Abstract
Please cite this paper as: Chao. (2012) Modelling the global transmission of antiviral‐resistant influenza viruses. Influenza and Other Respiratory Viruses 7(Suppl. 1), 58–62.
Background The mutations that confer resistance to antiviral agents are thought to be detrimental, or at best neutral, to influenza virus fitness. The fact that resistant influenza strains can circulate and sometimes replace sensitive strains is of great public health concern.
Objectives We used mathematical modeling to test various hypotheses about the transmission of antiviral‐resistant influenza viruses by comparing the model’s output with the observed rise in antiviral resistance of seasonal A(H1N1) influenza viruses between 2006 and 2009.
Methods We developed a mathematical model of the transmission of influenza among 321 cities around the globe. In the model, influenza strains resistant to antiviral agents competed with sensitive strains.
Results and Conclusions We found that a resistant strain of influenza could not displace the sensitive strain as rapidly as has been observed unless it was more transmissible than the sensitive strain in the general population. We believe that an antiviral‐resistant strain displaced the antiviral‐sensitive seasonal A(H1N1) virus by hitchhiking on an escape mutation. Because of the complex global patterns of influenza circulation, tracking the emergence and spread of antiviral resistance must be a coordinated global effort.
Keywords: Antiviral agents, influenza, mathematical modeling, neuraminidase inhibitors, resistance
Introduction
Antiviral agents are part of the initial response plans for pandemic influenza and play a key role in protecting high‐risk populations from seasonal influenza. 1 , 2 , 3 In recent years, the widespread circulation of seasonal A(H3N2) viruses resistant to the aminoadamantanes and seasonal A(H1N1) viruses resistant to the neuraminidase inhibitor oseltamivir has been observed. 4 , 5 , 6 The loss of effectiveness of antiviral agents is an important public health concern, and worldwide, surveillance has been established to monitor the situation. 7
Although influenza virus mutations that reduce the efficacy of antiviral agents often arise in those taking the drugs, the risk of transmission had been believed to be minimal because the mutations reduced viral fitness. 8 , 9 , 10 , 11 , 12 , 13 However, in certain genetic backgrounds, the mutations that confer resistance do not attenuate virus replication or transmission, as was found with oseltamivir resistance in seasonal H1N1. 14 , 15 , 16 , 17
To explore the conditions under which antiviral‐resistant influenza viruses can spread, we developed a mathematical model of influenza transmission (Figure 1). We used the model to test various hypotheses about how a resistant strain of influenza can displace a sensitive strain and offer suggestions on how to slow this process.
Figure 1.
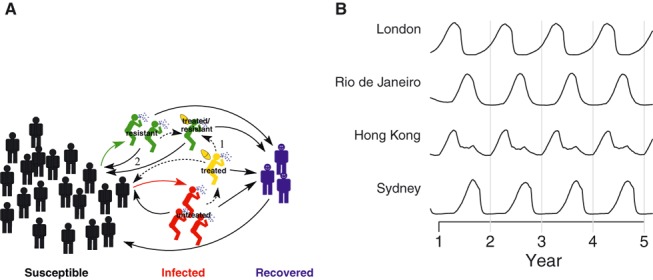
A mathematical model of influenza transmission. (A) Each city in the model is represented as a population of individuals susceptible to influenza. A susceptible individual can become infected by contact with infected individuals. Those infected may be treated with antiviral agent, which lowers their infectiousness. However, treated people may develop a resistant strain of influenza, which can co‐circulate with the sensitive strain. An individual becomes susceptible again after recovering from influenza. The numbered arrows highlight two pathways that generate more cases of resistant influenza: (1) treatment of infected individuals with antiviral agent and (2) transmission of resistant influenza virus. (B) Influenza season varies across the globe in the model. Generally, the influenza season is during the winter in the temperate Northern and Southern Hemispheres, and seasonality is less pronounced in the tropics. The plot shows the simulated relative prevalence of influenza in four representative cities in the model, one in the temperate Northern Hemisphere (London), one in the temperate Southern Hemisphere (Sydney), and two that are in the tropics (Brazil and Hong Kong).
