The kinesin KIF4A is locally activated by the central spindle pool of Aurora B and thereby preferentially controls anaphase spindle length.
Abstract
Anaphase central spindle formation is controlled by the microtubule-stabilizing factor PRC1 and the kinesin KIF4A. We show that an MKlp2-dependent pool of Aurora B at the central spindle, rather than global Aurora B activity, regulates KIF4A accumulation at the central spindle. KIF4A phosphorylation by Aurora B stimulates the maximal microtubule-dependent ATPase activity of KIF4A and promotes its interaction with PRC1. In the presence of phosphorylated KIF4A, microtubules grew more slowly and showed long pauses in growth, resulting in the generation of shorter PRC1-stabilized microtubule overlaps in vitro. Cells expressing only mutant forms of KIF4A lacking the Aurora B phosphorylation site overextended the anaphase central spindle, demonstrating that this regulation is crucial for microtubule length control in vivo. Aurora B therefore ensures that suppression of microtubule dynamic instability by KIF4A is restricted to a specific subset of microtubules and thereby contributes to central spindle size control in anaphase.
Introduction
Microtubules are highly versatile filaments formed by polymerization of the protein tubulin in the presence of GTP. In living cells the nucleation, polymerization, and depolymerization of microtubules is tightly controlled by a large number of specific protein complexes (Desai and Mitchison, 1997; Akhmanova and Steinmetz, 2008, 2010; Howard and Hyman, 2009). These factors are required to produce microtubule structures at the correct time, and with the correct mechanical properties and organization. This allows microtubules to carry out many diverse functions in dividing and nondividing cells. One of the most striking roles for microtubules is as part of the mitotic and anaphase spindle apparatus required for chromosome segregation and controlling the position of the cell division plane during cytokinesis (Wittmann et al., 2001). Formation of the anaphase central spindle requires the concerted action of a group of microtubule-associated proteins and kinesin motors (Eggert et al., 2006; Glotzer, 2009). Anaphase central spindle formation is controlled by a simple mechanism involving an anti-parallel microtubule-bundling factor PRC1 and the kinesin KIF4A, which suppresses microtubule dynamic instability (Bringmann et al., 2004; Kurasawa et al., 2004; Lee and Kim, 2004; Zhu and Jiang, 2005; Zhu et al., 2005; Gruneberg et al., 2006; Bieling et al., 2010; Ozlü et al., 2010; Subramanian et al., 2010; Hu et al., 2011). Previous studies have suggested that PRC1 is inhibited until the onset of anaphase by Cdk1–cyclin B phosphorylation, and this is a major factor delaying central spindle formation until the onset of anaphase (Jiang et al., 1998; Mollinari et al., 2002; Zhu et al., 2006; Neef et al., 2007). The idea that Cdk1 is the key negative regulator of central spindle formation has been questioned by a study indicating that PRC1 may be negatively regulated by Polo-like kinase 1 (Plk1; Hu et al., 2012). However, this would be inconsistent with a large body of literature showing that Plk1 activity is needed to promote initiation of cytokinesis in early anaphase (Niiya et al., 2006; Petronczki et al., 2007; Wolfe et al., 2009), and then plays a subsequent role in abscission timing (Fabbro et al., 2005; Bastos and Barr, 2010).
In addition to control by PRC1, the centralspindlin complex comprised of the MKlp1/KIF23 kinesin-like motor protein and the Cyk-4/MgcRacGAP Rho-family GTPase regulator is important for central spindle integrity (Mishima et al., 2002). Again, centralspindlin action is inhibited until the onset of anaphase by Cdk1–cyclin B phosphorylation (Mishima et al., 2004). MKlp1 is required for the bundling of anti-parallel microtubule overlaps, and this activity is promoted by Aurora B phosphorylation and binding of the 14-3-3 adaptor proteins (Mishima et al., 2002; Guse et al., 2005; Neef et al., 2006; Pavicic-Kaltenbrunner et al., 2007; Douglas et al., 2010). Aurora B localization to the central spindle is under the control of a second mitotic kinesin, MKlp2 (Gruneberg et al., 2004). This pathway creates a restricted pool of Aurora B activity on the central spindle specifically in anaphase cells (Fuller et al., 2008). In the absence of MKlp2 this Aurora B pool is lost and MKlp1 complexes fail to become phosphorylated, ultimately resulting in the failure of cytokinesis (Neef et al., 2006). In turn, the MAD2 spindle checkpoint pathway and Cdk1–cyclin B negatively regulate MKlp2 and the relocation of Aurora B to the central spindle, respectively (Hümmer and Mayer, 2009; Lee et al., 2010; Vázquez-Novelle and Petronczki, 2010). Together these regulatory events prevent the activity of central spindle factors before the onset of anaphase, and ensure that this structure forms at the right time and place. However, despite the central importance of KIF4A in anaphase spindle function, it remains unclear how its activity is controlled. Here we have investigated how the KIF4A–PRC1 system is regulated, focusing on the relationship between the key mitotic kinase Aurora B and KIF4A function.
Results
Association of KIF4A with the central spindle is Aurora B dependent
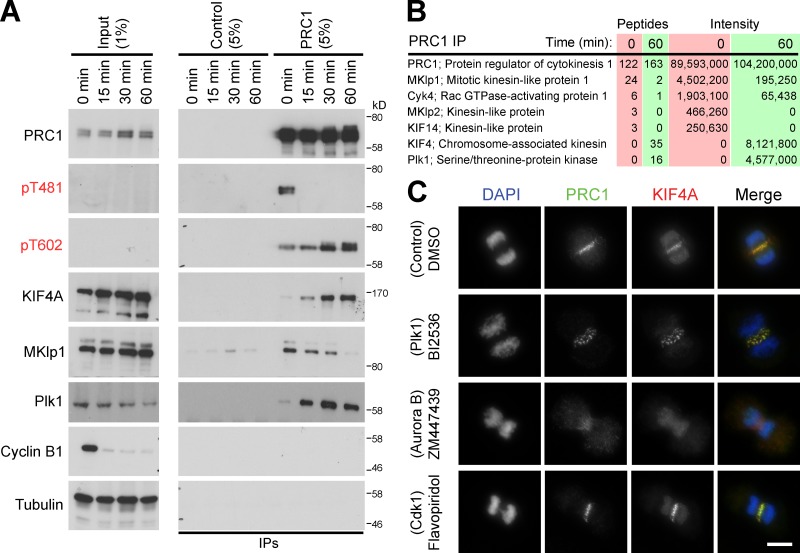
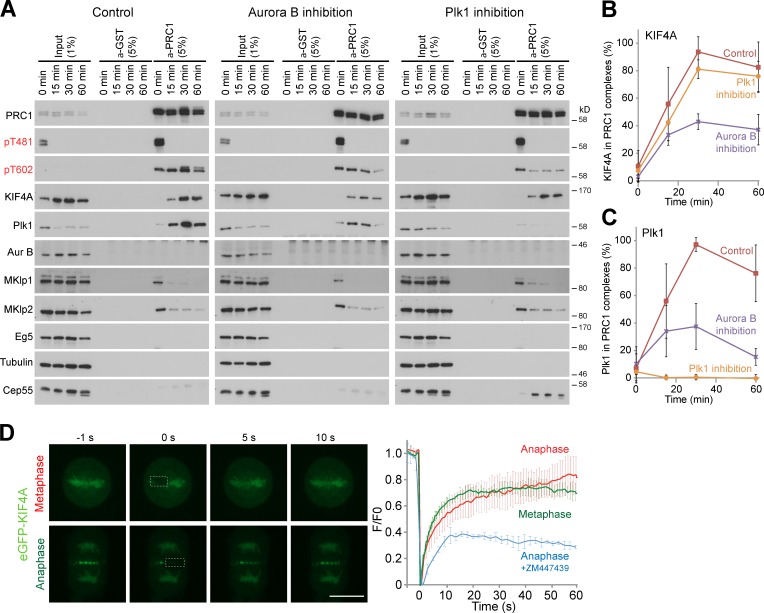
To analyze how the activities of KIF4A and PRC1 are regulated on the central spindle, PRC1 complexes were isolated from cells at different times in anaphase. Western blotting of these complexes revealed that PRC1 was phosphorylated by Cdk1 in metaphase samples on threonine 481 (pT481) and was then rapidly dephosphorylated as cells progress through anaphase (Fig. 1 A). Concurrently, PRC1 became phosphorylated on threonine 602 (pT602) and Plk1 accumulated in the complexes, reaching a maximum at 30–60 min (Fig. 1 A). KIF4A, like Plk1, was absent from metaphase PRC1 and then progressively accumulated throughout anaphase (Fig. 1 A). Mass spectrometry confirmed that KIF4A and Plk1 complexes accumulated as cells progressed from early to late anaphase (Fig. 1 B). This suggested that Plk1 activity might regulate the assembly of the KIF4A–PRC1 complex at the central spindle. To test this idea, cells were treated with kinase inhibitors specific for Cdk1, Plk1, and Aurora B. Surprisingly, Plk1 inhibition using BI2536 did not prevent PRC1 and KIF4A localization to the central spindle, although in some cells later in anaphase the staining pattern was fragmented into discrete punctae rather than the highly ordered structure seen in control cells (Fig. 1 C). By contrast, inhibition of Aurora B with ZM447439 resulted in the loss of KIF4A staining at the central spindle (Fig. 1 C). Treatment with the Cdk1 inhibitor flavopiridol had little effect on PRC1 or KIF4A staining, but did cause some defects in chromosome segregation (Fig. 1 C). These findings suggested that Aurora B rather than Plk1 regulates KIF4A localization in anaphase.
Figure 1.
Control of PRC1–KIF4A complexes by mitotic kinases. (A) PRC1 complexes were immunoprecipitated using specific antibodies from synchronized HeLa cells entering anaphase, at the time points indicated in the figure. Species-matched GST antibodies were used as a negative control (Control). The input and immunoprecipitated fractions (IPs) were analyzed by Western blotting with the antibodies shown in the figure, and (B) mass spectrometry. (C) Thymidine-synchronized HeLa cells were treated 8 h after the thymidine washout step for 5 min with DMSO, 1 µM BI2536, 5 µM flavopiridol, or 1 µM ZM447439. These cells were fixed, and then stained for DNA with DAPI, rabbit anti-PRC1, and sheep anti-KIF4A. Bar, 10 µm.
