Abstract
Mutant p53 (mtp53) gain of function (GOF) contributes to various aspects of tumor progression including cancer stem cell (CSC) property acquisition. A key factor of GOF is stabilization and accumulation of mtp53. However, the precise molecular mechanism of the mtp53 oncogenic activity remains unclear. Here, we show that ribophorin II (RPN2) regulates CSC properties through the stabilization of mtp53 (R280K and del126-133) in breast cancer. RPN2 stabilized mtp53 by inactivation of glycogen synthase kinase-3β (GSK3β) which suppresses Snail, a master regulator of epithelial to mesenchymal transition. RPN2 knockdown promoted GSK3β-mediated suppression of heat shock proteins that are essential for mtp53 stabilization. Furthermore, our study reveals that high expression of RPN2 and concomitant accumulation of mtp53 were associated with cancer tissues in a small cohort of metastatic breast cancer patients. These findings elucidate a molecular mechanism for mtp53 stabilization and suggest that RPN2 could be a promising target for anti-CSC therapy.
Recent studies show that some p53 mutations result in the loss of tumor-suppressing function (LOF) by the mutant allele and trans-dominant inactivation of the remaining wtp531. Importantly, the mutant p53 (mtp53) contributes to tumor progression. This mechanism is referred to as mtp53 gain of function (GOF). A key property of GOF is the stabilization and accumulation of mtp532. The mtp53 protein is rescued from degradation and contributes to malignant phenotypes such as invasion and metastasis or genomic instability by binding and inactivating p63 and Mre11, respectively3,4,5. In contrast to wild-type p53 (wtp53), the mtp53 protein adopts an aberrant conformation. Mtp53 forms stable complexes with heat shock proteins HSP90 and HSP70, MDM2, and the carboxyl terminus of HSP70-interacting protein (CHIP), which prevent mtp53 misfolding and aggregation6,7. In malignant cancer cells, the HSP90 and HSP70 chaperone machinery is upregulated and activated to protect mutated and overexpressed oncoproteins from degradation8,9. While several studies succeeded in identifying the molecular mechanisms that regulate mtp53 stability6,7 and the small molecules that induce mtp53 destabilization6,10, the mechanism that leads to mtp53 stabilization is not yet fully understood.
In breast cancer, p53 LOF or mutation induces epithelial to mesenchymal transition (EMT)11, which contributes to cancer progression and metastasis12,13. Several studies show a link between EMT and the acquisition of CSC properties11,14. Ectopic expression of EMT regulators, such as Twist and Snail, or shRNA-mediated knockdown of E-cadherin confer cancer stem cell (CSC) properties to mammary epithelial cells12. Snail expression is suppressed by glycogen synthase kinase-3β (GSK3β) at transcriptional and post-transcriptional levels15,16, and the inhibition of GSK3β by small molecules induces EMT and promotes CSC phenotypes in breast cancer17. Despite the critical role of GSK3β in the regulation of CSCs phenotypes, the physiological and molecular mechanisms underlying its function remain unclear.
Breast CSCs exhibit a CD44+CD24−/low antigenic phenotype with low expression of epithelial markers such as E-cadherin, and are characterized by high tumorigenicity and drug resistance14,18. Previously, we showed that ribophorin II (RPN2) is a novel regulator of drug resistance in breast cancer and affects docetaxel resistance by modulating the N-linked glycosylation of P-glycoprotein (ABCB1)19. To gain further insight into the regulation of mtp53 stability in CSCs, we screened for possible interactions between RPN2 and GSK3β. The present study suggests that the stabilization of mtp53 (R280K and del126-133) in breast cancer depends on RPN2 inhibition of GSK3β-mediated inactivation of HSP70 and HSP90 that are essential factors for the stabilization and oncogenic activities of mtp53.
Results
RPN2 is highly expressed in the CSC fraction
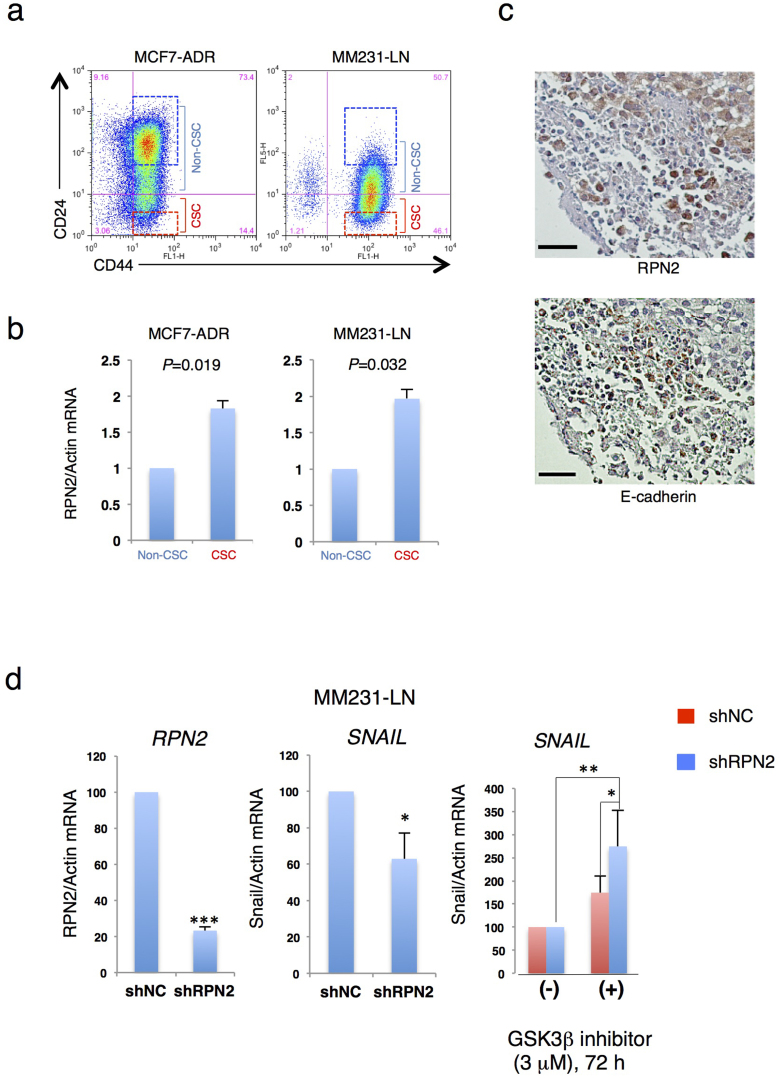
RPN2 was initially considered essential for the maintenance of CSCs because CSCs exhibit resistance to conventional chemotherapy18,20. To test this hypothesis, the expression of RPN2 in the CSC fraction was examined using two breast cancer cell lines, MCF7-ADR drug resistant human breast cancer cells and MDA-MB-231-D3H2-LN highly metastatic human breast cancer cells (MM231-LN). Since CSC fraction in breast cancer cells is reported to show the resistance to chemotherapy and the metastatic ability21,22, we selected these two cell lines. Flow cytometry analysis showed that MCF7-ADR and MM231-LN cells comprise approximately 15% and 50% of CSCs, respectively (Fig. 1a). In breast CSCs, the CD44high/CD24low fraction shows higher tumorigenicity than the CD44high/CD24high fraction23. Flow cytometry and quantitative reverse transcription PCR (qRT-PCR) of MCF7-ADR and MM231-LN cells showed that RPN2 was more highly expressed in the CSC fraction than in the non-CSC fraction (Fig. 1b). In addition, the CSC fraction of MM231-LN showed high tumorigenicity in an animal model (Suppl. Fig. S1). Immunostaining analysis showed inverse correlation between RPN2 and E-cadherin expression in MM231-LN xenograft tumors (Fig. 1c).
Figure 1. RPN2 is essential for the maintenance of the CSC fraction.
(a) The CSC fraction derived from MCF7-ADR and MM231-LN cells. (b) MCF7-ADR and MM231-LN cells were segregated by fluorescence-activated cell sorting (FACS) into CD44high/CD24low and CD44high/CD24high subsets; sorted subsets were then compared for RPN2 expression by quantitative real-time PCR (qRT-PCR). Each data point is the average of three experiments. (c) Immunohistochemistry (IHC) for RPN2 (Top panel) and E-cadherin (Bottom panel) in MM231-LN tumors. MM231-LN xenografts were grown for 5–6 weeks after fat pad injection. Sections are representative of at least four mice analyzed per group. Scale bar, 200 μm. (d) RPN2 knockdown suppressed Snail expression via GSK3β activation. In MM231-LN cells, RPN2 and Snail mRNA expression was monitored by qRT-PCR after 72 h of treatment with a GSK3β inhibitor (CHIR99021, 3 μM). (n = 3, *P < 0.05, **P < 0.01,***P < 0.001).
