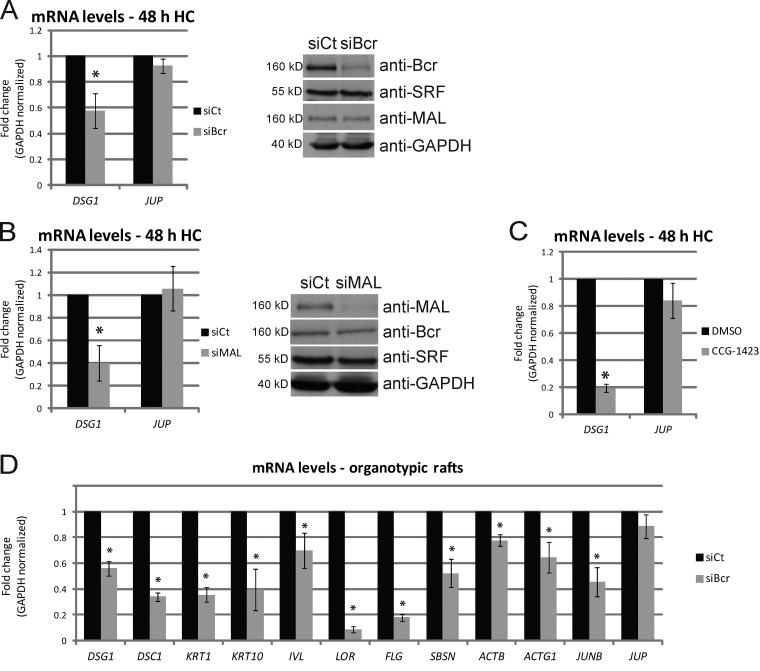
As shown above, loss of Bcr or MAL results in a reduction in protein levels of Dsg1, which has been shown to be required for proper differentiation (Getsios et al., 2009). DSG1 was among 72 mRNAs identified as altered in an array analysis of SRF-null mouse keratinocytes, indicating that this gene is under regulation of SRF (Luxenburg et al., 2011). We therefore analyzed whether KD of Bcr or MAL regulates DSG1 mRNA levels. Control or Bcr KD NHEKs were induced to differentiate for 2 d, after which total RNA was collected and mRNA levels analyzed by quantitative real-time PCR (qPCR). Bcr KD caused a significant reduction in the levels of DSG1 mRNA, but not of plakoglobin (JUP; Fig. 6 A). In addition, either KD of MAL or treatment with the MAL transcriptional inhibitor CCG-1423 caused a significant decrease in DSG1 mRNA (but not JUP; Fig. 6, B and C). Loss of SRF also recapitulated these data (Fig. S5 B). These data demonstrate for the first time that both Bcr and MAL are required for DSG1 mRNA expression during the process of differentiation. Bcr KD in rafts also resulted in decreased mRNA levels of all differentiation markers tested (Dsg1, Dsc1, keratin 1, keratin 10, involucrin, loricrin, filaggrin, and suprabasin), as well as other known SRF gene targets such as ActB, ActG1, and JunB (Fig. 6 D). Expression of the differentiation marker suprabasin was previously shown to be regulated by SRF (Park et al., 2002; Luxenburg et al., 2011). In addition, expression of JunB through MAL/SRF activity has been shown to be required for keratinocyte differentiation (Connelly et al., 2010).
Figure 6.
Loss of Bcr or MAL decreases mRNA transcript levels of the differentiation-specific desmosomal cadherin Dsg1. (A and B) Control NHEKs or those knocked down for (A) Bcr or (B) MAL were grown to confluence and subjected to high calcium to induce differentiation. 2 d after high calcium, total RNA was extracted from all samples, equalized, and cDNA prepared. qPCR analysis for DSG1 and PG (JUP) mRNA levels from 3–4 independent experiments are normalized to GAPDH and represented as fold-change over control. Error bars represent SD. *, P < 0.05. Duplicate samples prepared from a representative experiment were lysed and Western blots performed to analyze protein levels of Bcr, MAL, SRF, and GAPDH. (C) Identical experiments for analysis of DSG1 and JUP mRNA levels were performed in the presence of DMSO (vehicle) or CCG-1423 (600 nM) for 2 d after induction of differentiation by high calcium. In all cases, KD of Bcr, MAL, or inhibition of MAL/SRF signaling resulted in a decrease in DSG1 mRNA levels, but no significant change in levels of JUP mRNA. (D) Total RNA extracted from control and Bcr KD organotypic rafts at day 3 was used for real-time PCR analysis of a range of differentiation-specific and SRF target genes (DSG1, DSC1, KRT1, KRT10, IVL, LOR, FLG, SBSN, ACTB, ACTG1, JUNB, and JUP). Data from three independent rafts are normalized to GAPDH and represented as fold-change over control. Error bars represent SD. *, P < 0.05.