Local regulation of KIF4A by the central spindle pool of Aurora B
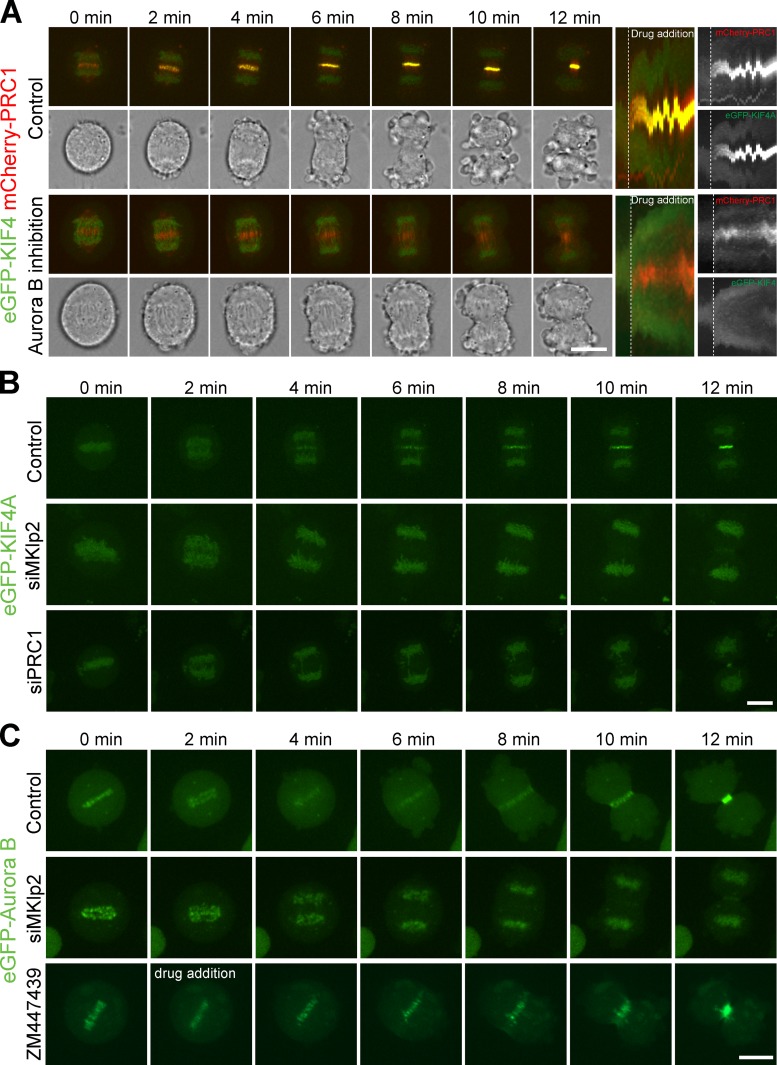
It has previously been shown that formation of a localized pool of Aurora B activity at the central spindle in anaphase requires the kinesin motor protein MKlp2 (Gruneberg et al., 2004; Hümmer and Mayer, 2009; Lee et al., 2010; Vázquez-Novelle and Petronczki, 2010). The formation of this pool of Aurora B is required for the phosphorylation of the Aurora B substrate MKlp1 at the central spindle (Neef et al., 2006). To test if this mechanism is also important for KIF4A targeting in anaphase, cell lines stably expressing eGFP-tagged KIF4A and mCherry-PRC1 were produced. Because it is difficult to study the events of early anaphase in fixed samples due to the short time of anaphase, 11 min in HeLa cells (Bastos and Barr, 2010), live-cell imaging was used to study the behavior of KIF4A and PRC1. First, cells entering anaphase were treated with the specific Aurora B inhibitor ZM447439 or a DMSO solvent control. In control cells KIF4A accumulated on the central spindle region 4–5 min after anaphase onset, ∼2 min after PRC1, which is present from 1–2 min (Fig. S1 A and Fig. 2 A). By contrast, Aurora B inhibition prevented recruitment of KIF4A to the central spindle, but, importantly, did not prevent PRC1 from targeting to the central spindle region (Fig. 2 B). Cells were then depleted of MKlp2 to specifically interfere with the activity of the central spindle pool of Aurora B (Gruneberg et al., 2004; Hümmer and Mayer, 2009; Lee et al., 2010; Vázquez-Novelle and Petronczki, 2010). Consistent with the effects of Aurora B inhibition using ZM447439, MKlp2 depletion resulted in retention of KIF4A on chromatin and the loss of the central spindle pool (Fig. 2 B and Fig. S1 B). After 12 min of anaphase a small amount of KIF4A was visible on the central spindle region (Fig. 2 B), possibly reflecting the activity of the delocalized pool of Aurora B in these cells. Depletion of PRC1 also resulted in a loss of KIF4A from the central spindle but not from the chromatin as cells progress into anaphase (Fig. 2 B and Fig. S1 B), consistent with the idea it is a major KIF4A binding partner in anaphase (Fig. 1, A and B; Kurasawa et al., 2004; Zhu and Jiang, 2005).
Figure 2.
The MKlp2–Aurora B pathway is required for KIF4A targeting to the central spindle in anaphase. (A) HeLa cells stably expressing eGFP-KIF4A and mCherry-PRC1 were treated with DMSO (control) or 1 µM ZM447439, then imaged every 30 s with a spinning disk confocal microscope. Maximum intensity–projected images of the merged eGFP and mCherry channels and a single brightfield image collected at the center plane of the image stack are shown. All times are relative to the drug addition (t = 0). A kymograph of the central spindle area is depicted. (B) HeLa cells stably expressing eGFP-KIF4A were treated with control, MKlp2, or PRC1 siRNA duplexes for 48 h, and then imaged every minute with a spinning disk confocal microscope. Maximum intensity–projected images of eGFP fluorescence are shown at the time indicated. (C) HeLa cells stably expressing eGFP–Aurora B were treated with control or MKlp2 siRNA duplexes for 48 h, and then imaged every minute with a spinning disk confocal microscope. Control cells were treated with 1 µM ZM447439 as indicated in the figure. Maximum intensity–projected images of eGFP fluorescence are shown at the time indicated. Bars, 10 µm.
To further support the idea that Aurora B regulates KIF4A, the timing of Aurora B recruitment to the central spindle was investigated. Cell lines stably expressing eGFP-tagged Aurora B were imaged as they passed into anaphase. This showed that Aurora B is initially retained on the centromeres for the first 2 min of anaphase, and then transitions to the central spindle after 4 min (Fig. 2 C). As expected, depletion of MKlp2 prevented the central spindle recruitment of Aurora B (Fig. 2 C and Fig. S1 C). Therefore, Aurora B and KIF4A are recruited to the central spindle at a similar time in anaphase, and share the same dependency for MKlp2. Inhibition of Aurora B using ZM447439 did not prevent Aurora B localization in anaphase (Fig. 2 C), providing further support for the view that Aurora B kinase activity rather than some other property is required for KIF4A regulation.
Together, these findings suggest that Aurora B rather than Plk1 is the key regulator of KIF4A targeting in anaphase. They also reveal a specific requirement for MKlp2 in this process. Because MKlp2 depletion does not affect the localization of Aurora B to centromeres, this indicates that the central spindle pool of Aurora B rather than its global activity regulates the timing of KIF4A recruitment.
Aurora B phosphorylates KIF4A at a conserved site
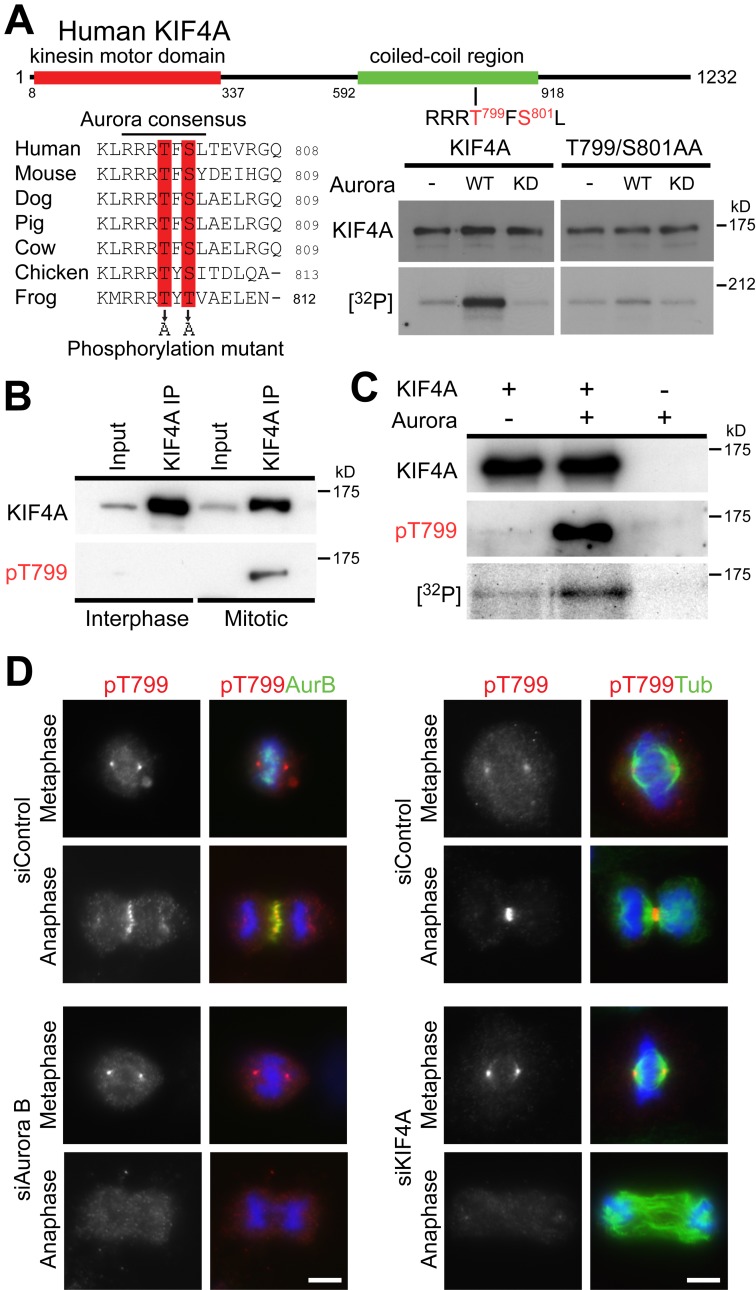
Aurora kinases predominantly phosphorylate a consensus sequence defined by a cluster of arginine and lysine residues followed by a patch of serine or threonine residues (Alexander et al., 2011; Kettenbach et al., 2011). Visual inspection of the vertebrate KIF4A sequences revealed the presence of a single such conserved sequence, RRRTFS, centered on threonine 799 and serine 801 in human KIF4A (Fig. 3 A). The KIF4A T799A/S801A Aurora site mutant showed strongly reduced levels of phosphorylation in vitro (Fig. 3 A), and specific phosphopeptide antibodies (pT799) were therefore raised against this site. Western blotting showed that the pT799 epitope was present on KIF4A complexes isolated from mitotic but not interphase cell populations (Fig. 3 B). In vitro kinase assays using recombinant full-length KIF4A showed that Aurora B phosphorylated KIF4A and created the pT799 epitope (Fig. 3 C). Furthermore, affinity-purified pT799 antibodies stained the central spindle in anaphase, and spindle poles in metaphase cells (Fig. 3 D). However, only the central spindle staining was lost after the depletion of Aurora B or KIF4A (Fig. 3 D), suggesting the spindle pole staining is not due to KIF4A. To provide evidence this staining requires Aurora B activity, specific kinase inhibitors were used. KIF4A pT799 staining was lost after inhibition of Aurora B, but not Plk1 or Cdk1 (Fig. S2 A). Depletion of MKlp2, which disregulates the central spindle pool of Aurora B, also resulted in a loss of pT799 staining (Fig. S2 B). Together, these results demonstrate that Aurora B phosphorylates the pool of KIF4A at the central spindle on threonine 799.
Figure 3.
KIF4A phosphorylation by Aurora B at T799. (A) A schematic depicting the position of the kinesin motor domain and central coiled-coil region of human KIF4A is shown at the top of the figure. A multiple sequence alignment shows the conserved Aurora phosphorylation consensus motif, comprising a patch of basic amino acids, in this case arginine, followed by serine or threonine residues at T799 and S801, which are predicted to be phosphorylated. In vitro kinase assays were performed using full-length wild-type KIF4A or KIF4A T799A/S801A mutant in the presence and absence of 50 ng Aurora kinase, 1 mM ATP, and 5 µCi γ-[32P]ATP per reaction. (B) KIF4A was isolated from asynchronous (Interphase) populations of HeLa cells, or cells released from a nocodazole block for 30 min. The input and immunoprecipitated fractions (IPs) were analyzed by Western blotting for KIF4A and KIF4A pT799. (C) In vitro kinase assays were performed using 1 µg full-length KIF4A or a buffer control in the presence and absence of 50 ng Aurora kinase, 1 mM ATP, and 5 µCi γ-[32P]ATP per reaction. (D) HeLa cells treated with control, Aurora B, and KIF4A siRNA duplexes for 48 h were fixed, and then stained for DNA with DAPI, mouse anti–Aurora B or mouse anti-tubulin, and rabbit anti-KIF4A pT799. Bars, 10 µm.