Methods
We developed a mathematical model of the global transmission of influenza. 18 Briefly, the model represents the populations of 321 cities across the globe. Each city is modeled as a well‐mixed population of susceptible, influenza‐infected, and recovered individuals (Figure 1A). A susceptible individual can become infected by contact with infected individuals in the same city. Soon after infection, an individual might be treated with an antiviral agent, which lowers that person’s infectiousness. Treated individuals may, with low probability, produce a drug‐resistant influenza strain, which could then co‐circulate with the sensitive strain. After recovery from influenza, a person is immune for a brief interval before becoming susceptible again. Infected individuals can travel between cities, with travel frequency based on airline transportation data, and can therefore spread influenza to other regions. Generally, influenza is most transmissible during the winter in the temperate Northern and Southern Hemispheres and is transmissible year‐round in the tropics. 19 , 20 To account for the different patterns of seasonality around the world, the transmissibility of influenza is raised during a region’s typical influenza season and lowered at other times in the model (Figure 1B). 21
Results
Using our mathematical model, we test various hypotheses that could explain the rapid spread of oseltamivir‐resistant seasonal H1N1 influenza viruses between 2006 and 2009. In our model, there are two explicit mechanisms that can cause the resistant strain of influenza to outcompete the sensitive one, as shown in Figure 1A: (i) Extensive drug use continuously generates a substantial number of resistant viruses and (ii) the resistant strain is inherently more transmissible than the sensitive strain, allowing it to sustain its own expansion once established in the general population. An alternative hypothesis is that random chance alone was responsible for the spread of resistance.
Did the use of antiviral agent drive the spread of resistant seasonal H1N1?
If a sufficient number of infected individuals are treated with antiviral agents, which are known to promote the development of resistant influenza viruses, then the overall proportion of resistance among circulating strains could rise (Figure 1A, pathway 1). We tested this hypothesis in our model by assuming that the same fraction of cases around the world are treated, resistance develops with a probability of 4% per day under antiviral treatment, 18 and that the resistant strain is exactly as transmissible as the sensitive strain among untreated individuals. We varied the fraction of cases treated, and compared the model output with the observed annual fraction of circulating seasonal H1N1 that was resistant to oseltamivir, which we estimated by identifying sequences from the Influenza Virus Resource and GISAID databases with the H275Y mutation. We found that approximately 30% of infected individuals worldwide needed to be treated with antiviral agent in order to match the observed rapid rise of oseltamivir‐resistant seasonal H1N1 (Figure 2A). We estimated that, in reality, <3% of influenza cases took oseltamivir in the parts of the world represented in our model. In fact, we suspect that the actual fraction of cases treated around the globe was even lower, because our model under‐represents the developing world by preferentially including cities with major airports.
Figure 2.
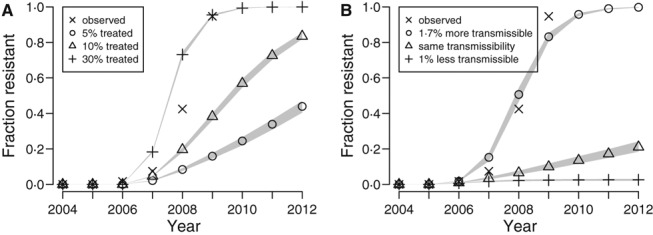
Exploring hypotheses of the spread of antiviral‐resistant influenza virus using a mathematical model. (A) We ran simulations to estimate what fractions of influenza cases around the world needed to take oseltamivir to drive the spread of resistance as quickly as was observed for seasonal H1N1, assuming that the resistant strain was exactly as transmissible as the sensitive strain among untreated individuals. The Xs show the observed proportion of circulating seasonal H1N1 strains that were resistant according to surveillance data. The other points represent the median values from 500 simulation runs, and the gray shaded areas cover the outcomes of 95% of the runs. We estimate that <3% of influenza cases around the world used oseltamivir. (B) We simulated the spread of a resistant strain of influenza that is 1% less transmissible, exactly as transmissible, or 1·7% more transmissible than the sensitive strain, and compared the results to the observed prevalence of resistant seasonal H1N1.
How transmissible was the antiviral‐resistant strain of seasonal H1N1 compared to the sensitive strain?
Once a resistant variant of influenza enters the general population, it may be transmissible enough to outcompete sensitive strains (Figure 1A, pathway 2). We used estimates of the number of infected individuals taking oseltamivir around 2006 7 , 18 , 22 , 23 and ran simulations in which the resistant strain was slightly more or less transmissible than the sensitive strain in untreated individuals.
