Abstract
The intracellular levels of many proteins are regulated by ubiquitin-dependent proteolysis. One of the best-characterized enzymes that catalyzes the attachment of ubiquitin to proteins is a ubiquitin ligase complex, Skp1-Cullin-F box complex containing Hrt1 (SCF). We sought to artificially target a protein to the SCF complex for ubiquitination and degradation. To this end, we tested methionine aminopeptidase-2 (MetAP-2), which covalently binds the angiogenesis inhibitor ovalicin. A chimeric compound, protein-targeting chimeric molecule 1 (Protac-1), was synthesized to recruit MetAP-2 to SCF. One domain of Protac-1 contains the IκBα phosphopeptide that is recognized by the F-box protein β-TRCP, whereas the other domain is composed of ovalicin. We show that MetAP-2 can be tethered to SCFβ-TRCP, ubiquitinated, and degraded in a Protac-1-dependent manner. In the future, this approach may be useful for conditional inactivation of proteins, and for targeting disease-causing proteins for destruction.
Degradation of cellular proteins is required for normal maintenance of cellular function, including proliferation, differentiation, and cell death. One of the major pathways to regulate proteins posttranslationally is ubiquitin-dependent proteolysis. Ubiquitination occurs through the activity of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin–protein ligases (E3), which act sequentially to catalyze the attachment of ubiquitin to lysine residues of substrate proteins (1). The E3s confer specificity to ubiquitination reactions by binding directly to substrate. Although the exact number of E3s cannot be determined with certainty from sequence data, there are probably >100 distinct F-box-containing E3s encoded within the human genome (2). One particular class of E3s, the heterotetrameric Skp1-Cullin-F box (SCF) complexes, consists of Skp1, a Cullin family member, the RING-H2 protein Hrt1 (also known as Roc1 or Rbx1), and an F box protein (3). These components are conserved from yeast to mammals. The mammalian F box protein, β-TRCP/E3RS, has been shown to bind IκBα, a negative regulator of NFκB (4). The SCFβ-TRCP complex promotes the ubiquitination and subsequent degradation of IκBα, which results in activation of NFκB during the inflammatory response (3).
The recruitment of IκBα to SCFβ-TRCP is mediated by a 10-aa peptide within IκBα, DRHDSGLDSM (4, 5). In response to diverse inflammatory signals, IκBα kinase (IKK) phosphorylates this motif on both serines, which triggers the binding of IκBα to β-TRCP. Because it is a well-defined ligand for a specific ubiquitin ligase, we sought to take advantage of this phosphopeptide to target an unrelated protein to SCFβ-TRCP for ubiquitination and degradation.
As proof of concept, we tested the ability of the IκBα phosphopeptide (IPP) to target methionine aminopeptidase-2 (MetAP-2) to SCFβ-TRCP. MetAP-2 catalyzes the cleavage of N-terminal methionine from nascent polypeptides (6) and seems to be the primary target of the potent angiogenesis inhibitors fumagillin and ovalicin (OVA; refs. 7 and 8). Both of these compounds inhibit MetAP-2 by covalently binding His-231 in the active site. The consequent reduction in MetAP-2 activity is thought to block endothelial cell proliferation by causing p53-dependent arrest in the G1 phase of the cell cycle (9). Importantly, MetAP-2 is not known to be ubiquitinated or a substrate for any SCF complex.
To determine whether MetAP-2 could artificially be targeted to SCFβ-TRCP, we synthesized proteolysis-targeting chimeric molecule 1 (Protac-1) that contained both the IPP and OVA. We hypothesized that the phosphopeptide moiety would bind β-TRCP, and the OVA moiety would bind MetAP-2, thereby recruiting MetAP-2 to SCFβ-TRCP for ubiquitination (Fig. 1A). We reasoned that this strategy might work because synthetic ligands that link distinct proteins have been shown to be capable of regulating signaling pathways in vivo (10). In this article, we report that Protac-1 indeed binds MetAP-2 to SCFβ-TRCP and thereby promotes MetAP-2 ubiquitination and degradation. Demonstrating that Protac-1 mediates the ubiquitination and degradation of a foreign substrate by SCF provides a basis to begin testing Protacs in vivo in addition to other targets known to promote disease.