Phosphorylation-defective mutants of KIF4A fail to support normal central spindle length control
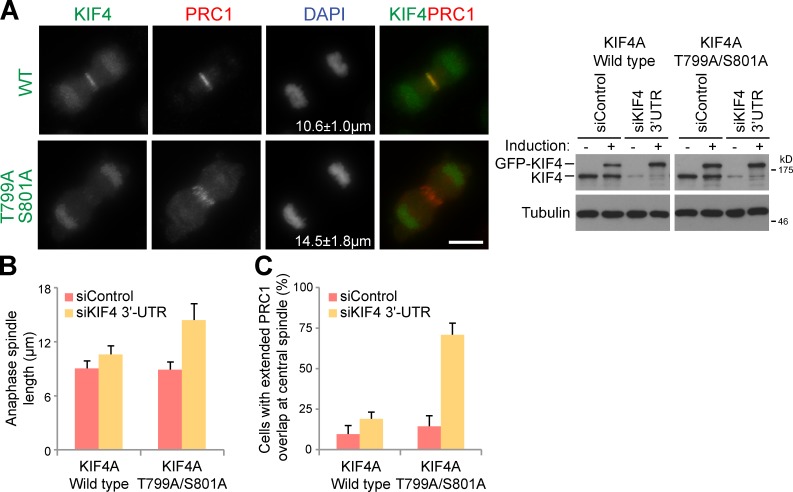
To investigate the role of threonine 799 phosphorylation in KIF4A targeting and function, cells stably expressing inducible eGFP-KIF4A or a KIF4T799A/S801A Aurora site mutant were created. These cells were then depleted of the endogenous KIF4A using siRNA directed to the 3′-UTR of the KIF4A mRNA or treated with a control siRNA. Replacement of the endogenous KIF4A with a wild-type GFP-tagged copy of KIF4A resulted in the expected pattern of localization to chromatin and central spindle (Fig. 4 A). Strikingly, the KIF4AT799A/S801A Aurora site mutant failed to target to the central spindle microtubules in anaphase cells (Fig. 4 A). Central spindle length in cells expressing the KIF4AT799A/S801A mutant was 14.5 ± 1.8 µm on average, compared with 10.6 ± 1.0 µm in cells expressing wild-type KIF4A (Fig. 4 B). This compared with 9.1 ± 1.1 µm in cells treated with control siRNA that expressed endogenous KIF4A, or 15.33 ± 1.5 µm in cells depleted of KIF4A but not rescued by induction of the transgene (4 independent experiments, >30 cells per experiment). Quantitation also revealed that the overlap region defined by PRC1 staining was extended in cells expressing the KIF4AT799A/S801A Aurora site mutant (Fig. 4 C). These highly penetrant phenotypes therefore support the view that phosphorylation of KIF4A at threonine 799 by Aurora B is crucial for its localization and function in controlling anaphase central spindle length.
Figure 4.
KIF4A phosphorylation at T799 regulates central spindle length. (A) HeLa cells stably expressing doxycycline-inducible eGFP-tagged KIF4A wild-type and KIF4AT799A/S801A Aurora site double mutants were treated with control or KIF4A-3′UTR siRNA duplexes for 24 h, then 1 µg/ml doxycycline was added. After a further 24 h the cells were fixed, and then stained for DNA with DAPI, mouse anti-tubulin (red), rabbit anti-PRC1 (red), and KIF4A-eGFP constructs (green). Bar, 10 µm. Equivalent samples were taken for Western blotting as shown in the figure to confirm KIF4A depletion and induction of eGFP-KIF4A. (B) Anaphase central spindle length and (C) the ability of PRC1 to focus on the overlapped region of the central spindle were measured. The values are plotted on the bar graphs for 4 independent experiments measuring >30 cells per experiment. Error bars show the standard deviation from the mean.
Regulation of the KIF4A–PRC1 interaction by Aurora B
To identify how Aurora B might regulate KIF4A, PRC1 complexes were isolated from synchronized cells treated with Aurora B or Plk1 inhibitors (Fig. 5 A). In control samples, KIF4A and Plk1 showed the expected pattern of behavior (Fig. 5, A–C), and bound to PRC1 only once cells had entered anaphase (Kurasawa et al., 2004; Neef et al., 2007; Santamaria et al., 2007). Western blotting revealed that the level of KIF4A in PRC1 complexes isolated from cells treated with the Aurora B inhibitor ZM447439 was reduced to 40% of the control value (Fig. 5, A and B), suggesting that Aurora B promotes the interaction between PRC1 and KIF4A. Plk1 inhibition with BI2536 had no effect on the amount of KIF4A present in PRC1 complexes (Fig. 5, A and B). By contrast, Plk1 interaction with PRC1 was lost and PRC1 phosphorylation at threonine 602 reduced in the samples treated with the Plk1 inhibitor BI2536 (Fig. 5, A and C), as published previously (Neef et al., 2007; Santamaria et al., 2007; Bastos and Barr, 2010). Investigation of KIF4A turnover at the central spindle using FRAP revealed that it had a short half-life in the range of 4–5 s at this structure, similar to the half-life on metaphase chromatin (Fig. 5 D). Inhibition of Aurora B immediately before bleaching reduced the recovery of KIF4A at the central spindle (Fig. 5 D). Aurora B is therefore required to maintain a highly dynamic pool of KIF4A at the central spindle. In part, this is by promoting the interaction with PRC1; however, this does not exclude further regulation of microtubule-binding and kinesin ATPase activities, and these possibilities were investigated further.
Figure 5.
Aurora B regulates the interaction of KIF4A and PRC1. (A) PRC1 complexes were immunoprecipitated using specific antibodies from synchronized HeLa cells entering anaphase treated with DMSO, 1 µM ZM447439, or 1 µM BI2536 at the time points indicated in the figure. Matched GST antibodies were used as a negative control (Control). The input and immunoprecipitated fractions (IPs) were analyzed by Western blotting with the antibodies shown in the figure. (B) The amount of PRC1-associated KIF4A and (C) Plk1 was measured using densitometry in ImageJ and is plotted in the graphs (n = 3). (D) FRAP analysis of eGFP-KIF4A. Fluorescence images of FRAP processes for eGFP-KIF4A for either metaphase chromatin or anaphase central spindle associated are shown. The square dashed line indicates areas of bleaching where fluorescence recovery was measured. Bar, 10 µm. Fluorescence recovery (fluorescence/prebleach fluorescence, F/F0) as a function of time for metaphase (green, n = 3), anaphase (red, n = 7), and ZM447439 treated (blue) is plotted.
KIF4A ATPase activity is increased by Aurora B phosphorylation
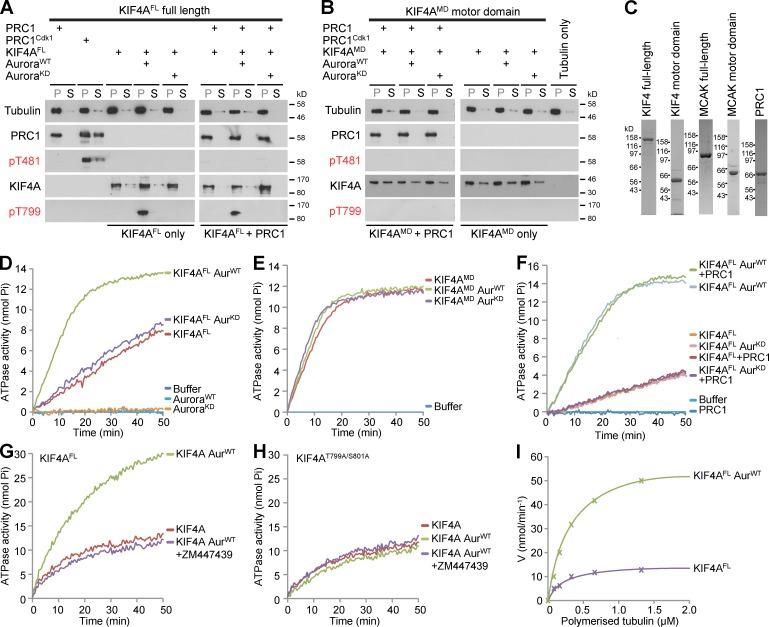
Previous studies have demonstrated that the Xenopus KIF4A orthologue XKlp1 has potent microtubule-stabilizing activity, and that this activity is directed toward anti-parallel microtubule overlaps created by PRC1 (Bieling et al., 2010; Subramanian et al., 2010). The potent microtubule-stabilizing action of KIF4A implies the need for regulation to restrict its function to the anaphase central spindle. However, these previous studies do not prove that PRC1 is the sole determinant limiting KIF4A activity to the central spindle. The data presented here suggest that Aurora B also helps restrict KIF4A activity to the central spindle. To investigate the roles of PRC1 and Aurora B in regulating the microtubule binding of KIF4A, purified recombinant PRC1 and KIF4A were therefore tested in microtubule-binding assays. Earlier work has shown that PRC1 is inhibited until the onset of anaphase by Cdk1–cyclin B phosphorylation (Mollinari et al., 2002; Neef et al., 2007), and this was therefore used as a control. As expected, PRC1 pelleted with microtubules and this was reduced for the Cdk1-phosphorylated form (Fig. 6 A). However, little difference was seen in the pelleting behavior of nonphosphorylated and threonine 799 Aurora-phosphorylated forms of full-length KIF4A in either the absence or presence of PRC1 (Fig. 6 A). Similarly, the KIF4A motor domain lacking the Aurora phosphorylation site bound directly to microtubules in a PRC1- and Aurora-independent fashion (Fig. 6 B). PRC1, KIF4A, and KIF4A motor domain pelleting was dependent on the presence of microtubules, indicating they were not precipitating under these conditions (Fig. S3 A). Titration of phosphorylated and nonphosphorylated KIF4A over a wide range of concentrations also failed to reveal any alteration in microtubule binding (Fig. S3 B). The purified proteins used for these assays are shown in Fig. 6 C.
Figure 6.
Phosphorylation by Aurora B increases KIF4A kinesin ATPase activity. (A) Microtubule-binding assays were performed as described in Materials and methods using 10 nM of the full-length KIF4A (KIF4FL), or (B) 33 nM of the KIF4A motor domain (KIF4MD) in the presence or absence of 10 nM PRC1 or Cdk1-phosphorylated PRC1 as indicated. Equivalent aliquots of the microtubule pellet fraction (P) and supernatant fractions (S) were analyzed by Western blotting. (C) Coomassie blue–stained gels of proteins used in the assays are shown. (D) Kinesin ATPase assays were performed using 50 nM KIF4FL (n = 5 independent experiments), (E) 75 nM KIF4MD (n = 2 independent experiments), or (F) KIF4FL in the presence or absence of 50 nM PRC1 (n = 2 independent experiments). For some conditions, KIF4A was phosphorylated using wild-type active Aurora kinase (AurWT) or an inactive “kinase-dead” mutant (AurKD). (G) Kinesin ATPase assays were performed using 50 nM KIF4FL or (H) KIF4S799A/T801A mutant left untreated or phosphorylated using wild-type active Aurora kinase (AurWT) in the presence or absence of 10 µM ZM447439 Aurora B inhibitor (n = 3 and n = 2 independent experiments, respectively). (I) Kinesin ATPases assays were performed using 50 nM full-length KIF4A or Aurora-phosphorylated KIF4A at different microtubule concentrations in 150 µl final volume for n = 2 independent experiments. Initial ATPase rates were plotted as a function of microtubule concentration. Assuming ATPase activity followed Michael-Menten kinetics, Kapp and Vmax were obtained.
The possibility that Aurora B phosphorylation promotes KIF4A motor activity was therefore tested using microtubule-stimulated ATPase assays. In the absence of microtubules, the ATPase activity of KIF4A was the same as ATP incubated in buffer alone. In the presence of microtubules, full-length KIF4A incubated with buffer or inactive kinase showed similar ATPase activities, whereas threonine 799 phosphorylated KIF4A had an ATPase activity nearly fourfold higher (Fig. 6 D). This effect was specific for the full-length motor because the motor domain alone was not sensitive to phosphorylation and showed high activity under all conditions tested (Fig. 6 E). Furthermore, the addition of PRC1 to assays performed using full-length phosphorylated and nonphosphorylated forms of KIF4A made no difference to the ATPase activity (Fig. 6 F). To confirm the effect of Aurora B was mediated by phosphorylation at the conserved T799/S801 site, this was mutated to alanine and recombinant protein tested in kinesin ATPase assays. As expected, wild-type KIF4A showed Aurora B stimulated ATPase activity prevented by inhibition of Aurora B using the specific inhibitor ZM447439 (Fig. 6 G). In contrast, the KIF4AT799/S801A mutant did not show Aurora B stimulation of ATPase activity (Fig. 6 H). Importantly, both wild-type KIF4A and the KIF4AT799/S801A mutant showed the same basal level of microtubule-stimulated ATPase activity before Aurora phosphorylation (Fig. 6, G and H).
ATPase assays were then performed under a range of microtubule concentrations to enable calculation of the apparent Km for microtubules and the maximal ATPase activity of KIF4A (Fig. 6 I). This revealed that phosphorylation did not alter Kapp MT, which was ∼0.2 µM in both cases, consistent with measurements of microtubule-binding activity. Microtubule-stimulated ATPase activity was increased from a kcat of 28.2 s−1 to 117.3 s−1 when KIF4A was prephosphorylated with Aurora B. In the absence of microtubules, ATPase activity was 5,000-fold lower. These values fall within the range reported for kinesins with high processivity (Friel and Howard, 2012).
