Abstract
Interpolation has become a default operation in image processing and medical imaging and is one of the important factors in the success of an intensity-based registration method. Interpolation is needed if the fractional unit of motion is not matched and located on the high resolution (HR) grid. The purpose of this work is to present a systematic evaluation of eight standard interpolation techniques (trilinear, nearest neighbor, cubic Lagrangian, quintic Lagrangian, hepatic Lagrangian, windowed Sinc, B-spline 3rd order, and B-spline 4th order) and to compare the effect of cost functions (least squares (LS), normalized mutual information (NMI), normalized cross correlation (NCC), and correlation ratio (CR)) for optimized automatic image registration (OAIR) on 3D spoiled gradient recalled (SPGR) magnetic resonance images (MRI) of the brain acquired using a 3T GE MR scanner. Subsampling was performed in the axial, sagittal, and coronal directions to emulate three low resolution datasets. Afterwards, the low resolution datasets were upsampled using different interpolation methods, and they were then compared to the high resolution data. The mean squared error, peak signal to noise, joint entropy, and cost functions were computed for quantitative assessment of the method. Magnetic resonance image scans and joint histogram were used for qualitative assessment of the method.
1. Introduction
1.1. Interpolation
One of the most important parts of designing a registration algorithm is choosing a good interpolation function in order to increase the accuracy of registration. Also, Interpolation is required if the fractional unit of the motion is not matched and located on high resolution (HR) grid. One of the ways by which we can help physicians in coming up with a better diagnosis and treatment is improving the resolution of images. One scheme for the interpolation step is shown in Figure 1. Here, a circle shows the reference HR image, and a diamond and a triangle represent a shifted HR pixel. For instance, if the image is downsampled by a factor of 4, a diamond has (0.25, 0.25) subpixel shift for the vertical and horizontal directions and a triangle has a shift that is less than (0.25, 0.25). In Figure 1, a triangle is not placed on the HR grid and it needs interpolation, but a diamond does not need interpolation.
Figure 1.
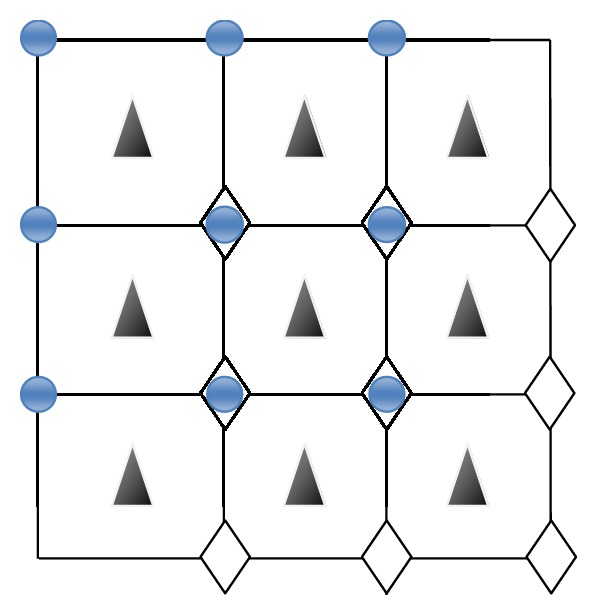
Scheme for interpolation. Straight line shows the original HR grid, circle shows the reference HR pixels, and a diamond and a triangle are shifted version of HR pixels.
Therefore, some interpolation approaches are proposed to overcome the problem of low resolution in medical imaging. Magnetic resonance imaging (MRI) is an invaluable modality in the medical field. Particularly, neuroimaging with MRI helps physicians to study the internal structure and functionality of the human brain. In these cases, high resolution and isotropic images are important because higher isotropic resolution could theoretically reduce partial volume artifacts, leading to better accuracy/precision in deriving volumetric measurement and decreasing considerable errors in the registration [1]. Clinically, acquiring a fully isotropic 3D image set is not feasible because of time, motion artifacts, and PSNR factors. Thus, typically, in 3D MR data, the in-plane direction has higher resolution than the slice direction (Z-axis). In this case, invaluable information will be lost in the latter direction. Our objective is to recover and fill in this missing information in order to enable the physicians to have a more accurate perspective of the underlying structure available in the data by optimizing the choice of interpolation techniques.
The study of interpolation approaches dates back to the 1980s [2]. In which a great diversity of techniques can be found in the literature. For example, B-splines were sometimes referred to as cubic splines [3], whereas cubic interpolation was also known as cubic convolution [4–6] and as high resolution spline interpolation [2]. Eight interpolation algorithms are reviewed in the following sections. We first present cubic Lagrangian, quintic Lagrangian, and heptic Lagrangian. Then, we explain a nearest neighbor interpolation approach which is associated with strong aliasing and blurring effect. Next, discussions of the trilinear interpolation approach as well as B-spline 3rd order, B-spline 4th order, and windowed Sinc are explained. Finally, we discuss and evaluate the performance of these interpolation algorithms in order to find the best interpolation method for upsampling of 3D MR images. Different 2D interpolation approaches exist in medical imaging [4]. However, in this paper, we compare the performance (quality and quantity) of eight common interpolation approaches on 3D data.
1.1.1. Lagrange Interpolation
Lagrange interpolation is a famous, classical technique for interpolation. The Lagrange interpolation is a way to pass a kernel of degree N − 1 through N × N points and is defined in X-direction (for 2D image, adds Y-direction, and for 3D image adds Y- and Z-directions) [5, 6, 8–13],
| (1) |
where i = j − (N/2) + 1 and n ∈ {−(N/2) + 1, −(N/2) + 2,…, N/2} are the Lagrange kernels. The Lagrange kernel for N = 1 equals the nearest neighbor interpolation. In this case, N = 2 equals the linear interpolation. The Lagrange kernels for N = 4 and N = 5 supporting points result in cubic and quartic polynomials, respectively, and are shown as follows
| (2) |
1.1.2. Nearest Neighbor Interpolation
Nearest neighbor interpolation (also known as zero-order interpolation) is the simplest method, and strong aliasing and blurring effects are associated with this interpolation [14]. The local 1-point Lagrange interpolation is equivalent to the nearest neighbor interpolation, defined by
| (3) |
The images when scaled up in size may look very blocky. Likewise, the local 2-point Lagrange interpolation is equivalent to the linear interpolation, defined by
| (4) |
1.1.3. Trilinear Interpolation
Trilinear interpolation calculates values placed between existing voxel values by linearly weighting the eight closest neighboring values. In other words, trilinear is the name given to the process of linearly interpolating points within a 3D box, given the values at the vertices of the box (see Figure 2) [15].
Figure 2.
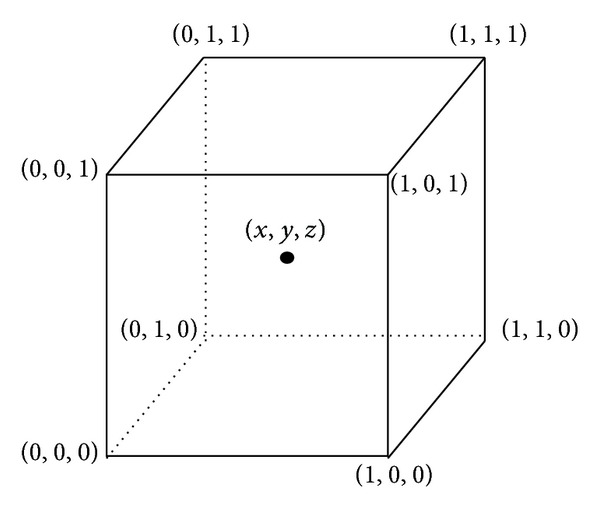
Trilinear interpolation computes values located between existing voxel values by linearly weighting the eight closest neighboring values (obtained from National Institutes of Health Center for Information Technology, Rockville, MD, USA).
The known values at each vertex are indicated as V000, V100, V010,…, V111, and the unknown value is calculated by merging the known corner values weighted by their distance from the point (x,y, z) within the cube.
1.1.4. B-Spline Interpolation
B-spline interpolation uses weighted voxel values in a wider neighborhood compared to trilinear interpolation, but both the B-spline and trilinear kernels are symmetrical and separable. The place of the neighboring points as control points relates to B-spline interpolation and combines the intensity values at these places using a set of polynomial basis according to (5) [16].
Equation (5) shows k-order B-spline with n + 1 control points (P1, P2,…, Pn),
| (5) |
In (5), N i,k are the polynomial functions of order k (degree k − 1), and n is the number of control points; k must be at least 2 (linear) and less than n + 1.
P(t) is validly defined for t min ≤ t < t max, where t min = t k and t max = t n+2. A knot vector (t 1, t 2,…, t k+(n+1)) must be determined. This specifies the values of t at which the pieces of curve join, like knots joining bits of string. It is important to note that the degree of the weighting polynomial (the order of the curve) is not dependent on the number of control points, n [17].
The weighting polynomial can be recursively defined by the following equation [18]:
| (6) |
In (6), t(i) represents an index that refers to the control points and t(i) are generally referred to as knot points (see Figure 3). The series of control point is defined as a control surface. This indexing scheme allows one to weight different control points more than other control points by using it once during the computation. Typically, the first and last control points are weighted more heavily than the internal points to give a smooth interpolating curve. Generally, the shape of the curve (N i,k) is specified by the relative spacing between the knots (t 0, t 1,…, t n). The sequence (t 0, t 1,…, t n) is called knot vector. Knot vectors are generally placed into one of three categories: uniform, nonuniform, and open uniform. Uniform knot vectors are the vectors for which t i+1 − t i = const, for example, [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]. Nonuniform knot vectors are a general case; the only constraint is the standard t i ≤ t i+1, for example, [0.2, 05, 0.8, 0.8, 0.8, 1.1, 1.1, 2.2, 2.7, 3.4]. The optimized automatic image registration (OAIR) method uses the open uniform knot vectors for computing B-spline. Open uniform knot vectors are uniform knot vectors which have k-equal knot values at each end as [3, 5, 9, 16–19]
| (7) |
for example, [0, 0, 0, 1, 2, 3, 4, 4, 4] (here k = 3, n = 5).
Figure 3.
