ABSTRACT
Does cell age matter in virulence? The emergence of persister cells during chronic infections is critical for persistence of infection, but little is known how this occurs. Here, we demonstrate for the first time that the replicative age of the fungal pathogen Cryptococcus neoformans contributes to persistence during chronic meningoencephalitis. Generationally older C. neoformans cells are more resistant to hydrogen peroxide stress, macrophage intracellular killing, and antifungal agents. Older cells accumulate in both experimental rat infection and in human cryptococcosis. Mathematical modeling supports the concept that the presence of older C. neoformans cells emerges from in vivo selection pressures. We propose that advanced replicative aging is a new unanticipated virulence trait that emerges during chronic fungal infection and facilitates persistence. Therapeutic interventions that target old cells could help in the clearance of chronic infections.
IMPORTANCE
Our findings that the generational age of Cryptococcus neoformans cells matters in pathogenesis introduces a novel concept to eukaryotic pathogenesis research. We propose that emerging properties of aging C. neoformans cells and possibly also other fungal pathogens contribute to persistence and virulence. Whereas the replicative life span of strains may not matter for virulence per se, age-related resilience and thus the generational age of individual C. neoformans cells within a pathogen population could greatly affect persistence of the pathogen population and therefore impact outcome.
Introduction
Cryptococcus neoformans is a fungal pathogen that causes disease worldwide predominantly in AIDS patients, resulting in more than 600,000 deaths per year due to cryptococcal meningoencephalitis (CME) (1). A hallmark of CME is the ability of fungal cells to persist and replicate in the cerebrospinal fluid (CSF) despite treatment with antifungal agents and appropriate antiretroviral therapy (ART). Most C. neoformans strains that are recovered from patients are susceptible to antifungal agents after in vitro cultivation (2). Recurrence of infection is caused by persistence of the initial infection (3, 4), and the extent of decrease in fungal burden in repeated lumbar punctures constitutes a better predictor of effective clearance than the minimum inhibitory concentrations (MICs) of C. neoformans isolates (5).
C. neoformans is ubiquitous in the environment, and human infection results from inhalation of aerosolized spores. Most environmental C. neoformans strains have reduced virulence (6), whereas clinical C. neoformans strains differ in virulence in murine models (7–9), suggesting strain-related differences in virulence traits, and these differences in clinical outcome can be at least partially attributed to strain-related variations (10, 11). Virulence traits that enhance survival in mammalian macrophages may be selected through interaction with environmental amoeboid predators (12). Some virulence traits, such as mating locus, are genetically encoded (13), whereas others, such as capsule induction, underlie complex epigenetic regulation (14, 15), which can also be passed onto progeny.
C. neoformans is a haploid fungus that can replicate sexually, but during human infection, C. neoformans populations expand predominantly clonally (16). Therefore, similar to Saccharomyces cerevisiae, Candida albicans and Schizosaccharomyces pombe (17, 18), we expected C. neoformans cells to undergo asymmetric mitotic divisions and cease division at the completion of their life span. The sum of these divisions determines their replicative life span (RLS) (19). Previous work indicates that old mother cells of C. neoformans strain RC-2 manifest phenotypic changes that render them more resistant to macrophage- and antifungal-mediated killing in vitro (20), which led to the hypothesis that accumulation of C. neoformans cells of advanced age that are otherwise exceedingly rare cells in a growing pathogen population (20) may in fact accumulate in vivo and facilitate persistence.
Here, we took advantage of a rat cryptococcosis model, for which pathogen and host conditions have been well characterized. This model closely mimics human CME (21, 22) and allows us to assess the generational age of C. neoformans cells in vivo. We correlated these results with those derived from C. neoformans cells obtained from patients with CME. Our results establish cellular age as a new factor in fungal virulence, and these findings have important implications for our understanding of how chronic fungal infections persist.
RESULTS
Features of replicative aging, advanced age, and death in C. neoformans strains.
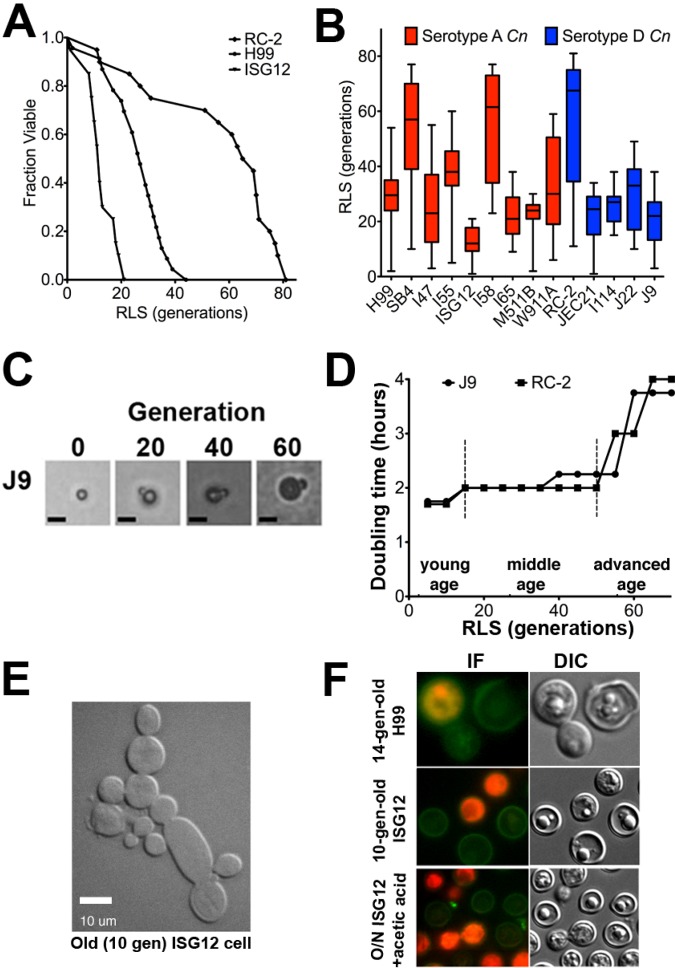
Recording of C. neoformans cell survival (Fig. 1A) resulted in RLS curves similar to those of S. cerevisiae (23). Notably, there was a wide range of RLSs for individual C. neoformans cells within a strain. The median RLS of 14 clinical C. neoformans strains (Fig. 1B) was 29.8 generations, and the RLS ranged from 12 to 67.5 generations among strains (P < 0.001 by log rank test). The median RLS of serotype A strains (33.4 generations) was comparable to that of serotype D strains (35.3 generations) (P = 0.845). In contrast to S. cerevisiae, the generational age of an individual C. neoformans cell could not be determined by vital stains for bud scars (see Fig. S1 in the supplemental material).
FIG 1 .