Together, these results suggest that phosphorylation by Aurora B at the central spindle is a major regulatory event controlling both the interaction of KIF4A with PRC1 and its kinesin ATPase activity. Furthermore, this stimulation of ATPase activity is a direct consequence of Aurora B phosphorylation at T799/S801, and does not require the interaction with PRC1.
Phosphorylated KIF4A reduces growth of PRC1-microtubule overlaps
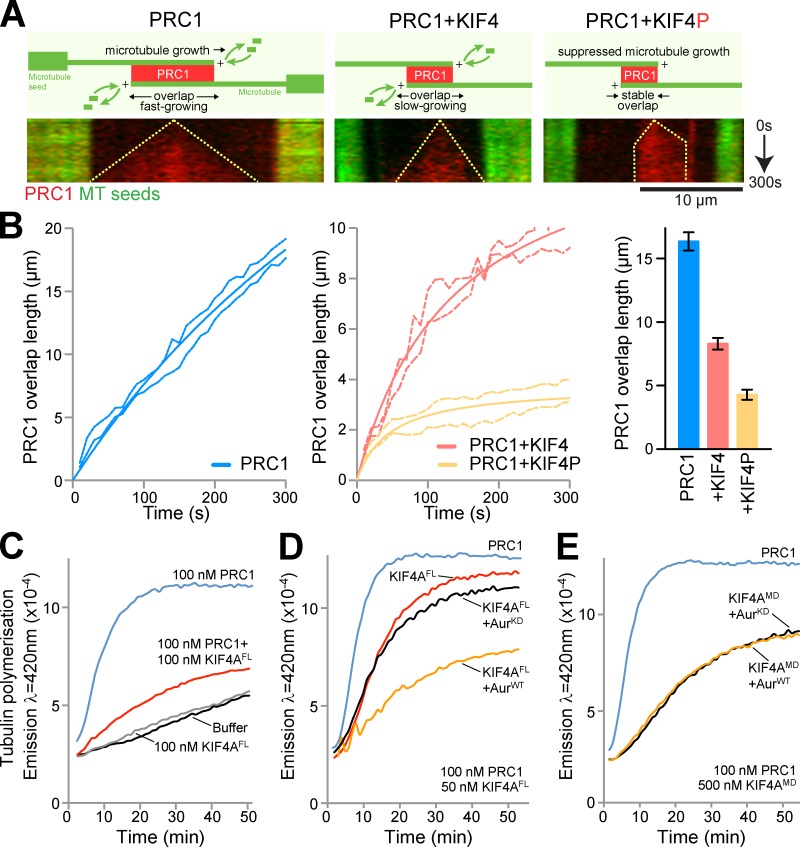
Anti-parallel microtubule overlaps were created in vitro using PRC1 and purified tubulin as described previously (Bieling et al., 2010). Imaging revealed that these PRC1–microtubule overlaps extended linearly to reach a mean length of 16 µm, and addition of KIF4A reduced this overlap length to 8–9 µm during the 5-min incubation period (Fig. 7, A and B). When Aurora B–phosphorylated KIF4A was used, the PRC1 overlap region extended for a short period at the start of the incubation and then stalled at a mean length of just under 5 µm.
Figure 7.
Reduced PRC1-microtubule overlap length in the presence of phosphorylated KIF4A. (A) Representative kymographs showing antiparallel overlaps formed by microtubules growing from GMP-CPP–stabilized microtubule seeds (green) in the presence of 14 nM mCherry-PRC1, 14 nM mCherry-PRC1 and 28.5 nM KIF4, or 14 nM mCherry-PRC1 and 28.5 nM KIF4P over 5 min. For the kymographs, t = 0 is when PRC1 localization is first observed, indicating establishment of an anti-parallel overlap. The growing overlap length is indicated by dotted lines. Bar, 10 µm. (B) Average length of anti-parallel microtubule overlaps in the presence of mCherry-PRC1 (left, n = 9), mCherry-PRC1 and KIF4A (middle, n = 15), or mCherry-PRC1 and KIF4P (middle, n = 20) as a function of time. Errors are shown by the dotted lines. Average overlap length after 5 min is plotted with standard error in the bar graph. (C) Microtubule polymerization assays were performed using 100 nM of PRC1 and 100 nM of full-length recombinant KIF4A (KIF4FL) for n = 3 independent experiments. (D) Full-length recombinant KIF4A was phosphorylated using wild-type active Aurora kinase (AurWT) or an inactive “kinase-dead” mutant (AurKD). Microtubule polymerization assays were then performed in the presence of 100 nM PRC1 and 50 nM of each form of KIF4A for n = 4 independent experiments. (E) Recombinant KIF4A motor domain was phosphorylated using wild-type active Aurora kinase (AurWT) or an inactive “kinase-dead” mutant (AurKD). Microtubule polymerization assays were then performed using 500 nM recombinant KIF4A motor domain (KIF4MD) in the presence of 100 nM PRC1 for n = 3 independent experiments.
Recombinant KIF4A motor domain and full-length KIF4A were then tested for their effects on microtubule polymerization from purified tubulin in solution. Under standard conditions where microtubule polymerization was limiting, 100 nM PRC1 promoted microtubule polymerization (Fig. 7 C). Addition of 100 nM full-length KIF4A reduced both the rate and extent of microtubule polymerization to a level that was intermediate to a buffer control or KIF4A alone (Fig. 7 C). Human KIF4A can therefore limit microtubule polymerization in the presence of PRC1. The effects of Aurora phosphorylation of KIF4A were then investigated. For this purpose the concentration of full-length KIF4A was reduced to 50 nM, a concentration at which it showed only a moderate effect on microtubule polymerization in the presence of 100 nM PRC1 (Fig. 7 D). Addition of 50 nM phosphorylated full-length KIF4A reduced both the rate and extent of microtubule polymerization, whereas KIF4A treated with inactive kinase behaved similarly to the untreated full-length motor protein (Fig. 7 D). As a further control, the effect of phosphorylation on the KIF4A motor domain, which lacks the Aurora B phosphorylation site, was tested. As expected, the effect of the KIF4A motor domain on microtubule polymerization was not altered by treatment with either wild-type or inactive kinase (Fig. 7 E). KIF4A therefore reduces microtubule polymerization in vitro, and this activity is enhanced by Aurora B phosphorylation.
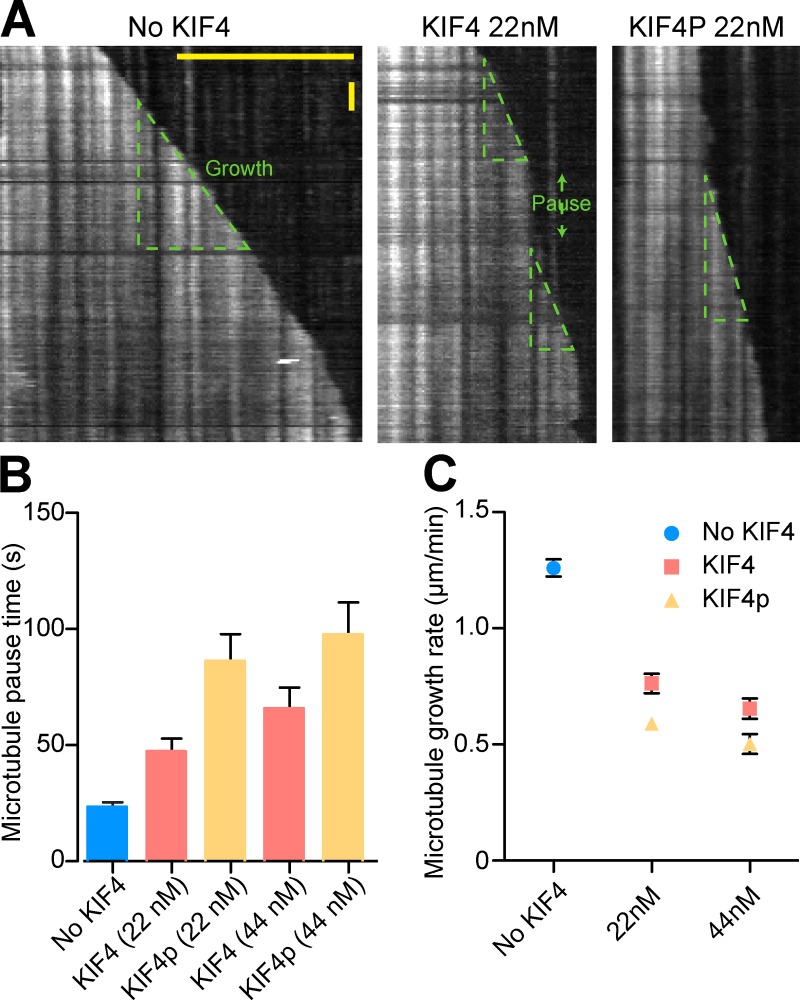
One explanation for these effects was that KIF4A has an ATP-dependent microtubule-destabilizing activity similar to MCAK (Walczak et al., 1996). This was tested using taxol-stabilized microtubules as a substrate (Fig. S4). As shown previously (Walczak et al., 1996, 2002), MCAK was able to depolymerize microtubules in an ATP-dependent fashion, and this activity was inhibited by Aurora B phosphorylation (Fig. S4, A and B) as expected (Andrews et al., 2004; Lan et al., 2004). Under the same conditions KIF4A had no effect on microtubule stability (Fig. S4, C and D), suggesting it is not an active microtubule-destabilizing kinesin. To further define how KIF4A reduces microtubule polymerization, single polymerizing microtubules were imaged over a period of 10 min (Fig. 8). In the absence of KIF4A, microtubules showed long periods of stable growth with only a few short pauses (Fig. 8, A and B). Under these conditions the mean growth rate was 1.3 µm/min (Fig. 8 C). Addition of 22 nM KIF4A altered this behavior such that the pause time increased to 50 s and the mean growth rate was reduced to 0.75 µm/min (Fig. 8, A–C). Aurora B–phosphorylated 22 nM KIF4A resulted in a doubling in the pause time to 100 s and an additional reduction in growth rate to 0.6 µm/min (Fig. 8, A–C). These effects were further increased when 44 nM KIF4A was used (Fig. 8, A–C). When taken together, these results support the view that KIF4A suppresses microtubule growth (Bieling et al., 2010), and demonstrate that this activity is enhanced by Aurora B phosphorylation. This provides a mechanism to restrict KIF4A activity to anaphase spindle microtubules.
Figure 8.
Suppression of microtubule dynamics by KIF4A is enhanced by Aurora B phosphorylation. (A) Kymographs of microtubule growth in the absence and presence of KIF4A and phosphorylated KIF4A (KIF4P). Microtubule growth was frequently interrupted by pauses indicated by green double-headed arrow. Microtubule growth rate is indicated by dotted triangles. Horizontal bar 10 µm; vertical bar, 0.5 min. (B) Average microtubule pause time was measured in the absence of KIF4A (n = 30), in the presence of 22 nM (n = 42) and 44 nM KIF4A (n = 29), and in the presence of 22 nM (n = 36) and 44 nM KIF4P (n = 32). (C) Average growth rate of microtubule in the absence of KIF4A (n = 36), in the presence of 22 nM (n = 42) and 44 nM KIF4A (n = 28), and in the presence of 22 nM (n = 35) and 44 nM KIF4P (n = 20). Error bars represent SEM.