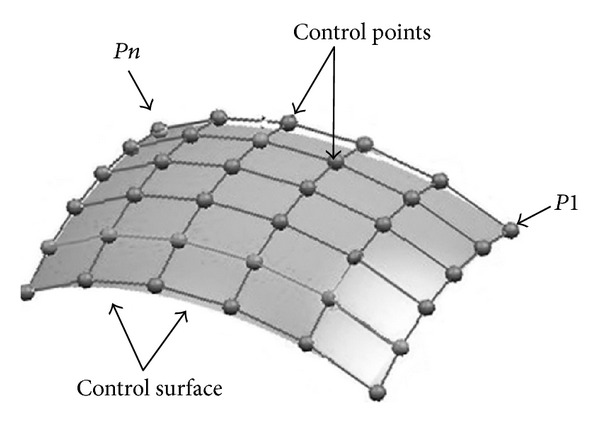
B-spline interpolation. There are n control points (P1, P2,…, Pn). The sequence of the control point is called a control surface (adapted from National Institutes of Health Center for Information Technology, Rockville, MD, USA).
1.1.5. Windowed Sinc Interpolation
This interpolation function has minimum aliasing artifacts in contrast to linear interpolation. Sinc function can be windowed more generally to yield [5, 10] the following
| (8) |
Think of an image data set comprising a 3D matrix voxel with intensities I(x, y, z), specified by integer position coordinates (x, y, z). If one wants to calculate the intensity value at an interior point defined by noninteger coordinates (x, y, z), this can be obtained by the following equation [5]:
| (9) |
For satisfying (9), two limiting conditions are required:
I(x, y, z) must be band limited. Put differently, the function must have Fourier transform F{I(x, y, z)} = I(f) = 0 for |f| > B for some maximum frequency B > 0;
the sampling rate—f s must be greater than twice the bandwidth, for example, f s > 2B.
The following section discusses the registration process and explains how interpolation is involved with registration.
1.2. Image Registration Algorithms
Image registration methods in medical imaging seek to align two or more images and can be applied in the same modality on the same patient for the purpose of monitoring and quantifying disease progression over time. Registration can also be applied across different modalities, which is useful for correction of different patient positions across scans, for instance, aligning positron emission tomography (PET) data to an MRI image. Also, image registration can be used on the different patients, which is useful for studies of variability between subjects. Image registration is classified into the following categories and depends on several factors: image modalities (MRI, PET, CT, etc.), the subject of registration (a single person or different persons), the object of registration (head or heart), the image dimensionality (e.g., 2D, 3D, and 4D), and geometrical transformation (affine, rigid, projective, etc.).
This study examines 3D affine registration of brain images using voxel intensities similarity measures such as normalized mutual information (NMI), normalized cross correlation (NCC), least squares (LS), and correlation ratio (CR). More explicitly, if a target image is resampled to match a reference image, the image intensities at each voxel should be similar in the two images. In fact, when utilizing an intensity-based cost function, it is essential to repeatedly resample one of the images to match the others at several various resolutions, while searching for the min cost function. This resampling process requires interpolation during the registration process [19]. In the optimized automatic image registration (OAIR) method, interpolation involves resampling of anisotropic voxels in the z-direction into isotropic cubic voxels. Also, it is important to note that in the OAIR method, the interpolation technique utilized for registration does not necessarily need to be the same interpolation technique used during registration to compute a final image using the optimal parameters.
In this paper, we are focusing on the effect of interpolation technique and cost function used for intensity-based registration. The following sections are organized as follows, we first give some background and provide a means of defining the critical components involved in image registration and establish a theoretical framework.
1.2.1. Geometric Transformation
When registering images, one should specify a geometric transformation that specially aligns one image to another. The common transformations can be classified as rigid, affine, and projective. Rigid transformation can be defined as a simple transformation that includes only translation and rotation. The projective transformation is the most general transformation and maps lines to lines (but does not necessarily preserve parallelism). An affine transformation includes scaling, rotation, translation, shearing, and reflection. There are several scanner-produced errors that can result in skewing or scaling terms, and affine transformations are applied to overcome these problems.
An affine transformation maps straight lines to straight lines and keeps the parallelism of lines, but not their lengths or their angles. Changing scaling and shearing factors for each image dimension will extend the degree of freedom (DOF, the number of independent pieces of information that go into the estimate of a parameter) of the rigid transformation [20–27]. Figure 4 shows the five basic components of affine transformations. The following matrices constitute the basic affine transforms in 3D, addressed in homogeneous form.
Figure 4.
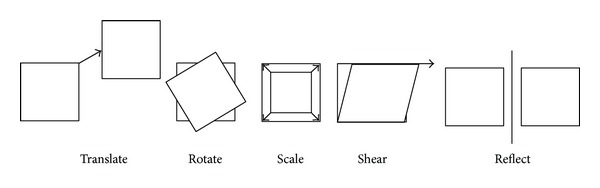
The five basic affine transformations are translate, rotate, scale, shear, and reflection. Translate moves a set of points a fixed distance in x- and y-directions, Rotate rotates a set of points about the origin, Scale scales a set of points up or down in x- and y-directions, and Shear offsets a set of points a distance proportional to their x- and y-coordinates. Reflection produces a mirror image of a set of points in x- or y-directions.
Translation. Translate a point in the xyz-plane to a new place by adding a vector (t x, t y, t z). x′ = x + t x, y′ = y + t y, and z′ = z + t z. P′ represents scaled matrices: P′ = T P, where
| (10) |
| (11) |
Scaling. Scaling is making the new scale of a coordinate direction p times larger. Scaling is applied to all axes, each with a different scaling factor (s x, s y, s z). x′ = s x × x, y′ = s y × y and z′ = s z × z. P′ represents of scaled matrices:
P′ = S P,
| (12) |
Rotation. If a point (x, y, z) is rotated an angle θ about the coordinate origin to become a new point (x′,y′, z′), the three basic rotations in 3D can be defined as follows:
Rotation about the x-axis:
| (13) |
Rotation about the y-axis:
| (14) |
Rotation about the z-axis:
| (15) |
There are several reasons for using homogeneous coordinates, including the ability to apply all four transformations multiplicatively. In view of the fact that transformation combinations (shearing, scaling, and rotation) are all multiplicative transforms, only translation is an additive transform.
Next, the following cost functions are defined and described: LS, NCC, CR, and NMI. Furthermore, an overview of the literature on their use in registration for medical applications is included. Among these cost functions, the NMI-based registration has become commonplace in many medical applications [28].
1.2.2. Cost Functions
The cost function or similarity measure evaluates the similarity between two images. In this section, the behaviors of four commonly used cost functions will be examined.
Least Squares (LS). The least squares method measures the average of the squared difference in image intensities [29]:
| (16) |
where R is the reference image, I is the input image, N is the number of values over which the sum is performed, and f is the least square. When two images differ only by Gaussian noise, the least squares will be the optimum cost function. Images of two different modalities such as MRI and PET will never differ by only Gaussian noise. Due to patient motion, even two images of the same modality, such as two MRI images, will rarely only differ by Gaussian noise. The effectiveness of LS will be extremely decreased by a small number of voxels having considerable intensity differences.
Correlation Ratio (CR). The main principle of the correlation ratio method is to calculate a “similarity measure” between a reference image and an input image and search for a spatial transformation T and an intensity mapping f such that by dis-replacing R and remapping its intensities, the resulting image f(R × T) can be seen as equivalent as possible to I. This can be obtained by minimizing the following CR function [30]:
| (17) |
which integrates over the voxel positions in the image I. The minimum and maximum values for the CR are 0 and 1, respectively. The CR can be applied in multimodal image registration involving positron emission tomography (PET), MRI, and computed tomography (CT) images, providing a good tradeoff between accuracy and robustness [31].
Normalized Cross-Correlation (NCC). The cross-correlation function works very well for aligning images of the same modality. Cross-correlation function is defined by the following equation:
| (18) |
where R is the reference image intensity, I is the input image intensity, and x and y represent the partials of images R and I in x and y-directions, respectively. The summation is taken over the region (u, v), where R and I overlap. When I(x, y) best matches R(x, y), CrossCorr(u, v) shows the maximum value.
Normalized Mutual Information (NMI). The algorithms of mutual information (MI) have been the most investigated measure for registration of medical image to date. The mutual information of images I and J is defined by the following [32, 33]:
| (19) |
where P i,j is the joint probability distribution image of I and J and p i and p j are the marginal probability distribution function of I and J, respectively. The minimum and maximum values for normalized mutual information are 0 and 1, respectively.
When images are correctly registered and aligned, there is maximal dependence between the gray values of the images, meaning that the amount of mutual information would be high. Misregistration will cause a decrease in the MI measure [34]. NMI has been used with success for a wide variety of combinations, including MR, CT, SPET, PET, and also time series images [35]. NMI can be found in a large number of studies [34, 36].
1.2.3. Optimized Automatic Image Registration 3D
OAIR is a robust image registration algorithm based on FLIRT (FLIRT stands for FMRIB's Linear Registration tool 1.3) [26, 37–39]. The OAIR technique specifies a transformation that minimizes a cost function, which represents the quality of alignment between two images. The method assesses the cost function at the number of different image resolutions, starting with the lowest resolution. Each step of increasing resolution uses the previously specified optimal transformation as the starting point and further refines its values. OAIR method usually works very well with the image of the same modality (e.g., MRI-MRI, CT-CT, and PET-PET). During the OAIR registration, the resampling process will influence the computed value of the cost function; therefore, choosing the best interpolation is important.
Outline of the OAIR Method. (1) The registration algorithm specifies the minimum resolution for each dimension of the target and reference images (they are subsampled by factors two, four, and eight). (2) The reference and target images are interpolated in order to create high resolution isotropic voxels. (3) The centers of mass (COM) for the reference and target images are then calculated and one translation level is implemented to align the COM.
The method uses the right-hand convention in 3D coordinate systems (X, Y, Z) in order to compute the COM. The image origin is generally at the corner of the image (the upper left-hand corner of the image). The axis directions are as follows the x-axis goes left to right, the y-axis goes top to bottom, and the z-axis goes into the image. To compute the COM, the characteristics function of an object in an image is defined by the following
| (20) |
Next, the area of the image is computed as
| (21) |
Figure 5 shows the COM.
Figure 5.