Clinical C. neoformans strains demonstrated variability in their life span. (A) Recording of RLS of individual cells of C. neoformans by generation of survival curves demonstrated short (ISG12), medium (H99), and long (RC-2) RLSs of strains that were statistically different from each other (P < 0.01 by Wilcoxon rank sum test). (B) The median RLSs (short horizontal lines in the boxes) of clinical serotype A and D C. neoformans strains were highly variable (20 to 60 cells per strain). The minimum and maximum values for all data are indicated by the ends of the whiskers of the box plots. (C) Increasing cell size was observed during the RLS shown for C. neoformans strain J9 at 0, 20, 40, and 60 replications. Bars, 10 µm. (D) The RLS of C. neoformans strains (J9 and RC-2) can be divided into young, middle, and advanced age based on changes in DT. (E) 10-generation-old C. neoformans cells (ISG12) manifested abnormal budding. Bar, 10 µm. (F) In order to investigate how cells of advanced age died, 14-generation-old H99 cells or 10-generation-old ISG12 cells were stained with annexin V. Cells undergoing apoptosis stained positive for annexin V (green), those undergoing necrosis stained for propidium iodine (PI) (red), and the cells that had suffered loss of membrane integrity stained for both (red with green boundary). Young cells (overnight [O/N] culture) were treated with acetic acid to induce apoptosis and stained similarly. The cells were examined by immunofluorescence (IF) and differential interference contrast (DIC) microscopy.
All strains demonstrated a gradual increase in cell body size with generational aging (Fig. 1C). On the basis of the observed doubling time (DT) and size-associated phenotypic changes, we divided the RLS of C. neoformans into three phases: young, middle, and advanced age (Fig. 1D). Generally, replications slowed significantly (>30% increase from the initial DT) to mark the beginning of advanced age after 70% completion of a strain’s respective RLS.
Analogous to S. cerevisiae, death was assumed if the old cell did not replicate for 24 h, but occasionally cells reemerged from this arrest to replicate a few more times. Incomplete release of buds was noted for all strains, including the short-lived ISG12 strain in approximately 15% (3 or 4 out of 20 to 40) of old cells in their final replications (Fig. 1E). Positive annexin V staining of older cells (14-generation-old H99 or 10-generation-old ISG12) (Fig. 1F) isolated by the gentle method of elutriation demonstrated that 15 to 20% died by apoptosis (data not shown).
RLS is regulated by phenotypic switching, but not by the mating locus, and selection of variants with altered RLS occurs during chronic infection.
Next, we investigated whether RLS is a stable or random trait in a C. neoformans strain. The majority of human-derived C. neoformans strains exhibit the MATα mating type (13) and are more virulent than congenic MATa strains (24). However, our data show that in serotype A and D strains, median RLSs were comparable between MATa and MATα congenic strains, indicating that RLS is a stable characteristic of a C. neoformans strain that is not regulated by the mating locus (Table 1).
TABLE 1 .
RLS was regulated and host passage selected for variants of RLS in some C. neoformans strains
| Strain | Type of comparison | Median RLS (no. of generations) | P valuea |
|---|---|---|---|
| Kn99a | Mating pair of serotype A strains | 33.0 | 0.502 |
| Kn99α | 22.0 | ||
| JEC20 | Mating pair of serotype D strains | 26.0 | 0.745 |
| JEC21 | 57.0 | ||
| SB4-S | Switch variants of serotype A | 11.0 | <0.001 |
| SB4-C | 30.0 | ||
| SB4-S-R | Revertant C to S | 67.5 | |
| RC-2-SM | Switch variants of serotype D | 31.0 | <0.001 |
| RC-2-MC | 25.0 | ||
| RC-2-SM-R | Revertant MC to SM | ||
| M7A | Sequential isolates; M7A collected on day 0 | 52.0 | 0.007 |
| M7E | Collected on day 15 | 34.0 | |
| M8A | Sequential isolates; M8A collected on day 0 | 9.5 | 0.514 |
| M8B | Collected on day 9 | 9.5 | |
| M9A | Sequential isolates; M9A collected on day 0 | 29.0 | 0.309 |
| M9B | Collected on day 17 | 35.0 | |
| M12A | Sequential isolates; M12A collected on day 0 | 99.0 | <0.001 |
| M12B | Collected on day 1 | 37.0 | |
| J9A | Sequential isolates; J9A collected on day 0 | 22.0 | 0.627 |
| J9B | Collected on day 7 | 16.5 | |
| J11A | Sequential isolates; J11A collected on day 0 | 35.5 | 0.337 |
| J11B | Collected on day 124 | 30.5 | |
| J22A | Sequential isolates; J22A collected on day 0 | 16.0 | 0.010 |
| J22B | Collected on day 150 | 24.5 | |
| J35A | Sequential isolates; collected on day 0 | 38.0 | 0.004 |
| J35B | Collected on day 1 | 27.5 |
The P value for the pair of strains or for the three strains.
In contrast, phenotypic switching in the previously described strains SB4 (25) and RC-2 (26) significantly changed RLS. In both strains, the hypervirulent variant (SB4-C and RC2-MC) exhibited a shortened RLS compared to the parent variant (SB4-S and RC2-SM) (Table 1). In strain RC-2, reversion of the MC variant back to SM (RC2-SM-R) did not correct this shortened RLS, despite reversion of the colony phenotype, whereas in strain SB4, reversion to S (SB4-S-R) extended RLS, but not to the extent of the original parent type.
As the RLSs of individual cells in a pathogen population exhibit a wide range, we hypothesized that C. neoformans cells with a longer or shorter RLS could be selected during chronic infection of the human host. The RLSs of sequential isolates (recovered 1 to 150 days apart) from 8 patients with CME were determined. Although all patients had CME, the strain, length of infection, and level of antifungal treatment were different for each patient. For 4 of the 8 patients (M7, M12, J22, and J35), the median RLS differed significantly between the early and late isolate (Table 1). Notably, in 3 of these 4 pairs, isolates with a significantly shortened RLS emerged during chronic infection. In 4 other patients (M8, M9, J9, and J11), the median RLSs of sequential isolates were not altered further after passage, supporting the notion that the median RLS of a population is a stable trait and not a random trait, and changes in RLS require selection of variants with altered RLSs in the host (Table 1).
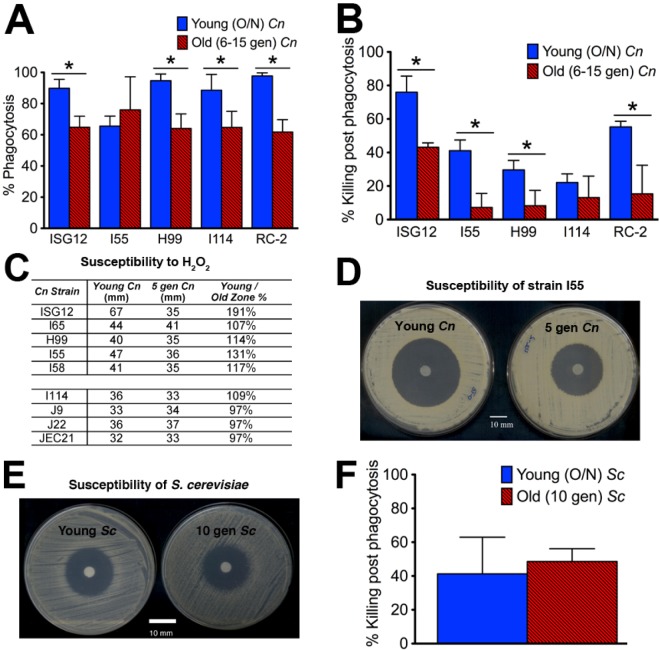
Aging promotes resistance to hydrogen peroxide (H2O2) and killing by macrophages.