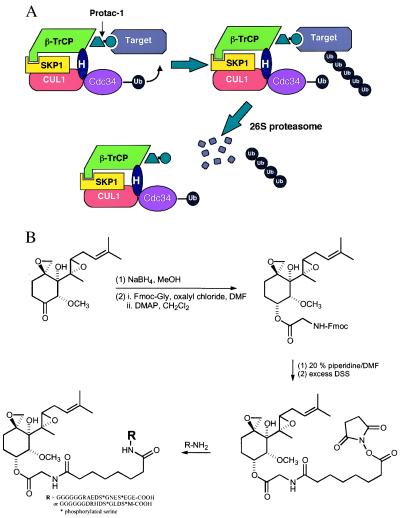
Figure 1.
(A) Protac-1 targets MetAP-2 to SCF. Protac-1 is a chimeric molecule that consists of a phosphopeptide moiety and a small molecule moiety that interacts with the protein target. Ub, ubiquitin; H, Hrt1. (B) The synthesis scheme for Protac-1 (see Materials and Methods). Fmoc, fluorenylmethoxycarbonyl; DMF, dimethylformamide; DMAP, dimethylaminopyridine; DSS, disuccinimidyl suberate.
Materials and Methods
Synthesis of IκBα-OVA Protac.
OVA (1.4 mmol) was dissolved in 10 ml of methanol at 0°C, and NaBH4 (3.0 mmol) was added slowly. After 30 min of stirring, methanol was removed under reduced pressure, and the resulting crude product was purified by flash column chromatography to yield ovalicinol (1.15 mmol, 82%). Fmoc-Gly was coupled to the ovalicinol to give Fmoc-Gly-ovalicinol. Specifically, dimethylformamide (DMF, 28 μl) was added to dichloromethane solution (30 ml) containing Fmoc-Gly-OH (3.56 mmol) and oxalyl chloride (7.12 mmol) at 0°C. After 3 hr of stirring at room temperature, dichloromethane was removed under nitrogen atmosphere. The resulting solid residue was redissolved in dichloromethane (10 ml) and was combined with ovalicinol (0.6 mmol) and dimethylaminopyridine (4.7 mmol) in dichloromethane (30 ml) at 0°C. The reaction mixture was stirred for 2 hr at room temperature. After dichloromethane was removed under reduced pressure, the resulting residue was flash-chromatographed to provide Fmoc-Gly-ovalicinol (0.39 mmol, 65%). Next, Fmoc-Gly-ovalicinol (0.09 mmol) was treated with 20% piperidine in DMF (2 ml) at room temperature for 10 min, and the DMF was removed under high vacuum. The resulting solid was redissolved in 2 ml of DMSO, and disuccinimidyl suberate (0.9 mmol) was added at room temperature. After overnight stirring, DMSO was removed under high vacuum, and the resulting crude product was flash-chromatographed to give monosuccinimidyl suberate-Gly-ovalicinol (0.06 mmol, 68%).
Monosuccinimidyl suberate-Gly-ovalicinol (12 μmol) in DMSO (0.6 ml) was added to DMSO solution (1 ml) containing IκBα peptide (3.67 μmol) and dimethylaminopyridine (11 μmol). After 20 min stirring at room temperature, the coupling reaction was completed, which was confirmed by a Kaiser test. DMSO was removed under high vacuum, and the resulting crude product was repeatedly washed with dichloromethane and methanol to remove excess monosuccinimidyl suberate-Gly-ovalicinol to give the final product, IκBα peptide-suberate-Gly-ovalicinol (5.8 mg, 2.59 μmol, 70%). The final product was characterized by electrospray (ES) mass spectrometry. ES-MS (M + H)+ for ovalicinol-Gly-suberate-IκBα peptide/dimethylaminopyridine was 2,231.56 Da. All other intermediates were characterized by 500-MHz 1H NMR spectroscopy.
MetAP-2–Protac-1 Coupling Assay.
MetAP-2 (9 μM) was incubated with increasing concentrations of Protac-1 (dissolved in water) at room temperature for 45 min. Reactions were supplemented with SDS loading dye, fractionated on an SDS/10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and immunoblotted with rabbit polyclonal anti-MetAP-2 antiserum (Zymed). Enhanced chemiluminescence was performed by using Amersham Pharmacia detection reagents.
Tissue Culture and Transfections.