Restoring Dsg1 expression is sufficient to rescue the differentiation defects seen upon loss of Bcr or MAL signaling
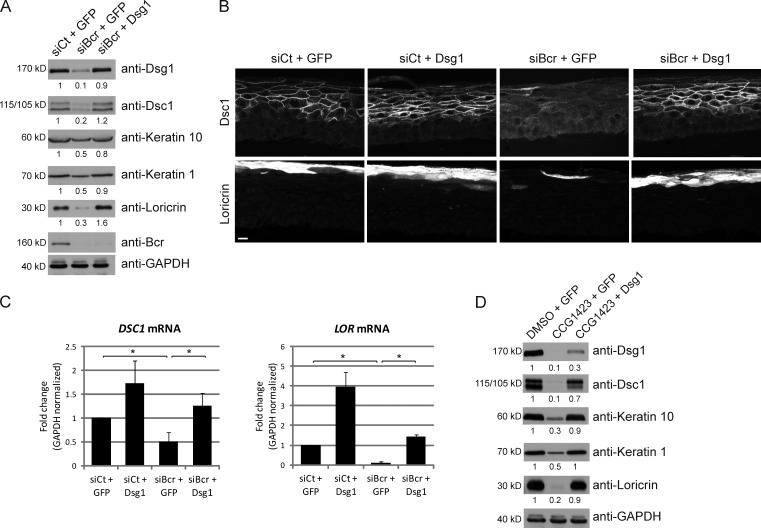
We have demonstrated that Bcr is required for differentiation and that Bcr and MAL can regulate mRNA levels of Dsg1. Considering the previously demonstrated importance of Dsg1 to differentiation, we hypothesized that regulation of Dsg1 expression is a crucial step involved in the mechanism of Bcr-induced differentiation. To test this idea, we rescued Dsg1 expression in NHEKs by retroviral infection of a full-length Dsg1 construct. Bcr KD resulted in a decrease in expression of Dsg1, keratin 1, keratin 10, and loricrin compared with control cells (Fig. 7 A). Ectopic rescue of Dsg1 in a Bcr KD background restored the expression of all these differentiation markers (Fig. 7 A). These data were also recapitulated in organotypic rafts, where expression of Dsg1 was able to restore both the proper localization (Fig. 7 B) and loss of mRNA levels (Fig. 7 C) of Dsc1 and loricrin seen in a Bcr KD condition. In addition, ectopic expression of Dsg1 was sufficient to rescue the differentiation defect observed upon inhibition of the MAL transcriptional program by CCG-1423 (Fig. 7 D). Taken together, these data demonstrate that regulation of Dsg1 expression by Bcr/MAL signaling is a crucial step in coordinating the onset of differentiation in NHEKs.
Figure 7.
Ectopic expression of Dsg1 is sufficient to restore the defects in differentiation seen upon Bcr KD or loss of MAL signaling. (A) NHEKs were infected with GFP (control) or Dsg1 expressing retrovirus as described in Materials and methods. 2 d after infection, control or Bcr siRNA were introduced by AMAXA electroporation. Cells were allowed to grow to confluence and differentiation induced by addition of high calcium for 1–2 d. Samples were then lysed, subjected to SDS-PAGE, and blotted for Dsg1, Dsc1, keratin 1, keratin 10, loricrin, Bcr, and GAPDH. Fold-change values over control quantified by densitometry are noted below blots. (B and C) Organotypic raft cultures were grown from siCt+GFP, siCt+Dsg1, siBcr+GFP, and siBcr+Dsg1 NHEKs, and processed for Dsc1 and loricrin staining (B; bar, 40 µm) or qPCR analysis (C) for DSC1 and LOR. Data from three independent rafts are normalized to GAPDH and represented as fold-change over control. Error bars represent SD. *, P < 0.05. (D) NHEKs were infected with GFP (control) or Dsg1 expressing retrovirus, and cells were allowed to grow to confluence. Differentiation was induced by addition of high calcium for 2 d, either in the presence of DMSO (vehicle) or the MAL transcriptional inhibitor CCG-1423 (600 nM). Samples were then lysed, subjected to SDS-PAGE, and blotted for Dsg1, Dsc1, keratin 1, keratin 10, loricrin, Bcr, and GAPDH. All Western blots shown are representative of three independent experiments. Ectopic expression of Dsg1 was sufficient to restore the defect in differentiation observed upon Bcr KD or CCG-1423 treatment.
Discussion
Rho GTPases have been established as important mediators of differentiation in keratinocytes, but little is known about specific Rho GEFs/GAPs that are responsible for coordinating Rho GTPase signaling during differentiation. We have identified the protein Bcr as a major regulator of RhoA activity, cytoskeletal organization, and differentiation in keratinocytes. We demonstrate further that loss of Bcr abrogates signaling through the MAL/SRF transcriptional nexus. KD of either Bcr or MAL reduces mRNA levels of the desmosomal cadherin Dsg1, restoration of which rescues the defects in differentiation seen upon loss of Bcr or MAL signaling. These data reveal the importance of Bcr in epidermal differentiation and establish the functional importance of a RhoA–SRF–Dsg1 pathway in this process.
Previous work has shown that RhoA activity increases in differentiating keratinocytes and is required along with its downstream effector PRK2 for maintenance of cell–cell adhesion (Calautti et al., 2002). Inhibition of either Rho or ROCK using pharmacological inhibitors (C3 and Y27632, respectively) decreases the ability of keratinocytes to differentiate (Sugai et al., 1992; McMullan et al., 2003; Ehrenreiter et al., 2009), while a constitutively active form of ROCK promotes differentiation (McMullan et al., 2003). RhoA/ROCK signaling has also been linked to cell compaction in the granular layer of the epidermis (Honma et al., 2006). RhoA is required for formation of cadherin-based cell–cell adhesion, and Rho/ROCK-mediated formation of a coordinated actomyosin network in keratinocytes is required for proper stratification of epidermal sheets in vitro (Braga et al., 1997; Vaezi et al., 2002). ROCKI and II are also required for inducing polarization and geometric cell shapes required for differentiation to occur (Kalaji et al., 2012). These studies therefore point to a positive role for RhoA/ROCK in regulating epidermal differentiation.
In contrast, a recent study concluded that conditional loss of RhoA in mouse skin resulted in no significant impairment to development of the epidermis. However, the lack of differentiation defects in RhoA-deficient skin might be due to overlapping functions of RhoB, whose expression in the epidermis is up-regulated fourfold in the absence of RhoA (Jackson et al., 2011). Other Rho family proteins such as Rnd3/RhoE also regulate differentiation. Unlike RhoA/Rac1, RhoE cannot hydrolyze GTP, and its functional activity is regulated by changes in expression (Chardin, 2006). RhoE expression was shown to increase upon differentiation, and loss of RhoE expression in keratinocytes results in hyperproliferation and an increase in integrin-mediated adhesion, both of which result in decreased differentiation (Liebig et al., 2009). Other studies have shown that although ROCK II promotes terminal differentiation in HaCaT cells, ROCK I has anti-differentiation effects (Lock and Hotchin, 2009). Taken together, these studies indicate that Rho-mediated signaling specificity in differentiation is tightly regulated and might even play divergent roles at different points during the process of epidermal morphogenesis. How different GEFs or GAPs might control Rho signaling specificity during the process of differentiation is unknown.