Re-wiring the Aurora B central spindle length control system
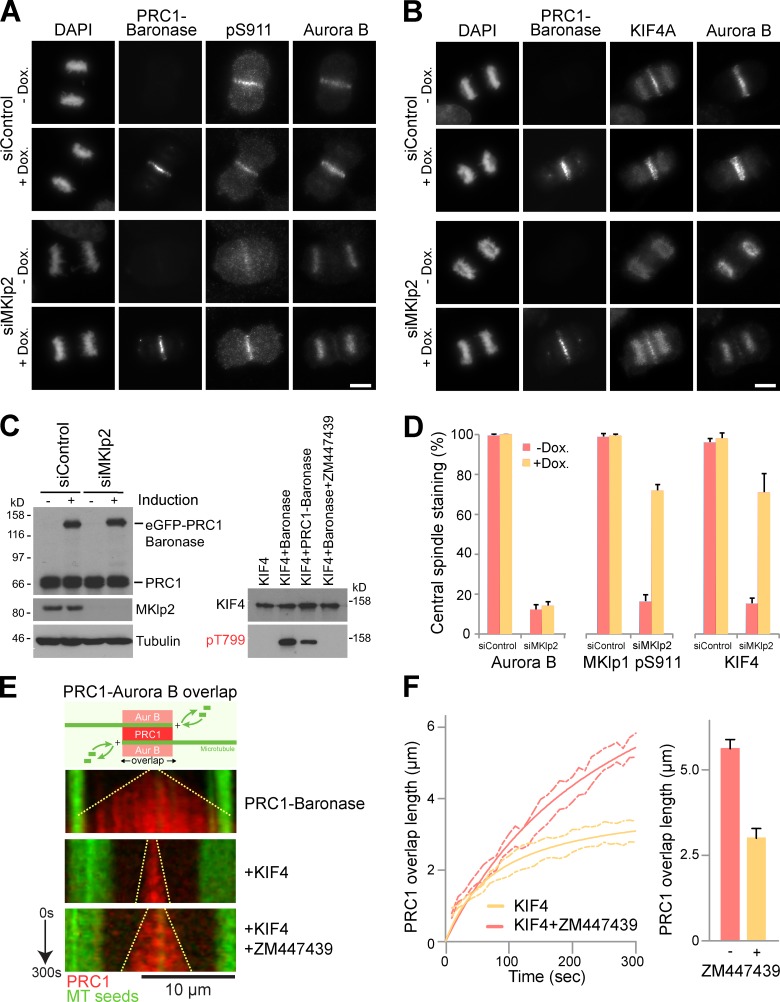
To further test the idea that phosphorylation at the anaphase central spindle is crucial for KIF4A function, the endogenous Aurora B targeting system was replaced by a synthetic construct. This used a kinase module comprising the activation region of INCENP joined to a truncated form of Aurora B (Sessa et al., 2005), coupled to the microtubule-targeting region of PRC1. This Baronase module (B-type Aurora kinase) exhibits high kinase activity toward both a model substrate and KIF4A and is sensitive to the Aurora B inhibitor ZM447439 (Fig. S5, A and B). A similar module, Aaronase (A-type Aurora kinase) was also created and tested and found to be sensitive to an Aurora A inhibitor but not used further here (Fig. S5 A). Stable cell lines expressing a doxycycline-inducible PRC1-Baronase were then created. When depleted of MKlp2 these cells retain Aurora B and KIF4A on the chromatin in anaphase, and Aurora B marker phosphorylation at MKlp1 pS911 is lost (Fig. 9, A–D). Induction of PRC1-Baronase rescues MKlp1 pS911 phosphorylation, and supports localization of KIF4A to the central spindle and KIF4A phosphorylation at threonine 799 (Fig. 9, A–D). Because Baronase was engineered to remove the epitope for the AIM1 monoclonal antibody, endogenous Aurora B could be detected independently of the transgene. This showed that although endogenous Aurora B was retained on the chromatin (Fig. 9, A and B), this did not interfere with KIF4A function.
Figure 9.
Reconstitution of Aurora B phosphorylation on PRC1-stabilized microtubule overlaps in vivo and in vitro. (A) HeLa cells stably expressing doxycycline-inducible eGFP-tagged PRC1-Baronase were treated with control or MKlp2-3′UTR siRNA duplexes for 24 h followed by thymidine synchronization for 20 h. Cells were washed with media and 1 µg/ml doxycycline (dox.) was either added or omitted. Cells were fixed 9 h after release, and then stained for DNA with DAPI, mouse anti–Aurora B, and either rabbit MKLp1 pS911or (B) sheep KIF4A. Bars, 10 µm. (C) Western blotting confirmed MKlp2 depletion and induction of eGFP-PRC1–Baronase construct. In vitro kinase assays were performed using full-length wild-type KIF4A in the absence or presence of Baronase kinase with and without ZM447439 treatment, as well as with the mCherry-PRC1–Baronase protein. Samples were analyzed by Western blotting for KIF4A and KIF4A pT799. (D) The presence of Aurora B, phosphorylated MKlp1 pS911, and KIF4A on the central spindle was scored. The values are plotted on the bar graphs for 3 independent experiments measuring >80 cells per experiment. Error bars show the standard deviation from the mean. (E) Kymographs show anti-parallel overlaps formed over 5 min by microtubules growing from GMP-CPP–stabilized seeds (green) in the presence of 14 nM mCherry-PRC1-Baronase, 14 nM mCherry-PRC1-Baronase and 28.5 nM KIF4, or 14 nM mCherry-PRC1-Baronase and 28.5 nM KIF4 with 10 µM ZM447439 to inhibit Aurora B kinase activity. For the kymographs, t = 0 is when mCherry-PRC1–Baronase is first observed, indicating establishment of an anti-parallel overlap. The growing overlap length is indicated by the dotted line. Bar, 10 µm. (F) Anti-parallel microtubule overlap length of in the presence of mCherry-PRC1 and KIF4A (n = 6), and mCherry-PRC1 and KIF4P (n = 8), is plotted as a function of time. Errors are shown by the dotted lines. Average overlap length at 5 min is plotted in the bar graph. Error bars show the standard error (n = 8 and n = 9).
This strategy also permits the reconstitution of Aurora B–decorated microtubule overlap structures in vitro. Anti-parallel microtubule overlaps were created in vitro using recombinant PRC1-Baronase and purified tubulin. Imaging revealed that these PRC1-Baronase microtubule overlaps extended linearly, similarly to those created using PRC1 (Fig. 9 E). Addition of KIF4A resulted in slow overlap length to 3 µm during the 5-min incubation period (Fig. 9, E and F). When Aurora B phosphorylation of KIF4A was prevented with ZM447439, the PRC1 overlap region extended more rapidly and achieved a mean length of 6 µm (Fig. 9, E and F). This system therefore recapitulates the regulation of KIF4A by a locally targeted pool of Aurora B at the microtubule overlap region of the central spindle.
Discussion
Suppression of microtubule dynamic instability by KIF4A and Aurora B
Here we have studied how the kinesin KIF4A and the anti-parallel microtubule-bundling factor PRC1 are regulated in anaphase (Bieling et al., 2010; Subramanian et al., 2010; Hu et al., 2011). We find that Aurora B directly phosphorylates KIF4A in the central coiled-coil stalk region and that this has two consequences. First, it enhances the binding of KIF4A to PRC1, and second, it increases the ATPase activity of KIF4A, thereby suppressing microtubule growth. Together, these two changes restrict KIF4A to the central spindle in anaphase. In the absence of Aurora B activity, KIF4A fails to localize to the central spindle, most likely due to a reduction in its affinity for PRC1. This results in an altered balance of microtubule polymerization events in this region, and over-extension of the anaphase central spindle and increased chromosome segregation. Interestingly, the results appear to eliminate a model whereby centromeric/chromatin Aurora B would promote KIF4A leaving the chromatin because cells depleted of MKlp2 retain both Aurora B and KIF4A on the chromatin. This chromatin-bound pool of KIF4A has a discrete function in lateral condensation of mitotic chromosomes (Samejima et al., 2012), and may therefore be functionally and spatially separated from the pool of KIF4A involved in central spindle formation.
The data presented here also show that PRC1 binding is necessary but not sufficient for full targeting and movement of KIF4A to the central spindle. This relocation also requires activation of the kinesin motor activity by Aurora B phosphorylation. A motor domain fragment lacking the stalk and coiled-coil region shows high constitutive ATPase activity unregulated by Aurora B. This suggests that full-length KIF4A is in a partially inhibited form perhaps with the head groups bound back on to a coiled-coil sequence in the stalk region similar to other motors (Hackney, 2007), or by the dual lockdown mechanism recently reported for kinesin-1 (Kaan et al., 2011). In both cases, phosphorylation of KIF4A in the stalk region might release auto-inhibition of the kinesin head domains and therefore increase activity. This may also be the structural basis for increased affinity to PRC1. However, more detailed analysis of the KIF4A ATPase cycle and single molecule measurements of movement and processivity will be needed to understand the molecular basis of this form of regulation. Although these parameters have not been measured here, the kcat of 28.2 s−1 for nonphosphorylated KIF4A assuming tight coupling of stepping to ATP turnover suggests a speed of 0.4 µm/s, consistent with axonemal gliding assays showing in vitro movement of 0.2 µm/s (Sekine et al., 1994). The increased ATPase activity seen after phosphorylation may therefore reflect increased processivity. This would increase the chance that after encountering a microtubule, KIF4A will reach the microtubule plus-end and suppress further microtubule growth or shrinkage.
To our knowledge, this is the first description of an activating role for Aurora B in the spatial and temporal regulation of a kinesin-like protein. Previous studies have implicated Aurora B in the inhibition of the microtubule-destabilizing kinesin-like protein MCAK important for destabilizing incorrect microtubule attachments to kinetochores during mitotic and meiotic spindle formation (Walczak et al., 1996, 2002; Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Tanenbaum et al., 2011). Interestingly, a recent report indicates that KIF4A may work together with MCAK to promote chromosome congression (Wandke et al., 2012), and our work hints that Aurora B may be a factor coordinating the relative activities of these two motors. Influential studies on the regulation of the KIF11/Eg5 motor protein required for centrosome separation during spindle formation have shown that Cdk1 phosphorylation at a conserved threonine residue is required for its spindle targeting and function (Blangy et al., 1995; Sawin and Mitchison, 1995), and thereby acts as a temporal control mechanism. Activating phosphorylations may therefore be a general mechanism controlling the temporal and spatial activity of kinesin-like motor proteins. However, neither in the case of KIF11/Eg5 nor KIF4A is this completely understood, and further structural studies will be necessary to validate these ideas.
Regulation of central spindle formation and size
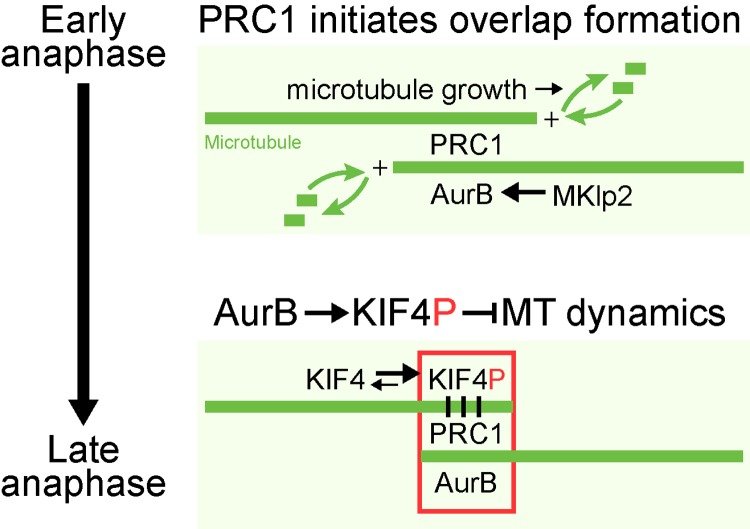
These results, taken together with previous findings, add to an emerging picture of how microtubule function at the central spindle is regulated in anaphase (Fig. 10). High Cdk1 and spindle checkpoint activity in metaphase cells is important for inhibiting the activity of PRC1 and other components of the central spindle (Mollinari et al., 2002; Mishima et al., 2004; Hümmer and Mayer, 2009; Lee et al., 2010; Vázquez-Novelle and Petronczki, 2010). Once the spindle checkpoint is satisfied, Cdk1 activity falls and PRC1 can start to organize anti-parallel microtubule bundles (Subramanian et al., 2010; Elad et al., 2011). PRC1 will therefore fail to promote microtubule organization in metaphase cells with high levels of Cdk1–cyclin B activity (Jiang et al., 1998; Mollinari et al., 2002; Zhu et al., 2006; Neef et al., 2007). Other factors not shown in the model, including the centralspindlin complex, also contribute to the formation of this anti-parallel overlapped structure and the formation of the midbody (Sellitto and Kuriyama, 1988; Nislow et al., 1992; Mishima et al., 2002; Douglas et al., 2010). KIF4A is then crucial for ensuring that these microtubule-stabilizing and -organizing factors are opposed, and a microtubule structure of the required size is created (Bringmann et al., 2004; Bieling et al., 2010; Hu et al., 2011). Here we explain how the activity of KIF4A in suppressing microtubule dynamics is restricted to the central spindle by a conserved activating phosphorylation at threonine 799 performed by the Aurora B kinase. Aurora B is delivered to the central spindle due to the activity of another kinesin-like motor protein, MKlp2, a process inhibited before the start of anaphase by Cdk1 and the spindle checkpoint machinery (Hümmer and Mayer, 2009; Vázquez-Novelle and Petronczki, 2010). Our data highlight the importance of the Aurora B chromosomal passenger complex for the correct ordering of events in anaphase after Cdk1 inactivation. It has been proposed that relocation of Aurora B away from the centromeres primarily ensures the spindle checkpoint remains silent upon sister chromatid separation once tension is lost, a condition that might reactivate the checkpoint if Aurora B remained active at the centromeres (Mirchenko and Uhlmann, 2010; Vázquez-Novelle and Petronczki, 2010). However, as shown here, human cell lines with both chromatin and central spindle–targeted Aurora B enter anaphase and carry out anaphase-specific phosphorylations (Fig. 2 C and Fig. 9, A–D). This suggests that the primary reason Aurora B relocates from the centromeres at the onset of anaphase is to ensure proper regulation of anaphase spindle dynamics by KIF4A and MKlp1 (Gruneberg et al., 2004; Guse et al., 2005; Fuller et al., 2008; Douglas et al., 2010). Together, these findings help explain how mitotic kinases and kinesin motor proteins work together in the temporal and spatial control of mitosis and cytokinesis.