293T cells were cultured in DMEM with 10% (vol/vol) FBS (GIBCO), penicillin (100 units/ml), streptomycin (100 mg/ml), and l-glutamine (2 mM). Cells were split 1:5 before the day of transfection and transiently transfected with 40 μg of plasmid. Cells were 60% confluent in 100-mm dishes on the day of transfection. DNA [20 μg of pFLAG-Cul1(RDB1347) and 20 μg of pFLAG-β-TRCP(RDB1189)] was added and the cells were transfected using calcium phosphate precipitation, as described (11). Cells were harvested 30 hr after transfection. Five micrograms of pGL-1, a plasmid containing the cytomegalovirus (CMV) promoter linked to the green fluorescent protein (GFP) cDNA, was cotransfected into cells at the same time to assess transfection efficiency. In all experiments, greater than 80% of the cells were GFP-positive at the time of harvesting.
Immunoprecipitation and Ubiquitination Assays.
293T cell pellets were lysed with 200 μl of lysis buffer (25 mM Tris⋅Cl, pH 7.5/150 mM NaCl/0.1% Triton X-100/5 mM NaF/0.05 mM EGTA/1 mM PMSF). Pellets from cells transfected with vector, pFLAG-β-TRCP, or pFLAG-Cul-1 were vortexed for 10 sec, then incubated on ice for 15 min. After centrifugation at 13,000 rpm in an Eppendorf microfuge (Germany) for 5 min at 4°C, the supernatant was added to 20 μl of FLAG M2 beads (Sigma), which were washed with lysis buffer three times before immunoprecipitation. Lysates were incubated with the beads on a rotator for 2 hr at 4°C, followed by one wash with buffer A (25 mM Hepes buffer, pH 7.4/0.01% Triton X-100/150 mM NaCl) and one wash with buffer B (the same buffer without the Triton X-100). For binding assays (Fig. 3), 50% (10 μl) of the 9 μM MetAP-2/50 μM Protac-1 mixture was loaded as input; the other 50% was added to the beads. After the addition of ligand, the beads were rotated at room temperature for 1 hr. The beads were washed once each with buffers A and B. After centrifugation at 13,000 rpm in an Eppendorf microfuge (Germany) for 1 min, half of the bead and supernatant fractions, representing bound and unbound, respectively, were evaluated by Western blotting as described above. For ubiquitination reactions, 4 μl of 18 μM MetAP-2, 4 μl of 100 μM Protac-1, 0.5 μl of 0.1 μg/μl purified mouse E1, 1 μl of 0.5 μg/μl human Cdc34 E2, and 1 μl of 25 mM ATP were added to 20 μl (packed volume) of washed FLAG-M2 beads. For competition experiments, the IPP (100 μM final) or OVA (100 μM final) was added simultaneously with the Protac-1. Reactions were incubated for 1 hr at 30°C in a thermomixer (Eppendorf) with constant mixing. SDS/PAGE loading buffer was added to terminate reactions, which were evaluated by Western blotting as described above.
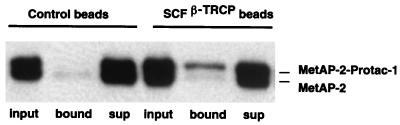
Figure 3.
Protac-1 recruits MetAP-2 to SCFβ-TRCP. Extracts from 293T cells transiently transfected with either control vector or plasmids expressing FLAG epitope-tagged Cul-1 and β-TRCP were subject to affinity purification on anti-FLAG resin to yield either control beads or SCFβ-TRCP beads. The matrices then were mixed with the preformed MetAP-2–Protac-1 complex (input), incubated, and separated into pellet (bound) and unbound (sup) fractions. Proteins were fractionated on an SDS/10% polyacrylamide gel, and immunoblotted with anti-MetAP-2 antiserum. MetAP-2 and MetAP-2–Protac-1 refer to free MetAP-2 and MetAP-2 complexed with Protac-1, respectively.
The experiments performed herein used two different preparations of MetAP-2. One preparation consisted primarily of a 47-kDa fragment that was generated either by a contaminating protease or by slow autoproteolysis. The second preparation consisted almost entirely of full-length 67-kDa MetAP-2. Essentially identical results were obtained with both preparations.
Degradation Experiments with Xenopus Extracts.