The concept of selection was then further investigated. First, we tested whether aging promoted resistance to reactive oxygen species (ROS)-mediated killing by host macrophages (27). We assessed the ability of older C. neoformans cells to evade phagocytosis and killing by murine macrophages. Phagocytosis indices of 5 (6- to 15-generation-old) C. neoformans strains showed that in 4 of 5 strains, older cells were significantly more resistant to phagocytic uptake by macrophages (Fig. 2A) than young cells (overnight culture). Older cells were also more resistant to killing by macrophages 1 to 3 h after phagocytosis (Fig. 2B). Only strain I114 did not demonstrate a difference in killing for older C. neoformans cells. Next, disc diffusion assays with H2O2 (Fig. 2C), demonstrated that older C. neoformans cells were more resistant to H2O2 in 6 out of 9 strains and consequently able to grow as demonstrated by a smaller zone of inhibition compared to the larger zone for young cells, which were susceptible to H2O2 (Fig. 2D). In the other 3 strains, even young C. neoformans cells demonstrated a high H2O2 resistance. In contrast to C. neoformans cells, 10-generation-old S. cerevisiae cells did not demonstrate enhanced resistance to H2O2 (Fig. 2E), nor did they resist macrophage killing (Fig. 2F).
FIG 2 .
Aging promoted resistance to killing by macrophages and H2O2. (A) Phagocytic uptake by macrophages was inhibited in most older C. neoformans compared to young C. neoformans cells. O/N, overnight; gen, generation. (B) Older cells were more resistant to killing 1 to 3 h after phagocytosis than young C. neoformans cells. The values in panels A and B that are significantly different (P < 0.001) are indicated by a bar and asterisk. (C) Accordingly, 5-generation-old C. neoformans cells were commonly more resistant to H2O2 than young cells. The susceptibility to H2O2 is indicated by the zone of growth inhibition. (D) H2O2 disc diffusion assay with strain I55 showed susceptibility of young cells as demonstrated by a larger zone of growth inhibition. (E) H2O2 assay showed a comparable zone of inhibition for young (36 mm) and 10-generation-old S. cerevisiae cells (34 mm). (F) 10-generation-old S. cerevisiae cells and young cells were comparably susceptible to macrophage killing.
Evidence for in vivo selection of cells of advanced age in rat CME.
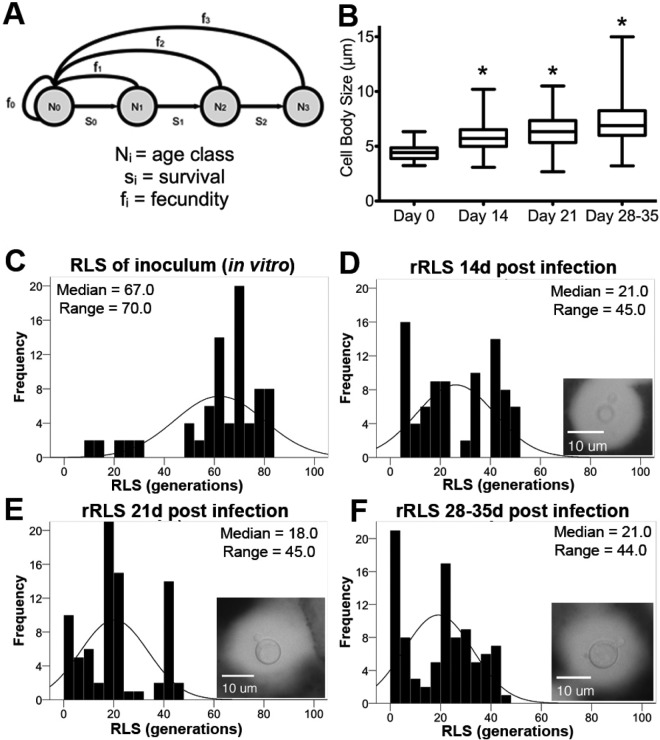
As outlined in our scheme (Fig. 3A), the ability of virgin cells (No) to age one generation (N1) depends on their survival (si) and on their fecundity (fi), which change with progression to each age class (generational age). In a clonally expanding population, we expect a 10-generation-old cell from a strain with a typical RLS (20 to 46 generations [see Fig. S2 in the supplemental material]) to occur approximately once in 1,024 cells. This representation is expected unless fecundity differs among generations and/or there is selection pressure that preferentially eliminates young cells, and thus allows cells of advanced age to accumulate. We hypothesized that such selection pressures are operative in vivo allowing older C. neoformans cells to accumulate. Using a Galleria mellonella infection model, we showed in vivo that S. cerevisiae cells derived from infected worms exhibited a higher number of bud scars than cells in the inoculum did (P < 0.01) (see Fig. S3 in the supplemental material). This indicated a selection toward older S. cerevisiae cells in worms even though S. cerevisiae is cleared in this model.
FIG 3 .
In vivo selection of older cells was observed in rat CME. (A) The ability of virgin cells (No) to age one generation (N1) depends on their survival (si) and on their fecundity (fi), which change with progression to each age class. In a replicating pathogen population, most cells are young (No to N2) and will live their full RLS. To investigate whether selection of older cells occurs in the host, rats were infected with RC-2 cells. (B) Cell body size significantly increased during infection as indicated by the asterisks (P < 0.001 by Kruskal-Wallis test). (C to F) rRLS was determined at 0, 14, 21, and 28 to 35 days postinfection and found to consistently decrease (P < 0.01 by Wilcoxon rank sum test). Note that the cells were budding (insets) and actively growing (increasing CFU [data not shown]). A total of 50 to 100 cells were microdissected in two independent experiments.
We then used a rat infection model that closely mimics human CME (21, 22) and found that the cell body size of CSF-derived C. neoformans cells (strain RC-2) increased significantly over 5 weeks of infection (Fig. 3B). Notably, the cell size of the overnight culture was comparable to that measured in vitro for 0-generation-old cells (4.80 µm), the size retrieved at 14 to 21 days of infection was comparable to that measured in vitro for 20-generation-old cells (5.93 µm), and the size retrieved at 28 to 35 days after infection was comparable to that measured in vitro for 40-generation-old cells (7.65 µm).
Next, the generational age of CSF-derived cells was assessed by immediately dissecting in vivo-derived cells (n ≥ 40 per spinal tap), thereby determining their remaining RLS (rRLS). In these experiments, we used the rRLS as a surrogate marker for generational age of the C. neoformans cells because unlike S. cerevisiae, bud scars do not adequately reflect the generational age of old C. neoformans cells (see Fig. S1 in the supplemental material). The rRLS was compared to the full RLS potential of virgin cells. The median rRLS of CSF-derived cells (Fig. 3D) was significantly shorter than the median RLS of cells in the inoculum (Fig. 3C). In addition, the rRLS continued to shorten in the course of chronic CNS infection (Fig. 3E and F).