To further investigate the role of RPN2 in CSCs, RPN2 knockdown experiments were performed in the two breast cancer cell lines, MCF7-ADR and MM231-LN, using lentivirus vectors expressing GFP and a small hairpin RNA against RPN2 (shRPN2-site2) (Suppl. Fig. S2A and S2B). RPN2 knockdown reduced the E-cadherin negative fraction in MM231-LN cells as detected by flow cytometry analysis (Suppl. Fig. S3A). We also found that RPN2 knockdown induced Snail suppression in MM231-LN cells (Fig. 1d). Compared with MM231-LN shNC, a 40% decrease in Snail expression was observed in MM231-LN shRPN2 (Fig. 1d). Moreover, the GSK3β inhibitor CH99021 caused Snail upregulation in MM231-LN shRPN2 (Fig. 1d).
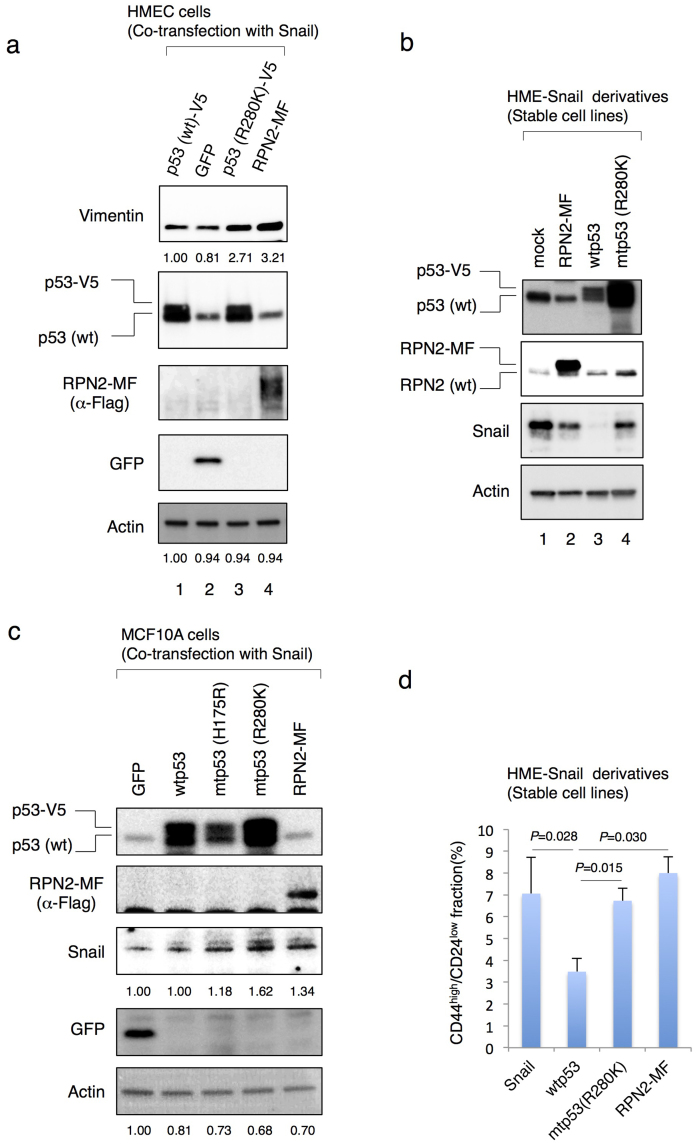
Consistent with the previous studies showing loss of epithelial phenotype by inactivation of p5311,14, we confirmed that the ectopic expression of Snail and a point mutant p53 (R280K) in human mammary epithelial cells (HME cells) promoted the expression of Vimentin which is one of the mesenchymal cell markers (Fig. 2a, lane 3). Co-expression of Snail and C-terminal Myc-Flag tagged RPN2 (RPN2-MF) also promoted the expression of Vimentin in HME cells (Fig. 2a, lane 4). Next we established HME-Snail cell line that shows predominant mesenchymal phenotype and contains the CD44high/CD24low fraction11 (Fig. 2b and 2d). Flow cytometry and western blot analysis revealed that while the expression of wtp53 suppressed Snail expression and reduced CSC fraction in HME-Snail cells, the expression of mtp53 or RPN2-MF did not alter the population of CSCs and Snail expression (Fig. 2b and 2d). More importantly, we also found that mtp53 (R280K) promoted the expression of RPN2 in HME-Snail cells (Fig. 2b lane 4). We also observed that mtp53 (R280K and R175H) promoted the protein stability of Snail and co-expression of RPN2-MF and mtp53 (R280K) induced the expression of N-cadherin in other human mammary epithelial cells (MCF10A cells) (Fig. 2c and Suppl. Fig. S3A). These results suggest that RPN2 plays an important role in the generation of CSC with EMT phenotype in breast cancer cells.
Figure 2. RPN2 plays the important roles in the generation of CSC fraction in breast cancer cells.
(a)–(c) Western blot analysis. Cell lysates were subjected to western blotting with anti-p53, anti-RPN2, anti-Snail, anti-Vimentin, anti-Flag, anti-GFP and anti-actin antibodies. (d) p53 status and RPN2 affected the population of CSCs in breast cancer cells. Flow cytometric analysis of CD44 and CD24 expression in HME-Snail cell line and its derivatives. Full-length gels and blots are shown in supplementary figure 10–12.
RPN2 regulates the tumorigenicity and metastasis of CSCs
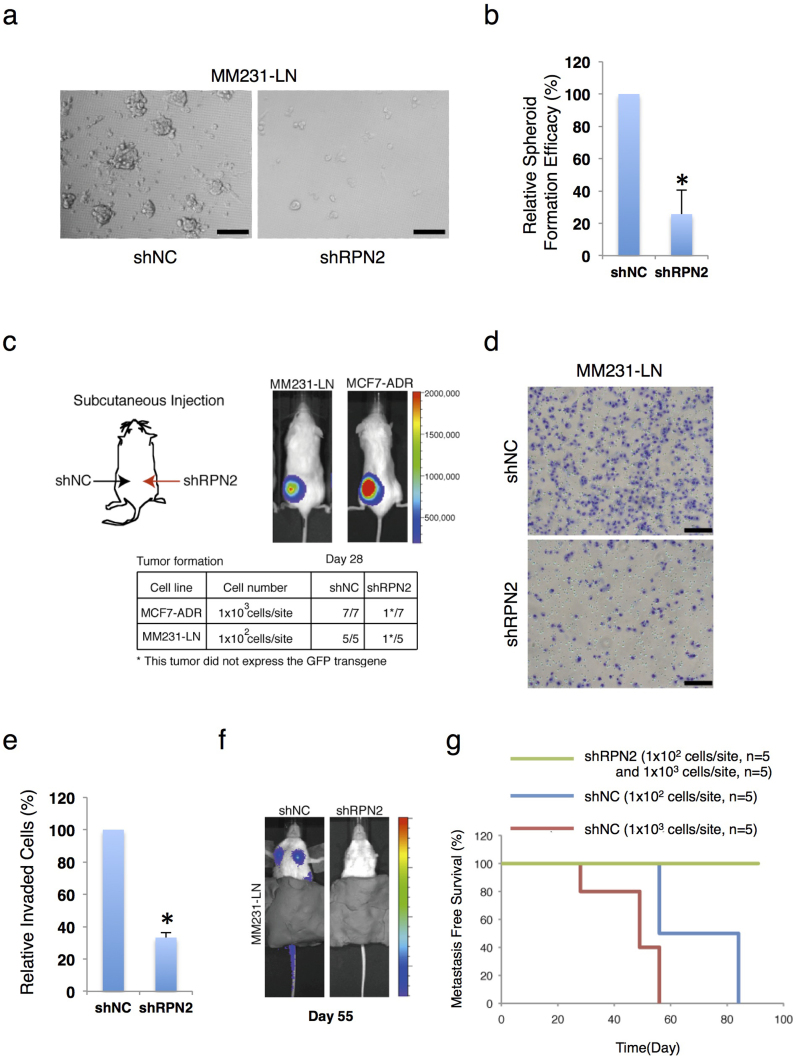
The tumorigenicity of RPN2 knockdown cell lines was then examined using a 3D spheroid culture system24. In several cancer cell lines, CSCs form spheroids, which are essential for tumor onset in immunodeficient mice14,24. MM231-LN CSCs exhibited high tumorigenicity in an animal model (Suppl. Fig. S1). Compared with the control CSC fraction (MM231-LN shNC), the RPN2-knockdown CSC fraction (MM231-LN shRPN2) formed very few spheroids (Fig. 3a and b). To evaluate tumor formation by in vivo imaging, a limiting-dilution assay was performed using 6-week-old NOD/SCID mice that had been injected in the hind legs with 102 cells from a CSC fraction (CD44high/CD24low/GFPhigh) derived from MM231-LN cells expressing firefly luciferase (Fig. 3c). The control CSC fraction formed tumors in all mice, whereas the RPN2-knockdown CSC fraction only formed one tumor from five injections (Fig. 3c and Suppl. Fig. S4A), and in that tumor, low RPN2 knockdown was confirmed by in vivo imaging (Suppl. Fig. S4B). Similar results were obtained with MCF7-ADR cells (Fig. 3c). The metastatic ability of RPN2 knockdown cell lines was examined next. In transwell migration assays, RPN2 knockdown reduced the invasiveness of CSCs in MM231-LN cells (Fig. 3d and e). After the transplantation of CSCs derived from MM231-LN shNC into the mammary fat pads of NOD/SCID mice, nodal and lung metastasis was observed in all mice (Fig. 3f). However, after transplantation of CSCs derived from MM231-LN shRPN2, nodal and lung metastasis was no longer observed (Fig. 3f and g), suggesting that RPN2 is essential for tumor formation and metastasis.