Here we show that Bcr regulates the activity of RhoA in epidermal keratinocytes (Fig. 1). The protein Bcr was originally identified in a group of leukemia patients who carried the Philadelphia translocation, where the resulting Bcr–Abl fusion protein is an oncogene characterized as being responsible for different types of leukemia, especially chronic myelogenous leukemia (Daley et al., 1990). Previous work has demonstrated that Bcr and Abr also regulate inflammatory responses and vestibular morphogenesis in mice (Kaartinen et al., 2002; Cunnick et al., 2009).
Among the ∼70 GEFs and 80 GAPs that have been identified for Rho GTPases, Bcr and Abr are unusual in having a GEF domain that can catalyze GTP exchange on RhoA, Rac, and Cdc42, and a GAP domain with intrinsic activity for Rac and Cdc42, but not RhoA (Heisterkamp et al., 1985, 1993; Chuang et al., 1995). Bcr therefore has the ability to regulate multiple GTPases, as evidenced from studies in fibroblasts demonstrating a dual ability to inactivate Rac1 and activate RhoA (Zheng et al., 2006). In contrast, other studies in macrophages and neurons have shown that Bcr seems to predominantly regulate Rac activity (Cho et al., 2007; Oh et al., 2010). To our knowledge, the functional activity of Bcr on different GTPases has not been studied in keratinocytes. Our analysis shows that whereas loss of Bcr dramatically reduces RhoA activity, it has a minimal effect on global Rac1 activity in keratinocytes, although local changes in Rac1 cannot be ruled out. These data raise the possibility that Rac1 activity in keratinocytes may be regulated by other GEF or GAPs, potentially Abr. It is also possible that Bcr activity may be targeted specifically to RhoA via a yet undiscovered scaffold, as has been demonstrated in other cell systems (Jaffe et al., 2005; García-Mata and Burridge, 2007; García-Mata et al., 2007).
We show that loss of Bcr reduces expression of desmosomal cadherins (Dsg1 and Dsc1), suprabasal keratins (keratin 1 and keratin 10), and cornified envelope proteins (loricrin), indicating the importance of this protein for the process of differentiation (Figs. 3 and 4). Loss of Abr does not affect differentiation, suggesting that Abr is not involved in this process (Fig. S1). The ability of Bcr to regulate differentiation depends on its GEF activity, as a GEF-dead mutant of Bcr was unable to rescue the differentiation defect seen upon Bcr KD (Fig. 3). To determine the mechanism via which Bcr regulates differentiation, we investigated signaling pathways downstream of Rho known to be important for differentiation. Specifically, SRF and its cofactor MAL have been shown to be required for shape-induced differentiation of keratinocytes on micropatterned collagen islands (Connelly et al., 2010), and a major role for SRF has been established in development of mouse epidermis (Koegel et al., 2009; Verdoni et al., 2010; Luxenburg et al., 2011). We demonstrate that loss of Bcr abrogates the nuclear translocation of MAL, a process that is required for activation of SRF via Rho-mediated cytoskeletal rearrangements (Fig. 5). We further show that loss of Bcr reduces SRF activity in differentiating keratinocytes, as well as the amount of junctional actin, a commonly studied target of SRF-mediated transcriptional activity (Koegel et al., 2009; Luxenburg et al., 2011). We also confirmed that like Bcr, RhoA KD reduces both stress fibers and SRF activity in keratinocytes (Fig. S1). Taken together, these data indicate that loss of Bcr causes a decrease in MAL/SRF-mediated signaling.
A recent study revealed the desmosomal cadherin Dsg1 as being among the mRNA transcripts that differ between control and SRF-null mouse keratinocytes (Luxenburg et al., 2011). We previously demonstrated that in addition to being critical for intercellular adhesion in the superficial layers of the epidermis, Dsg1 also promotes differentiation, including expression of Dsc1, suprabasal keratins, and loricrin (Getsios et al., 2009). Here we show that KD of Bcr, MAL, or inhibition of MAL signaling using the drug CCG-1423 causes a reduction in mRNA levels of Dsg1, indicating that its expression is regulated upstream by Bcr and MAL (Fig. 6). Transcriptional control of Dsg1 has previously been shown to be regulated by two different transcription factors, p63 and grainyhead-like 1 (Wilanowski et al., 2008; Ferone et al., 2013). Whether MAL and/or SRF regulate Dsg1 transcription directly or in conjunction with these other transcription factors is an area that awaits further investigation.
Considering the importance of Dsg1 for the proper differentiation of keratinocytes, we hypothesized that Dsg1 may be a key intermediary in the regulation of differentiation by a Bcr/MAL signaling pathway. Indeed, restoration of Dsg1 expression was sufficient to rescue the differentiation defect seen upon KD of Bcr or treatment with CCG-1423 (Fig. 7). These data suggest that Bcr regulates differentiation partially through expression of Dsg1. The desmosomal cadherins are targets of autoantibodies in pemphigus vulgaris and pemphigus foliaceus, which cause severe blistering of the skin in humans (Green and Simpson, 2007). Treatment of keratinocytes with pemphigus antibodies was shown to decrease RhoA activity, and reactivation of RhoA in pemphigus antibody–treated skin was shown to significantly rescue the blistering phenotype (Waschke et al., 2006). These studies demonstrate that RhoA activity can also be modulated by targeting desmogleins, suggesting the possibility of a feedback loop.
In summary, our study has implicated Bcr in the regulation of RhoA activity, MAL/SRF signaling, and epidermal differentiation in keratinocytes. These data highlight a novel pathway linking Bcr and MAL to Dsg1 expression during the process of differentiation. Taken together, these studies further our understanding of how Rho GTPase–mediated signaling pathways contribute to the process of epidermal differentiation.