Extracts from unfertilized Xenopus laevis eggs were prepared the day of the experiment, as described (12). The MetAP-2–Protac-1 mixture (4 μl of 9 μM MetAP-2 plus 50 μM Protac-1) or MetAP-2 alone was added to 10 μl of extract in addition to OVA (10 μM final), constitutively active IKK (IKK-EE, 0.4 μg), and okadaic acid (OA; 10 μM final). N-acetyl-leu-leu-norleucinal (LLnL, 50 μM final), epoxomicin (10 μM final), or DMSO vehicle were added to the indicated concentrations to inhibit degradation by the proteasome. The protease inhibitors chymostatin, pepstatin, and leupeptin (15 μg/ml final concentration) also were added to the extracts. Reactions were incubated for the indicated times at room temperature and terminated by adding 50 μl of SDS/PAGE loading buffer. Samples were evaluated by Western blotting as described above.
Results
MetAP-2 Specifically Binds Protac-1 in Vitro.
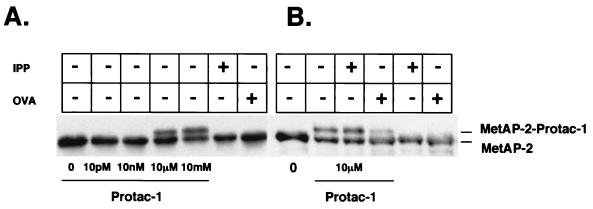
The IκBα-OVA chimera, Protac-1 (Fig. 1B), was synthesized as described in Materials and Methods. To demonstrate that purified MetAP-2 bound Protac-1, we incubated MetAP-2 (18 μM) with increasing concentrations of Protac-1 (Fig. 2A). A Western blot analysis was performed with anti-MetAP-2 antiserum. At Protac-1 concentrations of 10 μM and 10 mM, we observed two bands; the lower band represents unbound MetAP-2, and the upper band represents a complex of MetAP-2 bound to Protac-1. The addition of Protac-1 at higher concentrations did not increase the yield of MetAP-2–Protac-1 complexes, suggesting that only a fraction of the MetAP-2 molecules were active and able to bind Protac-1. Combining MetAP-2 with either free IPP or free OVA did not yield the doublet observed with Protac-1. We also tested the specificity of MetAP-2 interaction with Protac-1 in vitro. Free OVA, but not free IPP, inhibited the formation of the MetAP-2–Protac-1 complex (Fig. 2B). Therefore, our results demonstrate that MetAP-2 was specifically conjugated to the OVA moiety of Protac-1 in a concentration-dependent manner.
Figure 2.
MetAP-2 binds Protac specifically and in a concentration-dependent manner. (A) MetAP-2 (9 μM) was incubated with increasing concentrations of Protac-1 at room temperature for 45 min. The last two lanes depict MetAP-2 that was incubated with either free IPP (50 μM) or free OVA (50 μM), as indicated. After incubation, samples were supplemented with SDS/PAGE loading buffer, separated by SDS/PAGE, and immunoblotted with MetAP-2 antiserum. (B) Same as A, except MetAP-2 (9 μM) plus Protac-1 (10 μM) were supplemented with either IPP (50 μM) or OVA (50 μM), as indicated. Protac binding to MetAP-2 was inhibited by the addition of OVA, but not by the addition of IPP.
Protac-1 Recruits MetAP-2 to SCF.
Before testing the activity of Protac-1, we first adapted an approach to isolate and assay SCFβ-TRCP complexes in vitro as described (13). Lysates from 293T cells transfected with plasmids that encoded β-TRCP and Cul-1 proteins tagged with the FLAG epitope at the N terminus were immunoprecipitated with anti-FLAG antibody-conjugated resin. Immunoblot analysis confirmed that all components of SCFβ-TRCP were present in the anti-FLAG immunoprecipitate, including Skp1, Hrt1, and the transfected FLAGCul-1 and FLAGβ-TRCP (data not shown). Furthermore, control experiments confirmed that these immunoprecipitates promoted ubiquitination of IKK-phosphorylated glutathione S-transferase-IκBα in a manner that was inhibited by the IPP and Protac-1 (data not shown; ref. 7).