Figure 10.
A model for Aurora B regulation of central spindle size. In early anaphase, PRC1 promotes the formation of anti-parallel microtubule overlaps at the central spindle. At the same time Aurora B is transported to these overlap structures by Mklp2, establishing a local high concentration of Aurora B. Phosphorylated KIF4A (KIF4P) suppresses plus-end microtubule dynamics, and thereby limits growth of the central spindle. Motor activity of kinesin family proteins depends on its ability to alternate between ATP-bound and ADP-bound forms using its ATPase activity. The two forms allow kinesins to cycle between high affinity and low affinity toward microtubules, resulting in its movement along microtubules over time (Friel and Howard, 2012). The processivity of a motor (number of steps a motor takes upon encounter with a microtubule) is directly dependent on its ATPase activity (Vmax) as well as its affinity toward microtubules (Kapp MT). Phosphorylation of KIF4A by Aurora B increases its ATPase activity by ∼5,000 fold compared with its ATPase activity in the absence of microtubules. However, this increase in ATPase activity is not accompanied by increased affinity toward the microtubules. Alternatively, Aurora B mediated phosphorylation may enhance the processivity of KIF4A. Concentration of Aurora B activity at the central spindle during anaphase would allow increased processivity of KIF4A specifically at the central spindle, allowing the motor to preferentially reach microtubule plus-ends and suppress microtubule dynamics in this region.
Materials and methods
Reagents and antibodies
General laboratory chemicals and reagents were obtained from Sigma-Aldrich and Thermo Fisher Scientific. Sheep antibodies were raised to MKlp1 (aa 1–444), KIF4A (full-length protein expressed in insect cells), and MKlp2 (aa 63–193) (Neef et al., 2003, 2007; Bastos and Barr, 2010). Rabbit antibodies were raised to full-length recombinant PRC1, PRC1 pT602 peptide CILNS(pT)NIQS, KIF4A (aa 738–1323), KIF4A (aa 738–1232), KIF4A pT799 peptide LRRR(pT)FSLT, and MKlp1 pS911 peptide CRKRR(pS)STVA (Neef et al., 2003, 2006, 2007; Gruneberg et al., 2006). Specific antibodies were purified using the antigens conjugated to Affigel-15, eluted with 0.2 M glycine-HCl, pH 2.8, then dialyzed against PBS before storage at −80°C. Commercially available antibodies were used to α-tubulin (mouse DM1A; Sigma-Aldrich), Plk1 (mouse sc-17783; Santa Cruz Biotechnology, Inc.), PRC1 pT481 (sc-11768, Santa Cruz Biotechnology, Inc.; and EP1514Y, Epitomics, Inc.), the RGS-His tag epitope (QIAGEN), AIM1 (mouse 611083; BD), and cyclin B1 (mouse GNS3; EMD Millipore). Affinity-purified primary and secondary antibodies were used at 1 µg/ml final concentration. Secondary antibodies conjugated to HRP were obtained from Jackson ImmunoResearch Laboratories, Inc. Secondary antibodies conjugated to Alexa Fluor 488, 555, and 647 were obtained from Invitrogen. DNA was stained with DAPI (Sigma-Aldrich).
Kinase inhibitors were obtained from Sigma-Aldrich (flavopiridol 5 mM, 1,000× stock), Tocris Bioscience (ZM447439 10 mM, 1,000× stock), and Axon Medchem (BI2536 1 mM, 1,000× stock). Reagents for microtubule polymerization and kinesin ATPase assays were obtained from Cytoskeleton, Inc. A benchtop microfuge (5417R; Eppendorf) was used for all centrifugations unless otherwise indicated.
Molecular biology
Human MCAK was amplified by PCR from IMAGE clone #3909438 (Source Bioscience) using KOD polymerase (Takara Bio Inc.). Human KIF4A, Aurora A and B, INCENP, TPX2, and PRC1 splice variant 2 were amplified directly from human testis cDNA as described previously (Gruneberg et al., 2004; Klein et al., 2006; Neef et al., 2007). The KIF4AT799A/S801A phosphorylation site mutant was created using QuikChange mutagenesis according to the supplied instructions (Agilent Technologies). Mammalian expression constructs for N-terminally tagged KIF4A, Aurora B, and PRC1 were made using pcDNA4/TO and pcDNA5/FRT/TO vectors (Invitrogen) modified to encode the eGFP or mCherry reading frames (Bastos and Barr, 2010). The Aaronase and Baronase modules were designed from existing crystal structures (Bayliss et al., 2003; Sessa et al., 2005) and created using standard molecular biology procedures to fuse human INCENP aa 834–897 plus an additional glycine residue to the N terminus of Aurora B aa 44–344, or TPX2 aa 1–43 plus an additional glycine residue to Aurora A 100–403, creating BamHI–XhoI fragments. These were then ligated into pQE32 for bacterial expression, and in the case of Baronase fused to the C terminus of PRC1 for mammalian or insect cell expression. Recombinant baculovirus carrying hexahistidine-tagged full-length PRC1, KIF4A, KIF4AT799A/S801A, MCAK, mCherry-PRC1, or mCherry-PRC1-Baronase cloned into the pAcSG2 vector were produced using the Baculogold system (Invitrogen). Hexahistidine-tagged bacterial expression constructs for the motor domains of KIF4A (aa 1–512) and MCAK (aa 1–592) were made in pQE32 (QIAGEN). Mutagenesis was performed using the QuikChange method (Agilent Technologies). Primers were obtained from Metabion GmbH. The siRNA duplexes used were obtained from Thermo Fisher Scientific or QIAGEN and have been described previously (Gruneberg et al., 2006; Neef et al., 2007; Bastos and Barr, 2010). Target sequences were control 5′-CGTACGCGGAATACTTCGA-3′, PRC1 5′-GGCTTCTAGGCGTGAGGAG-3′, MKlp2 5′-CCACCTATGTAATCTCATG-3′, Aurora B 5′-CGCGGCACTTCACAATTGA-3′, and 5′-CAGGTCCAGACTACTACTC-3′ for the KIF4A 3′-UTR.
Human and insect cell culture
HeLa cells were cultured in DMEM containing 10% (vol/vol) bovine calf serum (Invitrogen) at 37°C and 5% CO2. For synchronization, cells were treated for 18 h with 2 mM thymidine, washed three times in PBS, and twice with growth medium. For plasmid transfection and siRNA transfection, Mirus LT1 (Mirus Bio LLC) and Oligofectamine (Invitrogen), respectively, were used. Stable HeLa cell lines with single copies of the desired transgene were created using the T-Rex doxycycline-inducible Flp-In system (Invitrogen). Sf-9 insect cells were grown in insect cell growth medium (TC100 containing 10% [vol/vol] bovine calf serum and Glutamax [Invitrogen]) at 27°C and atmospheric CO2.
Protein expression and purification
Recombinant baculoviruses encoding hexahistidine-tagged KIF4A, KIF4AT799A/S801A PRC1, MCAK, mCherry-PRC1, or mCherry-PRC1-Baronase were used to infect 4 × 107 insect cells in 20x 15-cm dishes with a multiplicity of infection of 10. To infect cells, the medium was removed, and the virus added in 3 ml of insect growth medium per dish. Dishes were gently rocked for 1 h; every 10 min the dishes were rotated by 90 degrees. After 1 h, 17 ml insect cell growth medium was added and the cells left for 60 h before harvesting by centrifugation at 200 g. To obtain M-phase Cdk-phosphorylated PRC1, the cells were treated for 3 h with 100 nM okadaic acid before harvesting. Cell pellets were washed once in ice-cold PBS, and then lysed in 10 ml of IMAC20 (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20 mM imidazole, and 0.2% [vol/vol] Triton X-100), and protease inhibitor cocktail (Sigma-Aldrich) for 20 min on ice. A cell lysate was prepared by centrifugation at 100,000 g in a TLA100.3 rotor (Beckman Coulter), then loaded onto a 1-ml HisTrap FF column (GE Healthcare) at 0.5 ml/min. The column was then washed with 30 ml of IMAC20, and eluted with a 20-ml linear gradient from 20 to 200 mM imidazole in IMAC20 collecting 1 ml fractions. Peak fractions judged by SDS-PAGE and absorbance at 280 nm were concentrated using Ultracel-10K centrifugal filters (EMD Millipore) according to the manufacturer’s instructions to a final volume of ∼1 ml. Samples were then buffer exchanged using 5 ml Zeba Desalt Spin columns (Perbio) into TND (20 mM Tris-HCl, pH 8, 300 mM NaCl, and 1 mM DTT). Protein samples were snap-frozen in 15-µl aliquots and stored at −80°C for further use. Aurora-phosphorylated proteins were prepared by incubating 4 µg purified KIF4A or MCAK with 0.4 µg of Aurora kinase and 1 mM Mg-ATP for 20 min at 37°C, then kept on ice until required. The Aurora B minikinase fusion, and motor domains of KIF4A and MCAK were expressed in Escherichia coli strain JM109 and purified in the same way, except that cell lysis was performed using an Emulsifex C5 cell breaker system (Avestin Europe GmbH).
Protein complex isolation from anaphase cells
Three 15-cm dishes of thymidine-synchronized HeLa S3 cells per condition were released for 3 h at 37°C, 100 ng/ml nocodazole added, and incubated further for 12–14 h. Mitotic cells were collected by shake-off and nocodazole was removed by washing three times with PBS and twice with growth medium warmed to 37°C. At each time point, specified in the figures and figure legends, cells were collected and washed three times with ice-cold PBS. Cell pellets were suspended in mitotic lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% [vol/vol] IGEPAL, 0.1% [wt/vol] sodium deoxycholate, 40 mM β-glycerophosphate, 10 mM NaF, 0.3 mM Na-vanadate, 100 nM okadaic acid, and protease and phosphatase inhibitor cocktails [Sigma-Aldrich]), left for 15 min on ice, and then clarified by centrifugation at 20,000 gav for 20 min at 4°C. Protein complexes were isolated from 3 mg of cell lysate using 3 µg rabbit antibodies against either GST as a matched negative control, or PRC1 bound to 20 µl protein A Sepharose by incubation for 2 h at 4°C. Isolated complexes were washed once with lysis buffer, then three times with 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% (vol/vol) IGEPAL. Samples were then analyzed by SDS-PAGE and Western blotting.