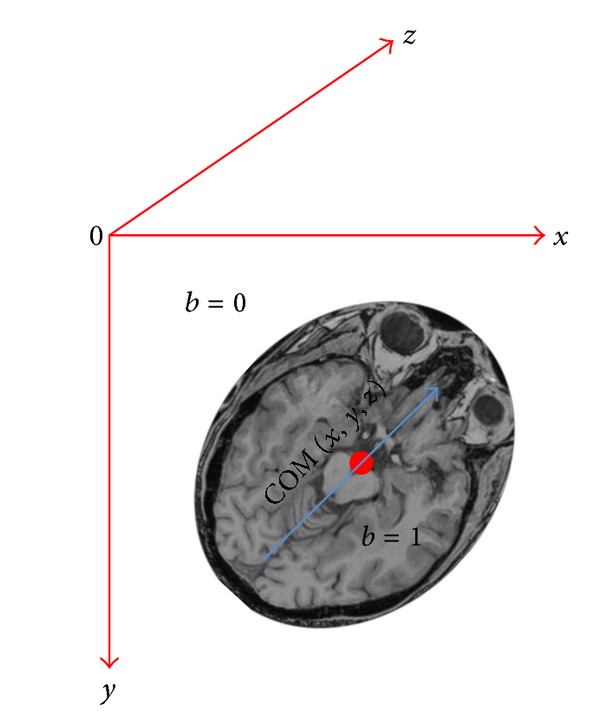
Calculating the COM in the image space; the image origin is in the upper left-hand corner of the image. The x-axis goes left to right, the y-axis goes top to bottom, and the z-axis goes into image.
The COM, indicated by (x com, y com, z com), is given by the first moments of the object:
| (22) |
(4) For each resampled image (which is 8, 4, 2, and 1 times) specifies the transform that minimizes the cost function.
Optimization Steps. The theoretical registration problem is completely determined by an interpolation method, a cost function, and a transformation space. However, in practice, an optimization method is needed to find the transformation that minimizes the cost function [26]. In general, all cost functions require global optimization. As a part of the transformation optimization process, the images are subsampled by several factors (e.g., eight, four, and two times) [38].
Levels Eight, Four, Two, and One Optimization. Reference and target images are interpolated and subsampled by eight, so each image is eight times smaller. The parameters corresponding to the minimum cost function are specified and used as the initial transformation. For the next level (level four) in the optimization, the reference and target images are interpolated and subsampled by four and, like in level eight, the transformation parameters corresponding to the minimum cost function are specified and used as the initial transformation for the next level (level two) in the optimization. For level two optimization, the process repeats, except that the reference and target images are first interpolated and subsampled by factor two. As mentioned above, the parameters of the transformation are systematically varied, and the cost function is assessed for each setting. For level one optimization, 1 mm interpolated images are used and the transformation is generalized to contain 12-DOF.
The merit of this multiresolution technique is that the initial optimization, at large n, has a noticeably reduced computational load, since the number of sample points is considerably less. Additionally, a large subsampling (n = 8) uses the lowest resolution image and coarse rotation angle, in which the large features of the image are dominate, and so the overall alignment is easier to find. The following sections explain in detail how each resampled image (which is 8, 4, 2, and 1 times) specifies the transform that minimizes the cost function.
(1) Level Eight Optimization. One of the most difficult tasks in image registration is finding the right orientation or rotation, and most of the erroneous registrations that have been examined have happened primarily because of an incorrect orientation. Thus, the search focuses on the rotational part of the transformation.
In level eight, the reference image is directed, interpolated, and the cost function is evaluated for coarse rotations angles (−30, −30, −30), (−30, −30, −15),…, (−30, −30, 30), where the values represent the amount of rotation in degrees about a, y, and z axes, respectively. In this case, there would be 125 possible angle configurations, since there are three rotation angles and each angle can contain five different values. A 4-DOF local optimization is also applied to find optimal translation and global scale for each angle configuration. The best 20% of the cost values and corresponding angle configurations (candidate local minima) are stored in vector of minima that is used as starting point for a further optimization, which uses a smaller step size over a narrow range of angles. For each parameter setting related to the top 20% of the cost function minima, the algorithm performs the minima over rotation as well as global scale and translation (previously, the algorithm had not optimized over rotation). For each of these sets of parameters, a 7-DOF optimization is then performed, storing the results of the transformation and cost before and after optimization in a vector of minima. A vector of parameters and top 20% of the min cost function values are considered for the next higher resolution (level four optimization) stage, because the relative costs of each candidate solution may change at higher resolutions. The algorithm also uses interpolation to transform images to this new orientation [26, 37–39].
(2) Level Four Optimization. The algorithm now calls level four with the interpolated images subsampled by 4 to specify the transformation that minimizes the cost function starting with the transformations determined in level eight. The optimization specifies a 7-DOF transformation that corresponds to minimum value of the cost function. This transformation is then perturbed and the cost function is calculated for these new settings. The perturbations correspond to six degrees for each rotation parameter and a global scaling factor of 0.8, 0.9, 1.0, 1.1, and 1.2. A vector of parameters and the top 20% of the cost function minima values are considered for the next step, which involves images subsampled by 2.
(3) Level Two Optimization. The algorithm uses the images interpolated and subsampled by 2 and computes the value of the cost function for each parameter setting obtained from the level four optimization. It finds the best minimum, and then optimizes it with the 7-DOF, then 9-DOF, and 12-DOF. The algorithm then returns the best minimum after optimization.
(4) Level One Optimization. The algorithm now level one to use the unsubsampled interpolated images and computes the value of the cost function for each parameter setting obtained from the level two optimization. In this step, one optimization run is performed, with the maximum allowable DOF, as determined by the user (max 12-DOF). The best answer is returned from level one and gives us the minimum cost of differences between the images.
2. Materials and Methods
A 3D spoiled gradient recalled (SPGR). MRI of the brain was acquired at Nationwide Children's Hospital of Columbus, Ohio, USA, using a 3T GE MR scanner from a 34-year-old participant. Interpolation techniques were performed on brain scans. Relevant imaging parameters are listed in Table 1. The first initial reference image, also the HR, dimensions were 512 × 512 × 120 (this is native scanner output) with voxel size of 0.5 × 0.5 × 1.3 mm3 and with slice thickness and spacing between slices of 1.3 mm (acquiring a fully isotropic 3D scan was not feasible because of time, motion artifact, and SNR factors). Because of interpolation and registration time, we simulated the new 3D HR images (simulated reference 2) with a resolution of 256 × 256 × 120 and with a voxel size of 1 × 1 × 1.3 mm3 and with slice thickness and spacing between slices of 1.3 mm. In the absence of gold standards, simulations are sometimes utilized to assess registration accuracy. A common tactic is to take real data and deform it using appropriate spatial transformation model (affine, rigid, and projective) and other factors that are thought to be relevant in limiting registration accuracy such as simulating the addition of noise and blurring.
Table 1.
Imaging parameters associated with 3D reference and low-resolution images.
| 3D images | No. of slices | Matrix size | Voxel size (mm3) |
|---|---|---|---|
| Reference 1 | 120 | 512 × 512 | 0.5 × 0.5 × 1.3 |
| Simulated reference 2 | 120 | 256 × 256 | 1 × 1 × 1.3 |
| Low resolution 1 | 60 | 512 × 512 | 0.5 × 0.5 × 2.6 |
| Low resolution 2 | 60 | 256 × 256 | 1 × 1 × 2.6 |
| Low resolution 3 | 120 | 256 × 128 | 1 × 2 × 1.3 |
| Low resolution 4 | 120 | 128 × 256 | 2 × 2 × 1.3 |
The first low resolution (LR) images were generated from the reference one, and the resolution was decreased (512 × 512 × 60 and with a voxel size of 0.5 × 0.5 × 2.6 mm3) along the slice direction by subsampling by a factor of 2. The second, third, and fourth LR images were generated from simulated reference 2, and they were subsampled by a factor of 2 in the x-, y-, and z directions. The second LR images were generated with a resolution of 256 × 256 × 60 (axial plane) and with a voxel size of 1 × 1 × 2.6 mm3. The third LR images were generated with resolution 256 × 128 × 120 (sagittal plane) and with voxel size of 1 × 2 × 1.3 mm3, and finally, the fourth LR images were generated with a resolution of 128 × 256 × 120 (coronal plane) and with a voxel size of 2 × 1 × 1.3 mm3. We rotated the LR images in x-direction by 5 degrees. Then, we translated the rotated image above in x by 2 mm and in y by 3 mm. The LR images are corrupted by Gaussian noise (10 standard deviation) and Gaussian blurring (horizontal 5 radius).
Afterward, we used these LR images as input to our interpolation algorithms (trilinear, cubic Lagrangian, quintic Lagrangian, heptic Lagrangian, windowed Sinc, B-spline 3rd order, and B-spline 4th order) to remap to a common size. They were upsampled and back to their original dimension (256 × 256 × 120), and then we compared them to the reference images in order to find the minimum interpolation error during upsampling. Image restoration (adaptive noise reduction and blind deconvolution techniques) was implemented upon the upsampled images to reduce blurring and noise. Adaptive noise reduction algorithm reduces noise without blurring the edges by replacing a pixel value with a weighted sum of all local pixels reached by following a path with small pixel intensity values between neighboring pixels, and blind deconvolution is a method, which allows recovering of the target object from a set of blurred images in the presence or a poorly specified or unknown point spread function (PSF) [40]. Restoration can be implemented by applying any deconvolution method that considers the presence of noise and blurring.
OAIR was applied on high resolution data set (simulated reference 2) with a resolution of 256 × 256 × 120 and with a voxel size of 1 × 1 × 1.3 mm3, and the transformed image with a resolution of 256 × 256 × 120 and with a voxel size of 1 × 1 × 1.3 mm3. Throughout the OAIR, when an optimal fit was achieved, the target image was reformatted using the transformation function and interpolations described above to match the reference image. For achieving a good registration (intensity-based cost function) between the fixed image (reference image) and the moving image (target image), the resampling was essential because the moving image did not necessarily have the same origin, spacing, and number of pixels as the fixed image. Therefore, the resampling process helped us to have the moving image in the grid of the fixed image.
The intensity-based registration method looked for the transformation that would give the smallest value of the cost function, which we assumed was the transformation that also gave the best alignment. During this registration for analyzing the effect of interpolation and cost function, we applied and tested various interpolations and cost functions. The cost functions which were performed in this method included
normalized mutual information (NMI);
normalized cross correlation (NCC);
least squares (LS);
correlation ratio (CR).
2.1. Image Assessment
There are various ways to evaluate the accuracy of registration technique. They can be divided into qualitative and quantitative methods. For qualitative and quantitative assessment of registered images, we proposed five ways to compare and evaluate the new transformation with the old; we needed to quantify the difference between the geometrically transformed source images with the target image.