Figure 3. RPN2 regulates the tumorigenicity and metastatic activity of CSCs.
(a) Phase-contrast images of spheroids seeded by MM231-LN shNC (right) and MM231-LN shRPN2 (left) cells. Scale bar, 50 μm. (b) Quantification of spheroid formation in MM231-LN shNC and MM231-LN shRPN2 cells. The data in b) represent three independent experiments, and values are means ± s.d. (n = 3, *P < 0.05). (c) CSC (CD44high/CD24low/GFPhigh) tumor formation in MM231-LN shNC (left) and MM231-LN shRPN2 (right) cells after subcutaneous injection. (d) and (e) Matrix invasion in MM231-LN shNC and MM231-LN shRPN2 cells (error bars = s.d., n = 3, *P < 0.05). (f) and (g) CSC (CD44high/CD24low/GFPhigh) tumor metastasis in MM231-LN shNC and MM231-LN shRPN2 cells after mammary fat pad injection and monitoring of survival (shNC, n = 5; shRPN2, n = 5).
RPN2 antagonizes GSK3β function via physical interaction
Several studies show that GSK3β suppresses Snail expression at transcriptional and post-transcriptional levels15,16, and the inhibition of GSK3β by small molecules induces EMT and promotes CSC phenotypes in breast cancer17. Our results also confirmed that the GSK3β inhibitor CH99021 suppressed E-cadherin expression in MCF7 cells (Suppl. Fig. S3B). In the present study, RPN2 knockdown reduced the E-cadherin negative fraction in MM231-LN cells via suppressing Snail expression (Fig. 1d and Suppl. Fig. S3C). These results indicate that RPN2 knockdown promotes GSK3β activation, and such activation is associated with the suppression of CSC phenotypes (Fig. 1 and 3).
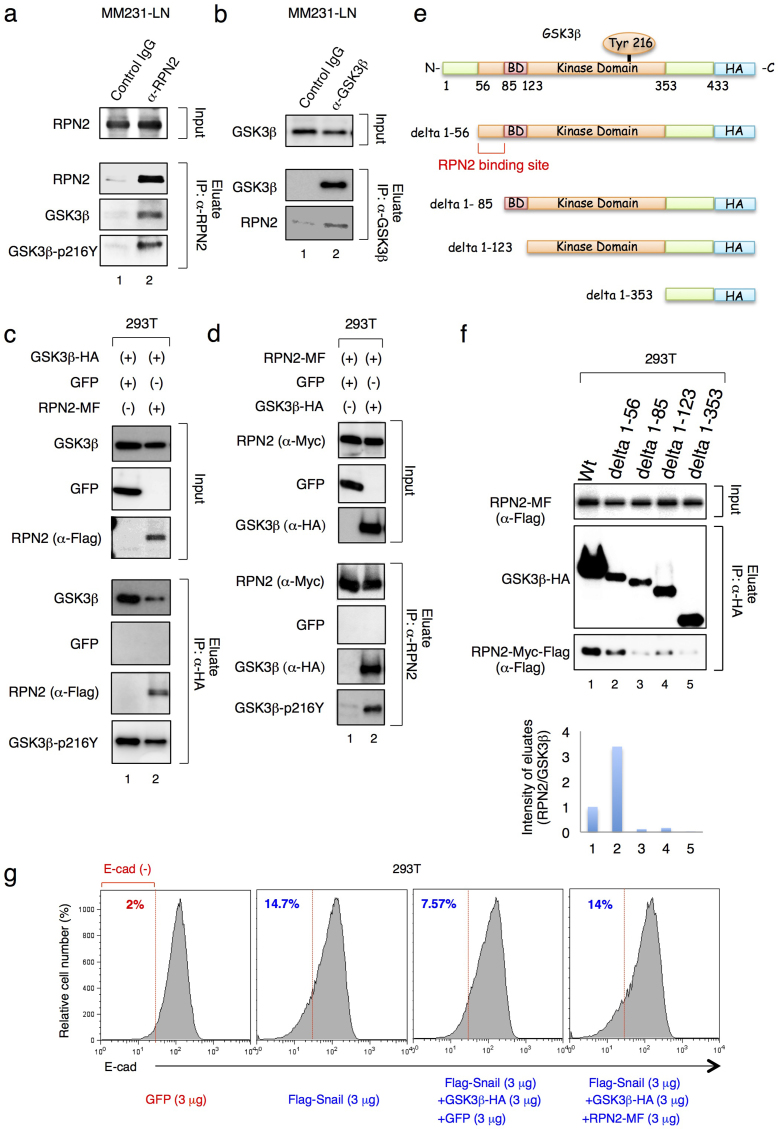
The results above indicate that RPN2 regulates GSK3β activity in MM231-LN cells (Fig. 1c and d and Suppl. Fig. S3C); however, the N-linked glycosylation of GSK3β has not been previously observed. We therefore examined the physical interaction between RPN2 and GSK3β in MM231-LN cells by immunoprecipitation of endogenous RPN2 from MM231-LN cells. Since the phosphorylation of GSK3β at Tyr216 (GSK3β-216Y) is essential for the nuclear localization and activation of GSK3β16, we examined the association of RPN2 with both Y216-phosphorylated and unphosphorylated GSK3β (Fig. 4a). Immunoprecipitation analysis showed that RPN2 associated with both Y216-phosphorylated and unphosphorylated GSK3β (Fig. 4a, lane 2). The physical interaction between RPN2 and GSK3β was also confirmed by using an anti-GSK3β antibody (Fig. 4b1, lane 2).
Figure 4. RPN2 antagonizes GSK3βfunction via physical interaction.
(a) and (b) RPN2 associated with GSK3β in MM231-LN cells. In the left panel, cell lysates were subjected to co-immunoprecipitation with an anti-RPN2 antibody and subjected to western blotting with anti-RPN2 (top), anti-GSK3β (middle) or anti-Phospho-GSK3β (Tyr216) antibodies (bottom). In the right panel, cell lysates were subjected to co-immunoprecipitation with an anti-GSK3β antibody and subjected to western blotting with anti-GSK3β (top and middle) and anti-RPN2 antibodies (bottom). (c) GSK3β associated with RPN2 in 293T cells. Cell lysates were subjected to co-immunoprecipitation with anti-HA Agarose and western blotting with anti-GSK3β (top), anti-GFP (middle), anti-RPN2 (bottom) or anti-Phospho-GSK3β (Tyr216) antibodies. (d) RPN2 associated with phosphorylated GSK3β in 293T cells. Cell lysates were subjected to co-immunoprecipitation with an anti-RPN2 antibody and western blotting with anti-RPN2 (top), anti-GFP (middle), anti-GSK3β (bottom) or anti-Phospho-GSK3β (Tyr216) antibodies. (e) Schematic representation of GSK3β and its deletion mutants. The binding domain (BD) designates the GSK3β-specific binding sites for its substrates and protein complexes. (f) RPN2-Myc-Flag, GSK3β-HA and the GSK3β deletion mutants were expressed in 293T cells and immunoprecipitated (IP) with an anti-HA antibody. Lysates (input) and immunoprecipitates were immunoblotted to detect co-immunoprecipitated RPN2. (g) Flow cytometry analysis and quantification of the percentage of E-cadherin-negative cells in 293T cells at 48 h after transfection. The amount of plasmid DNA used in each experiment was indicated in the figure. Full-length gels and blots are shown in supplementary figure 13 and 14.