We found that if the resistant strain is even slightly less transmissible than the sensitive strain, it would not spread, and even a strain that was exactly as transmissible would spread only slowly (Figure 2B). Thus, we conclude that a mutation associated with any attenuation in viral fitness could not spread. We find that the resistant strain needs to be 1–2% more transmissible in untreated individuals in order to spread as rapidly as was observed with seasonal H1N1.
The right genetic background might restore transmissibility of a resistant virus, 14 , 15 , 16 , 17 but resistance per se is unlikely to make a circulating influenza virus inherently more transmissible. It has been suggested that the resistance mutation “hitchhiked” on another mutation that made the virus more transmissible. Such hitchhiking is thought to be responsible for the spread of aminoadamantane resistance in seasonal H3N2 24 and oseltamivir resistance in seasonal H1N1. 6 , 16 , 25 , 26
Could antiviral‐resistant influenza viruses spread by random chance alone?
One could imagine that a resistant strain of influenza that is exactly as transmissible as a sensitive one could displace the sensitive one by chance. Our model is stochastic, meaning that it explicitly includes random effects. The results we present in Figures 2A,B show 95% of the range of outcomes from our stochastic simulations. One can see that when the resistant strain is exactly as transmissible as the sensitive one, the spread of resistance is usually negligible (Figure 2B). We conclude that random effects play only a minor role when considering the spread of viruses in a large population and that it is extremely unlikely that chance alone could cause a non‐competitive new influenza strain to displace circulating strains.
Reducing transmission from those treated with antiviral agents
Because surveillance efforts usually detect antiviral resistance mutations first among patients taking the drug, 1 these individuals are an obvious focus for resistance prevention strategies. If one could systematically reduce transmission from these potential sources of resistance, whether by more careful isolation of patients or better respiratory hygiene, one would reduce the rate of introduction of new resistant strains into the general population. If the resistant strain were no more transmissible than the sensitive strain, we find that this strategy slows the spread of resistance (Figure 3A). However, if the resistant strain is more transmissible, then this strategy does not slow the spread of resistance, although it might delay the onset of the rapid replacement of sensitive viruses (Figure 3B). In other words, once transmission of a resistant strain is sustained in the general population, controlling transmission from those treated with antiviral agent has little effect.
Figure 3.
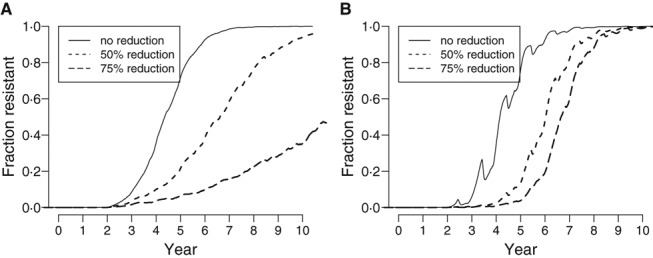
The simulated effects of reducing transmission from those treated with antiviral agent. We simulated the effect of reducing the transmission by 0%, 50%, and 75% from all individuals taking antiviral agent. (A) We assumed that 5% of all infected individuals worldwide took antiviral agent and that the resistant strain was exactly as transmissible as the sensitive strain. (B) We assumed that the fraction of infected individuals taking antiviral agent matched our 2006 estimates and that the resistant strain was 1·7% more transmissible than the sensitive strain. Each curve shows the results of a single stochastic simulation.
Conclusions
We used a mathematical model to explore the factors that would allow an antiviral‐resistant strain of influenza to displace a sensitive one. We show that aggressive use of antiviral agents, covering 30% of all cases, could drive the rapid spread of resistance, but this level of usage is not consistent with levels used in the past. The more likely explanation for the loss of antiviral susceptibility of seasonal H1N1 viruses between 2006 and 2009 is that the resistant virus was more transmissible than the sensitive ones.
Our model indicates that a strain of influenza that is only 1–2% more transmissible than other circulating strains can rapidly displace them. It may be difficult to detect such a small viral fitness difference experimentally. In addition, in vitro assays would be unable to detect the enhanced fitness of antigenic escape mutants. Immune pressure from prior exposure to influenza in the human population promotes the spread of escape mutants and is probably responsible for the frequent turnover of influenza strains every 2–8 years. 27 , 28 Therefore, the identification of the genetic background required to restore transmissibility to resistant influenza viruses in vitro is essential, but the actual transmissibility of a resistant strain “in the wild” is also affected by its antigenic profile and the current background of population‐level immunity to circulating strains.