2.1.1. Quantitative Assessment
For the quantitative assessment, we considered a mean squared error (MSE), peak signal to noise ratio (PSNR), and entropy. The MSE and PSNR measures are estimates of the quality of registration images, and entropy is also a suitable choice for quantitative assessment of the accuracy of registration method.
(1) Mean Square Error. MSE was computed between the original image (reference) and reconstructed image in order to measure the average of the squared difference in image intensities:
| (23) |
where i, j, and k represent the direct comparison of each coordinate location, R is the reference image, and I is the reconstructed image. The MSE was computed for 3D brain image in order to assign a value and compare the results.
| (24) |
where n, m, and l are the number of points in the x-, y-, z-directions, respectively, for the reconstructed volume.
(2) Peak Signal to Noise. PSNR in decibels (dB) between the original image and the registered image is defined by [41]
| (25) |
where MAX is the maximum pixel value of the image and RMSE is the square root of the MSE.
(3) Entropy. The desire for a measure of information (commonly termed entropy) of a message stems from communication theory [42]. Shannon introduced an adapted measure in 1984 [43], which weights the information per outcome by the probability of that outcome occurring. Given the events occurring with the probabilities, the Shannon entropy is defined as
| (26) |
where p = (histogram count in bin)/total count. The Shannon entropy can be applied and computed for an image and be used on the distribution of the gray values of the image. An image with a low entropy value has almost a single intensity; it contains very little information. An image with a high entropy value has more or less equal quantities of numerous different intensities; it contains a lot of information [42]. For instance, blurring an image reduces noise and high frequency and thus sharpens the images histogram, resulting in reduced entropy.
2.1.2. Qualitative Assessment
One way for qualitative assessment is the subtraction of the reference and registered images. Image subtraction techniques can be used to detect subtle changes that may reflect clinically important disease progression [21]. Another way to conduct a qualitative assessment is to create a joint histogram. The joint histogram is a functional tool for visualizing the relationship between the intensities of corresponding voxels in two or more images. Visual assessment is also considered for qualitative assessment.
(1) Joint Histogram. The joint histogram is two-dimensional for two grayscale images A and B and is created by plotting the intensity of each voxel in image A against the intensity of the corresponding voxel in image B. When two images of different modalities are produced, the spatial resolution is likely to be different (see Figure 6). Therefore, before calculating a joint histogram, it is essential to rescale the range of data of the first image to the range of data of the second image. When two images are perfectly aligned, the corresponding anatomical areas overlap, and their joint histogram is highly focused. In misaligned images, anatomical areas are not matched, and they are mixed up and their joint histogram is scattered. For example, the cerebrum region of one image overlaid onto the skull region of another image causes a more dispersed joint histogram. Note, joint histograms are commonly used for different modalities like MR-CT and PET-MR at different stages are investigated in some papers [7, 44]. We implemented the joint histogram technique on the registered images of the same modality for checking the effect of interpolations and cost functions on the accuracy of OAIR.
Figure 6.
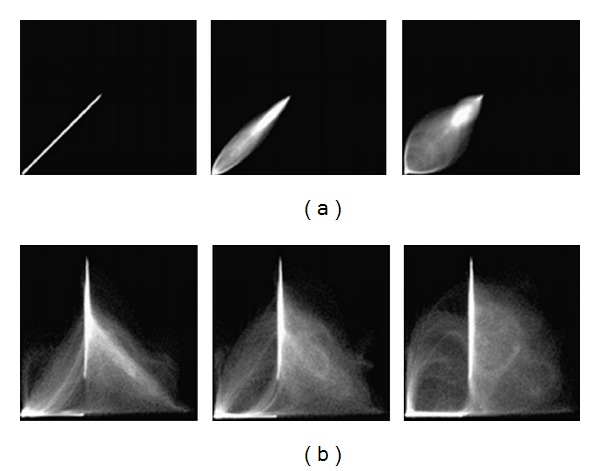
Example 2D histograms for (a) the same MR images of the head, (b) MR and CT images of the head. The left columns show two images when aligned, the middle columns show two image when translated by 2 mm, and the right columns show two images when translated by 5 mm. As can be seen, the joint histogram disperses with increasing mis-registration (obtained from Hill et al. (1994)).
3. Results and Discussion
In Figure 7, the interpolated images in axial, coronal, and sagittal views are shown for the first LR images. Part (a) illustrates the 3D image interpolated by the trilinear method; in part (b), the image generated by heptic Lagrangian and quintic Lagrangian interpolation images appears in part (c). Part (d), part (e), and part (f) show that 3D images are generated by windowed Sinc, cubic Lagrangian, and nearest neighbor interpolations, respectively. The B-spline 3rd and B-spline 4th interpolations are shown in parts (g) and (h), respectively. The 3D MSEs for 512-sized MRI images were computed in all three planes. To further test the interpolations in 3D, three typical matrix sizes were simulated, namely, 64, 128, and 256. The 3D MSEs of these matrix sizes were tabulated in Table 2. The MSE is inversely proportional to the 3D MRI images size. As a result, the trilinear method yielded more accurate (lower MSE) values than the discussed interpolations.
Figure 7.
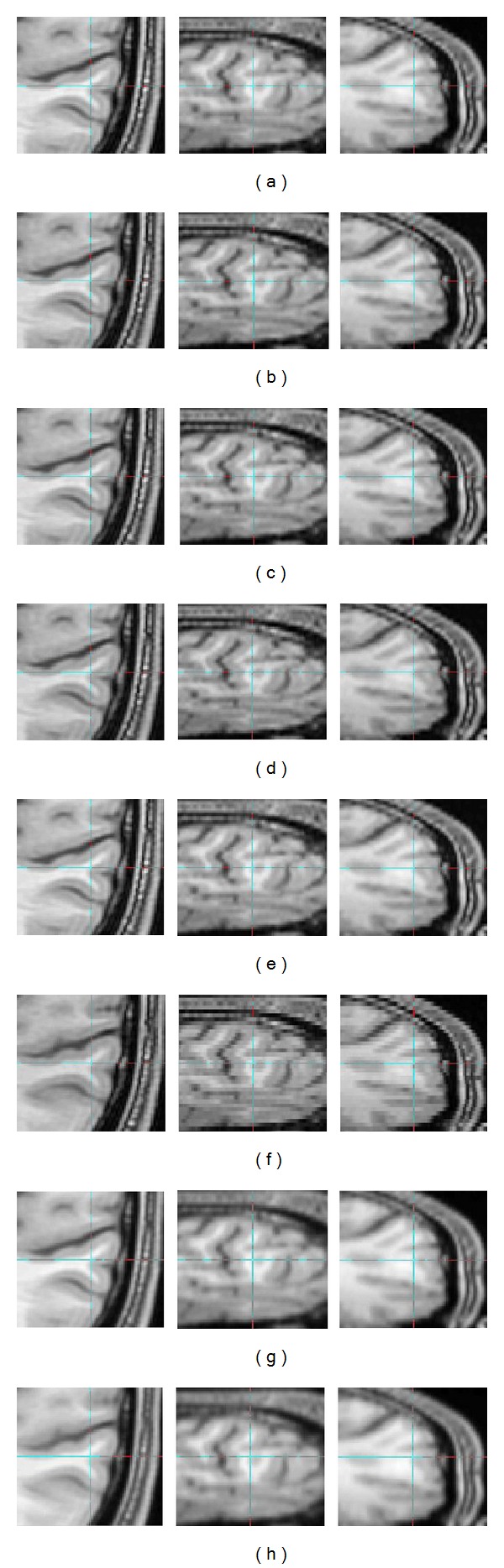
(a) Trilinear, (b) heptic Lagrange, (c) quintic Lagrange, (d) windowed Sinc, (e) cubic Lagrangian, (f) nearest neighbor, (g) B-spline 3rd order, (h) B-spline 4th order, axial view (left column), sagittal view (middle column), and coronal view (right column). These upsampled images are from images that were downsampled by a factor of two in z-direction.
Table 2.
3D MSE for MR images of 64–512 for interpolations of trilinear, nearest neighbor, B-spline 3rd order, B-spline 4th order, cubic Lagrangian, quintic Lagrangian, heptic Lagrangian, and windowed Sinc.
| MSE | ||||
|---|---|---|---|---|
| Matrix size | ||||
| Interpolation | 64 × 64 | 128 × 128 | 256 × 256 | 512 × 512 |
| Trilinear | 0.22683 | 0.03088 | 0.00282 | 0.00024 |
| Nearest neighbor | 0.75852 | 0.09615 | 0.01295 | 0.00135 |
| B-spline 3rd order | 0.50714 | 0.06435 | 0.00706 | 0.00095 |
| B-spline 4th order | 0.32088 | 0.05742 | 0.00618 | 0.00075 |
| Cubic Lagrangian | 0.24283 | 0.04956 | 0.00554 | 0.00027 |
| Quintic Lagrangian | 0.24204 | 0.03286 | 0.00552 | 0.00026 |
| Heptic Lagrangian | 0.24181 | 0.03284 | 0.00549 | 0.00025 |
| Windowed Sinc | 0.24338 | 0.05123 | 0.00559 | 0.00027 |
Also, the interpolated images were quantitatively evaluated by computing the PSNR, which is widely used in the evaluation of reconstructed images [11]. The 3D PSNRs for 64–512 sized MRI images were computed. The 3D PSNRs of these matrix sizes were tabulated in Table 3. The PSNR results for trilinear interpolation for matrix size of 64–512 were approximately 91 (dB), 100 (dB), 111 (dB), and 121 (dB), respectively. Based on Table 3, trilinear interpolation shows PSNR superiority against the other interpolation. In addition, the PSNR was found to slowly increase as the matrix size increased. The second LR images, the matrix size of 256 × 256 × 60 (axial view), the third LR images, the matrix size of 256 × 128 × 120 (sagittal view), and the fourth LR images, the matrix size of 128 × 256 × 120 (coronal view) were simply interpolated separately, and MSE was computed. The results are tabulated in Table 4.
Table 3.
3D PSNR for MR images of 64–512 for interpolations of trilinear, nearest neighbor, B-spline 3rd order, B-spline 4th order, cubic Lagrangian, quintic Lagrangian, heptic Lagrangian, and windowed Sinc.
| PSNR (dB) | ||||
|---|---|---|---|---|
| Matrix size | ||||
| Interpolation | 64 × 64 | 128 × 128 | 256 × 256 | 512 × 512 |
| Trilinear | 91.95 | 100.61 | 111.01 | 121.76 |
| Nearest neighbor | 86.78 | 95.76 | 104.47 | 114.29 |
| B-spline 3rd order | 87.62 | 96.57 | 106.17 | 114.73 |
| B-spline 4th order | 89.44 | 96.92 | 106.60 | 115.93 |
| Cubic Lagrangian | 91.73 | 98.63 | 108.14 | 121.29 |
| Quintic Lagrangian | 91.74 | 100.42 | 108.16 | 121.45 |
| Heptic Lagrangian | 91.75 | 100.44 | 108.19 | 121.61 |
| Windowed Sinc | 91.72 | 98.49 | 108.11 | 121.28 |
Table 4.