To determine whether Protac-1 could recruit MetAP-2 to SCFβ-TRCP, we first incubated MetAP-2 (18 μM) with Protac-1 (100 μM) for 45 min at room temperature. After isolation of SCFβ-TRCP complexes, the anti-FLAG beads were supplemented with the MetAP-2–Protac-1 mixture and rotated at room temperature for 1 hr. The beads and supernatant then were evaluated by Western blot analysis for the presence of MetAP-2. Anti-FLAG beads coated with SCFβ-TRCP, but not control beads preincubated with untransfected 293T cell lysates, specifically retained a fraction of the MetAP-2–Protac-1 complex and not the unliganded MetAP-2 (Fig. 3). These results demonstrate that Protac-1 specifically recruited MetAP-2 to SCFβ-TRCP.
Protac-1 Mediates the Ubiquitination of MetAP-2.
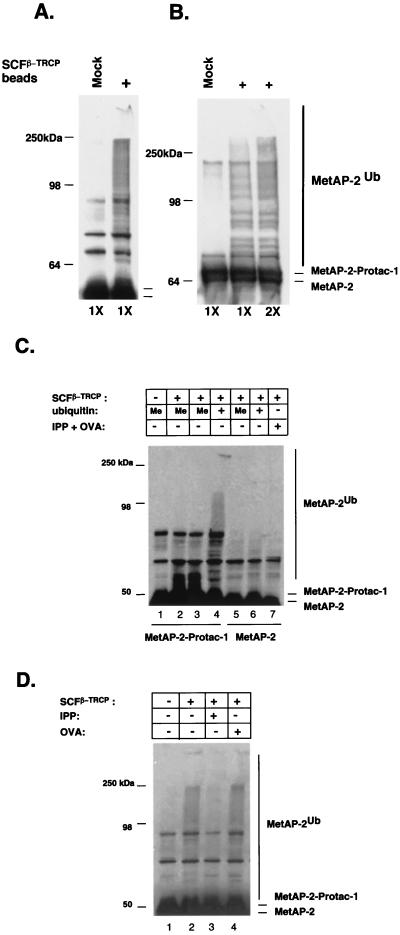
Because Protac-1 was able to tether MetAP-2 to SCFβ-TRCP, we next asked whether MetAP-2 could be ubiquitinated. To answer this question, we supplemented anti-FLAG beads coated with SCFβ-TRCP with ATP, MetAP-2–Protac-1, plus purified E1, E2 (human Cdc34), and ubiquitin. After incubation, Western blot analysis was performed with anti-MetAP-2 antiserum. This experiment was repeated with two different preparations of purified MetAP-2: one contained primarily a 47-kDa autocatalyzed cleavage product (Fig. 4A), and the other contained full-length 67-kDa protein (Fig. 4B). In both cases, MetAP-2–Protac-1 was extensively modified in the presence of SCFβ-TRCP-coated beads but not control beads (Mock). Substitution of the methyl ubiquitin, which acts as a chain-terminator of polyubiquitination, collapsed the high molecular weight forms of modified MetAP-2 to a series of 2–3 bands migrating directly above unmodified MetAP-2–Protac-1 complex (Fig. 4C, compare lanes 2 and 3 with lane 4), confirming that MetAP-2 was ubiquitinated by SCFβ-TRCP.
Figure 4.
Protac mediates MetAP-2 ubiquitination by SCF. (A) Ubiquitination of the 46-kDa fragment of MetAP-2. MetAP-2–Protac-1 mixture was added to either control (Mock) or SCFβ-TRCP beads (+) supplemented with ATP plus purified E1, E2 (Cdc34), and ubiquitin. The E2, UbcH5c (500 ng), was also tested in the reaction, which resulted in the same degree of ubiquitination as observed with Cdc34 (data not shown). Reactions were incubated for 1 hr at 30°C and were evaluated by SDS/PAGE followed by Western blotting with anti-MetAP-2 antiserum. (B) Ubiquitination of full-length (67-kDa) MetAP-2. Same as A, except that the 67-kDa preparation of MetAP-2 was used, and E1, E2, plus ubiquitin were either added at normal (1X) or 2-fold higher (2X) levels, as indicated. (C) Ubiquitination of MetAP-2 by SCFβ-TRCP depends on Protac-1. Same as A, except that methylubiquitin (Me) was substituted for ubiquitin, as indicated, and the reactions depicted in lanes 5–7 lacked Protac-1. In lane 7, unlinked IPP and OVA were added at 100 μM in place of Protac-1. (D) Protac-1-dependent ubiquitination of MetAP-2 is competitively inhibited by IPP. Same as A, except that reactions in lanes 3 and 4 were supplemented with 100 μM each IPP and OVA, respectively.