We entertained an alternative explanation for the shortened RLS and increased size, which could be affected by host conditions, such as temperature (28) or starvation stress (29, 30), both of which have been documented in S. cerevisiae to affect cell size and RLS. However, temperature is not a concern, because C. neoformans is a human pathogen, and all strains used are derived from patients and already adapted to 37°C. Cell size (data not shown) and RLS (see Fig. S4A in the supplemental material) of RC-2 cells was unaffected at 30°C or 37°C. To assess the effect of starvation stress, we grew RC-2 cells for a prolonged time under conditions mimicking human CSF, where we changed medium every 12 h. Under these conditions, cells divided every 12 h and retained a mean cell size of 5.0 ± 0.9 µm and a median RLS of 51 generations (Fig. S4B), both of which were comparable to the average cell size and RLS (Fig. 3B and C) of the inoculum that mostly contains young cells (overnight culture). Additionally, C. neoformans cells derived from CSF at all 3 time points continued to actively multiply and exhibited budding (Fig. 3D to F, insets) with indices of 18 to 20%, which was not the case in S. cerevisiae cells that underwent starvation stress (29). Rat CSF contained white blood cells and C. neoformans cells. Rats died eventually of CME with high brain CFUs (106 [data not shown]), further documenting ongoing pathogen replication.
Evidence for in vivo selection of cells of advanced age in human CME.
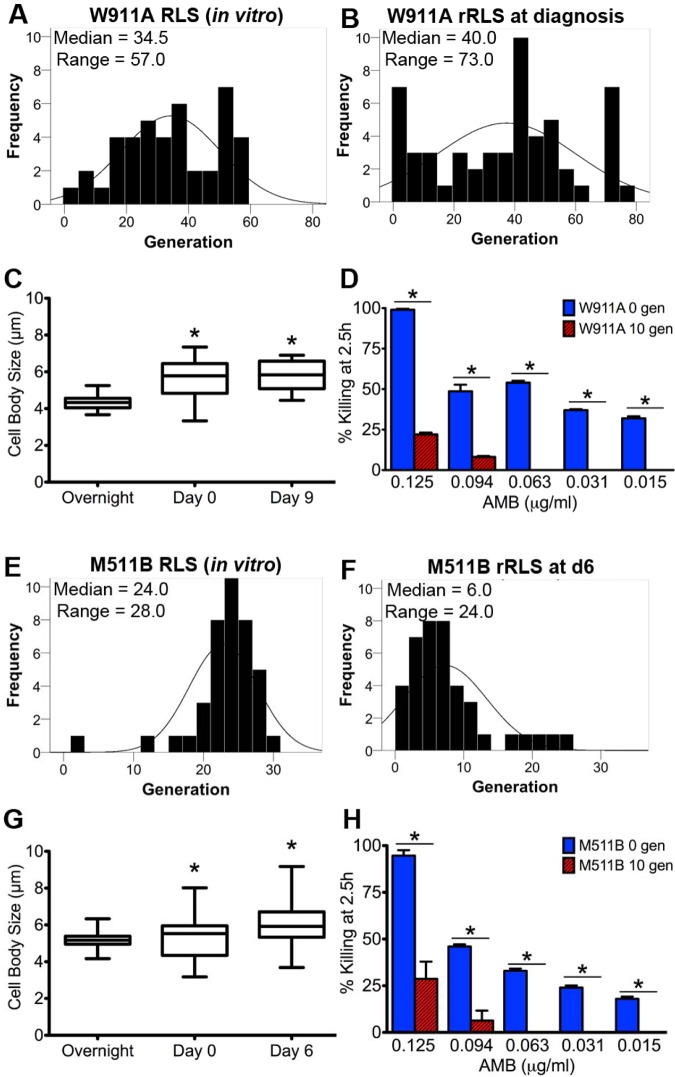
Next, the rRLS of C. neoformans cells directly derived from sequential lumbar punctures of two patients was determined. In both patients, CSF was isolated for initial diagnosis, and at 6 or 9 days as part of standard care. Both patients were treated with amphotericin B (AMB) only. Consistent with the accumulation of older cells, a significant increase in cell size was observed throughout the course of CME in cells derived from sequential isolates (Fig. 4C and G). In patient W911, C. neoformans cells from the first CSF sample collection on day 0 (W911A [Fig. 4B]) exhibited a median rRLS that was comparable to the median RLS of young W911A cells of an overnight culture (Fig. 4A) (P > 0.01 by Wilcoxon rank sum test). Notably, the rRLSs of these cells had a wider range than the overnight culture, and a trend suggesting the accumulation of older cells was apparent. C. neoformans cells were collected from CSF on day 9 after diagnosis (W911B), and cell size was measured (Fig. 4C). The rRLS could be determined on only 12 cells on this sample, of which 50% replicated 1 or 2 times and then stopped. C. neoformans cells from the first collection of CSF (M511A) from patient M511 arrived late, and although the cells were viable, transport was too long to determine rRLS accurately. C. neoformans cells from the second collection of CSF at day 6 (M511B) demonstrated a significantly shorter rRLS (Fig. 4F) than the overall RLS potential of the M511A strain (Fig. 4E) (P < 0.01 by Wilcoxon rank sum test), consistent with the accumulation of older cells after exposure to selection, namely, the host immune response and AMB treatment. We hypothesized that AMB would be less effective on older cells, thus leading to their selection, and tested this in vitro. As expected for both C. neoformans strains, in vitro killing assays with AMB demonstrated that 10-generation-old cells exhibited more resistance when exposed to AMB (Fig. 4D and H), and dissection of these cells confirmed the expected rRLS (approximately 10 generations shorter than that of the overnight culture [see Fig. S5A and S5B in the supplemental material]). This validated the accuracy of this assay and suggested that AMB itself does not shorten RLS. Importantly, enhanced AMB resistance of older cells was not documented for 10-generation-old S. cerevisiae cells, which showed enhanced sensitivity to AMB (see Fig. S5C in the supplemental material).
FIG 4 .
In vivo and in vitro selection of older cells was observed in human CME. (A and B) The rRLS of C. neoformans cells from the first lumbar puncture of patient W911 (W911A cells) showed a trend for selection of older cells with a shortened rRLS (40.0 versus 34.5 generations on average; P > 0.01 by Wilcoxon rank sum test). (C) Cell body size of in vivo cells from patient W911 derived at day 0 and day 9 was increased, suggesting that they were of advanced age (P < 0.001 by Kruskal-Wallis test). (D) To investigate whether antifungal treatment may explain the selection of older cells in the host, we exposed 0- or 10-generation-old W911A cells to AMB in vitro and found that they were more resistant to antifungal killing (P < 0.01) as indicated by the bars and asterisks. (E and F) Similarly, the rRLS of cells from the second lumbar puncture of patient M511 (M511B cells) was significantly reduced compared to its in vitro RLS (6.0 versus 24.0 generations on average; P < 0.01 by Wilcoxon rank sum test). Unlike the first patient, this patient had been on AMB when the cells were collected, which likely drove the selection of older cells. (G) The size of C. neoformans cells from patient M511 was increased relative to that of in vitro C. neoformans cells (P < 0.001 by Kruskal-Wallis test) as indicated by the asterisks. (H) AMB experiments confirmed that 10-generation-old M511B cells were more resistant to antifungal killing.