3D MSE for MR image of matrix sizes of 256 × 256 × 60, 256 × 128 × 120, and 128 × 256 × 120 for interpolations of trilinear, nearest neighbor, B-spline 3rd order, B-spline 4th order, cubic Lagrangian, quintic Lagrangian, heptic Lagrangian, and windowed Sinc (compared to 3D simulated reference 2 with resolution 256 × 256 × 120).
| MSE | |||
|---|---|---|---|
| Matrix size | |||
| Interpolation | 256 × 256 | 256 × 128 | 128 × 256 |
| Trilinear | 0.002793 | 0.002820 | 0.002983 |
| Nearest neighbor | 0.014754 | 0.016352 | 0.016544 |
| B-spline 3rd order | 0.010866 | 0.013058 | 0.012870 |
| B-spline 4th order | 0.006383 | 0.007063 | 0.007319 |
| Cubic Lagrange | 0.004519 | 0.005546 | 0.005213 |
| Quintic Lagrange | 0.004504 | 0.005530 | 0.005195 |
| Heptic Lagrange | 0.004132 | 0.005524 | 0.005192 |
| Windowed Sinc | 0.004605 | 0.005660 | 0.005285 |
As a result, in Table 4, the interpolated matrix size of 256 × 256 × 60 (axial view) yields more accurate (lower MSE) results than both matrix sizes of 256 × 128 × 120 (sagittal view), 128 × 256 × 120 (coronal view). In other words, the interpolated images in z-direction have more quality (lower MSE) than the interpolated images in x- and y-directions. However, the perceived quality of 128 × 256 × 120 (coronal view) was nearly as good as that of 256 × 128 × 120 (sagittal view).
3.1. Visual Quality of Interpolation Techniques
In Figure 8, we applied the trilinear algorithm on the second LR images, and the resolution in the axial view did not improve because the resolution in x- and y-directions was already high (256 × 256 × 60) with voxel size of 1 × 1 × 2.6 mm3, and the resolution in axial direction was constant. In contrast, the resolutions in sagittal and coronal views showed different results, and we saw enough improvement in both planes (interpolated images). As was mentioned before, the resolution in the slice-select direction is lower than plane direction, and we would like to improve the resolution in the slice-selection direction. In Figure 9, we applied a trilinear algorithm on the third LR images with a resolution of 256 × 128 × 120 and a voxel size of 1 × 2 × 1.3 mm3; the resolution in coronal view did not improve because the resolution in x-direction was high. However, the resolutions in sagittal and axial views were changed, and we saw enough improvement in their resolutions. The fourth LR images had a resolution of 128 × 256 × 120 and a voxel size of 2 × 1 × 1.3 mm3; those results are shown in Figure 10. The resolution in the sagittal view was constant because the resolution in y was high and just the sagittal resolution in axial and coronal views was changed. The downsampled results in Figure 8 (middle row) in the sagittal and the coronal views show significant jagged-edge distortion. The trilinear interpolation results in Figure 8 (bottom row) and the coronal views have smoother edges but somewhat blurred appearance overall. Also, the downsampled results in Figure 9 (middle row) in the axial and sagittal views are almost equivalent to the downsampled results in the sagittal and coronal views in Figure 9, and they showed noticeable jagged-edge distortion. The trilinear interpolation results in Figure 9 (bottom row) in the axial and sagittal views had smoother edges but blurred appearance slightly. One can see enough improvement in resolution of both planes (axial and sagittal), but the resolution of the coronal view was constant. The downsampled results in Figure 10 (middle row) were similar to the downsampled results in Figures 8 and 9, but with different perspective (axial and coronal). The trilinear interpolation results in Figure 10 (bottom row) in axial and coronal views showed enough improvement in their planes, but the sagittal view was constant.
Figure 8.
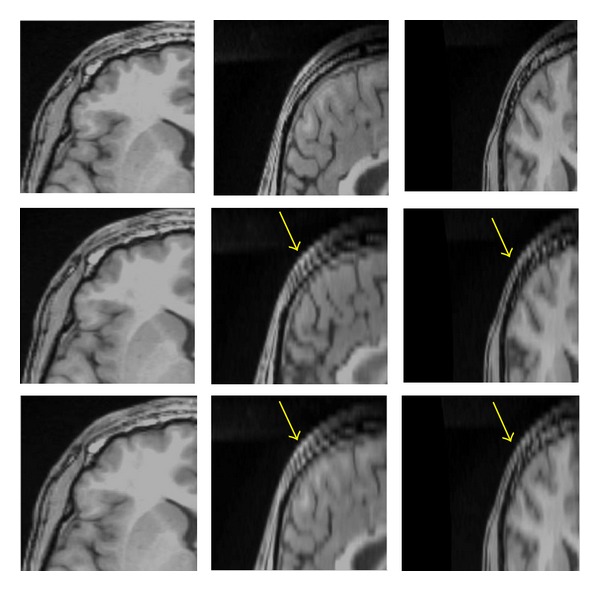
Reference with resolution 256 × 256 × 120 (top row), downsampled by a factor of 2 in the Z direction with resolution 256 × 256 × 60 (middle row), interpolated by trilinear (bottom row), axial view (left column), sagittal (middle column), and coronal view (right column). The yellow arrows in sagittal and coronal views show significant jagged-edge distortion.
Figure 9.
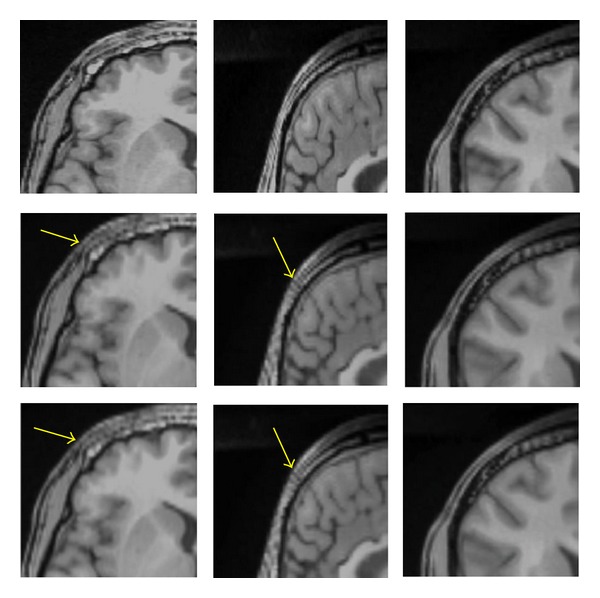
Reference with resolution 256 × 256 × 120 (top row), downsampled by a factor of 2 in the Y direction with resolution 256 × 128 × 120 (middle row), interpolated by trilinear (bottom row), axial view (left column), sagittal (middle column), and coronal views (right column). The yellow arrows in axial and sagittal views show significant jagged-edge distortion.
Figure 10.
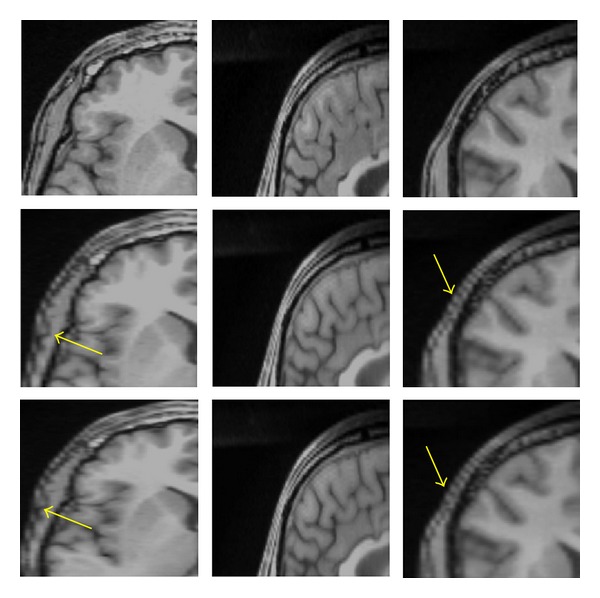
Reference with resolution 256 × 256 × 120 (top row), downsampled by a factor of 2 in the X direction with resolution 128 × 256 × 120 (middle row), interpolated by trilinear (bottom row), axial view (left column), sagittal (middle column), and coronal view (right column). The yellow arrows in axial and coronal views show significant jagged-edge distortion.
3.2. Runtime Measurement
The runtimes of the various interpolation schemes were computed on MR images with a resolution of 256 × 256 × 60 (axial view). In the axial view, the nearest neighbor was the fastest interpolation, with a runtime of 16.8 s, and the trilinear was a bit slower than the nearest neighbor with a runtime of 20.4 s. Cubic Lagrangian was fairly fast (35.7 s) and required less time than the quintic Lagrangian, heptic Lagrangian, and windowed Sinc, with 75.2 s, 159.6 s and 182.3 s, respectively.
Interpolation with the B-spline 3rd order and B-spline 4th order took about 48 and 185 times as long as nearest neighbor interpolation. This weak performance was caused by evaluation of the exponential function necessary to specify the weights and increasing the order of B-spline showed the interpolation drastically. The results of the run times are presented in Figure 11.
Figure 11.
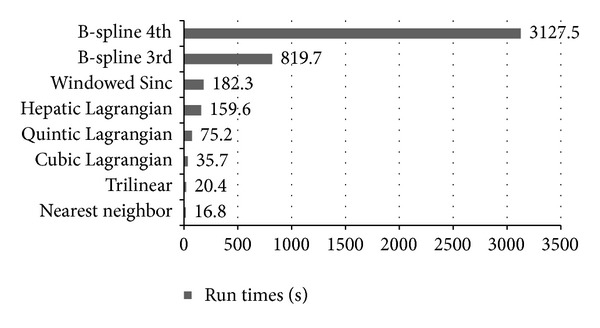
Run times measured on the Intel Xeon with 2.13 GHz 2 processor. Among the discussed interpolation techniques, the trilinear is one of the fastest interpolations and runs quickly, with 20.4 s.