Materials and methods
Growth and maintenance of cells
The SCC9 cell line (gift of L. Hudson, University of New Mexico, Albuquerque, NM) was maintained in DMEM/F12 medium (Corning) supplemented with 10% FBS (Atlanta Biologicals) and penicillin/streptomycin solution (Sigma-Aldrich). NHEKs were regularly obtained from the Northwestern University Skin Disease Research Center, where they are isolated from neonatal human foreskin as described in Halbert et al. (1992). In brief, foreskins (n = 3) were incubated overnight in 2.4 U/ml dispase (Roche) at 4°C to separate the epidermis from the dermis. Epidermal sheets are trypsinized, and keratinocytes released from the tissue by mechanical dispersion and passage through a 40-µm nylon sieve (BD). The cells were grown in medium 154 containing human keratinocyte growth supplement, gentamycin/amphotericin B solution (Invitrogen), and 0.07 mM CaCl2.
Calcium switch and induction of differentiation in submerged and organotypic raft cultures
Calcium switch experiments in SCC9s were performed by switching the cells to low calcium DMEM (0.05 mM CaCl2) for 4–16 h, followed by reintroduction of SCC9 growth media to induce formation of cell–cell junctions (Godsel et al., 2010). For NHEKs, analysis of cell–cell contact–induced cytoskeletal changes was performed by switching cells to high Ca2+ medium 154 (containing 1.2 mM CaCl2) for 1 or 4 h. To induce differentiation of submerged cultures, NHEKs were grown to confluence and switched to medium 154 with 1.2 mM CaCl2 for a period of 1–4 d. Organotypic raft cultures were grown according to previously published protocols (Meyers and Laimins, 1994). In brief, J2-3T3 fibroblasts are suspended in a mixture of collagen I (BD) and DMEM, and allowed to polymerize at 37°C. NHEKs are seeded on top of the collagen plugs, and 48 h later, lifted to an air–medium interface and grown for a period of 3–6 d.
DNA constructs, siRNA, and chemical reagents
The retroviral expression constructs LZRS-GFP and C-terminally tagged LZRS-Dsg1-Flag have been described in Getsios et al. (2009). N-terminally tagged pECFP-Bcr (WT) and pECFP-Bcr (N689A/E690A) expression constructs were a gift of N. Heisterkamp (University of Southern California, Los Angeles, CA). siRNA refractory silent mutations were introduced into these constructs using the QuikChange mutagenesis kit (Agilent Technologies). The pLPCX retroviral construct for expression of C-terminally tagged HA-MAL was a gift from G. Posern (Max Planck Institute of Biochemistry, Martinsried, Germany). The adenoviral pAd-CMV-shRhoA knockdown construct (Aghajanian et al., 2009) was a gift from K. Burridge (University of North Carolina at Chapel Hill, Chapel Hill, NC). Control siRNA oligonucleotides and those specific for KD of Bcr were purchased from Invitrogen. A pool of three siRNA oligonucleotides were used for >80% KD of Bcr in NHEKs (target sequences: siBcr#1, 5′-CCTGGAGGTGGATTCCTTTGGGTAT-3′; siBcr#2, 5′-TCCGCATGATCTACCTGCAGACGTT-3′; siBcr#3, 5′-CAGAAGAAGTGTTTCAGAAGCTTCT-3′), whereas individual oligonucleotides were sufficient for KD of Bcr in SCC9 cells. KD of Abr in NHEKs was also performed using a pool of three siRNA oligonucleotides (target sequences: 5′-GAGGAGGTTGGCATCTACAGGATAT-3′, 5′-GAGGTCAGAGTGGAGAGAAGCAATT-3′, and 5′-TACAAAGCGTTTGTCGATAACTATA-3′). KD of SRF was performed with two different siRNA oligonucleotides (target sequences: siSRF#1, 5′-CCTGGCACCAGTGTCTGCTAGTGTC-3′; and siSRF#2, 5′-GCAGGTCCCAGTGCAGGCCATTCAA-3′), and the same was achieved for MAL (target sequences: siMAL#1, 5′-GGACAGAGGACTATCTCAAACGGAA-3′; and siMAL#2, 5′-TCGATGGCCATGATTTGCAGCTGCA-3′). siRNA delivery in SCC9s was achieved by transfection with DharmaFECT (Thermo Fisher Scientific) and by AMAXA nucleoporation (Lonza) in NHEKs, both according to the manufacturers’ instructions. EGF (CN02) was purchased from Cytoskeleton, Inc. The MAL/SRF transcriptional inhibitor CCG-1423 was purchased from Cayman Chemical and used at the concentrations described during induction of differentiation in NHEKs with 1.2 mM Ca2+ medium 154.
Virus production and infection of keratinocytes
The phoenix packaging cell line was grown in DMEM supplemented with 10% FBS and penicillin/streptomycin solution. For production of GFP, Dsg1, and MAL-HA retrovirus, phoenix cells were transfected with the appropriate DNA constructs and placed in 1 µg/ml puromycin selection media 48 h after transfection. After drug selection, phoenix cells were placed at 32°C for 16–24 h for collection of retroviral supernatant. Fresh supernatant with 4 µg/ml polybrene was then used to infect cells for 90 min at 32°C. After retroviral infection, cells were washed twice with 1× PBS and returned to 37°C with fresh growth media. SRE-luciferase lentivirus was produced by the Northwestern University Skin Disease Research Center using a construct obtained from L. Shea (Northwestern University, Chicago, IL). Luciferase activity was measured using a standard luciferase reporter assay (Promega).
Quantitative real-time PCR
For measurement of mRNA transcript levels using quantitative real-time PCR, RNA was isolated using the RNeasy Mini kit (QIAGEN), according to the manufacturer’s instructions. Total RNA concentrations were equalized between samples, and cDNA prepared using the Superscript III First Strand kit (Invitrogen). Quantitative PCR was performed using SYBR Green PCR master mix (Applied Biosystems) and gene-specific primers in a StepOnePlus instrument (Applied Biosystems). Calculations for relative mRNA levels were performed using the ΔΔCt method, normalized to GAPDH and represented as fold-change values compared with control siRNA samples. Statistical analysis was performed using a student two-tailed t test.