To determine the binding site of GSK3β on RPN2, in vitro binding assays were performed using extracts from 293T cells expressing RPN2-MF and C-terminal HA-tagged GSK3β (GSK3β-HA). Co-immunoprecipitation analysis revealed that GSK3β interacted with RPN2 in this extract (Fig. 4c and d, lane 2). Next, the binding region for GSK3β to RPN2 was examined using 293T cell extracts expressing RPN2-MF and GSK3β deletion mutants (Fig. 4e). Co-immunoprecipitation analysis revealed that the N-terminal amino acid region between 56–85 of GSK3β was critical for RPN2 binding (Fig. 4f, lane3).
To further investigate whether RPN2 antagonizes GSK3β activity via physical interaction, we examined the E-cadherin expression in 293T cells. In a control experiment, we confirmed that ectopic expression of N-terminal Flag-tagged Snail (Flag-Snail) induced E-cadherin suppression (Fig. 4g) and that expression of GSK3β-HA inhibited the Snail-mediated suppression of E-cadherin (Fig. 4g). Therefore, we examined whether ectopic expression of RPN2-MF with GSK3β-HA restores the Snail activity. Flow cytometry analysis showed that the expression of RPN2-MF restored the Snail-mediated suppression of E-cadherin (Fig. 4g). These results indicate that RPN2 inhibits the nuclear localization and activation of GSK3β via a physical interaction with its N-terminal region, and partially provide a molecular mechanism by which E-cadherin negative fraction acquires drug resistance.
RPN2 knockdown promoted GSK3β−mediated inactivation of heat shock proteins
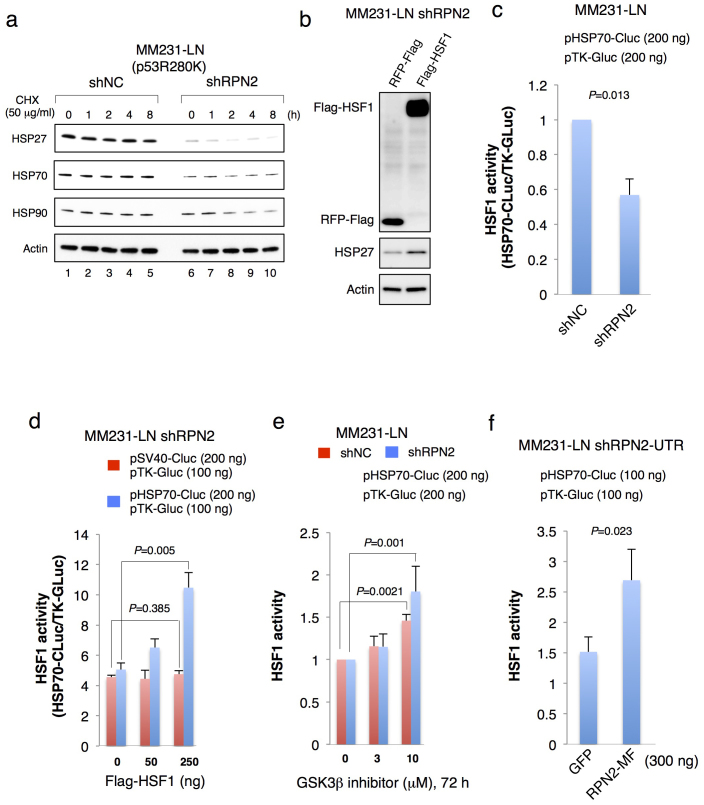
Recent studies show that different types of mtp53 form stable complexes with MDM2, CHIP, HSP90 and HSP7025,26,27 and that HSP70 is transcriptionally regulated by heat shock transcription factor1 (HSF1)28,29. HSF1 transcriptional activity is negatively regulated by GSK3β30, which suggests that RPN2 knockdown may promote GSK3β−induced downregulation of HSP70 (Suppl. Fig. S5A). Immunoblot analysis revealed that RPN2 knockdown reduced the expression of HSP70 (Fig. 5a, lanes 1 and 6). We also confirmed the downregulation of HSP27 that is a transcriptional target of HSF131,32 and contributes the maintenance of breast CSCs through the regulation of EMT and NF-κB activity33 (Fig. 5a, lane 1 and 6). Importantly, HSP70 protein also functions as co-chaperones for HSP909. Therefore, we examined the effect of HSP70 downregulation on the destabilization of HSP90 by blocking protein synthesis with cycloheximide (CHX) treatment (Fig. 5a). While the effect on HSP70 protein stability was minimal, RPN2 knockdown reduced the stability and half-life of HSP90 (Fig. 5a and Suppl. Fig. S6). We also confirmed that the expression of N-terminal Flag-tagged HSF1 (Flag-HSF1) restored HSP27 expression in MM231-LN shRPN2 cells (Fig. 5b).
Figure 5. RPN2 knockdown suppressed the heat shock proteins via GSK3β activation.
(a) RPN2 knockdown suppressed the expression of HSP27 and HSP70 and reduced HSP90 protein stability. MM231-LN shNC and MM231-LN shRPN2 cells were treated with cycloheximide (50 μg/ml) for 0–8 h. Cell lysates were subjected to western blotting with anti-HSP27, anti-HSP70, anti-HSP90 and anti-actin antibodies. (b) HSF1 restored the HSP27 expression in MM231-LN shRPN2 cells. After transfection of pFlag-HSF1 into MM231-LN shRPN2 cells, Cell lysates were subjected to western blotting with anti-Flag (top), anti-HSP27 (middle), anti-actin (bottom) antibodies. (c) HSF1 transcriptional activity in RPN2 knockdown cell line. HSP70 promoter-driven cypridina luciferase activity (HSP70-CLuc) was normalized to the thymidine kinase promoter-driven gaussia luciferase activity (TK-GLuc). MM231-LN shNC or MM231-LN shRPN2 cells were transfected with 200 ng of pHSP70-Cluc in combination with 200 ng of pTK-Gluc. Transfection and luciferase assays were performed as described in experimental procedure. (d) HSF1 restored the HSP70 expression in MM231-LN shRPN2 cells. MM231-LN shRPN2 cells were transfected with pFlag-HSF1 with 200 ng of pHSP70-Cluc, or pSV40-Cluc in combination with 100 ng of pTK-Gluc, and luciferase activity was measured at 48 h after transfection. (e) GSK3β inhibitor restored HSF1 transcriptional activity. Six hours after transfection, cells were exposed to CHIR99021 (3, 10 μM) or an equivalent volume of dimethyl sulfoxide as a control added to the growth medium (CHIR99021, 0 μM). Treatments lasted for 72 h, after which the luciferase activity was examined. (f) Rescue experiment using MM231-LN shRPN2-UTR cells. RPN2-MF restored the HSP70 activity in MM231-LN shRPN2-UTR cells. pHSP70-Cluc was transfected into MM231-LN shRPN2-UTR cells with 300 ng of pCMV-RPN2-MF, or pEGFP-N1 in combination with 100 ng of pTK-Gluc, and luciferase activity was measured at 48 h after transfection. Full-length gels and blots are shown in supplementary figure 15.
Next, we examined the HSF1 transcriptional activity by using the expression vector of HSP70 promoter-driven secreted cypridina luciferase (HSP70-Cluc, Fig. 5c). Luciferase assays showed that RPN2 knockdown induced a 50% repression of HSF1 activity (Fig. 5c). When Flag-HSF1 was transfected with HSP70-Cluc in MM231-LN shRPN2 cells, luciferase activity was markedly enhanced 1.5- to 3-fold compared with those transfected with SV40 promoter-driven secreted cypridina luciferase (SV40-Cluc, Fig. 5d). We also confirmed that GSK3β inhibitor restored the HSF1 transcriptional activity in MM231-LN shRPN2 cells (Fig. 5e).