Mathematical modeling explains experimental data in rat and human CME.
Finally, mathematical modeling was used to validate our hypothesis. A Leslie matrix model was employed to determine whether selection could explain the trend toward accumulation of older cells, which are otherwise rare in a growing population. Instead of assuming a clonal expansion, the model accounted for dynamics of an age-structured population by considering the average offspring (fecundity) of each age class (generation) and its respective survival rate (fitness) (see Fig. S6 in the supplemental material). A scenario with no selection and an exponentially decaying fecundity was suitable as a null model for the in vitro behavior of C. neoformans cells. The age distributions obtained under these assumptions indicate that finding old cells is unlikely (<1% [data not shown]). Then, when a Gaussian function was used for parameterizing fecundity, an age distribution mimicking the ones observed in vivo after selection was obtained. It is not unreasonable to consider that the fecundity of the younger generations was effectively reduced in vivo due to the higher selective pressure or higher clearance of younger cells. Thus, this “effective fecundity” Gaussian function, different from the exponential one observed in vitro, could be considered to result from selection not accounted by the fitness function of the model, which included implicit DT differences. Under these new fecundity conditions, using a constant fitness function, the minimum chi-square (a measure of the difference between the observed and predicted age class distribution) obtained was high (Fig. 5A), and thus, the distributions did not show a bias toward the older generations (Fig. 5B). When a fitness function that shifted toward the older generations (whereby older cells are fitter/more resilient) was used, the minimum chi-square improved substantially (Fig. 5C), obtaining distributions that showed a bias toward the older generations (Fig. 5D). This reproduced the qualitative behavior of the experimental data in rat and human CME (Fig. 3D to F and Fig. 4B and F). Therefore, to explain the observed experimental data, our model suggests a fitness regimen where older cells are selected because they are fitter (more resilient) than younger cells in the host environment.
FIG 5 .
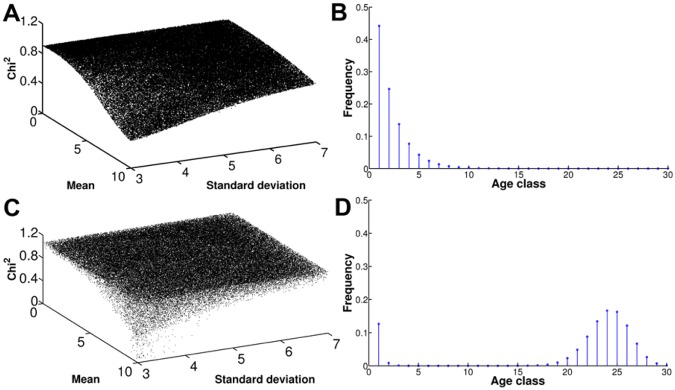
Mathematical modeling explained experimental data showing accumulation of older C. neoformans cells. (A and B) In a neutral scenario that assumed a constant fitness function, the minimum chi-square obtained was high (A), and the distribution did not show a bias toward the older age classes (B). (C and D) When selection was added to the scenario, the minimum chi-square improved substantially (C), obtaining distributions that showed a bias toward the older age classes (D).
DISCUSSION
This study for the first time examined RLS in C. neoformans and established that RLS is a reproducible but highly variable trait among C. neoformans strains, which is similar to the situation in S. cerevisiae (31). The RLSs of the two serotypes of C. neoformans were comparable, and all strains tested had similar phenotypic features. Specifically, a gradual increase in cell size and constant doubling times for the first two-thirds of life was followed by a rather abrupt transition to advanced age with prolonged doubling time and impaired fitness. We found that cells of advanced age were smaller than previously described giant cells, which emerge in a comparably shorter time (15, 32, 33). Our data indicated that cells of advanced age die by apoptosis analogous to S. cerevisiae (34). It remains unclear whether C. neoformans cells die only by apoptotic death and whether this mechanism of death in the CSF relates to the commonly observed paucity of inflammatory response in patients with high fungal burden (35, 36).
Being an accidental but successful pathogen with a broad host range, C. neoformans has adapted its regulatory pathways to survive in multiple hosts (37). Host selection pressure can also promote the emergence of variants with genetic and epigenetic changes. Therefore, it was of particular interest to investigate generational aging in the setting of host selection pressure. Although our data clearly demonstrated that advanced generational age in C. neoformans cells conveys resistance to macrophage and antifungal killing, one cannot necessarily deduce from these data alone that older C. neoformans cells would be selected in vivo and thus matter for virulence.
The phenotype of an older cell may convey resistance similar to genetic mutants or phenotypic switch variants; however, it is not inherited by the daughter cell, except in progeny derived from very old mothers as found in S. cerevisiae (38). Consequently, old cells should not statistically dominate in a replicating pathogen population, as was predicted in our mathematical model; in fact, they are expected to be exceedingly rare unless there is overwhelming selection for that specific phenotype, resulting in a shift to a population composed of older generations. Because bud scar stains cannot identify old C. neoformans cells, we devised an indirect method to demonstrate that in fact, older cells accumulate during chronic infection. We used a well-characterized rat infection model (21, 22), which allowed us to closely mimic human CME and institute appropriate controls. This model avoids confounders of drug treatment or patient status, as is the case with the human data, and has the advantage of a larger sample size. Rats infected intracisternally with a small number of young C. neoformans cells underwent weekly CSF collections, and cells were immediately measured to determine their size or microdissected to determine their rRLS. These serial collections demonstrated that C. neoformans cells increased in size and displayed significantly shorter rRLS during chronic infection. We concluded that C. neoformans cells of advanced age accumulate and thus complete fewer replications than virgin cells. The results of our mathematical modeling were consistent with our conclusion and suggest a fitness regimen where older C. neoformans cells are selected because they are fitter than young C. neoformans cells in the host environment.
Several other explanations for loss of RLS were entertained. One was switching of SM to MC, which would result in a variant with a shortened RLS. This was not observed and usually occurs later in the course of chronic infection (39). Another alternative explanation for the shorter RLS was that it was due to starvation stress in the host, which results in a shorter RLS in S. cerevisiae (29). Because bud scar count cannot be used to determine generational age of C. neoformans, we cannot completely rule out this possibility. However, our data do not support this conclusion. First, in rats, the percentage of dead cells (cells without RLS) was much smaller and only slowly rose over 1 month of infection to a maximum of 12% compared to 50% in S. cerevisiae (29). Second, C. neoformans cells actively grew in the CSF as evidenced by budding indices and 4-log-unit increases in CFU (21, 22). Third, the shortened RLS observed in the in vivo-derived C. neoformans cells was not reproduced in cells exposed to maximal starvation stress during in vitro growth. Finally, the cell size of CSF-derived cells was consistent with their estimated generational age in vitro, and in vitro starvation did not increase cell size on its own. It is noteworthy that larger C. neoformans cells have also been observed in electron microscopy pictures of tissue derived from chronically infected mice (40). All these findings support our conclusion that the shortened RLS and the increased cell size do in fact reflect a change in the generational distribution of the pathogen population because young cells are killed more readily and older cells persist. This may also explain why MICs, which are done with young cells, commonly do not correlate with clinical outcome in fungal infections (5, 41).