We next tested whether MetAP-2 ubiquitination depended on Protac-1. As shown in Fig. 4C, MetAP-2 was not ubiquitinated in the absence of either SCFβ-TRCP (lane 1) or Protac-1 (lanes 5 or 6; for some experiments, methyl ubiquitin was used in place of ubiquitin to simplify detection of ubiquitin conjugates). Moreover, unlinked OVA (50 μM) plus IPP (50 μM) were not able to substitute for the OVA–IPP conjugate (lane 7). Protac-1-dependent ubiquitination of MetAP-2 was specific; it was readily competed for by free IPP (Fig. 4D, lane 3 vs. lane 2). In contrast, free OVA did not compete (Fig. 4D, lane 4), presumably because it was unable to displace the Protac-1 that previously was linked covalently to MetAP-2. Taken together, these observations indicate that Protac-1 specifically elicited ubiquitination of MetAP-2 by SCFβ-TRCP, and that successful targeting required that the two components of Protac-1 be bound together as a chimeric molecule.
MetAP-2–Protac-1 Is Degraded in Xenopus Egg Extracts.
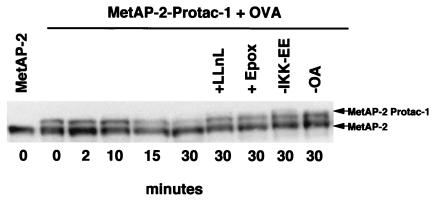
The experiments described above demonstrated that MetAP-2 was ubiquitinated in a Protac-1-dependent manner by highly purified SCFβ-TRCP. However, the key issues are whether Protac-1 can specifically activate MetAP-2 degradation, and whether targeted degradation can be achieved by endogenous ubiquitin/proteasome pathway components at typical intracellular concentrations. To address these questions, we preincubated MetAP-2 with Protac-1 to allow the complexes to form, and then added the mixture to Xenopus egg extract supplemented with IKK-EE (14), OA, and OVA. The addition of IKK-EE and OA was intended to sustain phosphorylation of the IκBα peptide moiety of Protac-1, whereas OVA was added to prevent the further linkage of Protac-1 to MetAP-2. Remarkably, MetAP-2–Protac-1 complex (top band) but not MetAP-2 alone (bottom band) was mostly degraded after 30 min (Fig. 5). Degradation of MetAP-2–Protac-1 was attenuated in extracts supplemented with the proteasome inhibitors LLnL or epoxomicin (15), but not by other protease inhibitors (chymotrypsin, pepstatin, and leupeptin) added to the reaction. Moreover, addition of both IKK-EE and OA was required for optimal degradation of MetAP-2–Protac-1. Similarly, we have seen specific turnover of the MetAP-2–Protac-1 complex, but not free MetAP-2, in three independent experiments. Because the IPP does not have lysines and the OVA does not have free amino groups, it is unlikely that Protac itself serves as a target for ubiquitin-dependent proteolysis. Taken together, these results suggest that Protac-1 targeted MetAP-2 for degradation by means of the proteasome. MetAP-2 turnover seemed to be very specific, in that maximal degradation required agents predicted to sustain phosphorylation of the IκBα peptide.
Figure 5.
MetAP-2-Protac is degraded in Xenopus extracts. The MetAP-2–Protac-1 mixture or MetAP-2 alone was added to Xenopus egg extract fortified with OVA (100 μM), IKK-EE (0.4 μg), and OA (10 μM). Where indicated, reactions were either deprived of IKK-EE or OA, or were further supplemented with 50 μM LLnL or 10 μM epoxomicin (Epox). Reactions were incubated for the indicated times at room temperature, terminated by adding SDS/PAGE loading buffer, and evaluated by SDS/PAGE followed by Western blotting with anti-MetAP-2 antiserum.