We also examined whether HSF1 activity correlated with RPN2 expression using MM231-LN cells expressing HSP70 promoter-driven GFP (MM231-LN HSP70-GFP) (Suppl. Fig. S5B). Flow cytometry and qRT-PCR analysis of MM231-LN HSP70-GFP cells showed that HSP70 and RPN2 were more highly expressed in the GFP high cell fraction than in the GFP low cell fraction (Suppl. Fig. S5C and 5D).
Finally, to exclude the off-target effects of shRNA against RPN2, we performed the rescue experiment using MM231-LN cells expressing shRNA against the 3′-untranslated region of RPN2 (Suppl. Fig.5E, MM231-LN shRPN2-UTR) to replace endogenous RPN2 with ectopically expressed RPN2-MF. Compared to the co-expression of shRPN2-UTR with GFP, co-expression of shRPN2-UTR with RPN2-MF restored the HSF1 activity in MM231-LN shRPN2-UTR cells (fig. 5f). These results strongly suggest that the formation of RPN2/GSK3β protein complexes inhibits the function of GSK3β, and that RPN2 knockdown induces the destabilization of mtp53 by GSK3β−mediated inactivation of heat shock proteins, leading to CSC phenotypic suppression.
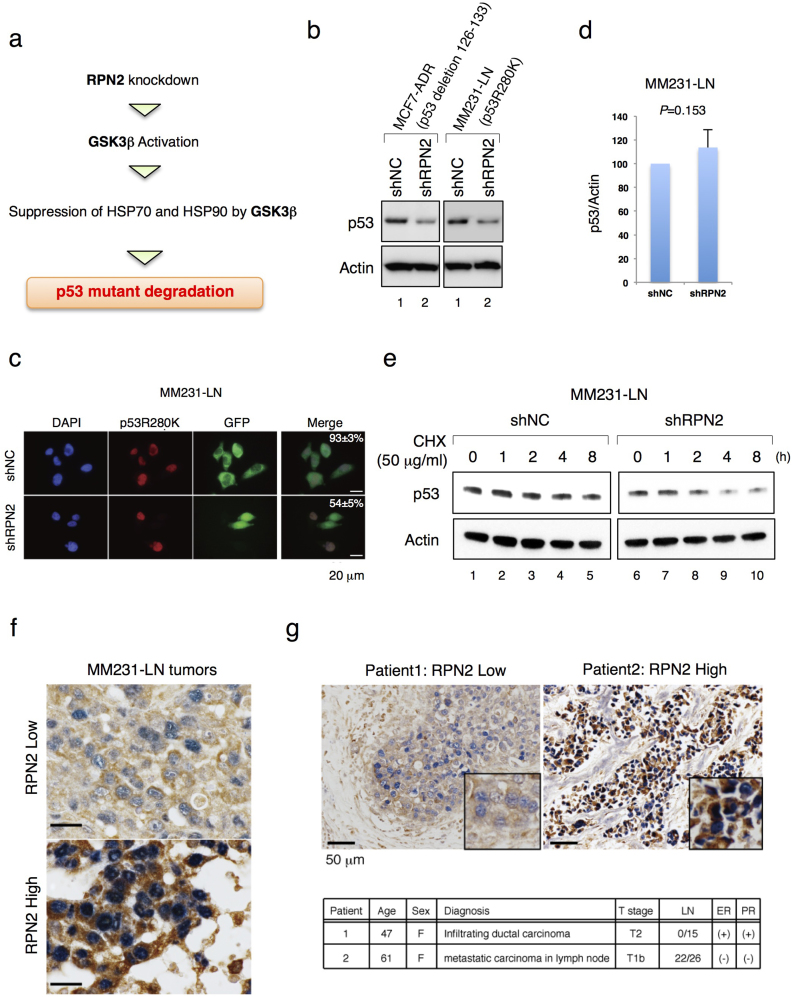
RPN2 knockdown induces mtp53 degradation
Since RPN2 knockdown promoted GSK3β−mediated inactivation of heat shock proteins that are essential for mtp53 stabilization6,25, we examined the expression and stability of mtp53 in MCF7-ADR and MM231-LN cells that express a p53 deletion mutant (p53 deletion 126–133)34 and a p53 point mutant (p53R280K)5, respectively (Fig. 6a and b). Immunoblot analysis with MCF7-ADR and MM231-LN cells showed that RPN2 knockdown induced mtp53 downregulation (Fig. 6b). As the lentivirus vector expressing shRPN2 also expressed the reporter gene GFP, mtp53 expression was examined in GFP-positive cells. Immunofluorescence staining with anti-p53 and anti-GFP antibodies revealed that mtp53 levels were considerably reduced in cells expressing shRPN2 compared with those expressing shNC (Fig. 6c). QRT-PCR revealed that p53 mRNA levels were not altered in MM231-LN shRPN2 cells (Fig. 6d). Therefore, the protein stability of mtp53 was examined in RPN2 knockdown cells by cycloheximide (CHX) treatment (Fig. 6e). Immunoblot analysis showed that RPN2 knockdown reduced the half-life of mtp53 compared to the control (Fig. 6e and Suppl. Fig. S6). These findings were confirmed by RPN2 and mtp53 immunohistochemical staining in mouse MM231-LN tumors. Accumulation of mtp53 was observed in primary tumors with strong RPN2 expression but was reduced in primary tumors with low RPN2 expression (Fig. 6f). Immunohistochemical staining for RPN2 and mtp53 expression in breast cancer tissues yielded similar results to those obtained using tissues from subjects with a high incidence of lymph node metastasis (Fig. 6g and Suppl. Fig. S7).
Figure 6. RPN2 knockdown induces mtp53 degradation.
(a) Working model for GSK3β-mediated downregulation of mtp53 in RPN2 knockdown cells. (b) and (d) Expression of mutant p53 (mtp53) in MCF7-ADR and MM231-LN cells. (c) Expression of mtp53 in MM231-LN shNC and MM231-LN shRPN2 cells. Immunofluorescence staining of mtp53 (Red) and GFP (Green) and merged images are shown. Nuclei are shown in blue (DAPI). Scale bar, 20 μm. (e) RPN2 knockdown reduced mtp53 protein stability. MM231-LN shNC and MM231-LN shRPN2 cells were treated with cycloheximide (50 μg/ml) for 0–8 h. Cell lysates were subjected to western blotting with anti-p53 and anti-actin antibodies. (f) Expression of RPN2 and mtp53 in MM231-LN tumors in mice. Panels show representative immunohistochemistry results for RPN2 (Brown) and mtp53 (Blue) in MM231-LN tumors. Scale bar, 20 μm. (g) The status of RPN2 (Brown) and mtp53 (Blue) in breast cancer tissues. The tissue sections obtained from patients were classified by the extent of lymph node metastasis (LN). ER, Estrogen Receptor; PR, Progesterone Receptor. Scale bar, 50 μm. Full-length gels and blots are shown in supplementary figure 16.