Whether a shortened or extended RLS confers a benefit to a C. neoformans strain cannot be conclusively determined from these data. A simple genetic approach to fulfill Koch’s postulate is difficult to design, because the loss of genes known to affect longevity commonly affect growth, and the interpretation of survival experiments with such null mutants would be impossible (42, 43). Also, multiple virulence traits can be identified in clinical strains (44, 45), making associations between RLS of a strain and virulence challenging. Both hypervirulent switch variants manifest considerably shortened life spans and seem to have traded longevity for virulence. Also, most of the sequential pairs of C. neoformans strains demonstrate a shortened RLS rather than an extended one. Investigations on long-lived S. cerevisiae mutants indicate that prolonged longevity comes at the cost of fitness early in life (46), which could be detrimental for a pathogen that has to establish an infection in a defensive host environment. Recent genomic studies on a pair of sequential C. neoformans isolates identified genetic changes that affect transcriptional regulators (47). Our data may indicate that a strain with a shortened life span may acquire the beneficial “old” phenotype earlier, and thus would be more resilient under host and antifungal attack. What causes the RLS to shorten is unclear. The fact that the shortened RLS is not reverted or only partially reverted in the switch variants suggests potentially complex epigenetic regulation or permanent loss of genes.
Although aging in prokaryotic and eukaryotic organisms is fundamentally different and cannot be compared, recent studies of Mycobacterium smegmatis indicate that older bacterial cells are also more resistant to antimicrobial agents (48). Thus, the concept of generational phenotypes that promote their selection in human hosts treated with antimicrobial agents seems to also be suggested in this study and may be relevant for other fungal pathogens that can infect the human host, such as Candida albicans and Candida glabrata, which replicate similarly. For C. neoformans cells with a relatively short RLS, a midlife yeast phenotype may strike the best balance to maintain chronic infection, having acquired considerable resistance while maintaining good replication at virtually the same DT. Alternatively, strains with an extended RLS could conceivably have a different biological benefit, as they may be more likely to sustain growth of the yeast population over a long time in vivo and promote latency. Although the slower DT of older cells may protect them against antifungal action as is the case with cancer therapies that target the fastest growing cells (49), we do not believe that it is the main mechanism of the observed resistance. In the macrophage- and antifungal-mediated killing assays, differences in DT between young and older cells were controlled for (see Text S1 in the supplemental material). Additionally, 15-generation-old cells do not grow slower than 0-generation-old cells, because for most strains, 15 generations is before their median RLS. These cells are not senescent, and they can still replicate and establish infection. Our earlier data suggest that the cell wall size is markedly increased in older C. neoformans cells (20), which could result in decreased penetration of antifungal agents.
It is noteworthy that although this study established common phenotypic characteristics, such as increased cell size and variability in RLS in S. cerevisiae and C. neoformans cells, it also delineated important differences between these fungi, especially with respect to the resilience of older S. cerevisiae and C. neoformans cells. Older C. neoformans cells are resistant to macrophage, H2O2, and AMB killing, whereas older S. cerevisiae cells are not. Most likely, the scattered budding of S. cerevisiae cells weakens the cell wall more than in C. neoformans, where bud scars seem to heal. This observation highlights the possibility that resilience and potential selection of older fungal cells may be affected by coevolution in the host environment, and thus, data derived from S. cerevisiae cannot necessarily be applied to human fungal pathogens.
Finally, our results encourage research on cellular aging in fungal pathogens, as age is an emerging natural trait that contributes to virulence, and targeting the process of aging could constitute a novel therapeutic approach in antifungal therapy.
MATERIALS AND METHODS
Strains.
S. cerevisiae or C. neoformans strains and media used in this study are listed in Text S1 and in Tables S1 and S2, respectively, in the supplemental material.
RLS and rRLS determination.
The RLS of C. neoformans strains (Table 1) was determined by microdissection as previously published for S. cerevisiae (23) with some modifications. Briefly, 20 to 40 cells of each strain of C. neoformans were arrayed on a yeast extract-peptone-dextrose (YPD) agar plate maintained at 37°C. The first bud of each cell was identified as the virgin mother cell, which then grew in size with every budding event and could easily be distinguished. New buds from the mother cell were separated at the end of each division (1 to 2 h) using a 50-µm fiber optic needle (Cora Styles) on a tetrad dissection Axioscope A1 microscope (Zeiss) at a magnification of ×10. The plate was returned to the incubator after each separation or to 4°C overnight to prevent excessive budding. The study was terminated when cells had failed to divide for 24 h, and then the plates were kept and incubated for an additional week to ensure that the failure to divide was from death, not cell cycle arrest. The RLS or rRLS of each cell was the sum of the total buds until cessation of division. The rRLS was determined the same way on C. neoformans cells derived directly from rat cisternal punctures (21) (see Text S1 in the supplemental material) or on C. neoformans cells (M511A, M511B, W911A, and W911B) derived directly from lumbar punctures of patients at hospitals affiliated with the Albert Einstein College of Medicine. Human experiments were carried out with the approval of the Albert Einstein College of Medicine Institutional Review Board.
Isolation of cells of advanced age.
0-generation-old C. neoformans cells of newly budded cells were isolated by counterflow centrifugal elutriation (20), labeled with Sulfo-NHS-LC-Biotin (Thermo Scientific) (sulfosuccinimidyl-6-[biotin-amido]hexanoate) and then grown for 5 or 6 DT in SD+ medium (see Table S2 in the supplemental material). Larger cells were separated by elutriation, labeled with streptavidin-conjugated magnetic microbeads (Miltenyi Biotec, Auburn, CA), and then isolated on an LS magnetic column (Miltenyi Biotec). The purity of older cells was confirmed on a sample by fluorescein isothiocyanate (FITC)-conjugated streptavidin labeling, then the cells were regrown in new medium to prevent stationary growth as confirmed by increasing cell number, and the procedure was repeated to obtain older cells of interest. Cells that washed off from the column when the cells were on the magnet were used as a control for young cells (referred to as overnight [O/N] culture), as they had undergone the same manipulations used to obtain the older cells with the exception of biotin or streptavidin conjugate labeling. Older S. cerevisiae cells were isolated by an initial round of elutriation, followed by magnetic bead selection only.
Apoptosis stain.
14-generation-old H99, 10-generation-old ISG12, or young (O/N culture) cells of each strain were stained with FITC-conjugated annexin V (stain green) and propidium iodide (stain red) in annexin V binding buffer per the instructions of the kit (Abcam). For a control, an O/N culture was incubated in 175 mM acetic acid for 200 min to induce apoptosis.
Mathematical modeling.
Briefly, the Leslie matrix of an age-structured population was built as previously described (50) and used to model the aging phenomenon, being defined by the average offspring or fecundity (fi) of each age class and their respective survival rates (si). See Text S1 in the supplemental material for additional details.
Additional methods.