Discussion
Because degradation of ubiquitinated proteins occurs rapidly in cells, we reasoned that ubiquitin-dependent proteolysis might provide an effective means to modulate the phenotype of normal and diseased cells. Thus, we sought to develop a method to target proteins, at will, to the ubiquitin/proteasome pathway. The linchpin of the strategy described here is the development of chimeric molecules—referred to as Protacs—that link a desired target protein to a ubiquitin ligase. A related method for regulated activation and termination of signaling pathways with synthetic ligands has been described (10). As a target protein for a proof of principle experiment, we chose MetAP-2, which covalently binds the angiogenesis inhibitor OVA. For the ubiquitin ligase, we chose the SCFβ-TRCP complex, for reasons described in more detail below. Although MetAP-2 has 36 lysines, it has not been reported to be an unstable protein in vivo, and it was not clear whether it would serve as a substrate for SCFβ-TRCP, or whether it would be degraded by the ubiquitin/proteasome pathway (16). Our results suggest that Protac-1 in fact can recruit MetAP-2 to SCFβ-TRCP for ubiquitination. In addition, we report that Protac-1 specifies degradation of MetAP-2 by the endogenous ubiquitin/proteasome pathway in Xenopus egg extracts.
We selected SCFβ-TRCP as a candidate ubiquitin ligase for the development of Protac technology for two reasons. First, the apparent constitutive activity of SCF complexes (3) enables a general strategy to manipulate normal or diseased cells. Second, the mechanism underlying substrate selection is well understood for SCFβ-TRCP. Pioneering work by Ben-Neriah and coworkers (4, 5) demonstrated that a 10-aa internal phosphopeptide mediates ubiquitination and degradation of IκBα. Subsequently, it was established that β-TRCP is the receptor that links this phosphopeptide to the ubiquitin/proteasome pathway (4, 17). Furthermore, while our work was underway, Zhou, Howley, and colleagues (18) demonstrated that engineered SCF complexes can be used to target heterologous proteins for destruction. Zhou et al. fused the human papillomavirus E7 protein to the F-box proteins Cdc4 and β-TRCP to create chimeras, which assemble to form hybrid SCFCdc4-E7 and SCFβ-TRCP-E7 complexes. E7 binds tightly to retinoblastoma protein (Rb), and the F-box protein/E7 chimeras stimulate turnover of Rb by means of the SCF pathway in both yeast and mammalian cells. A limitation of the F-box fusion approach is that it depends on gene transfer of the chimeric F-box protein; thus, its use is limited to transgenic organisms. Its potential use as a therapeutic strategy awaits the development of safe and reliable gene therapy protocols.
We propose that Protacs may be useful research tools for manipulating the phenotype of cells by means of the targeted elimination of specific proteins, or as useful therapeutic agents for targeting the elimination of disease-promoting proteins. An obstacle to realizing these goals, however, is that the phosphopeptide-containing Protac-1 described here is unlikely to penetrate cells. For future applications, Protacs will need to be modified to enhance delivery to cells. For example, attachment of the tat peptide (19) may promote transduction of Protac-1 into cells, or a β-TRCP-binding peptide that is derived from the HIV Vpu protein (20, 21), and that is phosphorylated by constitutively active casein kinase II may allow delivery of an unphosphorylated Protac to cells. The ultimate goal is to identify small molecules that can substitute for the E3 targeting activity of the IPP.
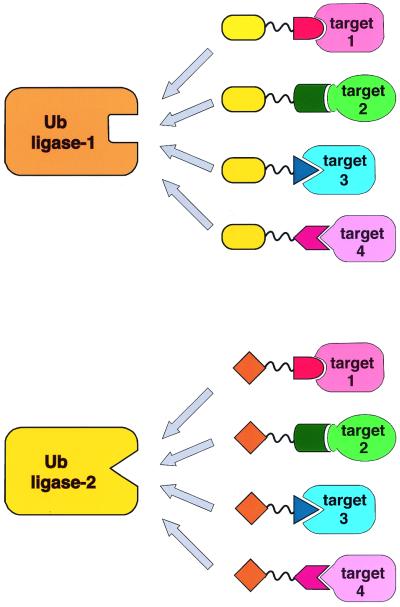
We envision the potential to develop an entire suite of Protac compounds (Fig. 6). Several approaches have been developed for identifying small molecules that bind to any target protein of interest (22–24). Candidates emerging from such screens would serve as platforms for the production of new Protacs, regardless of the topology of their interaction with the target protein. Because the human genome encodes a large number of RING- and HECT-based ubiquitin ligases, it should be possible to develop multiple Protacs that target a single protein to allow for fine-tuning its rate of degradation. Moreover, because some ubiquitin ligases are likely to exhibit restricted patterns of expression, it even may be possible to develop Protac-based drugs whose effects are limited to specific tissues.