Western blotting
To analyze protein expression levels, cells from submerged or raft cultures were washed briefly in phosphate-buffered saline (PBS) and lysed in urea sample buffer (8 M deionized urea, 1% SDS, 10% glycerol, 60 mM Tris, pH 6.8, and 5% β-mercaptoethanol) and equalized for total protein concentration. Samples were subjected to SDS-PAGE on 7.5 or 15% polyacrylamide gels, followed by transfer to polyvinylidene fluoride (PVDF) or nitrocellulose membranes (EMD Millipore). Membranes were probed with specific primary and secondary antibodies (as described below) and visualized using enhanced chemiluminescence and x-ray film (Thermo Fisher Scientific). Western blots were quantified by standard densitometry analysis using ImageJ software (National Institutes of Health, Bethesda, MD). All Western blots shown are representative data obtained from three independent experiments.
Immunofluorescence and immunohistochemical analysis
For immunofluorescence, cells grown on coverslips were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in PBS. Primary and secondary antibody incubations were performed at room temperature for 1 h, interspaced by multiple washes in PBS, and followed by mounting coverslips in polyvinyl alcohol (Sigma-Aldrich). Fixed cells were visualized with a microscope (model DMR; Leica) fitted with a 40× objective (PL APO, NA 1.32). Images were captured with an ORCA-100 CCD camera (model C4742-95; Hamamatsu Photonics) and MetaMorph 6.1 imaging software (Molecular Devices). Confocal imaging was performed at the Northwestern University Cell Imaging Facility. For quantification of stress fibers, cells were counted based on having either prominent or reduced/no stress fibers. More than 500 cells were counted from randomly imaged fields for each cell condition, and the data represented as percentage of the total cells counted on a bar graph. For quantification of MAL localization, cells from randomly imaged fields were scored for nuclear staining of ectopically expressed HA-tagged MAL, and the data represented as percentage of cells counted.
Organotypic raft cultures were fixed in 10% neutral-buffered formalin and embedded in paraffin. Antigen retrieval for paraffin-embedded sections was performed by heating samples to 95°C in 0.01 M citrate buffer. Sections were blocked in 10% normal goat serum, and incubated with primary and secondary antibodies for 1 h at 37°C. After mounting in polyvinyl alcohol, sections were visualized as described above. Hematoxylin and eosin staining from paraffin-embedded sections was performed according to established protocols. Quantification of cortical actin staining and E-cadherin was performed by measuring fluorescence intensity per area in regions of interest formed by tracing cell–cell borders in randomly imaged fields. Statistical analysis was performed using a student two-tailed t test.
Antibodies
The following primary antibodies were used: 26C4/anti-RhoA, anti-RhoC, anti-Cdc42, anti-Bcr (N-20), anti-SRF, anti-MAL, PY99/anti-phosphotyrosine (Santa Cruz Biotechnology, Inc.), anti-Rac1, anti–β-catenin (BD), 27B2/anti-Dsg1, anti-GFP (Invitrogen), U100/anti-Dsc1 (RDI), 1407/anti-PG (Aves Laboratories), anti-actin, anti-Ki67 (EMD Millipore), anti-GAPDH, hVin1/anti-Vinculin, anti-HA (Sigma-Aldrich), HECD1/anti–E-cadherin (Takara Bio Inc.), anti-PKP2 (Progen Biotechnik), anti-DP (Angst et al., 1990), and 12G10/anti-tubulin (provided by J. Frankel and E.M. Nelson from the Developmental Studies Hybridoma Bank under the auspicies of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA). The following antibodies were gifts of J. Segre (National Human Genome Research Institute, National Institutes of Health): rabbit polyclonal anti–mouse keratin 1, rabbit polyclonal anti–mouse keratin 10, rabbit polyclonal anti–mouse involucrin, and rabbit polyclonal anti–mouse loricrin. Secondary antibodies for Western blotting included HRP-conjugated goat anti–mouse, –rabbit, and –chicken antibodies from Kirkegaard & Perry Laboratories. Secondary antibodies for immunocytochemistry/histochemistry included Alexa Fluor 488– or 568–conjugated goat anti–mouse, –rabbit, and –chicken antibodies from Invitrogen. Alexa Fluor 488–conjugated phalloidin was used to visualize F-actin (Invitrogen).
Rho, Rac, and Cdc42 activity assays
Construction of the pGEX4T-1 prokaryotic expression construct containing Rhotekin Rho-binding domain (GST-RBD) has been described previously (Liu and Burridge, 2000). Purification of GST-RBD and pull-down assays for active Rho GTPases was performed as described previously (Noren et al., 2000; Dubash et al., 2007). In brief, expression of the fusion proteins in Escherichia coli was induced with 100 µM IPTG for 12–16 h at room temperature. Bacterial lysate collected in 50 mM Tris, pH 7.6, 50 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 1 mM DTT, and protease inhibitors (Complete; Roche) was incubated with glutathione Sepharose 4B beads (GE healthcare) for 1 h at 4°C, followed by washes in lysis buffer. For GTPase pull-downs, cells were lysed in 50 mM Tris, pH 7.6, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate, 10 mM MgCl2, and protease inhibitors. Lysates were clarified by centrifugation, equalized for total protein concentration, and incubated with 30–60 µg of GST-RBD for 30 min at 4°C. Bead pellets were washed in 50 mM Tris, pH 7.6, 150 mM NaCl, 1% Triton X-100, 10 mM MgCl2, and protease inhibitors, and further processed for SDS-PAGE and Western blotting. G-LISA assays for RhoA, Rac1, and Cdc42 activity were performed according to the manufacturer’s instructions (Cytoskeleton, Inc.), and absorbance measurements for GTPase activity was obtained using a Synergy 2 plate reader (BioTek Instruments, Inc.).
Online supplemental material
Fig. S1 shows defects in stress fiber formation, SRF activity, and keratinocyte differentiation upon RhoA KD. Fig. S2 highlights localization of cell–cell junction proteins upon Bcr KD. Fig. S3 demonstrates no change in differentiation upon Abr KD. Fig. S4 shows Ki67 staining upon Bcr KD. Fig. S5 indicates differentiation defects and loss of Dsg1 mRNA upon SRF KD. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304133/DC1.