For immunofluorescence image capture, cell size measurements, H2O2 disc diffusion assay, phagocytosis index, macrophage- or antifungal-mediated killing assays, animal infection models, and detailed elutriation protocol, see Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download
Calcofluor stains successfully quantitated bud scars in S. cerevisiae, but not in C. neoformans. Calcofluor reliably stained chitin for 6-generation-old S. cerevisiae cells, but not 6-generation-old C. neoformans cells, where a wheat germ agglutinin lectin stain against N-acetylglucosamine residues had more success, but was not accurate. Download
Clinical C. neoformans strains demonstrate a wide distribution in RLS. RLS of each cell from 14 clinical C. neoformans was pooled to plot a wide range (whiskers) and a RLS that typically fell between 20–46 generations (box). Download
In vivo selection of older S. cerevisiae cells was documented in a G. mellonella infection model. (A) S. cerevisiae cells with a high number of bud scars were found at day 3 after infection in G. mellonella compared to the inoculum (day 0). (B) 28% of the inoculum contained cells with 2 or more bud scars compared to 65% of the cells collected ex vivo, which indicated that there was a selection for older S. cerevisiae cells. Download
Shortened RLS in rat CME was not a result of stress. rRLS was dissected on RC-2 cells grown for 7 d in in vitro conditions that mimicked CSF. After maximal stress, these cells had transitioned to stationary phase and exhibited only a mildly reduced rRLS (51.0 generations on average). Download
Older C. neoformans cells, not S. cerevisiae cells, were more resistant to antifungal killing. (A) To confirm that the effect of amphotericin B (AMB) was on a pure population of 0- or 10-generation-old W911A cells, rRLS was determined validating that older cells (patterned box) completed fewer replications. (B) 10-generation-old M511B cells also completed fewer replications compared to 0-generation-old cells. (C) As opposed to the enhanced AMB resistance seen in C. neoformans cells, 10-generation-old S. cerevisiae cells exhibit enhanced sensitivity to AMB compared to young cells (overnight culture). Download
Fitness and fecundity assumptions were made in a selective model. (A) Fitness was assumed to be variable with each age class, but higher for older age classes. (B) Fecundity was assumed to be higher for the younger age classes. Download
C. neoformans strains used in this study.
Media used in this study.
ACKNOWLEDGMENTS
We thank Arturo Casadevall for 18B7 antibody, Sue Jaspersen for strain Fy86, and George Sutphin for technical advice. We thank Arturo Casadevall, Josh Nosanchuk, and Marion Schmidt for critical reading of the manuscript.
B.C.F. is supported by R21 A1087564, RO1 AI059681, and the Center for AIDS Research at the Albert Einstein College of Medicine and Montefiore Medical Center (NIH AI-51519). T.B. is supported by the Institutional AIDS Training Grant, “HIV, AIDS and Opportunistic Infections” (T32 AI007501). X.P. is supported by a Fulbright grant (grantee ID no. 15120166). A.B. is supported by RO1 CA164468.
Footnotes
Citation Bouklas T, Pechuan X, Goldman DL, Edelman B, Bergman A, Fries BC. 2013. Old Cryptococcus neoformans cells contribute to virulence in chronic cryptococcosis. mBio 4(4):e00455-13. doi:10.1128/mBio.00455-13.
REFERENCES
- 1. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 2. Casadevall A, Spitzer ED, Webb D, Rinaldi MG. 1993. Susceptibilities of serial Cryptococcus neoformans isolates from patients with recurrent cryptococcal meningitis to amphotericin B and fluconazole. Antimicrob. Agents Chemother. 37:1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manosuthi W, Sungkanuparph S, Thongyen S, Chumpathat N, Eampokalap B, Thawornwan U, Foongladda S. 2006. Antifungal susceptibilities of Cryptococcus neoformans cerebrospinal fluid isolates and clinical outcomes of cryptococcal meningitis in HIV-infected patients with/without fluconazole prophylaxis. J. Med. Assoc. Thai. 89:795–802 [PubMed] [Google Scholar]
- 4. Spitzer ED, Spitzer SG, Freundlich LF, Casadevall A. 1993. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet 341:595–596 [DOI] [PubMed] [Google Scholar]
- 5. Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, Taseera K, Rebe K, Loyse A, Jarvis J, Bekker LG, Wood R, Limmathurotsakul D, Chierakul W, Stepniewska K, White NJ, Jaffar S, Harrison TS. 2009. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin. Infect. Dis. 49:702–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Litvintseva AP, Mitchell TG. 2009. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect. Immun. 77:3188–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fries BC, Casadevall A. 1998. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J. Infect. Dis. 178:1761–1766 [DOI] [PubMed] [Google Scholar]
- 8. Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, Kronstad JW, Perfect JR. 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2(3):e00103-11. 10.1128/mBio.00103-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rzaieva OM, Varbanets’ LD, Hudzenko OV. 2011. Optimization of cultivation conditions of Cryptococcus albidus—producers of alpha-l-rhamnosidase. Mikrobiol. Z. 73:10–16 (In Ukrainian.) [PubMed] [Google Scholar]
- 10. McFadden DC, Fries BC, Wang F, Casadevall A. 2007. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot. Cell 6:1464–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alanio A, Desnos-Ollivier M, Dromer F. 2011. Dynamics of Cryptococcus neoformans-macrophage interactions reveal that fungal background influences outcome during cryptococcal meningoencephalitis in humans. mBio 2(4):e00158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steenbergen JN, Shuman HA, Casadevall A. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. U. S. A. 98:15245–15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kwon-Chung KJ, Bennett JE. 1978. Distribution of alpha and alpha mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337–340 [DOI] [PubMed] [Google Scholar]
- 14. Zaragoza O, Chrisman CJ, Castelli MV, Frases S, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2008. Capsule enlargement in Cryptococcus neoformans confers resistance to oxidative stress suggesting a mechanism for intracellular survival. Cell. Microbiol. 10:2043–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. García-Rodas R, Casadevall A, Rodríguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. 2011. Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS One 6:e24485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J. Clin. Microbiol. 43:556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu XH, Meng FL, Hu Y, Zhou JQ. 2008. Candida albicans, a distinctive fungal model for cellular aging study. Aging Cell 7:746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roux AE, Chartrand P, Ferbeyre G, Rokeach LA. 2010. Fission yeast and other yeasts as emergent models to unravel cellular aging in eukaryotes. J. Gerontol. A Biol. Sci. Med. Sci. 65:1–8 [DOI] [PubMed] [Google Scholar]
- 19. Kaeberlein M. 2010. Lessons on longevity from budding yeast. Nature 464:513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jain N, Cook E, Xess I, Hasan F, Fries D, Fries BC. 2009. Isolation and characterization of senescent Cryptococcus neoformans and implications for phenotypic switching and pathogenesis in chronic cryptococcosis. Eukaryot. Cell 8:858–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldman DL, Casadevall A, Cho Y, Lee SC. 1996. Cryptococcus neoformans meningitis in the rat. Lab. Invest. 75:759–770 [PubMed] [Google Scholar]