Mass spectrometry of protein complexes
Protein samples for mass spectrometry were separated on 4–12% gradient NuPAGE gels, then stained using a colloidal Coomassie blue stain. Gel lanes were cut into 12 slices, and then digested with trypsin (Wilm et al., 1996). The resulting tryptic peptide mixtures in 0.05% trifluoracetic acid were then analyzed by online LC-MS/MS with a nanoAcquity UPLC (Waters) and Orbitrap XL ETD mass spectrometer (Thermo Fisher Scientific) fitted with a Proxeon nano-electrospray source. Peptides were loaded onto a 5-cm × 180-µm BEH-C18 Symmetry trap column (part no. 186003514; Waters) in 0.1% (volv/vol) formic acid at 15 µl/min, and then resolved using a 25-cm × 75-µm BEH-C18 column (part no. 186003815; Waters) in 99–37.5% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid at a flow rate of 400 nl/min. The mass spectrometer was set to acquire an MS survey scan in the Orbitrap (R = 30,000) and then perform MS/MS on the top five ions in the linear quadrupole ion trap after fragmentation using collision ionization (30 ms, 35% energy). A 90-s rolling exclusion list with n = 3 was used to prevent redundant analysis of the same ions. Maxquant and Mascot (Matrix Science) were then used to compile and search the raw data against the human IPI database (Cox and Mann, 2008; Cox et al., 2009). Protein group and peptide lists were sorted in Excel (Microsoft). MS and MS/MS spectra were manually inspected using Xcalibur Qualbrowser (Thermo Fisher Scientific).
Fixed- and live-cell microscopy
For fixed-cell imaging samples were prepared and imaged exactly as described previously (Bastos et al., 2012). In brief, cells grown on no. 1.5 glass coverslips (Menzel-Gläser; Thermo Fisher Scientific) were washed twice with 2 ml of PBS, and fixed with 2 ml of 3% (wt/vol) paraformaldehyde in PBS for 15 min. Fixative was removed and the cells quenched with 2 ml of 50 mM NH4Cl in PBS for 10 min. Coverslips were washed three times in 2 ml PBS before permeabilization in 0.2% (vol/vol) Triton X-100 for 5 min. In all cases primary and secondary antibody staining was performed in PBS for 60 min at room temperature. Affinity-purified antibodies were used at 1 µg/ml, whereas commercial antibodies were used as directed by the manufacturers. DAPI was added to the secondary antibody staining solution at 0.3 µg/ml. Coverslips were mounted in Mowiol 4-88 mounting medium (EMD Millipore). Fixed samples on glass slides were imaged using a 60×/NA 1.35 oil immersion objective on an upright microscope (BX61; Olympus) with filtersets for DAPI; GFP/Alexa Fluor 488, 555, 568, and 647 (Chroma Technology Corp.); a CoolSNAP HQ2 camera (Roper Scientific); and MetaMorph 7.5 imaging software (Molecular Dynamics, Inc.). Illumination was provided by a Lumen 200-Watt metal halide light source (Prior Scientific Instruments, Ltd). Image stacks of 12–35 planes with a spacing of 0.2–0.4 µm through the cell volume were taken. Image stacks were maximum intensity projected and then merged to create 24-bit RGB TIFF files in MetaMorph. Images in 24-bit RGB format were then cropped in Photoshop CS3 and placed into Illustrator CS3 (Adobe Systems, Inc.) to produce the figures.
For live-cell imaging using spinning disk confocal microscopy, cells were plated in 35-mm dishes with a 14-mm no. 1.5 thickness coverglass window in the bottom (MatTek Corporation). For imaging, the dishes were placed in a 37°C and 5% CO2 environment chamber (Tokai Hit) on the microscope stage. Imaging was performed at 37°C in 5% CO2 using an inverted microscope (IX81; Olympus) with a 60×/1.42 NA oil immersion objective coupled to a spinning disk confocal system (Ultraview Vox; PerkinElmer) fitted with an EM-CCD camera (C9100-13; Hamamatsu Photonics). Exposure times were 30 ms for GFP-KIF4A, eGFP-Aurora B, and mCherry-PRC1 using 4% laser power. Image stacks of 24 planes spaced 0.6 µm apart were taken at 1–4 stage positions every minute for up to 6 h. A brightfield reference image was also taken to visualize cell shape. Maximum intensity projection of the fluorescent channels was performed in Volocity to create 24-bit RGB TIFF files. For FRAP prebleach, 5 image stacks of 4 planes with 0.4-µm spacing were acquired at 1-s intervals. Bleach was performed using an UltraVIEW PK Device with the 488-nm laser set at 10% with the following settings: cycles 5, step size 1, spot period 10, stop period 10, spot cycles 1, small spot size and no attenuation. Recovery images were acquired for 60 time points every second and then a further 60 time points every 2 s. Quantification and analysis of the FRAP data were performed using the Volocity FRAP plug-in. Images in 24-bit RGB TIFF format were then placed into Adobe Illustrator CS3 to produce the figures.
Microtubule polymerization assays
Standard microtubule polymerization assays were performed using porcine tubulin and a commercial fluorescence-based assay system (Cytoskeleton, Inc.). Porcine tubulin stock solution was in GTB (80 mM Pipes, pH 6.9, 2 mM MgCl2, and 0.5 mM EGTA). A tubulin master mix was created on ice from 243 µl of BP01 (80 mM Pipes-KOH, pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, and 10 µM 4’,6-diamidino-2-phenylindole), 112 µl of TGB (80 mM Pipes-KOH, pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, and 60% [vol/vol] glycerol), 85 µl of 10 mg/ml porcine tubulin, 4.44 µl of 100 mM GTP, and 4.44 µl of 100 mM ATP. Reactions were set up in 96-well plates by mixing the protein in a total volume of 7.5 µl TND on ice. Proteins were tested in the range of 50–500 nM final assay concentration. To start the assay, 50 µl of tubulin master mix was added to each well while on ice, to give a total assay volume of 57.5 µl. Final assay composition was: 80 mM Pipes-KOH, pH 6.9, 39 mM NaCl, 2 mM MgCl2, 0.5 mM EGTA, 0.87 mM GTP, 0.87 mM ATP, 13% (vol/vol) glycerol, and 2.2 µM 4’,6-diamidino-2-phenylindole. This was then rapidly transferred to a plate reader (Tristar LB 941; Berthold Technologies) set to 340 nm excitation and 420 nm emission, and 37°C. Readings were acquired every minute for one hour. Because reactions were pipetted on ice, condensation on the plates formed as they were transferred into the plate reader and interfered with the measurements. After an initial period of equilibration after which the plate comes to the set temperature and this condensation clears (defined as t = 0 min), values were plotted as a function of time.
Microtubule pelleting assays
Taxol-stabilized microtubules were produced by incubating 45 µl 7.4 mg/ml porcine tubulin, 4.5 µl TGB, and 0.55 µl 100 mM GTP at 37°C for 20 min. To stabilize the microtubules, 8 µl of 2 mM paclitaxel in 750 µl GTB was added to the microtubule polymerization reaction. This tubulin stock mix (3.75 µM) was stored at room temperature until further use. Proteins to be tested were diluted in GTB to give the final concentrations defined in the figure legends. For experiments involving titration, 3.125–100 nM of either KIF4A or KIF4A prephosphorylated by Aurora B was incubated with 175 nM polymerized tubulin to give a final volume of 80 µl 80 mM Pipes-KOH, pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, and 20 µM paclitaxel. Samples were centrifuged for 10 min at 80,000 rpm in a TLA-100 rotor. The supernatant was removed and mixed with 20 µl 5× SDS-PAGE sample buffer. The pellet was resuspended in 100 µl of 1× SDS-PAGE sample buffer. Equal amounts of both supernatant and pellet fractions were analyzed by SDS-PAGE and Western blotting.
Microtubule destabilization assays
Taxol-stabilized microtubules were produced by incubating 45 µl 7.4 mg/ml porcine tubulin, 4.5 µl TGB, and 0.55 µl 100 mM GTP at 37°C for 20 min. To stabilize the microtubules, 8 µl of 2 mM paclitaxel in 750 µl GTB was added to the microtubule polymerization reaction. This 3.75 µM tubulin stock mix was stored at room temperature until further use. An 80-µl reaction mixture was assembled containing 1.4 µM polymerized microtubules, 25 µg/ml BSA, 250 nM MCAK, or 200 nM KIF4A full-length proteins or motor domains. For some reactions MCAK and KIF4A were prephosphorylated using Aurora B. Final buffer composition was 80 mM Pipes, pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, and 9 µM paclitaxel. The reaction mix was incubated in the absence or presence of 1 mM ATP added from a 100-mM stock for 20 min at 37°C. Samples were centrifuged and analyzed as described for microtubule pelleting assays.
Kinesin motor ATPase assays
A commercial enzyme–linked inorganic phosphate assay was used to measure kinesin ATPase activity (Cytoskeleton, Inc.). In brief, a microtubule premix was created at room temperature by mixing 1 ml of reaction buffer (15 mM Pipes-KOH, pH 7, and 5 mM MgCl2), 10 µl of 2 mM paclitaxel, 80 µl of preassembled microtubules (1 mg/ml tubulin, 15 mM Pipes-KOH, pH 7, 5 mM MgCl2, 1 mM GTP, and 20 µM paclitaxel), 240 µl of 1 mM 2-amino-6-mercapto-7-methylpurine riboside, and 12 µl of 0.1 U/µl purine nucleoside phosphorylase. Phosphorylated KIF4FL or KIF4MD were prepared by preincubating the motor protein with 10 nM of either wild-type Aurora kinase or the inactive “kinase-dead” mutant in a final volume of 7.5 µl TND containing 1 mM Mg-ATP for 30 min at 30°C. Reactions were set up in 96-well plates by mixing the protein of interest in a total volume of 7.5 µl TND with 147.5 µl of the microtubule premix at room temperature. To start the assay, 10 µl of 10 mM ATP was added to each well. Final assay volume was 165 µl of 12 mM Pipes-KOH, pH 7, 4 mM MgCl2, 0.61 mM ATP, and 14.5 mM NaCl. This was then rapidly transferred to a 37°C plate reader (Tristar LB 941; Berthold Technologies) set to read absorbance at 360 nm. Readings were acquired every 30 s for 1 h. An inorganic phosphate standard curve was created in the same assay buffer and used to convert absorbance to nmol ATP hydrolyzed.
Dynamic microtubule overlap assays
Porcine brain tubulin was purified and used to produce double-cycled GMPCPP-stabilized microtubule seeds using published methods (Castoldi and Popov, 2003; Gell et al., 2010). In brief, 10 µl of 2 µM biotin-labeled tubulin (Cytoskeleton, Inc.), 2 µM HiLyte-488 tubulin (Cytoskeleton, Inc.), 16 µM unlabeled tubulin in BRB80 (80 mM Pipes-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA), and 1 mM Mg-GMPCPP (Jena Bioscience) were incubated for 30 min at 37°C and pelleted for 10 min at 80,000 rpm in a TLA-100 rotor. The microtubule seeds were resuspended in 7.2 µl of ice-cold BRB80 and incubated on ice for 20 min to promote disassembly. The once-cycled tubulin was then supplemented with 1 mM GMP-CPP, incubated for 30 min at 30°C, and pelleted. The double-cycled microtubule seeds were resuspended in 100 µl BRB80 and stored in the dark at 37°C. Imaging chambers were prepared using silanized coverslips separated by double-sided tape (Gell et al., 2010). To immobilize biotinylated microtubule seeds, imaging chambers were first incubated with 50 µg/ml Neutravidin (Thermo Fisher Scientific) at room temperature for 5 min followed by 1% (vol/vol) Pluronic F127 (Sigma-Aldrich) at room temperature for 5 min. Biotinylated, GMPCPP-stabilized microtubule seeds were then bound to the imaging chambers by flowing the microtubule seeds through the chambers and incubating for 15 min at 37°C. KIF4A was phosphorylated in vitro by incubating 445 nM KIF4A with 55 nM Baronase and 1 mM ATP for 30 min at 30°C. For experiments where the effect of KIF4A was studied to compare with phosphorylated KIF4, 445 nM of KIF4A was incubated with 1 mM ATP for 30 min at 30°C in the absence of Baronase. Microtubule growth was initiated by flowing in tubulin reaction mixture containing 17.5 µM unlabeled tubulin, 1 mM GTP, 1 mM ATP, and 14 nM mCherry-PRC1 to fill the chamber. KIF4A or in vitro–phosphorylated KIF4A was added to the tubulin reaction mixture to study its effect on microtubule overlaps as defined in the figure legends. The imaging chambers were sealed with nail polish and immediately placed on the spinning disk confocal microscope for imaging. Imaging was performed at 37°C using an inverted microscope (IX81; Olympus) with a 60×/1.42 NA oil immersion objective coupled to a spinning disk confocal system (Ultraview Vox; PerkinElmer) fitted with an EM-CCD camera (C9100-13; Hamamatsu Photonics). The temperature of the objective was maintained at 37°C throughout the experiment. Exposure times were 30 ms for HiLyte-488 tubulin and mCherry-PRC1 using 4% laser power. The images were taken every 10 s for 20 min. The ability of PRC1-Baronase to form anti-parallel microtubule overlaps was studied by replacing mCherry-PRC1 with mCherry-PRC1-Baronase in the tubulin reaction mixture in the absence and presence of 10 µM ZM447439.