Supplementary Material
Acknowledgments
We would like to thank V. Todorovic and S. Getsios for critical reading of the manuscript, along with all other members of the Green laboratory for their technical assistance and support. We would like to thank J. Segre for her gift of multiple antibodies used in this study (keratin 1, keratin 10, loricrin, and involucrin), G. Posern for his gift of the HA-MAL construct, and K. Burridge for the RhoA knockdown adenovirus. We are very grateful to N. Heisterkamp for her gift of WT and GEF-dead Bcr constructs, and also to L. Shea for providing the SRE-luciferase lentivirus. We would also like to thank D. Quach for his assistance with qPCR (Northwestern University). Keratinocytes were obtained from the Keratinocyte Core of the Northwestern University Skin Disease Research Center, and histological analysis was conducted either by the Mouse Phenotyping Core of the R.H. Lurie Comprehensive Cancer Center (supported by NIH/NCI; P30 CA060553-159026) or by the Pathology Core of the Northwestern University Skin Disease Research Center, Chicago, IL (5P30AR057216-02), with support from the NIH/NIAMS. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Northwestern University Skin Disease Research Center or the NIH/NIAMS. Imaging work was performed in part at the Northwestern University Cell Imaging Facility (generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center).
This work was supported by NIH grant R01 AR041836 and AR43380, with partial support from CA122151, to K. Green. Additional support was provided by the J.L. Mayberry endowment to K. Green. A. Dubash is supported by an American Heart Association post-doctoral fellowship (11POST7380001).
Footnotes
Abbreviations used in this paper:
- Bcr
- breakpoint cluster region
- DP
- desmoplakin
- Dsc1
- desmocollin-1
- Dsg1
- desmoglein-1
- E-cad
- E-cadherin
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- KD
- knockdown
- NHEK
- normal human epidermal keratinocyte
- PG
- plakoglobin
- PKP
- plakophilin
- ROCK
- Rho kinase
- SRF
- serum response factor
- WT
- wild type
References
- Aghajanian A., Wittchen E.S., Campbell S.L., Burridge K. 2009. Direct activation of RhoA by reactive oxygen species requires a redox-sensitive motif. PLoS ONE. 4:e8045 10.1371/journal.pone.0008045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai M., Stanley J.R. 2012. Desmoglein as a target in skin disease and beyond. J. Invest. Dermatol. 132:776–784 10.1038/jid.2011.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst B.D., Nilles L.A., Green K.J. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257 [DOI] [PubMed] [Google Scholar]
- Asselineau D., Prunieras M. 1984. Reconstruction of ‘simplified’ skin: control of fabrication. Br. J. Dermatol. 111(Suppl 27):219–222 10.1111/j.1365-2133.1984.tb15608.x [DOI] [PubMed] [Google Scholar]
- Bass-Zubek A.E., Godsel L.M., Delmar M., Green K.J. 2009. Plakophilins: multifunctional scaffolds for adhesion and signaling. Curr. Opin. Cell Biol. 21:708–716 10.1016/j.ceb.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitah S.A., Frye M., Glogauer M., Watt F.M. 2005. Stem cell depletion through epidermal deletion of Rac1. Science. 309:933–935 10.1126/science.1113579 [DOI] [PubMed] [Google Scholar]
- Bishop A.L., Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241–255 10.1042/0264-6021:3480241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter E., Garcia-Mata R., Guilluy C., Dubash A., Rossi G., Brennwald P.J., Burridge K. 2010. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 12:477–483 10.1038/ncb2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga V.M., Machesky L.M., Hall A., Hotchin N.A. 1997. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell–cell contacts. J. Cell Biol. 137:1421–1431 10.1083/jcb.137.6.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. 2004. Rho and Rac take center stage. Cell. 116:167–179 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- Busche S., Descot A., Julien S., Genth H., Posern G. 2008. Epithelial cell-cell contacts regulate SRF-mediated transcription via Rac-actin-MAL signalling. J. Cell Sci. 121:1025–1035 10.1242/jcs.014456 [DOI] [PubMed] [Google Scholar]
- Calautti E., Grossi M., Mammucari C., Aoyama Y., Pirro M., Ono Y., Li J., Dotto G.P. 2002. Fyn tyrosine kinase is a downstream mediator of Rho/PRK2 function in keratinocyte cell–cell adhesion. J. Cell Biol. 156:137–148 10.1083/jcb.200105140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P. 2006. Function and regulation of Rnd proteins. Nat. Rev. Mol. Cell Biol. 7:54–62 10.1038/nrm1788 [DOI] [PubMed] [Google Scholar]
- Cho Y.J., Cunnick J.M., Yi S.J., Kaartinen V., Groffen J., Heisterkamp N. 2007. Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol. Cell. Biol. 27:899–911 10.1128/MCB.00756-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang T.H., Xu X., Kaartinen V., Heisterkamp N., Groffen J., Bokoch G.M. 1995. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc. Natl. Acad. Sci. USA. 92:10282–10286 10.1073/pnas.92.22.10282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.T., Gautrot J.E., Trappmann B., Tan D.W., Donati G., Huck W.T., Watt F.M. 2010. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat. Cell Biol. 12:711–718 10.1038/ncb2074 [DOI] [PubMed] [Google Scholar]
- Cunnick J.M., Schmidhuber S., Chen G., Yu M., Yi S.J., Cho Y.J., Kaartinen V., Minoo P., Warburton D., Groffen J., Heisterkamp N. 2009. Bcr and Abr cooperate in negatively regulating acute inflammatory responses. Mol. Cell. Biol. 29:5742–5750 10.1128/MCB.00357-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q., Van Etten R.A., Baltimore D. 1990. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 247:824–830 10.1126/science.2406902 [DOI] [PubMed] [Google Scholar]