- 22. Fries BC, Lee SC, Kennan R, Zhao W, Casadevall A, Goldman DL. 2005. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect. Immun. 73:1779–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steffen KK, Kennedy BK, Kaeberlein M. 2009. Measuring replicative life span in the budding yeast. J. Vis. Exp. 2009:e1209. 10.3791/1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK, Jr, Giles SS, Mitchell TG, Casadevall A, Perfect JR, Heitman J. 2005. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 73:4922–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldman DL, Fries BC, Franzot SP, Montella L, Casadevall A. 1998. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc. Natl. Acad. Sci. U. S. A. 95:14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. 1999. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect. Immun. 67:6076–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, Perfect JR. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai CY, Jaruga E, Borghouts C, Jazwinski SM. 2002. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics 162:73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ashrafi K, Sinclair D, Gordon JI, Guarente L. 1999. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 96:9100–9105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qin H, Lu M. 2006. Natural variation in replicative and chronological life spans of Saccharomyces cerevisiae. Exp. Gerontol. 41:448–456 [DOI] [PubMed] [Google Scholar]
- 32. Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6:e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6:e1000945. 10.1371/journal.ppat.1000945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fröhlich KU, Madeo F. 2001. Apoptosis in yeast: a new model for aging research. Exp. Gerontol. 37:27–31 [DOI] [PubMed] [Google Scholar]
- 35. Bicanic T, Brouwer AE, Meintjes G, Rebe K, Limmathurotsakul D, Chierakul W, Teparrakkul P, Loyse A, White NJ, Wood R, Jaffar S, Harrison T. 2009. Relationship of cerebrospinal fluid pressure, fungal burden and outcome in patients with cryptococcal meningitis undergoing serial lumbar punctures. AIDS 23:701–706 [DOI] [PubMed] [Google Scholar]
- 36. Darras-Joly C, Chevret S, Wolff M, Matheron S, Longuet P, Casalino E, Joly V, Chochillon C, Bédos JP. 1996. Cryptococcus neoformans infection in France: epidemiologic features of and early prognostic parameters for 76 patients who were infected with human immunodeficiency virus. Clin. Infect. Dis. 23:369–376 [DOI] [PubMed] [Google Scholar]
- 37. Casadevall A, Pirofski LA. 2007. Accidental virulence, cryptic pathogenesis, Martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot. Cell 6:2169–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kennedy BK, Austriaco NR, Jr, Guarente L. 1994. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J. Cell Biol. 127:1985–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fries BC, Taborda CP, Serfass E, Casadevall A. 2001. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Invest. 108:1639–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feldmesser M, Kress Y, Casadevall A. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147:2355–2365 [DOI] [PubMed] [Google Scholar]
- 41. Huang YC, Kao HT, Lin TY, Kuo AJ. 2001. Antifungal susceptibility testing and the correlation with clinical outcome in neonatal candidemia. Am. J. Perinatol. 18:141–146 [DOI] [PubMed] [Google Scholar]
- 42. Cruz MC, Goldstein AL, Blankenship J, Del Poeta M, Perfect JR, McCusker JH, Bennani YL, Cardenas ME, Heitman J. 2001. Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob. Agents Chemother. 45:3162–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waugh MS, Nichols CB, DeCesare CM, Cox GM, Heitman J, Alspaugh JA. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191–201 [DOI] [PubMed] [Google Scholar]
- 44. Ma H, May RC. 2009. Virulence in cryptococcus species. Adv. Appl. Microbiol. 67:131–190 [DOI] [PubMed] [Google Scholar]
- 45. Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, Geddes JM, Griffiths EJ, Choi J, Cadieux B, Caza M, Attarian R. 2012. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot. Cell 11:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delaney JR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. 2011. Quantitative evidence for early life fitness defects from 32 longevity-associated alleles in yeast. Cell Cycle 10:156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ormerod KL, Morrow CA, Chow EW, Lee IR, Arras SD, Schirra HJ, Cox GM, Fries BC, Fraser JA. 2013. Comparative genomics of serial isolates of Cryptococcus neoformans reveals gene associated with carbon utilization and virulence. G3 3(4):675-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collado M, Blasco MA, Serrano M. 2007. Cellular senescence in cancer and aging. Cell 130:223–233 [DOI] [PubMed] [Google Scholar]
- 50. Leslie PH. 1945. On the use of matrices in certain population mathematics. Biometrika 33:183–212 [DOI] [PubMed] [Google Scholar]
- 51. Kwon-Chung KJ, Edman JC, Wickes BL. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jain N, Wickes BL, Keller SM, Fu J, Casadevall A, Jain P, Ragan MA, Banerjee U, Fries BC. 2005. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J. Clin. Microbiol. 43:5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68:4225–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cotter G, Doyle S, Kavanagh K. 2000. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 27:163–169 [DOI] [PubMed] [Google Scholar]
- 55. Meyer CD. 2000. Matrix analysis and applied linear algebra. Miscellaneous titles in applied mathematics series. Society for Industrial and Applied Mathematics (SIAM), Philadelphia, PA. [Google Scholar]
- 56. Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. 1953. Equations of state calculations by fast computing machines. J. Chem. Phys. 21:1087–1092 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download
Calcofluor stains successfully quantitated bud scars in S. cerevisiae, but not in C. neoformans. Calcofluor reliably stained chitin for 6-generation-old S. cerevisiae cells, but not 6-generation-old C. neoformans cells, where a wheat germ agglutinin lectin stain against N-acetylglucosamine residues had more success, but was not accurate. Download
Clinical C. neoformans strains demonstrate a wide distribution in RLS. RLS of each cell from 14 clinical C. neoformans was pooled to plot a wide range (whiskers) and a RLS that typically fell between 20–46 generations (box). Download
In vivo selection of older S. cerevisiae cells was documented in a G. mellonella infection model. (A) S. cerevisiae cells with a high number of bud scars were found at day 3 after infection in G. mellonella compared to the inoculum (day 0). (B) 28% of the inoculum contained cells with 2 or more bud scars compared to 65% of the cells collected ex vivo, which indicated that there was a selection for older S. cerevisiae cells. Download
Shortened RLS in rat CME was not a result of stress. rRLS was dissected on RC-2 cells grown for 7 d in in vitro conditions that mimicked CSF. After maximal stress, these cells had transitioned to stationary phase and exhibited only a mildly reduced rRLS (51.0 generations on average). Download
Older C. neoformans cells, not S. cerevisiae cells, were more resistant to antifungal killing. (A) To confirm that the effect of amphotericin B (AMB) was on a pure population of 0- or 10-generation-old W911A cells, rRLS was determined validating that older cells (patterned box) completed fewer replications. (B) 10-generation-old M511B cells also completed fewer replications compared to 0-generation-old cells. (C) As opposed to the enhanced AMB resistance seen in C. neoformans cells, 10-generation-old S. cerevisiae cells exhibit enhanced sensitivity to AMB compared to young cells (overnight culture). Download
Fitness and fecundity assumptions were made in a selective model. (A) Fitness was assumed to be variable with each age class, but higher for older age classes. (B) Fecundity was assumed to be higher for the younger age classes. Download
C. neoformans strains used in this study.
Media used in this study.