Single-microtubule polymerization assay
Flow chambers were prepared as for the dynamic microtubule overlap assay. The tubulin reaction mixture was prepared using 15.75 µM tubulin, 1.75 µM Hilyte-488 tubulin, 1 mM GTP, 1 mM ATP, anti-blinking cocktail (500 nM glucose oxidase, 64 nM catalase, 40 mM d-glucose, 1% [vol/vol] β-mercaptoethanol), and 0.1% methyl cellulose (4000 cP, Sigma-Aldrich). KIF4A or in vitro–phosphorylated KIF4A at the concentrations defined in the figure legends were added to the tubulin reaction mixture to study their effects on microtubule dynamics. Imaging chambers were filled with tubulin reaction mixture and then sealed with nail polish. TIRF imaging was performed at 30°C with a super resolution microscope (OMX V3; Applied Precision) using the 60×/1.49 NA oil immersion objective and an sCMOS camera (PCO AG). The images were taken using a 488-nm laser and the 10% neutral density filter every 3 s for 10 min with a 50-ms exposure time.
Online supplemental material
Fig. S1 shows additional characterization of the eGFP-PRC1 and KIF4A stable cell lines. Fig. S2 confirms the specificity of the KIF4A pT799 antibody. Figs. S3 and S4 show the effects of KIF4A and MCAK with and without Aurora B phosphorylation on microtubule binding and microtubule destabilization. Fig. S5 describes the characterization of the Baronase module. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201301094/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201301094.dv.
Supplementary Material
Acknowledgments
We thank Ian Dobbie (Micron Advanced Bioimaging Unit) for technical assistance with TIRF microscopy.
This study was funded by a Cancer Research UK program award (C20079/A9473) to F.A. Barr. U. Gruneberg was supported by a Cancer Research UK career development fellowship (C24085/A8296). E.A. Nigg acknowledges supports from the Swiss National Science Foundation and the Swiss Cancer League. A Royal College of Surgeons/Freemasons Research Fellowship and the Liverpool CRUK center support R.D. Baron. Parts of this work originated when the authors were located at the Max Planck Institute of Biochemistry, Martinsried, Germany and supported by the Max Planck Society.
Footnotes
Abbreviation used in this paper:
- Plk1
- Polo-like kinase 1
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Steinmetz M.O. 2010. Microtubule +TIPs at a glance. J. Cell Sci. 123:3415–3419 10.1242/jcs.062414 [DOI] [PubMed] [Google Scholar]
- Alexander J., Lim D., Joughin B.A., Hegemann B., Hutchins J.R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E.A., et al. 2011. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4:ra42 10.1126/scisignal.2001796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Bastos R.N., Barr F.A. 2010. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J. Cell Biol. 191:751–760 10.1083/jcb.201008108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R.N., Penate X., Bates M., Hammond D., Barr F.A. 2012. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J. Cell Biol. 198:865–880 10.1083/jcb.201204107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R., Sardon T., Vernos I., Conti E. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bieling P., Telley I.A., Surrey T. 2010. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 142:420–432 10.1016/j.cell.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Blangy A., Lane H.A., d’Hérin P., Harper M., Kress M., Nigg E.A. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 83:1159–1169 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Bringmann H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., Surrey T. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522 10.1126/science.1094838 [DOI] [PubMed] [Google Scholar]
- Castoldi M., Popov A.V. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. 2009. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4:698–705 10.1038/nprot.2009.36 [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- Douglas M.E., Davies T., Joseph N., Mishima M. 2010. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr. Biol. 20:927–933 10.1016/j.cub.2010.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert U.S., Mitchison T.J., Field C.M. 2006. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75:543–566 10.1146/annurev.biochem.74.082803.133425 [DOI] [PubMed] [Google Scholar]
- Elad N., Abramovitch S., Sabanay H., Medalia O. 2011. Microtubule organization in the final stages of cytokinesis as revealed by cryo-electron tomography. J. Cell Sci. 124:207–215 10.1242/jcs.073486 [DOI] [PubMed] [Google Scholar]
- Fabbro M., Zhou B.B., Takahashi M., Sarcevic B., Lal P., Graham M.E., Gabrielli B.G., Robinson P.J., Nigg E.A., Ono Y., Khanna K.K. 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell. 9:477–488 10.1016/j.devcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Friel C.T., Howard J. 2012. Coupling of kinesin ATP turnover to translocation and microtubule regulation: one engine, many machines. J. Muscle Res. Cell Motil. 33:377–383 10.1007/s10974-012-9289-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C., Bormuth V., Brouhard G.J., Cohen D.N., Diez S., Friel C.T., Helenius J., Nitzsche B., Petzold H., Ribbe J., et al. 2010. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 95:221–245 10.1016/S0091-679X(10)95013-9 [DOI] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E.A., Barr F.A. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167–172 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Li X., Chan E.H., Chalamalasetty R.B., Nigg E.A., Barr F.A. 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172:363–372 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Mishima M., Glotzer M. 2005. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 15:778–786 10.1016/j.cub.2005.03.041 [DOI] [PubMed] [Google Scholar]
- Hackney D.D. 2007. Jump-starting kinesin. J. Cell Biol. 176:7–9 10.1083/jcb.200611082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J., Hyman A.A. 2009. Growth, fluctuation and switching at microtubule plus ends. Nat. Rev. Mol. Cell Biol. 10:569–574 10.1038/nrm2713 [DOI] [PubMed] [Google Scholar]
- Hu C.K., Coughlin M., Field C.M., Mitchison T.J. 2011. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21:815–824 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.K., Ozlü N., Coughlin M., Steen J.J., Mitchison T.J. 2012. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell. 23:2702–2711 10.1091/mbc.E12-01-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümmer S., Mayer T.U. 2009. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 19:607–612 10.1016/j.cub.2009.02.046 [DOI] [PubMed] [Google Scholar]
- Jiang W., Jimenez G., Wells N.J., Hope T.J., Wahl G.M., Hunter T., Fukunaga R. 1998. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol. Cell. 2:877–885 10.1016/S1097-2765(00)80302-0 [DOI] [PubMed] [Google Scholar]
- Kaan H.Y., Hackney D.D., Kozielski F. 2011. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 333:883–885 10.1126/science.1204824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach A.N., Schweppe D.K., Faherty B.K., Pechenick D., Pletnev A.A., Gerber S.A. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4:rs5 10.1126/scisignal.2001497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U.R., Nigg E.A., Gruneberg U. 2006. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol. Biol. Cell. 17:2547–2558 10.1091/mbc.E05-12-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y., Earnshaw W.C., Mochizuki Y., Dohmae N., Todokoro K. 2004. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23:3237–3248 10.1038/sj.emboj.7600347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Lee S.H., McCormick F., Saya H. 2010. Mad2 inhibits the mitotic kinesin MKlp2. J. Cell Biol. 191:1069–1077 10.1083/jcb.201003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.M., Kim W. 2004. Kinesin superfamily protein member 4 (KIF4) is localized to midzone and midbody in dividing cells. Exp. Mol. Med. 36:93–97 10.1038/emm.2004.13 [DOI] [PubMed] [Google Scholar]
- Mirchenko L., Uhlmann F. 2010. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr. Biol. 20:1396–1401 10.1016/j.cub.2010.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54 10.1016/S1534-5807(01)00110-1 [DOI] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Grüneberg U., Nigg E.A., Glotzer M. 2004. Cell cycle regulation of central spindle assembly. Nature. 430:908–913 10.1038/nature02767 [DOI] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.P., Jiang W., Schoehn G., Hunter T., Margolis R.L. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175–1186 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Preisinger C., Sutcliffe J., Kopajtich R., Nigg E.A., Mayer T.U., Barr F.A. 2003. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162:863–875 10.1083/jcb.200306009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Klein U.R., Kopajtich R., Barr F.A. 2006. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr. Biol. 16:301–307 10.1016/j.cub.2005.12.030 [DOI] [PubMed] [Google Scholar]
- Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., Barr F.A. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Niiya F., Tatsumoto T., Lee K.S., Miki T. 2006. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene. 25:827–837 10.1038/sj.onc.1209124 [DOI] [PubMed] [Google Scholar]
- Nislow C., Lombillo V.A., Kuriyama R., McIntosh J.R. 1992. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 359:543–547 10.1038/359543a0 [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T.J. 2004. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 15:2895–2906 10.1091/mbc.E04-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlü N., Monigatti F., Renard B.Y., Field C.M., Steen H., Mitchison T.J., Steen J.J. 2010. Binding partner switching on microtubules and aurora-B in the mitosis to cytokinesis transition. Mol. Cell. Proteomics. 9:336–350 10.1074/mcp.M900308-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavicic-Kaltenbrunner V., Mishima M., Glotzer M. 2007. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol. Biol. Cell. 18:4992–5003 10.1091/mbc.E07-05-0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Glotzer M., Kraut N., Peters J.M. 2007. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 12:713–725 10.1016/j.devcel.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Samejima K., Samejima I., Vagnarelli P., Ogawa H., Vargiu G., Kelly D.A., de Lima Alves F., Kerr A., Green L.C., Hudson D.F., et al. 2012. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J. Cell Biol. 199:755–770 10.1083/jcb.201202155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A., Neef R., Eberspächer U., Eis K., Husemann M., Mumberg D., Prechtl S., Schulze V., Siemeister G., Wortmann L., et al. 2007. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell. 18:4024–4036 10.1091/mbc.E07-05-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., Mitchison T.J. 1995. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA. 92:4289–4293 10.1073/pnas.92.10.4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Okada Y., Noda Y., Kondo S., Aizawa H., Takemura R., Hirokawa N. 1994. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J. Cell Biol. 127:187–201 10.1083/jcb.127.1.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto C., Kuriyama R. 1988. Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J. Cell Biol. 106:431–439 10.1083/jcb.106.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., Musacchio A. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Subramanian R., Wilson-Kubalek E.M., Arthur C.P., Bick M.J., Campbell E.A., Darst S.A., Milligan R.A., Kapoor T.M. 2010. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 142:433–443 10.1016/j.cell.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macurek L., van der Vaart B., Galli M., Akhmanova A., Medema R.H. 2011. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr. Biol. 21:1356–1365 10.1016/j.cub.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Vázquez-Novelle M.D., Petronczki M. 2010. Relocation of the chromosomal passenger complex prevents mitotic checkpoint engagement at anaphase. Curr. Biol. 20:1402–1407 10.1016/j.cub.2010.06.036 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Mitchison T.J., Desai A. 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 84:37–47 10.1016/S0092-8674(00)80991-5 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Gan E.C., Desai A., Mitchison T.J., Kline-Smith S.L. 2002. The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr. Biol. 12:1885–1889 10.1016/S0960-9822(02)01227-7 [DOI] [PubMed] [Google Scholar]
- Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., et al. 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 198:847–863 10.1083/jcb.201110060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 379:466–469 10.1038/379466a0 [DOI] [PubMed] [Google Scholar]
- Wittmann T., Hyman A., Desai A. 2001. The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 3:E28–E34 10.1038/35050669 [DOI] [PubMed] [Google Scholar]
- Wolfe B.A., Takaki T., Petronczki M., Glotzer M. 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7:e1000110 10.1371/journal.pbio.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Jiang W. 2005. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA. 102:343–348 10.1073/pnas.0408438102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J.D., Bossy-Wetzel E., Fan J.B., Abraham R.T., Jiang W. 2005. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 16:3187–3199 10.1091/mbc.E05-02-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Lau E., Schwarzenbacher R., Bossy-Wetzel E., Jiang W. 2006. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc. Natl. Acad. Sci. USA. 103:6196–6201 10.1073/pnas.0506926103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.