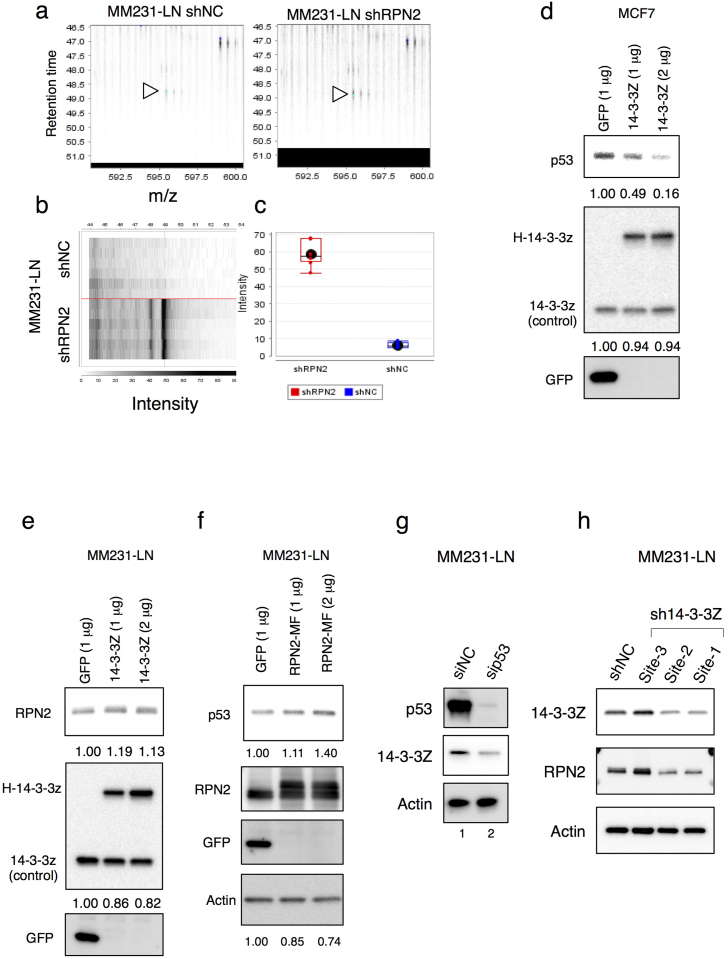
To further elucidate the role of RPN2 in tumor initiation and metastasis, we performed label-free proteomic analysis, two-dimensional image-converted analysis of liquid chromatography and mass spectrometry (2DICAL)35, and identified proteins differentially expressed between MM231-LN shRPN2 and MM231-LN shNC cells (Fig. 7a–c and Suppl. Table S1). Although several proteins were differentially expressed, we focused on 14-3-3zeta upregulation, which has been implicated in breast cancer pathogenesis36,37. Despite the suppression of CSC phenotypes by RPN2 knockdown (Fig. 1–6), 14-3-3zeta was upregulated in RPN2 knockdown cells, which is a typical feature of the early stages of breast cancer wherein 14-3-3zeta upregulation promotes wtp53 degradation via phosphorylation of the MDM2 E3 ligase, and thus, cancer progression36,37. Although the peptide specificity was very low, 2DICAL also showed that RPN2 knockdown induced the downregulation of mtp53 (Suppl. Table S1). Consistent with previous reports36,37, we confirmed that N-terminal Halo-tagged 14-3-3zeta (H-14-3-3zeta) induced downregulation of wtp53 in MCF7 cells (Fig. 7d). Determination of the effect of 14-3-3zeta upregulation on the mtp53 protein stability revealed that H-14-3-3zeta expression partially induced RPN2 expression in MM231-LN cells (Fig. 7e). We also observed that RPN2 overexpression induced mtp53 upregulation in MM231-LN cells (Fig. 7f) and that mtp53 knockdown strongly reduced the expression of 14-3-3zeta in MM231-LN cells (Fig. 7g). Moreover, 14-3-3zeta knockdown reduced the expression of RPN2 in MM231-LN cells (Fig. 7h and Suppl. Fig. S18). These results suggest that 14-3-3zeta-mediated RPN2 upregulation contributes to the stability and accumulation of mtp53 and that RPN2 knockdown inhibits the 14-3-3zeta-dependent feedback regulation of mtp53 protein stability (Suppl. Fig S8).
Figure 7. 14-3-3zeta-mediated RPN2 upregulation is required for the mtp53 stability.
(a) Detected peptides that differed between RPN2 knockdown cell line (MM231-LN shRPN2) and control (MM231-LN shNC). MS peak of 14-3-3Zeta (1433Z) in MM231-LN shNC (Right) and MM231-LN shRPN2 (Left) (indicated by arrows). (b) Gel-like views of MS peaks with retention time (RT) along the vertical axes (top) and distribution of the mean peak intensity of duplicates (bottom) across 6 samples. (c) Spot intensity of 14-3-3Z in MM231-LN shNC and MM231-LN shRPN2. (d) 14-3-3zeta reduced wtp53 expression. Lysates from MCF7 cells, transfected as indicated, were immunoblotted as shown. (e) and (f) RPN2 stabilized mtp53 in MM231-LN cells. Lysates from MM231-LN cells, transfected as indicated, were immunoblotted as shown. (g) and (h) 14-3-3zeta-dependent feedback regulation of mtp53 protein stability. Full-length gels and blots are shown in supplementary figure 17 and 18.
Discussion
LOF or mtp53 accumulation is associated with acquisition of the EMT phenotype and development of high-grade tumors in breast cancers11. The half-life of wtp53 is < 30 min, whereas mtp53 is more stable, with a half-life of several hours34,38. Although the molecular mechanism of mtp53 stabilization is not completely understood, recent studies demonstrated that the contribution of mtp53 to tumor progression and its promotion of EMT onset account for the development of CSC properties11,14. Several pathways such as Wnt/β-catenin, TGF-β, Notch and Hedgehog signaling are critical for the acquisition of CSC properties20. In the present study, we provide evidence that the expression of RPN2 is regulated by p53 mutant (R280K and del126-133) which is associated with the acquisition of EMT phenotype in breast cancer and that RPN2 regulates CSC phenotypes via stabilization of mtp53 (R280K and del126-133). We demonstrated that shRNA-mediated knockdown of RPN2 significantly suppresses the CSC phenotype in vitro and in vivo and promotes GSK3β-mediated destabilization of mtp53.
While GSK3β promotes ubiquitin-proteasome pathway-mediated β-catenin degradation39 and suppresses Snail expression at transcriptional and post-transcriptional levels15,16, it is unclear how GSK3β suppresses CSC tumorigenesis and metastasis. Our results demonstrate, for the first time, a novel role for GSK3β in mtp53 destabilization and indicate that its activity is regulated by RPN2 (Fig. 4,5,6). We also demonstrated that RPN2 antagonized GSK3β function via physical interaction, and identified N-terminal amino acids 56–85 of GSK3β as important for its interaction with RPN2 (Fig. 4). Our previous study showed that RPN2 affects docetaxel resistance by modulating the N-linked glycosylation of P-glycoprotein19. Ribophorin-1 (RPN1) is also a component of the oligosaccharide transferase (OST) complex40,41. However, a recent study showed that RPN1 regulates the cell surface localization of μ-opioid receptor (MOR) via direct interaction with MOR42. Therefore, our data suggest that, in addition to being a component of the OST complex, RPN2 may play an important role in tumor onset and metastasis.
HSP90 is a promising therapeutic target for cancer therapy because the HSP90 chaperone machinery is upregulated and activated in malignant cancers and inhibition of this single protein causes the simultaneous degradation of multiple oncoproteins8,9. The stabilization and accumulation of mtp53 is associated with the formation of stable protein complexes between HSP90 and various types of mtp53, and the formation of such complexes inhibits the constitutive E3 ligase activity of MDM2 and CHIP6,25. Therefore, the identification and development of HSP90 inhibitors has been a focus of several studies8,43. One of main problems associated with the inhibition of HSP90 is the HSF1-dependent induction of HSP70 and HSP27 in response to HSP90 inhibitor treatment8,9. Considering that HSP70 chaperones also act as co-chaperones for HSP90 and have a well-documented antiapoptotic function that is independent of their interaction with HSP909,44, our finding that RPN2 knockdown suppresses HSP70 expression (Fig. 6) is important not only for understanding the regulation of mtp53 stability, but also for developing ways to overcome the side effects of HSP90 inhibitors.
We also found that 14-3-3zeta involved in the stabilization and accumulation of mtp53 (Fig. 7 and Suppl. Fig S8). Since forced expression of 14-3-3zeta showed only a modest effect on RPN2 expression in MM231-LN (Fig. 7e), it might be necessary to consider the other molecules and pathways that regulate RPN2 expression. In a clinical study, the upregulation of 14-3-3zeta via amplification of the chromosome region where it is located (8q22) promoted chemoresistance to anthracyclines and metastatic recurrence of breast cancer45. However, the molecular mechanism underlying the chemoresistance and tumor recurrence has not been addressed. A recent study also showed that anthracyclines increase the amount of mtp53 mRNA and protein46. Therefore, the present study provides further insight into these mechanisms by showing that 14-3-3zeta promotes breast cancer malignancy.
TP53 mutations are present in 80% of triple-negative breast cancers (TNBC), which are characterized by the lack of estrogen and progesterone receptors and the absence of human epidermal growth factor receptor 247. Patients with TNBC show resistance to conventional chemotherapies and are in need of effective therapeutic agents to prevent recurrence and improve survival48. As shown in Figures 6g and S7, high expression of RPN2 and accumulation of mtp53 were observed in clinical samples of breast cancer tissues associated with lymph node metastasis. Moreover, our results are supported by a recent clinical study that showed that HSP70 expression correlates significantly with metastasis in TNBC patients49. Therefore, the concomitant expression of RPN2 and mtp53 could be a novel diagnostic marker for malignant progression and poor prognosis in TNBC. This is currently being investigated in a large cohort of TNBC patients at our NCC hospital and it would reveal that which types of p53 mutants are associated with RPN2 expression.