- Delva E., Tucker D.K., Kowalczyk A.P. 2009. The desmosome. Cold Spring Harb. Perspect. Biol. 1:a002543 10.1101/cshperspect.a002543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubash A.D., Wennerberg K., García-Mata R., Menold M.M., Arthur W.T., Burridge K. 2007. A novel role for Lsc/p115 RhoGEF and LARG in regulating RhoA activity downstream of adhesion to fibronectin. J. Cell Sci. 120:3989–3998 10.1242/jcs.003806 [DOI] [PubMed] [Google Scholar]
- Dusek R.L., Godsel L.M., Green K.J. 2007. Discriminating roles of desmosomal cadherins: beyond desmosomal adhesion. J. Dermatol. Sci. 45:7–21 10.1016/j.jdermsci.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Ehrenreiter K., Kern F., Velamoor V., Meissl K., Galabova-Kovacs G., Sibilia M., Baccarini M. 2009. Raf-1 addiction in Ras-induced skin carcinogenesis. Cancer Cell. 16:149–160 10.1016/j.ccr.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2002. Rho GTPases in cell biology. Nature. 420:629–635 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Evelyn C.R., Wade S.M., Wang Q., Wu M., Iñiguez-Lluhí J.A., Merajver S.D., Neubig R.R. 2007. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol. Cancer Ther. 6:2249–2260 10.1158/1535-7163.MCT-06-0782 [DOI] [PubMed] [Google Scholar]
- Ferone G., Mollo M.R., Thomason H.A., Antonini D., Zhou H., Ambrosio R., De Rosa L., Salvatore D., Getsios S., van Bokhoven H., et al. 2013. p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum. Mol. Genet. 22:531–543 10.1093/hmg/dds464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata R., Burridge K. 2007. Catching a GEF by its tail. Trends Cell Biol. 17:36–43 10.1016/j.tcb.2006.11.004 [DOI] [PubMed] [Google Scholar]
- García-Mata R., Dubash A.D., Sharek L., Carr H.S., Frost J.A., Burridge K. 2007. The nuclear RhoA exchange factor Net1 interacts with proteins of the Dlg family, affects their localization, and influences their tumor suppressor activity. Mol. Cell. Biol. 27:8683–8697 10.1128/MCB.00157-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getsios S., Simpson C.L., Kojima S., Harmon R., Sheu L.J., Dusek R.L., Cornwell M., Green K.J. 2009. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J. Cell Biol. 185:1243–1258 10.1083/jcb.200809044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel L.M., Dubash A.D., Bass-Zubek A.E., Amargo E.V., Klessner J.L., Hobbs R.P., Chen X., Green K.J. 2010. Plakophilin 2 couples actomyosin remodeling to desmosomal plaque assembly via RhoA. Mol. Biol. Cell. 21:2844–2859 10.1091/mbc.E10-02-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.J., Simpson C.L. 2007. Desmosomes: new perspectives on a classic. J. Invest. Dermatol. 127:2499–2515 10.1038/sj.jid.5701015 [DOI] [PubMed] [Google Scholar]
- Halbert C.L., Demers G.W., Galloway D.A. 1992. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66:2125–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsell S., Cowin P. 2001. Deconstructing desmoplakin. Nat. Cell Biol. 3:E270–E272 10.1038/ncb1201-e270 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. 2007. Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesion? Biochim. Biophys. Acta. 1773:69–77 10.1016/j.bbamcr.2006.04.009 [DOI] [PubMed] [Google Scholar]
- Heisterkamp N., Stam K., Groffen J., de Klein A., Grosveld G. 1985. Structural organization of the bcr gene and its role in the Ph’ translocation. Nature. 315:758–761 10.1038/315758a0 [DOI] [PubMed] [Google Scholar]
- Heisterkamp N., Kaartinen V., van Soest S., Bokoch G.M., Groffen J. 1993. Human ABR encodes a protein with GAPrac activity and homology to the DBL nucleotide exchange factor domain. J. Biol. Chem. 268:16903–16906 [PubMed] [Google Scholar]
- Hennings H., Michael D., Cheng C., Steinert P., Holbrook K., Yuspa S.H. 1980. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 19:245–254 10.1016/0092-8674(80)90406-7 [DOI] [PubMed] [Google Scholar]
- Honma M., Benitah S.A., Watt F.M. 2006. Role of LIM kinases in normal and psoriatic human epidermis. Mol. Biol. Cell. 17:1888–1896 10.1091/mbc.E05-12-1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B., Peyrollier K., Pedersen E., Basse A., Karlsson R., Wang Z., Lefever T., Ochsenbein A.M., Schmidt G., Aktories K., et al. 2011. RhoA is dispensable for skin development, but crucial for contraction and directed migration of keratinocytes. Mol. Biol. Cell. 22:593–605 10.1091/mbc.E09-10-0859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A., Schmidt A. 2005. Association of CNK1 with Rho guanine nucleotide exchange factors controls signaling specificity downstream of Rho. Curr. Biol. 15:405–412 10.1016/j.cub.2004.12.082 [DOI] [PubMed] [Google Scholar]
- Kaartinen V., Nagy A., Gonzalez-Gomez I., Groffen J., Heisterkamp N. 2002. Vestibular dysgenesis in mice lacking Abr and Bcr Cdc42/RacGAPs. Dev. Dyn. 223:517–525 10.1002/dvdy.10071 [DOI] [PubMed] [Google Scholar]
- Kalaji R., Wheeler A.P., Erasmus J.C., Lee S.Y., Endres R.G., Cramer L.P., Braga V.M. 2012. ROCK1 and ROCK2 regulate epithelial polarisation and geometric cell shape. Biol. Cell. 104:435–451 10.1111/boc.201100093 [DOI] [PubMed] [Google Scholar]
- Koegel H., von Tobel L., Schäfer M., Alberti S., Kremmer E., Mauch C., Hohl D., Wang X.J., Beer H.D., Bloch W., et al. 2009. Loss of serum response factor in keratinocytes results in hyperproliferative skin disease in mice. J. Clin. Invest. 119:899–910 10.1172/JCI37771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke M.D., Delva E., Kowalczyk A.P. 2006. The desmosome: cell science lessons from human diseases. J. Cell Sci. 119:797–806 10.1242/jcs.02888 [DOI] [PubMed] [Google Scholar]