Among the interpolations techniques discussed, the trilinear method was one of the fastest techniques and had the smallest interpolation error. The nearest neighbor had a strong point in which the original voxel intensities were preserved, but the resulting image was degraded significantly and had a blocky appearance. Our experiments showed that the heptic Lagrangian technique had smaller error than the quintic Lagrangian and the cubic Lagragian. The windowed Sinc had smaller error than the nearest neighbor, B-spline 3rd order, and B-spline 4th order. The main drawback of windowed Sinc interpolation was that, it generated significant ripple artifacts in the surrounding of image edges. The B-spline 3rd order and B-spline 4th order were the slowest techniques in this study, and B-spline 3rd order produced one of the worst results in terms of similarity to the original image. These results demonstrated that the increment of the order in B-spline will not significantly improve the interpolation quality, and this will just magnify the edge effects and the degree of blurriness, which already noticeable when compared to trilinear and Lagrangian methods. The theory and application of B-spline were analyzed by researchers [45, 46], and they found the third-order B-spline interpolator to be sufficient for some specific practical applications [47]. Currently, we believe that the trilinear can offer the best compromise between speed and accuracy in upsampling.
3.3. Analyzing the Effect of Interpolation Techniques on Accuracy of Cost Functions-Based OAIR Algorithm
We implemented OAIR 3D described in Section 1 to perform registration between images, and seven interpolation techniques using similarity measures NCC, LS, CR, and NMI. We computed MSE and PSNR of our results, and the experimental results are listed in Table 5. It is important to note that the interpolation error during upsampling (before registration) is different than the interpolation error of geometric transformation (during registration). For instance, the interpolation algorithm, which has remarkable performance in upsampling process, may have insufficient performance in geometric transformation [48]. Statistical analysis of Table 5 showed that there was insignificant difference between the sets of image registered using CR, LS, NCC, and NMI (P value > 0.9994 for all cost functions). However, the effect of interpolation was considerable, and we observed significant difference between the sets of image registered using different interpolations (P-value < 0.0001 for all interpolations). For instance, sets of images registered using windowed Sinc interpolation were significantly better than sets of images registered using B-spline 3rd-order interpolation with similar cost functions (lower MSE and higher PSNR). For qualitative assessment, we investigated the accuracy of registered results using intensity-based cost functions (CR, LS, NCC, NMI). Windowed Sinc and B-spline 3rd-order interpolations were used during registration (other interpolations schemes can also be used if more investigation is desired). Figure 12 shows axial slices from two registered 3D MRI volumes with their subtractions. The panels show axial slices from two data sets (3D simulated images with a resolution of 256 × 256 × 120 and with a voxel size of 1 × 1 × 1.3 mm3) after registration of the three-dimensional volumes using an intensity-based CR (first column), LS (second column), NCC (third column), and NMI (fourth column). Windowed Sinc (panel one) and B-spline 3rd-order (panel two) interpolations were used as resampling. In the second row of the panels, after registration, the pixel intensities of the reference and target images were roughly identical and different images were considerably smooth. In both panels, although differences of registered 3D MRI volumes using an intensity-based CR (first column), LS (second column), NCC (third column), and NMI (fourth column) were difficult to see by visual inspection, changing the interpolation showed that significant differences between two panels exist. In the panel two, where B-spline 3rd order was used during registration, the boundary of the skull, which was masked out of the images during registration, could still be observed easily in the difference images, whereas in the panel one, where windowed Sinc was used during registration, the skull in different images was not easily detected by eye. A possible reason for checking the effect of interpolations and cost functions for registration can be seen by visual inspection of the joint histogram in Figure 13, which contains several histograms of 3D MRI using an intensity-based CR (first column), LS (second column), NCC (third column), and NMI (fourth column). In the top and bottom rows, windowed Sinc and B-spline 3rd-order interpolations were used during registration, respectively. The top row in Figure 13 showed the joint histogram for 3D images at registration using windowed interpolation and with small amount of misregistration, and there was a diagonal in the distribution with the small dispersion. However, in the bottom row, the B-spline 3rd-order interpolation led to large mis-registration of the image, and increased off-diagonal entries started to appear, and the distribution became more dispersed. The distribution of the bottom row is nonsymmetric, and so the linear relationship is not preserved. In general, the intense inhomogeneity will noticeably change for different interpolations; this is one of the reasons that will induce nonsymmetric dispersion and a nonlinear relationship between intensities. However, the change in the appearance of the histograms for these 3D MRI volumes using CR, LS, NCC, and NMI is insignificant. We used these joint histograms to better understand the effect of different interpolations and cost functions during registration. We also measured joint entropy of the registered images using various interpolations and cost functions. Because joint entropy is directly related, the joint probability distribution described the statistical relationship of corresponding voxel intensities. Entropy increased with increasing mis-registration as can be seen in visual appearance of the joint histogram (see Figure 13). High dispersion of the joint histogram is equivalent to high joint entropy [49]. The joint entropy results are shown in Table 6, and there are no significant differences between the entropies of registered images using CR, LS, NCC, and NMI (P-value ≥ 0.9999 for all cost functions), which means that they have the similar dispersion in the joint histogram, but there were significant differences between the entropies of registered images using different interpolations (P-value < 0.0001 for all interpolations). Also, we computed costs for different voxel similarity cost functions that were used in registration with different interpolations and cost functions. We used inverse cost functions; thus, the minimum cost function corresponded to better registration. The results are shown in Table 7, and the registered images using CR, LS, NCCI, and NMI yielded very close results (P-value ≥ 0.9999 for all cost functions), whereas the registered images using different interpolations yielded different results (P-value ≤ 0.0041).
Table 5.
The 3D MSE and PSNR for registered image of 256 × 256 × 120 using interpolations of trilinear, B-spline 3rd order, B-spline 4th order, cubic Lagrange, quintic Lagrange, heptic Lagrangian, windowed Sinc, and cost functions of CR, LS, NCC, and NMI. The affine transformation contains 12 DOF and was implemented during registration.
| Interpolation | Cost function | MSE | PSNR (dB) |
|---|---|---|---|
| Trilinear | Correlation ratio | 0.023505 | 115.32 |
| Trilinear | Least squares | 0.023591 | 115.16 |
| Trilinear | Normalized cross-correlation | 0.023597 | 115.10 |
| Trilinear | Normalized mutual information | 0.023623 | 115.09 |
| B-spline 3rd order | Correlation ratio | 0.065592 | 106.30 |
| B-spline 3rd order | Least squares | 0.065732 | 106.27 |
| B-spline 3rd order | Normalized cross-correlation | 0.064311 | 106.34 |
| B-spline 3rd order | Normalized mutual information | 0.065588 | 106.31 |
| B-spline 4th order | Correlation ratio | 0.037540 | 111.37 |
| B-spline 4th order | Least squares | 0.037597 | 111.36 |
| B-spline 4th order | Normalized cross-correlation | 0.037631 | 111.35 |
| B-spline 4th order | Normalized mutual information | 0.037604 | 111.36 |
| Cubic Lagrange | Correlation ratio | 0.020723 | 116.35 |
| Cubic Lagrange | Least squares | 0.020887 | 116.28 |
| Cubic Lagrange | Normalized cross-correlation | 0.020891 | 116.27 |
| Cubic Lagrange | Normalized mutual information | 0.020793 | 116.32 |
| Quintic Lagrange | Correlation ratio | 0.019982 | 116.76 |
| Quintic Lagrange | Least squares | 0.020108 | 116.71 |
| Quintic Lagrange | Normalized cross-correlation | 0.020082 | 116.72 |
| Quintic Lagrange | Normalized mutual information | 0.019979 | 116.76 |
| Heptic Lagrange | Correlation ratio | 0.019588 | 116.86 |
| Heptic Lagrange | Least squares | 0.019770 | 116.78 |
| Heptic Lagrange | Normalized cross-correlation | 0.019736 | 116.79 |
| Heptic Lagrange | Normalized mutual information | 0.019639 | 116.83 |
| Windowed Sinc | Correlation ratio | 0.019119 | 117.06 |
| Windowed Sinc | Least squares | 0.019190 | 117.02 |
| Windowed Sinc | Normalized cross-correlation | 0.019329 | 116.96 |
| Windowed Sinc | Normalized mutual information | 0.019160 | 117.04 |
Figure 12.
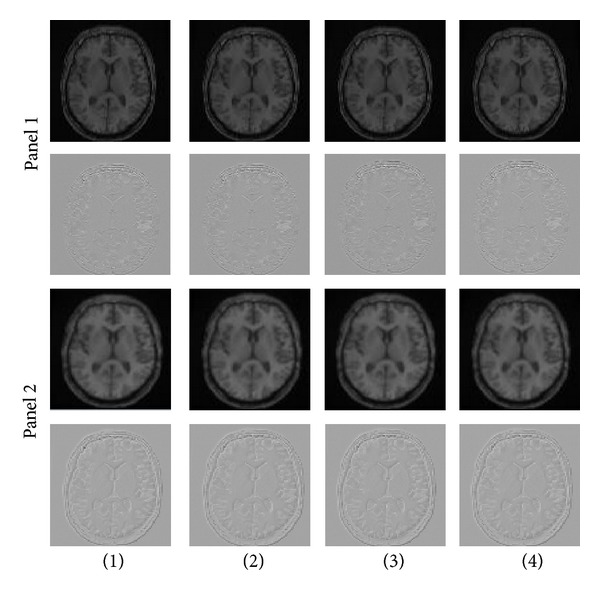
In the top panel one, axial slices from two unregistered 3D MRI volumes using an intensity-based cost function (CR (first column), LS (second column), NCC (third column), and NMI (fourth column)) are shown. Windowed Sinc interpolation was used during registration for the panel one. The second row of the panel one shows subtraction of the images (registered and reference images). The panel two shows similar axial slices from the same two data sets after registration of the full 3D volumes using the same intensity-based cost function (CR (first column), LS (second column), NCC (third column), and NMI (fourth column)). B-spline 3rd-order interpolation was used during registration for the panel two. The second row of the panel two shows subtraction of the images. Although differences of the first rows of panel one and panel two are not easily observed by eye, subtraction of the images shows that differences are present, as seen on the second rows of panel one and panel two.
Figure 13.