In summary, the present results indicate that inhibition of the RPN2/mtp53 regulatory network is a promising approach for overcoming the progression of malignant breast tumors and suppressing the CSC phenotype. Furthermore, our data suggest a novel mechanism for the modulation of nucleocytoplasmic proteins in cancer biology. Given the diverse biological roles of RPN2, further investigation aimed at understanding the role of RPN2 in processes associated with tumor progression is warranted.
Methods
Plasmids
N-terminal Flag-tagged Snail (pFlag-Snail), N-terminal Flag-tagged HSF1 (pFlag-HSF1), pBabe-puro-Snail, C-terminal V5-tagged wtp53 (pLenti6/V5-p53), C-terminal V5-tagged mtp53 (R280K, pLenti6/V5-p53R280K) and C-terminal HA-tagged GSK3β (pcDNA3-GSK3β-HA) were purchased from Addgene. To obtain C-terminal Myc- and Flag-tagged RPN2 constructs (pLenti-RPN2-Myc-Flag), cDNA encoding full-length human RPN2 (GenBank accession number Y00282) was amplified by polymerase chain reaction (PCR) using cDNA pools from MDA-MB231-D3H2-LN cells (MM231-LN). PCR fragments were inserted into EcoRI- and NotI-treated pLenti-C-Myc-DDK (Origene: PS100064). To obtain a HSP70 promoter-driven GFP (pHSP70-GFP), HSP70 promoter region was amplified by PCR using genomic DNA from MM231-LN cells (GenBank accession number M11717). The PCR fragment was inserted into BglII- and SalI-treated pEGFP-1 (Clontech). To obtain a HSP70 promoter-driven secreted cypridina luciferase (pHSP70-CLuc), HSP70-GFP was digested with KpnI and NotI and the fragment containing GFP was replaced with cypridina luciferase gene derived from pCLuc-Basic2 (NEB). All constructs were verified by DNA sequencing.
Cell culture
MCF7, MCF7-ADR, MCF7-ADR-Luc, MM231-LN, 293T, MCF10A and HME cells have been described previously14,19. To establish MM231-LN cells expressing HSP70-GFP, pHSP70-GFP containing the neomycin resistance gene was transfected with lipofectamine LTX (Invitrogen). The transfected cells were cultivated under selective growth medium including G418 (0.6 mg/ml). For spheroid culture, cells were plated on NanoCulture plates (Scivax) and cultured for 3 days.
Antibodies
The primary antibodies and dilutions were anti-RPN2 (1:2000, sc-166421, Santa Cruz Biotechnology), anti-p53 (1:2000, sc-126, Santa Cruz Biotechnology), anti-V5 (1:1000, V8137, Sigma), anti-Vimentin (1:2000, 550513, BD Pharmingen), anti-HA (1:2000, #3724, CST), anti-HSP27(1:2000, #2402, CST), anti-Myc (1:1000, 2278, CST), anti-Flag (1:1000, M185-7, MBL), anti-HA agarose (#3956S, CST), anti-HSP70 (1:1000, 610607, BD Transduction Laboratories), anti-HSP90 (1:1000, 610418, BD Transduction Laboratories), anti-GSK3β (1:1000, 610201, BD Transduction Laboratories), anti-Phospho-GSK3β (Tyr216) (1:1000, 612312, BD Transduction Laboratories), anti-N-cadherin (1:2000, #4061S, CST) and anti-actin antibodies (1:5000, MAB1501, Millipore). An anti-p53 antibody (Abcam, Pab240) was used for the detection of mtp53 by IHC. Staining was visualized using Alexa 488 or Alexa 594 (Molecular Probes). Immunofluorescence-stained cells were observed by fluorescence microscopy or confocal fluorescence microscopy (Leica). The signal intensity in immunoblot analysis was quantified using ImageJ software (http://rsbweb.nih.gov/ij/).
Lentiviral shRNA transduction
Cell lines stably expressing RPN2 shRNA, 14-3-3zeta shRNA or control non-target shRNA were established using a vector-based shRNA technique. Human RPN2 shRNA targets 5′-GGAGGAGATTGAGGACCTTGT-3′ (shRPN2-site1), 5′-GCCACTTTGAAGAACCCAATC-3′ (shRPN2-site2), 5′-TCCAGATTGTAGTTATACTTC-3′ (shRPN2-UTR), human 14-3-3zeta shRNA targets 5′-GCAGAGAGCAAAGTCTTCTAT-3′ (sh14-3-3zeta-site1), 5′-GCAATTACTGAGAGACAACTT-3′ (sh14-3-3zeta-site2), 5′-GCTCGAGAATACAGAGAGAAA-3′ (sh14-3-3zeta-site3), 5′-GAGAGGAATCTTCTCTCAGTT-3′ (sh14-3-3zeta-site4), 5′-CTCTGTGTTCTATTATGAGAT-3′ (sh14-3-3zeta-site5) and control shRNA targets 5′-GAAATGTACTGCGCGTGGAGAC-3′. Briefly, each fragment was subcloned into pGreenPuro (System Biosciences). Recombinant lentiviruses were produced according to the manufacturer's instructions. In knockdown experiments, MCF7-ADR and MM231-LN cells were infected with recombinant lentiviruses expressing control shRNA (shNC) or shRNA against RPN2 (shRPN2).
Matrigel invasion assay
The matrigel invasion assay was performed using the Matrigel Invasion Chamber (BD Bioscience) according to the manufacturer's protocol. In brief, 5 × 104 cells were plated in the upper chamber in serum-free media. The bottom chamber contained RPMI media with 10% FBS. After 24–48 h, the bottom of the chamber insert was fixed and stained with Diff-Quick stain. Cells on the stained membrane were counted under a dissecting microscope. Each membrane was divided into four quadrants and an average from all four quadrants was calculated. The matrigel invasion assays were performed with biological triplicates.
Dual luciferase assay
MM231-LN shNC or MM231-LNshRPN2 cells (2 × 104 cells) were seeded in a 24-well plate and cotransfected with HSF1 expression vector (pFlag-HSF1) in combination with reporter constructs of HSP70 promoter (pHSP70-Cluc), and SV40 promoter (pSV40-Cluc) using Lipofectamine 2000 (Invitrogen). The amount of plasmid DNA used in each experiment is indicated in the figure legends. A thymidine kinase promoter-driven secreted gaussia luciferase (pTK-Gluc, NEB) was mixed in a DNA–liposome complex as an internal control. Luciferase activity was quantified by a dual-luciferase assay system (NEB) and relative transactivation was calculated according to the manufacturer's instructions. All experiments were repeated at least 3 times.
Fluorescence-activated cell sorting
FITC or APC-conjugated anti-CD44 antibody (BD Bioscience, clone G44-26), PE-conjugated anti-CD24 antibody (Biolegend, clone LM5), APC-conjugated anti-E cadherin antibody (Biolegend, clone 67A4) and propidium iodide (5 μg/ml) were used for fluorescence-activated cell sorting (FACS) analysis using JSAN in accordance with the manufacturer's protocols. Data were processed by FlowJo software.
Immunoprecipitation
MM231-LN cells were lysed using immunoprecipitation buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol, 1 mM DTT, protease inhibitor cocktail, phosphatase inhibitor). After brief sonication, the lysates were cleared by centrifugation at 4°C. Supernatants were incubated with anti-RPN2 or anti- anti-GSK3β antibodies for 4 h and protein A/G-Sepharose beads (Invitrogen) for 4 h at 4°C. The immunocomplexes were washed four times, boiled in sample buffer and immunoblotted with anti-GSK3β, anti-Phospho-GSK3β (Tyr216) and anti-RPN2 antibodies.
Co-immunoprecipitation analysis
Extracts of 293T cells were obtained using immunoprecipitation buffer as described above. Supernatants were incubated with anti-HA Agarose (CST) under rotation for 4 h at 4°C. After washing four times with immunoprecipitation buffer, the immunoprecipitated protein complex bound to the beads was eluted with the HA peptide (Wako). The eluates from the immunoprecipitation and cell lysates were immunoblotted with anti-GSK3β, anti-Phospho-GSK3β (Tyr216) and anti-GFP antibodies.
Real-time reverse transcription PCR
Total RNA was isolated from cells and tumor tissues with an RNeasy Mini Kit and an RNase-Free DNase Set (Qiagen), and cDNA was produced with an ExScript RT reagent Kit (Takara). The cDNA samples were subjected to real-time PCR with SYBR Premix Ex Taq (Invitrogen) and specific primers (Supplementary Methods). All reactions were performed in a Light Cycler (Applied Biosystems). Gene expression levels were normalized to those of β-actin.