- Liebig T., Erasmus J., Kalaji R., Davies D., Loirand G., Ridley A., Braga V.M. 2009. RhoE Is required for keratinocyte differentiation and stratification. Mol. Biol. Cell. 20:452–463 10.1091/mbc.E07-11-1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.P., Burridge K. 2000. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol. Cell. Biol. 20:7160–7169 10.1128/MCB.20.19.7160-7169.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock F.E., Hotchin N.A. 2009. Distinct roles for ROCK1 and ROCK2 in the regulation of keratinocyte differentiation. PLoS ONE. 4:e8190 10.1371/journal.pone.0008190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxenburg C., Pasolli H.A., Williams S.E., Fuchs E. 2011. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat. Cell Biol. 13:203–214 10.1038/ncb2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey L.M., Macara I.G. 2011. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 21:727–735 10.1016/j.tcb.2011.06.005 [DOI] [PubMed] [Google Scholar]
- McGrath J.A. 2005. Inherited disorders of desmosomes. Australas. J. Dermatol. 46:221–229 10.1111/j.1440-0960.2005.00188.x [DOI] [PubMed] [Google Scholar]
- McGrath J.A., McMillan J.R., Shemanko C.S., Runswick S.K., Leigh I.M., Lane E.B., Garrod D.R., Eady R.A. 1997. Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nat. Genet. 17:240–244 10.1038/ng1097-240 [DOI] [PubMed] [Google Scholar]
- McMullan R., Lax S., Robertson V.H., Radford D.J., Broad S., Watt F.M., Rowles A., Croft D.R., Olson M.F., Hotchin N.A. 2003. Keratinocyte differentiation is regulated by the Rho and ROCK signaling pathway. Curr. Biol. 13:2185–2189 10.1016/j.cub.2003.11.050 [DOI] [PubMed] [Google Scholar]
- Meyers C., Laimins L.A. 1994. In vitro systems for the study and propagation of human papillomaviruses. Curr. Top. Microbiol. Immunol. 186:199–215 10.1007/978-3-642-78487-3_11 [DOI] [PubMed] [Google Scholar]
- Moon S.Y., Zheng Y. 2003. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 13:13–22 10.1016/S0962-8924(02)00004-1 [DOI] [PubMed] [Google Scholar]
- Müller E.J., Williamson L., Kolly C., Suter M.M. 2008. Outside-in signaling through integrins and cadherins: a central mechanism to control epidermal growth and differentiation? J. Invest. Dermatol. 128:501–516 10.1038/sj.jid.5701248 [DOI] [PubMed] [Google Scholar]
- Nikolova E., Mitev V., Minner F., Deroanne C.F., Poumay Y. 2008. The inhibition of the expression of the small Rho GTPase Rac1 induces differentiation with no effect on cell proliferation in growing human adult keratinocytes. J. Cell. Biochem. 103:857–864 10.1002/jcb.21455 [DOI] [PubMed] [Google Scholar]
- Noren N.K., Liu B.P., Burridge K., Kreft B. 2000. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J. Cell Biol. 150:567–580 10.1083/jcb.150.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D., Han S., Seo J., Lee J.R., Choi J., Groffen J., Kim K., Cho Y.S., Choi H.S., Shin H., et al. 2010. Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J. Neurosci. 30:14134–14144 10.1523/JNEUROSCI.1711-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., Nordheim A. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353–365 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.T., Lim S.E., Jang S.I., Morasso M.I. 2002. Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor. J. Biol. Chem. 277:45195–45202 10.1074/jbc.M205380200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G., Treisman R. 2006. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16:588–596 10.1016/j.tcb.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Simpson C.L., Patel D.M., Green K.J. 2011. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol. 12:565–580 10.1038/nrm3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai M., Hashimoto K., Kikuchi A., Inoue S., Okumura H., Matsumoto K., Goto Y., Ohgai H., Moriishi K., Syuto B., et al. 1992. Epidermal cell differentiation inhibitor ADP-ribosylates small GTP-binding proteins and induces hyperplasia of epidermis. J. Biol. Chem. 267:2600–2604 [PubMed] [Google Scholar]
- Thomason H.A., Scothern A., McHarg S., Garrod D.R. 2010. Desmosomes: adhesive strength and signalling in health and disease. Biochem. J. 429:419–433 10.1042/BJ20100567 [DOI] [PubMed] [Google Scholar]
- Tunggal J.A., Helfrich I., Schmitz A., Schwarz H., Günzel D., Fromm M., Kemler R., Krieg T., Niessen C.M. 2005. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 24:1146–1156 10.1038/sj.emboj.7600605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaezi A., Bauer C., Vasioukhin V., Fuchs E. 2002. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell. 3:367–381 10.1016/S1534-5807(02)00259-9 [DOI] [PubMed] [Google Scholar]
- Vaughan E.M., Miller A.L., Yu H.Y., Bement W.M. 2011. Control of local Rho GTPase crosstalk by Abr. Curr. Biol. 21:270–277 10.1016/j.cub.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoni A.M., Ikeda S., Ikeda A. 2010. Serum response factor is essential for the proper development of skin epithelium. Mamm. Genome. 21:64–76 10.1007/s00335-009-9245-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschke J., Spindler V., Bruggeman P., Zillikens D., Schmidt G., Drenckhahn D. 2006. Inhibition of Rho A activity causes pemphigus skin blistering. J. Cell Biol. 175:721–727 10.1083/jcb.200605125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilanowski T., Caddy J., Ting S.B., Hislop N.R., Cerruti L., Auden A., Zhao L.L., Asquith S., Ellis S., Sinclair R., et al. 2008. Perturbed desmosomal cadherin expression in grainy head-like 1-null mice. EMBO J. 27:886–897 10.1038/emboj.2008.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A., Hatzfeld M. 2010. A role of plakophilins in the regulation of translation. Cell Cycle. 9:2973–2978 10.4161/cc.9.15.12446 [DOI] [PubMed] [Google Scholar]
- Zheng X., Güller S., Beissert T., Puccetti E., Ruthardt M. 2006. BCR and its mutants, the reciprocal t(9;22)-associated ABL/BCR fusion proteins, differentially regulate the cytoskeleton and cell motility. BMC Cancer. 6:262 10.1186/1471-2407-6-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.