It may be tempting to focus surveillance efforts in regions where antiviral agents are used the most and where one might expect resistance to arise. However, influenza strains might not persist between influenza seasons, as each season a region’s circulating strains are imported from around the globe. 29 Therefore, the regional prevalence of antiviral resistance might not correlate with regional usage of antiviral agents. 6 , 23 , 30
Surveillance of antiviral resistance must therefore be a coordinated global effort. 7 The first weeks of the 2009 H1N1 pandemic demonstrated that new strains of influenza travel along major international airline routes. 31 , 32 The airline transportation network is the backbone of the global spread of person‐to‐person transmitted infectious diseases. 33 , 34 , 35 However, it is not enough to simply follow the flow of people. The different patterns of influenza seasonality around the world determine when new strains of influenza can take root in a region. The interplay between human migration and influenza seasonality creates the complex global patterns of influenza circulation, which many groups are beginning to describe. 36 , 37 , 38 , 39 Our mathematical model represents our efforts to capture these patterns, and further research is needed to build systems to help us predict the spread of antiviral resistance.
Conflicts of interest
The author has no conflicts of interest to declare.
Acknowledgements
This work was partially supported by the National Institute of General Medical Sciences MIDAS grant U01‐GM070749. The author would like to thank the International Society for Influenza and other Respiratory Virus Diseases (isirv) for their inaugural “Influenza Antivirals: Efficacy and Resistance” conference, where many fruitful discussions took place.
References
- 1. Ward P, Small I, Smith J, Suter P, Dutkowski R. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J Antimicrob Chemother 2005; 55:i5–i21. [DOI] [PubMed] [Google Scholar]
- 2. Schünemann HJ, Hill SR, Kakad M et al. WHO Rapid Advice Guidelines for pharmacological management of sporadic human infection with avian influenza A (H5N1) virus. Lancet Infect Dis 2007; 7:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM. Antiviral agents for the treatment and chemoprophylaxis of influenza — recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2011; 60:1–24. [PubMed] [Google Scholar]
- 4. Weinstock DM, Zuccotti G. The evolution of influenza resistance and treatment. JAMA 2009; 301:1066–1069. [DOI] [PubMed] [Google Scholar]
- 5. Moscona A. Global transmission of oseltamivir‐resistant influenza. N Engl J Med 2009; 360:953–956. [DOI] [PubMed] [Google Scholar]
- 6. Baranovich T, Saito R, Suzuki Y et al. Emergence of H274Y oseltamivir‐resistant A(H1N1) influenza viruses in Japan during the 2008–2009 season. J Clin Virol 2010; 47:23–28. [DOI] [PubMed] [Google Scholar]
- 7. Monto AS, McKimm‐Breschkin JL, Macken C et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 2006; 50:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carr J, Ives J, Kelly L et al. Influenza virus carrying neuraminidase with reduced sensitivity to oseltamivir carboxylate has altered properties in vitro and is compromised for infectivity and replicative ability in vivo. Antiviral Res 2002; 54:79–88. [DOI] [PubMed] [Google Scholar]
- 9. Herlocher ML, Carr J, Ives J et al. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res 2002; 54:99–111. [DOI] [PubMed] [Google Scholar]
- 10. Bouvier NM, Lowen AC, Palese P. Oseltamivir‐resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J Virol 2008; 82:10052–10058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ives JA, Carr JA, Mendel DB et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res 2002; 55:307–317. [DOI] [PubMed] [Google Scholar]
- 12. Herlocher ML, Truscon R, Elias S et al. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J Infect Dis 2004; 190:1627–1630. [DOI] [PubMed] [Google Scholar]
- 13. Abed Y, Goyette N, Bovin G. A reverse genetics study of resistance to neuraminidase inhibitors in an influenza A/H1N1 virus. Antivir Ther 2004; 9:577–581. [PubMed] [Google Scholar]
- 14. Rameix‐Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog 2008; 4:e1000103 Available from: http://dx.doi.org/10.1371%2Fjournal.ppat.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baz M, Abed Y, Simon P, Hamelin ME, Boivin G. Effect of the neuraminidase mutation H274Y conferring resistance to oseltamivir on the replicative capacity and virulence of old and recent human influenza A(H1N1) viruses. J Infect Dis 2010; 201:740–745. [DOI] [PubMed] [Google Scholar]