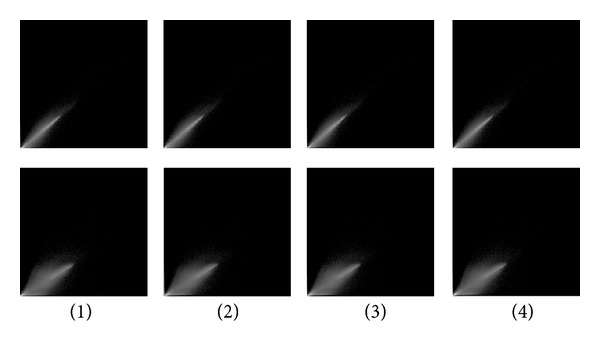
Joint histogram for registered 3D MRI volumes using an intensity-based cost function (CR (first column), LS (second column), NCC (third column), and NMI (fourth column)) are shown. The top row is generated from the images when registered using windowed Sinc interpolation, and the bottom row is generated from the images when registered using B-spline 3rd-order interpolation.
Table 6.
The joint entropy for the registered images of matrix size of 256 × 256 × 120 using interpolations of trilinear, B-spline 3rd order, B-spline 4th order, cubic Lagrange, quintic Lagrange, heptic Lagrange, windowed Sinc, and cost functions of CR, LS, NCC, and NMI. Their entropies were achieved using mutual information.
| Interpolation | Cost function | Entropy |
|---|---|---|
| Trilinear | Correlation ratio | 2.3723 |
| Trilinear | Least squares | 2.3718 |
| Trilinear | Normalized cross-correlation | 2.3704 |
| Trilinear | Normalized mutual information | 2.3787 |
| B-spline 3rd order | Correlation ratio | 2.4499 |
| B-spline 3rd order | Least squares | 2.4486 |
| B-spline 3rd order | Normalized cross-correlation | 2.4489 |
| B-spline 3rd order | Normalized mutual information | 2.4529 |
| B-spline 4th order | Correlation ratio | 2.3874 |
| B-spline 4th order | Least squares | 2.3889 |
| B-spline 4th order | Normalized cross-correlation | 2.3891 |
| B-spline 4th order | Normalized mutual information | 2.3881 |
| Cubic Lagrange | Correlation ratio | 2.3649 |
| Cubic Lagrange | Least squares | 2.3613 |
| Cubic Lagrange | Normalized cross-correlation | 2.3615 |
| Cubic Lagrange | Normalized mutual information | 2.3636 |
| Quintic Lagrange | Correlation ratio | 2.3527 |
| Quintic Lagrange | Least squares | 2.3559 |
| Quintic Lagrange | Normalized cross-correlation | 2.3534 |
| Quintic Lagrange | Normalized mutual information | 2.3544 |
| Heptic Lagrange | Correlation ratio | 2.3549 |
| Heptic Lagrange | Least squares | 2.3561 |
| Heptic Lagrange | Normalized cross-correlation | 2.3511 |
| Heptic Lagrange | Normalized mutual information | 2.3517 |
| Windowed Sinc | Correlation ratio | 2.3425 |
| Windowed Sinc | Least squares | 2.3452 |
| Windowed Sinc | Normalized cross-correlation | 2.3434 |
| Windowed Sinc | Normalized mutual information | 2.3443 |
Table 7.
The CR, MI, NMI, and NCC of reference and registered images. The 3D registered images using trilinear, B-spline 3rd order, B-spline 4th order, cubic Lagrange, quintic Lagrange, heptic Lagrange, windowed Sinc interpolations, and cost functions of CR, LS, NCC, and NMI.
| Registration | Pixel similarity cost functions | ||||
|---|---|---|---|---|---|
| Interpolation | Cost function | Correlation ration | Mutual information | Normalized mutual information | Normalized cross-correlation |
| Trilinear | Correlation Ratio | 0.030921 | 0.572625 | 0.758732 | 0.016443 |
| Trilinear | Least square | 0.030943 | 0.575558 | 0.758843 | 0.016472 |
| Trilinear | Normalized cross-correlation | 0.030978 | 0.577968 | 0.758742 | 0.016498 |
| Trilinear | Normalized mutual information | 0.030874 | 0.571644 | 0.758756 | 0.016452 |
| B-spline 3rd | Correlation ratio | 0.091192 | 0.906945 | 0.789837 | 0.047106 |
| B-spline 3rd | Least square | 0.091371 | 0.905995 | 0.790105 | 0.047204 |
| B-spline 3rd | Normalized cross-correlation | 0.089311 | 0.905822 | 0.788936 | 0.046135 |
| B-spline 3rd | Normalized mutual information | 0.091554 | 0.906234 | 0.789860 | 0.047101 |
| B-spline 4th | Correlation ratio | 0.047523 | 0.645413 | 0.764792 | 0.026291 |
| B-spline 4th | Least square | 0.047664 | 0.647675 | 0.764965 | 0.026334 |
| B-spline 4th | Normalized cross-correlation | 0.047837 | 0.646529 | 0.764938 | 0.026347 |
| B-spline 4th | Normalized mutual information | 0.047741 | 0.646291 | 0.764879 | 0.026328 |
| Cubic Lagrange | Correlation ratio | 0.027451 | 0.557755 | 0.738121 | 0.014453 |
| Cubic Lagrange | Least square | 0.027678 | 0.559016 | 0.738517 | 0.014556 |
| Cubic Lagrange | Normalized cross-correlation | 0.027751 | 0.557785 | 0.738253 | 0.014558 |
| Cubic Lagrange | Normalized mutual information | 0.027632 | 0.558993 | 0.734889 | 0.014486 |
| Quintic Lagrange | Correlation ratio | 0.026226 | 0.533578 | 0.731717 | 0.013931 |
| Quintic Lagrange | Least square | 0.026307 | 0.538433 | 0.730597 | 0.014006 |
| Quintic Lagrange | Normalized cross-correlation | 0.026227 | 0.533604 | 0.731881 | 0.013986 |
| Quintic Lagrange | Normalized mutual information | 0.026234 | 0.532218 | 0.731289 | 0.013912 |
| Heptic Lagrange | Correlation ratio | 0.025983 | 0.537712 | 0.729622 | 0.013651 |
| Heptic Lagrange | Least square | 0.026274 | 0.532524 | 0.731221 | 0.013767 |
| Heptic Lagrange | Normalized cross-correlation | 0.026273 | 0.531087 | 0.730581 | 0.013741 |
| Heptic Lagrange | Normalized mutual information | 0.026162 | 0.531199 | 0.730681 | 0.013672 |
| Windowed Sinc | Correlation ratio | 0.024201 | 0.263132 | 0.720291 | 0.013392 |
| Windowed Sinc | Least square | 0.025253 | 0.260997 | 0.720884 | 0.013445 |
| Windowed Sinc | Normalized cross-correlation | 0.024727 | 0.266973 | 0.720306 | 0.013544 |
| Windowed Sinc | Normalized mutual information | 0.024819 | 0.262372 | 0.730032 | 0.013426 |
Statistical analysis of Tables 5, 6, and 7 showed that interpolations had a significant effect on the registration accuracy, whereas cost functions had no effect on the registration accuracy.
4. Conclusion
Interpolation techniques play a critical role in the improvement and deterioration of the quality of the image as the resolution changes. Thus, the interpolation error is crucial in assessing the interpolation techniques. The interpolation error depends on features such as geometric deformation and the content of the image; therefore, only one evaluation method would not be adequate to evaluate all properties of an algorithm, and a variety of methods should be applied. The comparison is performed by visual quality assessment, quantitative interpolation error determination, and run time measurement. In this study, the results of the algorithms showed that the trilinear method had the smallest interpolation error and the highest PSNR and was one of the fastest techniques, making it appropriate for upsampling in 3D MR images and super resolution. Although super computers are able to compute a huge amount of data in real time, fast methods might be required for online resampling of image sequences or films [50]. The resulting images for trilinear interpolation were less smooth and blocky than other interpolated images. Nevertheless, trilinear interpolation has the effect of losing some high frequency information from the image [51].
Also, the effect of cost functions (LS, NMI, NCC, and CR), and interpolations (trilinear, cubic Lagrange, quintic Lagrange, heptic Lagrange, windowed Sinc, B-spline 3rd order, and B-spline 4th order) for OAIR of 3D brain images was examined, and our experimental results showed that interpolations can effectively decrease or increase the failure possibility of the registration algorithm, and the robustness of method was not due to the choice of cost function, but the choice of interpolation was critical on the robustness of registration. In addition, each component of the optimization method was also necessary to achieve the accurate registrations. Studying the precise effect of transformation on registration is an active research area [52] but is beyond the scope of this paper.
We noted that the study presented in the tables and figures relied on the same modality (MRI); in addition, these results are not representative of different modality combinations. Perhaps other conclusions would be obtained by the use of different modality combinations or transformations, for instance, MRI-PET or CT-MRI.
Also, OAIR was explained in detail, and it was a powerful image registration algorithm. This algorithm was a fully automated algorithm and proposed various resampling interpolation methods combined with CR, LS, NCC, and NMI as a cost function. One of the advantages of this method was using a feature detector (corners are used as the features) to automatically choose a large number of potentially matchable feature points in both images. The algorithm is able to detect identical features in all projections of the scene regardless of the particular image deformation. This method has direct potential for registering clinical MRI images. We have validated this method quantitatively and qualitatively, on the simulated and real data, respectively. Also, there are many excellent sources for more in-depth discussions of image registration that the reader may wish to read and learn [53–56].
5. Future Work
We would like to study the precise effect of transformation on registration, and we are also interested in combining the information of three MRI plane orientations using brain images in order to increase the resolution of 3D brain image based on super resolution reconstruction (SRR) technique.