Figure 6.
General application of Protacs. A schematic of how different disease-promoting proteins might be recruited to different ubiquitin ligases for ubiquitination and degradation by unique Protacs.
Acknowledgments
We thank the members of the Deshaies lab for their helpful suggestions and Zhen-Quiang Pan for providing reagents and valuable input on the reconstitution of SCFβ-TRCP activity. This work was supported by the University of California Los Angeles Jonsson Comprehensive Cancer Center, CapCURE (K.M.S., R.J.D., and C.M.C.), the Gates Grubstake Fund (R.J.D.), and the Howard Hughes Medical Institute (R.J.D.). K.M.S. is a Scholar of the Leukemia and Lymphoma Society of America.
Abbreviations
- E1
ubiquitin-activating enzymes
- E2
ubiquitin-conjugating enzymes
- E3
ubiquitin–protein ligases
- SCF
Skp1–Cullin–F box
- IKK
IκBα kinase
- IPP
IκBα phosphopeptide
- MetAP-2
methionine aminopeptidase-2
- OVA
ovalicin
- Protac
proteolysis targeting chimeric
- IKK-EE
constitutively active IKK
- OA
okadaic acid
References
- 1.Ciechanover A, Orian A, Schwartz A L. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 2.Winston J F, Koepp D M, Zhu C, Elledge S J, Harper J W. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- 3.Deshaies R J. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 4.Yaron A, Hatzubai A, Davis M, Lagoon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Ben-Neriah Y. Nature (London) 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 5.Yaron A, Gonen H, Alakalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben-Neriah Y. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Chang Y-H. Proc Natl Acad Sci USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sin N, Meng L, Wang M Q, Wen J J, Bornmann W G, Crews C M. Proc Natl Acad Sci USA. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith E C, Su Z, Turk B E, Chen S, Chang Y H, Wu Z, Biemann K, Liu J O. Chem Biol. 1997;4:461–471. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 9.Yeh J-R J, Mohan R, Crews C M. Proc Natl Acad Sci USA. 2000;97:12782–12787. doi: 10.1073/pnas.97.23.12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyapina S A, Correll C C, Kipreos E T, Deshaies R J. Proc Natl Acad Sci USA. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray A W. Methods Cell Biol. 1991;3:581–605. [PubMed] [Google Scholar]
- 13.Tan P, Fuchs S Y, Chen A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Mol Cell. 1999;3:327–333. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 14.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennet B L, Li J W, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 15.Meng L, Mohan R, Kwok B H B, Elofsson M, Sin N, Crews C M. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Widom J, Kemp C W, Crews C M, Clardy J. Science. 1998;282:324–327. doi: 10.1126/science.282.5392.1324. [DOI] [PubMed] [Google Scholar]
- 17.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 18.Zhou P, Bogachi R, McReynolds L, Howley P M. Mol Cell. 2000;6:751–756. doi: 10.1016/s1097-2765(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 19.Nagahara H, Vocero-Akbani A M, Snyder E L, Ho A, Latham D G, Lissy N A, Becker-Hapak M, Ezhevsky S A, Dowdy S F. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 20.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 21.Schubert U, Antón L C, Bacík I, Cox J H, Bour S, Bennink J R, Orlowski M, Strebel K, Yewdell J W. J Virol. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borchardt A, Liberles S D, Biggar S R, Crabtree G R, Schreiber S L. Chem Biol. 1997;4:961–968. doi: 10.1016/s1074-5521(97)90304-5. [DOI] [PubMed] [Google Scholar]
- 23.MacBeath G, Koehler A N, Schreiber S L. J Am Chem Soc. 1999;121:7967–7968. [Google Scholar]
- 24.You A J, Jackman R J, Whitesides G M, Schreiber S L. Chem Biol. 1997;4:969–975. doi: 10.1016/s1074-5521(97)90305-7. [DOI] [PubMed] [Google Scholar]