- 16. Matsuzaki Y, Mizuta K, Aoki Y et al. A two‐year survey of the oseltamivir‐resistant influenza A(H1N1) virus in Yamagata, Japan and the clinical effectiveness of oseltamivir and zanamivir. Virol J 2010; 7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 2010; 328:1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chao DL, Bloom JD, Kochin B, Antia R, Longini IM Jr. The global spread of drug‐resistant influenza. J R Soc Interface 2012; 9:648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lofgren E, Fefferman NH, Naumov YN, Gorski J, Naumova EN. Influenza seasonality: underlying causes and modeling theories. J Virol 2007; 81:5429–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tamerius J, Nelson MI, Zhou SZ, Viboud C, Miller MA, Alonso WJ. Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ Health Perspect 2011; 119:439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenah E, Chao DL, Matrajt L, Halloran ME, Longini IM Jr. The global transmission and control of influenza. PLoS ONE 2011; 6:e19515 Available from: http://dx.doi.org/10.1371%2Fjournal.pone.0019515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. F Hoffmann‐La Roche Ltd . Media release: Roche announces further progress in Tamiflu production expansion; 2005. http://www.roche.com/med-cor-2005-11-07, accessed on October 4, 2010.
- 23. Kramarz P, Monnet D, Nicoll A, Yilmaz C, Ciancio B. Use of oseltamivir in 12 European countries between 2002 and 2007–lack of association with the appearance of oseltamivir‐resistant influenza A(H1N1) viruses. Euro Surveill 2009; 14:19112. [DOI] [PubMed] [Google Scholar]
- 24. Simonsen L, Viboud C, Grenfell BT et al. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 2007; 24:1811–1820. [DOI] [PubMed] [Google Scholar]
- 25. Zaraket H, Saito R, Suzuki Y et al. Genetic makeup of amantadine‐resistant and oseltamivir‐resistant human influenza A/H1N1 viruses. J Clin Microbiol 2010; 48:1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niman HL. Emergence and fixing of antiviral resistance in influenza A via recombination and hitch hiking. Nature Precedings. 2009; Available at http://hdl.handle.net/10101/npre.2009.2832.1.
- 27. Plotkin JB, Dushoff J, Levin SA. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc Natl Acad Sci USA 2002; 99:6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith DJ, Lapedes AS, de Jong JC et al. Mapping the antigenic and genetic evolution of influenza virus. Science 2004; 305:371–376. [DOI] [PubMed] [Google Scholar]
- 29. Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog 2007; 3:1220–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meijer A, Lackenby A, Hungnes O et al. Oseltamivir‐resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis 2009; 15:552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khan K, Arino J, Hu W et al. Spread of a novel influenza A (H1N1) virus via global airline transportation. N Engl J Med 2009; 361:212–214. [DOI] [PubMed] [Google Scholar]
- 32. Fraser C, Donnelly CA, Cauchemez S et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009; 324:1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rvachev LA, Longini IM Jr. A mathematical model for the global spread of influenza. Math Biosci 1985; 75:3–22. [Google Scholar]
- 34. Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci USA 2004; 101:15124–15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Balcan D, Colizza V, Gonçalves B, Hu H, Ramasco JJ, Vespignani A. Multiscale mobility networks and the spatial spreading of infectious diseases. Proc Natl Acad Sci USA 2009; 106:21484–21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell CA, Jones TC, Barr IG et al. The global circulation of seasonal influenza A (H3N2) viruses. Science 2008; 320:340–346. [DOI] [PubMed] [Google Scholar]
- 37. Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature 2008; 453:615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bedford T, Cobey S, Beerli P, Pascual M. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog 2010; 6:e1000918 Available from: http://dx.doi.org/10.1371%2Fjournal.ppat.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bahl J, Nelson MI, Chan KH et al. Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc Natl Acad Sci USA 2011; 108:19359–19364. [DOI] [PMC free article] [PubMed] [Google Scholar]