References
- 1.Sajjad M, Khattak N, Jafri N. Image magnification using adaptive interpolation by pixel level data-dependent geometrical shapes. International Journal of Computer Science and Engineering. 1(2) [Google Scholar]
- 2.Parker JA, Kenyon RV, Troxel DE. Comparison of interpolating methods for image resampling. IEEE Transactions on Medical Imaging. 1983;MI-2(1):31–39. doi: 10.1109/TMI.1983.4307610. [DOI] [PubMed] [Google Scholar]
- 3.Hou HS, Andrews HC. Cubic splines for image interpolation and digital filtering. IEEE Trans Acoust Speech Signal Process. 1978;26(6):508–517. [Google Scholar]
- 4.Maeland E. On the comparison of interpolation methods. IEEE Transactions on Medical Imaging. 1988;7(3):213–217. doi: 10.1109/42.7784. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann TM, Gönner C, Spitzer K. Survey: interpolation methods in medical image processing. IEEE Transactions on Medical Imaging. 1999;18(11):1049–1075. doi: 10.1109/42.816070. [DOI] [PubMed] [Google Scholar]
- 6.Keys RG. Cubic convolution interpolation for digital image processing. IEEE Transactions on Acoustics, Speech, and Signal Processing. 1981;29(6):1153–1160. [Google Scholar]
- 7.Hill DL, Studholme C, Hawkes DJ. Voxel similarity measures for automated image registration. In: Richard Robb A, editor. Visualization in Biomedical Computing; 1994; pp. 205–216. [Google Scholar]
- 8.Göçeri E, Loménie N. Interpolation approaches and spline based resampling for MR images. Proceedings of the 5th International Symposium on Health Informatics and Bioinformatics (HIBIT '10); April 2010; pp. 137–143. [Google Scholar]
- 9.Hill DLG, Batchelor P. Medical image registration. In: Joseph Hajnal V, Derek Hill LG, David Hawkes J, editors. Registration Methodology: Concepts and Algorithms. chapter 3. CRC Press; 2001. pp. 40–70. [Google Scholar]
- 10.Woods RP. Handbook of medical image processing and analysis. In: Isaac N, editor. Within-Modality Registration Using Intensity-Based Cost Functions. chapter 33. Academic Press; 2000. pp. 529–553. [Google Scholar]
- 11.Hildebrand FB. Introduction to Numerical Analysis. 2nd edition. New York, NY, USA: Dover; 1974. [Google Scholar]
- 12.Jean-Jacques F, Delyon B. Min-max interpolators and Lagrange interpolation formula. Proceedings of the IEEE International Symposium on Circuits and Systems; May 2002; pp. IV/429–IV/432. [Google Scholar]
- 13.Chang Y, Lee C-D. Algebraic decoding of a class of binary cyclic codes via Lagrange interpolation formula. IEEE Transactions on Information Theory. 2010;56(1):130–139. [Google Scholar]
- 14.Bourke P. Interpolation Methods. December 1999.
- 15.Trilinear interpolation. July 1997.
- 16.Thévenaz P, Blu T, Unser M. Interpolation Revisited. IEEE Transactions on Medical Imaging. 2000;19(7) doi: 10.1109/42.875199. [DOI] [PubMed] [Google Scholar]
- 17.Du H, Mei W. Image resizing using exponential B-spline functions. Proceedings of the 2nd International Congress on Image and Signal Processing (CISP '09); October 2009; pp. 1–5. [Google Scholar]
- 18.Farin G. Curves and Surfaces for CAGD: A Practical Guide. 5th edition. Academic Press; [Google Scholar]
- 19.Woods RP. Handbook of medical image processing and analysis. In: Isaac Bankman N, editor. Within-Modality Registration Using Intensity-Based Cost Functions. chapter 33. Academic Press; 2000. pp. 529–536. [Google Scholar]
- 20.Radau PE, Slomka PJ, Julin P, Svensson L, Wahlund L-O. Evaluation of linear registration algorithms for brain SPECT and the errors due to hypoperfusion lesions. Medical Physics. 2001;28(8):1660–1668. doi: 10.1118/1.1388894. [DOI] [PubMed] [Google Scholar]
- 21.Blackall JM, Rueckert D, Maurer Jr CR, Penney GP, Hill DLG, Hawkes DJ. Medical Image Computing and Computer-Assisted Intervention. Vol. 1935. Berlin, Germany: Springer; 2000. An image registration approach to automated calibration for freehand 3D ultrasound; pp. 462–471. [Google Scholar]
- 22.Erdi YE, Rosenzweig K, Erdi AK, et al. Radiotherapy treatment planning for patients with non-small cell lung cancer using positron emission tomography (PET) Radiotherapy and Oncology. 2002;62(1):51–60. doi: 10.1016/s0167-8140(01)00470-4. [DOI] [PubMed] [Google Scholar]
- 23.Van Laere K, Koole M, D’Asseler Y, et al. Automated stereotactic standardization of brain SPECT receptor data using single-photon transmission images. Journal of Nuclear Medicine. 2001;42(2):361–375. [PubMed] [Google Scholar]
- 24.Mangin JF, Poupon C, Clark C, Le Bihan D, Bloch I. Eddycurrent distortion correction and robust tensor estimation for MR diffusion imaging. In: Niessen WJ, Viergever MA, editors. Medical Image Computing and Computer-Assisted Intervention. Vol. 2208. Berlin, Germany: Springer; 2001. pp. 186–194. (Lecture Notes in Computer Science). [Google Scholar]
- 25.Viola P, Wells WM., III Alignment by maximization of mutual information. International Journal of Computer Vision. 1997;24(2):137–154. [Google Scholar]
- 26.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 27.Mangin JF, Poupon C, Clark C, Le Bihan D, Bloch I. Eddycurrent distortion correction and robust tensor estimation for MR diffusion imaging. In: Niessen WJ, Viergever MA, editors. Medical Image Computing and Computer-Assisted Intervention. Vol. 2208. Berlin, Germany: Springer; 2001. pp. 186–194. (Lecture Notes in Computer Science). [Google Scholar]
- 28.Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Physics in Medicine and Biology. 2001;46(3):R1–R45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- 29.Woods RP. Handbook of medical image processing and analysis. In: Isaac N, editor. Within-Modality Registration Using Intensity-Based Cost Functions. chapter 33. Bankman, Academic Press; 2000. pp. 529–553. [Google Scholar]
- 30.Roche A, Pennec X, Rudolph M, et al. Generalized Correlation Ratio for Rigid Registration of 3D Ultrasound with MR Images. Institut National de Recherche en Informatique et en Automatique; 2000. [Google Scholar]
- 31.Roche A, Malandain G, Ayache N. The Correlation Ratio as a New Similarity Measure for Multimodal Image Registartion. Vol. 1496. Springer; 1998. (Lecture Notes in Computer Science). [Google Scholar]
- 32.Kostelec PJ, Periaswamy S. Image Registration for MRI. Vol. 46. Modern Signal Processing, MSRI Publications; 2003. [Google Scholar]
- 33.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32(1):71–86. [Google Scholar]
- 34.Denton ERE, Holden M, Christ E, et al. The identification of cerebral volume changes in treated growth hormone-deficient adults using serial 3D MR image processing. Journal of Computer Assisted Tomography. 2000;24(1):139–145. doi: 10.1097/00004728-200001000-00025. [DOI] [PubMed] [Google Scholar]
- 35.Pluim JPW, Maintz JBAA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Transactions on Medical Imaging. 2003;22(8):986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 36.Erdi YE, Rosenzweig K, Erdi AK, et al. Radiotherapy treatment planning for patients with non-small cell lung cancer using positron emission tomography (PET) Radiotherapy and Oncology. 2002;62(1):51–60. doi: 10.1016/s0167-8140(01)00470-4. [DOI] [PubMed] [Google Scholar]
- 37.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 38.Jenkinson M, Smith S. Optimization in Robust Linear Registartion of Brain Images. FMRIB. (TR00MJ2)
- 39.Smith S. FMRIB. TR00SMS2b. Headington, UK: Oxford Centre for Functional Magnetic Resonance Imaging of the Brain), Department of Clinical Neurology, Oxford University, John Radcliffe Hospital; BET: ‘Brain Extraction Tool’. [Google Scholar]
- 40.Holmes TJ. Blind deconvolution of quantum-limited incoherent imagery: maximum-likelihood approach. Journal of the Optical Society of America A. 1992;9(7):1052–1061. doi: 10.1364/josaa.9.001052. [DOI] [PubMed] [Google Scholar]
- 41.Sung M-M, Kim H-J, Kim E-K, Kwak J-Y, Yoo J-K, Yoo H-S. Clinical evaluation of JPEG2000 compression for digital mammography. IEEE Transactions on Nuclear Science. 2002;49(3):827–832. [Google Scholar]
- 42.Pluim JPW, Maintz JBAA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Transactions on Medical Imaging. 2003;22(8):986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 43.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- 44.Hill DLG, Hawkes DJ, Harrison NA, Ruff CF. A strategy for automated multimodality image registration incorporating anatomical knowledge and imager characteristics. Information Processing in Medical Imaging. 1993;687:182–196. [Google Scholar]
- 45.Unser M, Aldroubi A, Eden M. Polynomial spline signal approximations: filter design and asymptotic equivalence with Shannon’s sampling theorem. IEEE Transactions on Information Theory. 1992;38(1):95–103. [Google Scholar]
- 46.Unser M, Aldroubi A, Eden M. B-spline signal processing. Part II. Efficient design and applications. IEEE Transactions on Signal Processing. 1993;41(2):834–848. [Google Scholar]
- 47.Unser M, Aldroubi A, Eden M. Fast B-spline transforms for continuous image representation and interpolation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1991;13(3):277–285. [Google Scholar]
- 48.Amanatiadis A, Andreadis I. A survey on evaluation methods for image interpolation. Measurement Science and Technology. 2009;20(10)104015 [Google Scholar]
- 49.Holden M, Hill DLG, Denton ERE, et al. Voxel similarity measures for 3-D serial MR brain image registration. IEEE Transactions on Medical Imaging. 2000;19(2):94–102. doi: 10.1109/42.836369. [DOI] [PubMed] [Google Scholar]
- 50.Evans OD, Kim Y. Efficient implementation of image warping on a multimedia processor. Real-Time Imaging. 1998;4(6):417–428. [Google Scholar]
- 51.Ashburner J, Penny KFW. Human Brain Function. 2nd edition. chapter 2 [Google Scholar]
- 52.Van den Elsen PA, Pol EJD, Viergever MA. Medical image matching—a review with classification. IEEE Engineering in Medicine and Biology Magazine. 1993;12(1):26–39. [Google Scholar]
- 53.Brown LG. Survey of image registration techniques. ACM Computing Surveys. 1992;24(4):325–376. [Google Scholar]
- 54.Maintz JBA, Viergever MA. A survey of medical image registration. Medical Image Analysis. 1998;2(1):1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- 55.Maurer CR, Fitzpatrick JM. American Association of Neurological Surgeons. Park Ridge, Ill, USA: 1993. A review of medical image registration, chapter in interactive image-guided neurosurgery. [Google Scholar]
- 56.Maes F. Segmentation and registration of multimodal medical images: from theory, implementation and validation to a useful to tool in clinical practice [Ph.D. thesis] Leuven, Belgium: Catholic University of Leuven; 1998. [Google Scholar]