Bioluminescence imaging
Animal experiments were performed in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. Female non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice (NOD.CB17-Prdkcscid/J, CLEA Japan, Shizuoka, Japan) aged 4–6 weeks were anaesthetized by exposure to 3% isoflurane on day zero and subsequent days. Images were analyzed with Living Image software (Xenogen, part of Caliper Life Sciences). Bioluminescent flux (photonss−1 sr−1 cm−2) was determined for the primary tumors, lungs or lymph nodes (upper abdomen, region of interest).
Mammary fat pad xenografts
MCF7-ADR or MM231-LN cells were suspended in a PBS/Matrigel (Sigma) mixture (1:1) and injected into the mammary fat pad in a 50 μl volume (n = 5 each and 102–106 cells per animal).
Tissue arrays
The tissue arrays of breast cancer samples were purchased from Super Bio CHIP. Immunohistochemical staining of RPN2 and mutant p53 was performed with DAB peroxidase and alkaline phosphatase substrate kits (Vector Laboratories).
Statistical analysis
Data are presented as mean ± standard error of the mean (s.e.m.) or mean ± standard deviation (s.d.). Statistical significance was determined by Student's two-tailed t-test unless otherwise noted. A P value < 0.05 was considered statistically significant.
Author Contributions
R.T. and K.H. designed the experiments and analysed the data. R.T. and F.T. performed the experiments. K.K. provided human breast cancer pathology information. M.O. performed proteome analysis by 2DICAL. R.T. and T.O. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary figures and tables
Acknowledgments
We thank Dr S. Koizumi for providing the human mammary carcinoma cell lines MCF7, MCF7-ADR and MDA-MB-231-D3H2-LN, Dr Y. Yamamoto for helpful discussions and Miss. A. Inoue for her excellent technical assistance. This study was supported in part by a grant-in-aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control of Japan, a grant-in-aid for Scientific Research on Priority Areas Cancer from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation of Japan, and supported by a Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) from the Japan Society for the Promotion of Science (JSPS).
References
- Shaulian E., Zauberman A., Ginsberg D. & Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol 12, 5581–5592 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo E. et al. Mutant p53 attenuates the SMAD-dependent transforming growth factor beta1 (TGF-beta1) signaling pathway by repressing the expression of TGF-beta receptor type II. Mol Cell Biol 27, 8228–8242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P. A. et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 (2009). [DOI] [PubMed] [Google Scholar]
- Song H., Hollstein M. & Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 9, 573–580 (2007). [DOI] [PubMed] [Google Scholar]
- Adorno M. et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 137, 87–98 (2009). [DOI] [PubMed] [Google Scholar]
- Li D., Marchenko N. D. & Moll U. M. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ 18, 1904–1913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Chen L., Li C., Lu W. & Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem 276, 40583–40590 (2001). [DOI] [PubMed] [Google Scholar]
- Trepel J., Mollapour M., Giaccone G. & Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 10, 537–549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. V., Clarke P. A. & Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14, 250–262 (2008). [DOI] [PubMed] [Google Scholar]
- Yan W. et al. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. J. et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 13, 317–32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T. T. et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 68, 3645–3654 (2008). [DOI] [PubMed] [Google Scholar]
- Oft M., Akhurst R. J. & Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol 4, 487–494 (2002). [DOI] [PubMed] [Google Scholar]
- Mani S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelder R. E., Yoon S. O., Franci C., de Herreros A. G. & Mercurio A. M. Glycogen synthase kinase-3 is an endogenous inhibitor of Snail transcription: implications for the epithelial-mesenchymal transition. J Cell Biol 168, 29–33 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook J. I. et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 8, 1398–1406 (2006). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res 16, 2580–2590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P. B. et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138, 645–659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma K. et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med 14, 939–948 (2008). [DOI] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 17, 313–319 (2011). [DOI] [PubMed] [Google Scholar]
- Yin H. & Glass J. The phenotypic radiation resistance of CD44+/CD24(-or low) breast cancer cells is mediated through the enhanced activation of ATM signaling. PloS one 6, e24080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno A. M. et al. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. Journal of the National Cancer Institute 102, 1637–1652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J. & Clarke M. F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M. et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 1, 389–402 (2007). [DOI] [PubMed] [Google Scholar]
- Li D. et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 9, 577–588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P., Hrstka R., Coomber D., Lane D. P. & Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene 27, 3371–3383 (2008). [DOI] [PubMed] [Google Scholar]
- Esser C., Scheffner M. & Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem 280, 27443–27448 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. HSF1-dependent upregulation of Hsp70 by sulfhydryl-reactive inducers of the KEAP1/NRF2/ARE pathway. Chem Biol 18, 1355–1361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W. C., Ching T. T., Lee H. C., Mousigian C. & Hsu A. L. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148, 322–334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier I. J. et al. Glycogen synthase kinase 3beta negatively regulates both DNA-binding and transcriptional activities of heat shock factor 1. J Biol Chem 275, 29147–29152 (2000). [DOI] [PubMed] [Google Scholar]
- Kim D., Kim S. H. & Li G. C. Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem Biophys Res Commun 254, 264–268 (1999). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. p27 suppresses arsenite-induced Hsp27/Hsp70 expression through inhibiting JNK2/c-Jun- and HSF-1-dependent pathways. J Biol Chem 285, 26058–26065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L. et al. Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial-mesenchymal transition and nuclear factor-kappaB. Breast Cancer Res 13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B. & Safa A. R. Expression of the mutated p53 tumor suppressor protein and its molecular and biochemical characterization in multidrug resistant MCF-7/Adr human breast cancer cells. Oncogene 14, 499–506 (1997). [DOI] [PubMed] [Google Scholar]
- Ono M. et al. Prolyl 4-hydroxylation of alpha-fibrinogen: a novel protein modification revealed by plasma proteomics. J Biol Chem 284, 29041–29049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal C. L. & Yu D. 14-3-3zeta as a prognostic marker and therapeutic target for cancer. Expert Opin Ther Targets 14, 1343–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danes C. G. et al. 14-3-3 zeta down-regulates p53 in mammary epithelial cells and confers luminal filling. Cancer Res 68, 1760–1767 (2008). [DOI] [PubMed] [Google Scholar]
- Zambetti G. P. & Levine A. J. A comparison of the biological activities of wild-type and mutant p53. FASEB J 7, 855–865 (1993). [DOI] [PubMed] [Google Scholar]
- Cohen P. & Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol 2, 769–776 (2001). [DOI] [PubMed] [Google Scholar]
- Crimaudo C., Hortsch M., Gausepohl H. & Meyer D. I. Human ribophorins I and II: the primary structure and membrane topology of two highly conserved rough endoplasmic reticulum-specific glycoproteins. EMBO J 6, 75–82 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. M., Roebuck Q. & High S. Ribophorin I regulates substrate delivery to the oligosaccharyltransferase core. Proc Natl Acad Sci U S A 105, 9534–9539 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Loh H. H. & Law P. Y. mu-Opioid receptor cell surface expression is regulated by its direct interaction with Ribophorin I. Mol Pharmacol 75, 1307–1316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P. P. et al. Effective targeting of triple-negative breast cancer cells by PF-4942847, a novel oral inhibitor of Hsp 90. Clin Cancer Res 17, 5432–5442 (2011). [DOI] [PubMed] [Google Scholar]
- Yiu C. C. et al. Down-regulation of heat-shock protein 70 (HSP-70) correlated with responsiveness to neoadjuvant aromatase inhibitor therapy in breast cancer patients. Anticancer Res 30, 3465–3472 (2010). [PubMed] [Google Scholar]
- Li Y. et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 16, 214–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug M. & Dobbelstein M. Anthracyclines induce the accumulation of mutant p53 through E2F1-dependent and -independent mechanisms. Oncogene (2011). [DOI] [PubMed] [Google Scholar]
- Cleator S., Heller W. & Coombes R. C. Triple-negative breast cancer: therapeutic options. Lancet Oncol 8, 235–244 (2007). [DOI] [PubMed] [Google Scholar]
- Gluz O. et al. Triple-negative breast cancer--current status and future directions. Ann Oncol 20, 1913–1927 (2009). [DOI] [PubMed] [Google Scholar]
- Sun B. et al. Identification of metastasis-related proteins and their clinical relevance to triple-negative human breast cancer. Clin Cancer Res 14, 7050–7059 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables