Abstract
Adeno-associated virus (AAV)–mediated RNA interference shows promise as a therapy for chronic hepatitis B virus (HBV) infection, but its low efficacy and hepatotoxicity pose major challenges. We have generated AAV vectors containing different promoters and a panel of HBV-specific short hairpin RNAs (shRNAs) to investigate factors that contribute to the efficacy and pathogenesis of AAV-mediated RNA interference. HBV transgenic mice injected with high doses of AAV vectors containing the U6 promoter produced abundant shRNAs, transiently inhibited HBV, but induced severe hepatotoxicity. Sustained HBV suppression without liver toxicity can be achieved by lowering the dose of AAV-U6 vectors. AAVs containing the weaker H1 promoter did not cause liver injury, but their therapeutic efficacy was highly dependent on the sequence of the shRNA. Mice treated with the toxic U6-promoter-driven shRNA showed little change in hepatic microRNA levels, but a dramatic increase in hepatic leukocytes and inflammatory cytokines and chemokines. Hepatotoxicity was completely absent in immunodeficient mice and significantly alleviated in wild-type mice depleted of macrophages and granulocytes, suggesting that host inflammatory responses are the major cause of liver injury induced by the overexpressed shRNAs from AAV-U6 vectors. Our results demonstrate that selection of a highly potent shRNA and control its expression level is critical to achieve sustained HBV suppression without inducing inflammatory side effects.
Sun and colleagues generate a panel of AAV vectors containing various promoters and hepatitis B virus (HBV)-specific short hairpin RNAs (shRNAs) to investigate factors that contribute to the efficacy and pathogenesis of AAV-mediated RNA interference. They show that injecting HBV transgenic mice with high doses of AAV vectors containing the U6 promoter produce abundant shRNAs, transiently inhibit HBV, but induce severe hepatotoxicity. By contrast, AAVs containing the weaker H1 promoter do not cause liver injury, but their therapeutic efficacy is highly dependent on the sequence of the shRNA.
Introduction
Hepatitis B virus (HBV) is a major human pathogen that causes acute liver disease and chronic infection. Although an effective vaccine is available to decrease the incidence of HBV infection, more than 350 million people worldwide are estimated to be chronically infected with HBV and at high risk of developing liver failure, cirrhosis, and hepatocellular carcinoma (Ganem and Prince, 2004). Current anti-HBV therapies, including IFN-α and nucleoside and nucleotide analogs, have limited effectiveness in complete elimination of viral DNA templates, and their use is usually accompanied by selection of drug-resistant mutations and a high rate of relapse when treatment is discontinued (Kwon and Lok, 2011).
RNA interference (RNAi) represents an alternative therapy for chronic HBV (Giladi et al., 2003; Klein et al., 2003; McCaffrey et al., 2003; Shlomai and Shaul, 2003; Morrissey et al., 2005a, b; Uprichard et al., 2005; Kim et al., 2007; Carmona et al., 2009; Ivacik et al., 2011). However, because of the high viral burden in these patients, successful RNAi treatment requires the combined use of potent short hairpin RNAs (shRNAs) and a highly efficient vector system that can transduce most of the infected cells and produce effective and sustained levels of shRNAs to eliminate the viral targets. Adeno-associated virus (AAV)–based vectors represent a promising delivery system for RNAi-based therapy because of their ability to infect both dividing and nondividing cells, to achieve extensive target organ transduction in vivo, and to direct long-term gene expression in these tissues (Mingozzi and High, 2011; Van Der Laan et al., 2011). Using a transgenic mouse line carrying a complete HBV genome and producing high-serum viral titers (>108 genome copies per ml), we previously reported that intravenous (i.v.) injection of a double-stranded AAV serotype 8 vector encoding a HBV-targeting shRNA markedly reduced the serum HBV titer and liver levels of HBV DNA, mRNA, and protein (Chen et al., 2007). The antiviral effect could be prolonged by consecutive injections of alternative AAV serotypes (AAV7 and AAV9) encoding the same anti-HBV shRNA, leading to a significant reduction in liver cell injury, hepatic regeneration, and incidence of liver tumor (Chen et al., 2009, 2012), which develop spontaneously in these HBV transgenic mice. Animals receiving shRNA-expressing AAV vectors did not show liver damage or mortality during the 1-year observation period (Chen et al., 2012). These results suggest that AAV-delivered shRNA has potential application in the treatment of chronic HBV and HBV-associated liver cancers.
However, in other reports, similar shRNA-encoding AAV8 vectors targeting HBV and a variety of other genes caused dose-dependent acute hepatitis and lethality in the majority of treated mice (Grimm et al., 2006, 2010). The proposed mechanism for the shRNA-mediated toxicity involves shRNA interference with endogenous microRNA processing and function (Grimm et al., 2006). The factors causing the difference in therapeutic efficacy and liver toxicity between our study and other studies have not been clearly defined. In the present study, we directly compared the influence of different promoters (H1 vs. U6) and HBV-targeting shRNAs on hepatic toxicity and therapeutic efficacy of AAV-mediated RNAi therapy in a chronic HBV model. Our results showed that at high doses AAV-U6 vectors produced abundant shRNAs and had only a transient effect on HBV suppression because of extensive liver injury and subsequent loss of AAV vectors and shRNA expression. The hepatotoxicity was caused by immune-mediated inflammatory responses, with saturation of endogenous microRNA synthesis pathways having only a little effect. Safe, effective, and sustained HBV suppression was achieved using the weaker H1 promoter and a highly potent shRNA or the stronger U6 promoter at lower doses. A better understanding of these mechanisms may help increase the safety and therapeutic benefits of AAV-based RNAi therapy in HBV patients.
Methods
Animals
All experimental procedures were reviewed and approved by the Animal Care and Use Committee of the Academia Sinica, Taipei, Taiwan. The C57BL/6J and FVB mice were purchased from the National Laboratory Animal Breeding and Research Center (Taipei, Taiwan) and the ICR mice from BioLASCO (Ilan, Taiwan). The ICR/HBV transgenic mouse line Tg[HBV1.3]24-3, which produces high and stable HBV titers, has been reported previously (Chen et al., 2007). The NOD.Cg-Prkdcscid IL2Rγtm1Wjl/SzJ (Nod-scid/IL2Rγ−/−), C57BL/6-I-Aβ−/− (I-Aβ−/−), and C57BL/6-CD8αtm1Mak (CD8α−/−) mice were originally obtained from the Jackson Laboratory. CD1d−/− mice were kindly provided by Dr. Chyung-Ru Wang (Northwestern University Feinberg School of Medicine, Chicago, IL). The various immunodeficient mouse lines were maintained as small breeding colonies in a specific pathogen-free environment in the animal facilities of the Institute of Biomedical Sciences, Academia Sinica.
Construction and production of the pseudotyped AAV8 vectors
The construction of the pAAVEMBL-H1/HBV-S1 and pAAVEMBL-H1/GL2 plasmids, which contain the H1 promoter and the shRNA coding sequence, has been reported previously (Chen et al., 2007). The oligonucleotide fragment containing the sAg19 or sAg25 sequence (Fig. 1a) was similarly cloned into the pAAVEMBL-H1 plasmid to generate pAAVEMBL-H1/sAg19 and pAAVEMBL-H1/sAg25, respectively. To generate shRNAs driven by the U6 promoter, the oligonucleotide fragment containing the GL2, HBV-S1, sAg19, or sAg25 sequence was cloned into the intermediate plasmid, pSilencer (Invitrogen/Life Technologies, Carlsbad, CA), containing the U6 promoter. The HindIII/EcoRI fragment containing the U6 promoter and the shRNA coding sequence was then released from the various pSilencer plasmids and used to replace the corresponding fragment in pAAVEMBL-H1/GL2 to generate plasmids pAAVEMBL-U6/GL2, pAAVEMBL-U6/HBV-S1, pAAVEMBL-U6/sAg19, and pAAVEMBL-U6/sAg25, respectively. The various pAAVEMBL-H1 and pAAVEMBL-U6 plasmids were then used to produce, respectively, the pseudotyped AAV8-H1 and AAV8-U6 vectors using the triple transfection method as described previously (Chen et al., 2009).
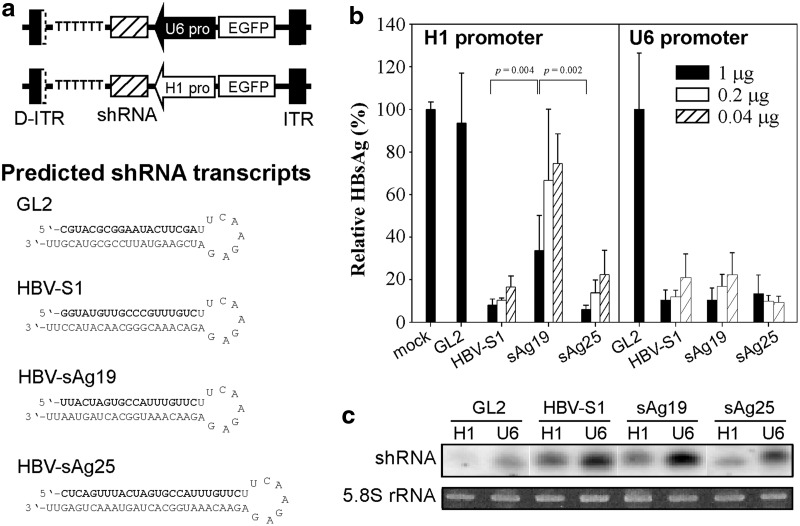
FIG. 1.
In vitro inhibition of HBV gene expression by H1- or U6-driven shRNAs. (a) Schematic representation of the shRNA-encoding pAAVEMBL plasmids containing the H1 or U6 promoter. The predicted structures and sequences of the three HBV-specific shRNAs and the control GL2 shRNA are depicted, with the sense strand shown in bold. (b) Inhibition of HBsAg expression by shRNAs. Huh-7 cells were transfected with pHBV1.3 alone or together with the indicated amounts of shRNA-expressing pAAVEMBL-H1 or pAAVEMBL-U6 plasmids for 72 hr, and then HBsAg levels in the culture supernatant were analyzed by enzyme-linked immunosorbent assay. The data are presented as a percentage of that produced by cells transfected with pHBV1.3 alone (mock transfection, mean±SD by three wells per condition). (c) Small RNAs from cells transfected with 1 μg of the different pAAVEMBL-H1 or pAAVEMBL-U6 plasmids transfected were detected by Northern blot using radiolabeled probes identical in sequence to the antisense strand of the corresponding HBV or luciferase shRNAs. 5.8S rRNA was stained with ethidium bromide as a loading control. HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; ITR, inverted terminal repeat; SD, standard deviation; shRNA, short hairpin RNA.
Co-transfection in vitro
Huh-7 cells in 24-well plates were co-transfected with 2 μg of pHBV1.3 plasmid containing a 1.3-fold overlength HBV genome (Chou et al., 2005) and different amounts (0.04, 0.2, or 1 μg) of various pAAVEMBL-H1 or pAAVEMBL-U6 plasmids using lipofectamine 2000 (Invitrogen) and the amount of hepatitis B surface antigen (HBsAg) in the supernatants analyzed 3 days later by enzyme-linked immunosorbent assay.
Animal experiments
Male mice at 8–12 weeks of age were used. The ICR/HBV mice used were selected from those with a serum HBV titer >108 genome copies per ml. Each mouse received a single i.v. injection of 1012 vector genomes (vg) of various AAV8-H1 or AAV8-U6 vectors as indicated in the figure legend. In some experiments, lower doses of AAV-U6/HBV-S1 at 2×1011, 6.6×1010, or 8×109 vg were intravenously injected into each mouse. For depletion of NK cells or granulocytes, C57BL/6 mice were injected intraperitoneally, respectively, with 20 μl of rabbit anti-asialo-GM1 antiserum (Wako Pure Chemical, Osaka, Japan) at days −2, 0, and 3 or with 0.5 mg of rat anti-Gr-1 monoclonal antibody (mAb) (RB6-8C5) at day −2 and with 0.25 mg at days 0 and 3. Mice treated with the same dose and schedule of normal rabbit serum (BioWest, Nuaille, France) or a monoclonal normal rat immunoglobulin G (IgG; HAA), derived from a hybridoma clone isolated from a naive rat, were included as controls. For Kupffer cell depletion, C57BL/6 mice were injected intravenously with 200 μl of clodronate liposome (ClodronateLiposomes, Amsterdam, The Netherlands) or 30 mg/kg of gadolinium chloride (GdCl3; Sigma-Aldrich, St. Louis, MO) for five consecutive days from day −1. Serum and liver samples were collected at different times for analysis.
HBV DNA and HBsAg analysis
Serum HBV DNA was quantified by hybridization probe-based real-time polymerase chain reaction (PCR; LightCycler FastStart; Roche Diagnostics GmbH, Mannheim, Germany) as described previously (Chen et al., 2009). HBsAg was measured using an Elecsys Systems electrochemiluminescence kit and a Cobas analyzer (Roche Diagnostics GmbH).
Small RNA Northern blot analysis
GL2 and HBV shRNAs (HBV-S1, sAg19, or sAg25), miR-122, and 5S rRNA were quantified by Northern blot analysis as described previously (Chen et al., 2009). The probes contained 32P-labeled oligonucleotides corresponding to the antisense strand of the various shRNAs, miR-122, or 5S rRNA. For in vitro study, equal RNA loading was assessed by ethidium bromide staining of 5S rRNA. ImageQuant software (Molecular Dynamics, Sunnyvale, CA) was used to quantify small RNA signals, as described previously (Chen et al., 2009).
AAV DNA Southern blot analysis
AAV DNA in the liver was quantified by Southern blot analysis as described previously (Chen et al., 2009). The probe contained a 32P-labeled DNA fragment corresponding to the green fluorescent protein coding sequence, which is present in all pAAVEMBL-H1 or pAAVEMBL-U6 plasmids used in this study and the intensity of the signal was quantified using ImageQuant software.
Measurement of serum alanine aminotransferase activity and albumin levels
Serum alanine aminotransferase (ALT) activity and albumin levels were measured using Vitros Chemistry Products ALT slides or albumin slides, respectively, and a Vitros 950 chemical analyzer (Johnson & Johnson, Rochester, NY).
Histology
Liver sections were fixed in 4% paraformaldehyde in phosphate-buffered saline, embedded in paraffin, sectioned (5 μm), and stained with hematoxylin and eosin, and then were mounted and observed by light microscopy.
Flow cytometric analysis of liver leukocyte populations
Hepatic mononuclear cells were isolated on a Percoll density gradient and analyzed by flow cytometric analysis as described previously (Chang et al., 2010). Cells were preincubated with anti-CD16/32 mAb (2.4G2; ATCC) to block nonspecific binding and then were incubated with the following mAbs: NK1.1 (PK136), CD3ɛ (145-2C11), CD8 (53-6.7), CD4 (RM4-5), CD19 (6D5), CD11c (HL3), CD11b (M1/70), Gr-1 (RB6-8C5), and the viability dye 7-AAD. In the multiple staining combinations, CD4+ and CD8+ cells were gated from the NK1.1−CD3+ population and CD11c+, CD11b+, and CD19+ cells were gated from the NK1.1−CD3− population. All mAbs were purchased from either BD Biosciences Pharmingen (San Jose, CA) or BioLegend (San Diego, CA). The stained cells were analyzed on a BD LSRII (BD Biosciences, San Diego, CA) and the data processed using FlowJo V.7.6.5 software (Treestar, Inc., Ashland, OR).
Cytokine and chemokine arrays
Liver inflammatory proteins were detected using a Mouse Cytokine Array Panel A (R&D Systems, Inc., Minneapolis, MN) according to the manufacturer's instructions. Briefly, liver tissues were extracted and homogenized in phosphate-buffered saline containing protease inhibitors (Complete Mini Protease Inhibitor Cocktail Tablets; Roche Diagnostics GmbH) and the samples frozen at −80°C until use. The total protein concentration of the samples was measured using the BCA assay (BioRad, Hercules, CA), and then 200 μg of liver tissue lysate protein was mixed with a cocktail of biotinylated detection antibodies and the mixture incubated overnight at 4°C with the Mouse Cytokine Array, a nitrocellulose membrane spotted with capture antibodies for the multianalyte profiling of 40 cytokines and chemokines. The array was then incubated with horseradish-peroxidase-conjugated streptavidin, the signals were developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA), and the intensity of the signals was quantified using ImageQuant software. For each spot, the net optical density level was determined by subtracting the background optical level from the total raw optical density level.
Cytokine and chemokine PCR
One microgram of total liver RNA was transcribed using Expand Reverse Transcriptase (Roche Diagnostics GmbH) and used in a quantitative real-time PCR (FastStart SYBR Green Master; Roche Diagnostics GmbH) with the specific primer sets listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/hum). All samples were run in duplicate along with negative reverse transcriptase controls and water blanks. mRNA levels were assessed from the cycle number at which the cytokine or chemokine amplification exceeded the threshold crossing point (ct) and these values were standardized against the GAPDH value obtained for the same sample. Cytokine and chemokine mRNA levels in the liver of AAV8-U6/sAg19- or AAV8-H1/sAg19-injected mice were expressed as the fold-change compared with levels in the saline control mice, where fold-change=2(saline control ct − experimental ct).
Statistical analysis
Results are presented as the mean±standard deviation (SD). The Student's t-test was used to analyze statistical differences between experimental groups of animals; p<0.05 was considered as statistically significant.
Results
Comparison of the HBV-suppressive effects of shRNAs driven by the H1 or U6 promoter
In this study, we directly compared the influence of different promoters and target sequences of the shRNA on the hepatotoxicity and therapeutic efficacy of AAV-mediated RNAi therapy against HBV. We designed an shRNA expression cassette under the control of the H1 or U6 promoter in such a way that an identical shRNA transcript was produced by these AAV vectors (Fig. 1a). We utilized the pAAVEMBL plasmid, which was designed to produce double-stranded AAV (Wang et al., 2003), and three previously reported HBV-specific shRNAs, HBV-S1, sAg19, and sAg25 (Grimm et al., 2006; Chen et al., 2007), to construct the corresponding pAAVEMBL-H1 and pAAVEMBL-U6 plasmids. Our previous studies showed that HBV-S1 shRNA under the control of the H1 promoter has a potent suppressive effect on HBV, leading to a 3–4 log10 decrease in HBV load in HBV transgenic mice and, importantly, exhibiting no detectable toxicity (Chen et al., 2007, 2009, 2012). The use of sAg19 and sAg25 shRNAs was reported by Grimm et al. (2006), who used similar double-stranded AAV8 vectors and the U6 promoter and showed that sAg19 was nontoxic and highly efficient in HBV suppression in mice, while sAg25 caused severe liver injury and lethality. We also cloned firefly-luciferase-targeting GL2 shRNA into the pAAVEMBL-H1 and pAAVEMBL-U6 plasmids to generate control AAV vectors. These pAAVEMBL plasmids are expected to produce 19- or 25-mer hairpin sequences with a 9-nucleotide loop (Fig. 1a).
To evaluate the HBV suppressive potency of these U6- and H1-driven shRNAs, Huh-7 cells were co-transfected with a replication-competent HBV plasmid, pHBV1.3, and various amounts (1, 0.2, and 0.04 μg) of shRNA-expressing pAAVEMBL plasmids, and HBsAg levels in culture supernatants were measured 3 days later. As shown in Figure 1b (bottom panel), compared with mock transfection, transfection with the pAAVEMBL-U6 plasmids encoding the three HBV-specific shRNAs (HBV-S1, sAg19, or sAg25) greatly reduced HBsAg secretion at all doses tested (between 78% and 91% reduction), whereas transfection with the control pAAVEMBL-U6/GL2 plasmid, even at the highest dose of 1 μg, had no substantial effect on HBsAg levels. When expressed under the control of the H1 promoter, HBV-S1 and sAg25 were still effective in HBsAg suppression, whereas sAg19 was significantly less effective [mean of 66%±16% reduction at the dose of 1 μg versus 92%±3% for HBV-S1 (p=0.004) or 94%±2% for sAg25 (p=0.002)] (Fig. 1b, top panel). Northern blot analysis of total RNAs extracted from the transfected Huh-7 cells showed that much higher levels of shRNA transcripts were generated from the pAAVEMBL-U6 plasmids than from the corresponding pAAVEMBL-H1 counterparts (Fig. 1c). These results demonstrate that HBV-S1 and sAg25 are more potent shRNAs than sAg19, and for the weaker sAg19 a high shRNA transcript level is required for effective HBV suppression.
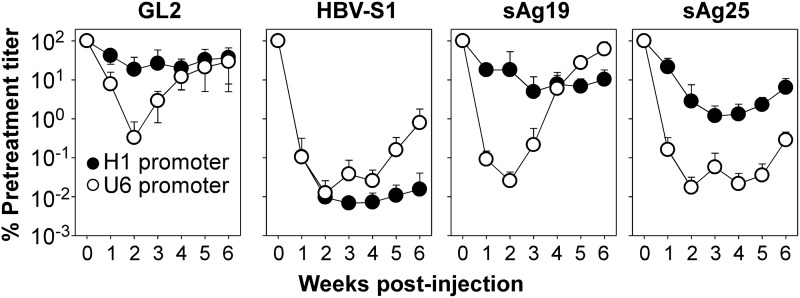
We then packaged these pAAVEMBL-H1 and pAAVEMBL-U6 plasmids in AAV8 vectors and tested their in vivo therapeutic efficacy in ICR/HBV transgenic mice, which produce sustained high titers of HBV (>108 genome copies/ml) and thus represent a clinically relevant animal model for chronic HBV infection (Chen et al., 2007). Mice (n=7) were injected intravenously with 1012 vg per mouse of AAV8-U6 or AAV8-H1 vectors encoding HBV-S1, sAg19, or sAg25 shRNAs, while controls were injected with the same amount of GL2 shRNA-encoding AAV8-U6 or AAV8-H1 vector. Serum samples from each group were collected weekly after AAV administration and HBV DNA quantified by real-time PCR. As shown in Figure 2, of the AAV8-H1 vectors, HBV-S1 was far more effective than sAg19 and sAg25 in HBV suppression, with a peak reduction of 14,700-fold in serum HBV DNA levels at week 3 and a 6,450-fold reduction at week 6 compared with a 20-fold reduction at week 3 and 10-fold reduction at week 6 for sAg19 and an 84-fold reduction at week 3 and 16-fold reduction at week 6 for sAg25. Treatment with the control AAV8-H1/GL2 vector had little effect on the amount of serum HBV DNA. In mice treated with AAV8-U6 vectors, the greatest reduction in serum HBV DNA levels (8,000-fold reduction for HBV-S1, 3,910-fold reduction for sAg19, and 5,810-fold reduction for sAg25) was seen at week 2 and was followed by a rapid rebound. At week 6 after AAV administration, the AAV8-U6/HBV-S1 and AAV8-U6/sAg25 vectors showed only a partial effect on HBV suppression, with, respectively, a 126-fold and 350-fold reduction in HBV DNA levels, whereas AAV8-U6/sAg19 showed no anti-HBV effect. Interestingly, treatment with the control AAV8-U6/GL2 vector also resulted in a significant HBV reduction of 305-fold at week 2, suggesting that the transient HBV suppression by the U6-promoter-driven shRNAs was, at least in part, caused by target sequence-independent mechanisms.
FIG. 2.
In vivo HBV DNA inhibition by AAV8-H1- or AAV8-U6-encoded shRNAs. Groups of ICR/HBV mice (n=7) were injected intravenously with 1012 vg per mouse of AAV8-H1 or AAV8-U6 vectors encoding HBV-specific shRNAs (HBV-S1, sAg19, or sAg25) or control GL2 shRNA, and then serum samples were collected weekly for analysis of HBV DNA levels and the results displayed as a percentage of the pretreatment titer for each group (mean±SD). The experiment was repeated twice with similar results.
Hepatotoxicity is induced by U6-promoter-driven, but not H1-promoter-driven, shRNAs
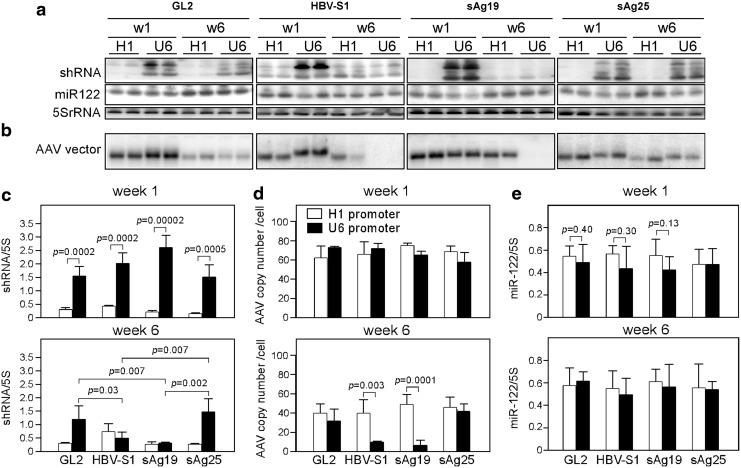
To determine the cause of the rapid loss of HBV suppression in mice treated with AAV8-U6 vectors, ICR/HBV mice were injected intravenously with 1012 vg per mouse of various AAV8-U6 or AAV8-H1 vectors and euthanized 1 or 6 weeks later. We first measured the expression of each processed shRNA in the liver by Northern blot analysis using a specific isotope-labeled oligonucleotide probe normalized to that of the internal control 5S rRNA. Figure 3a shows one representative blot and Figure 3c shows the summarized results for two independent experiments. Except for the AAV8-H1/HBV-S1 vector, which produced more shRNA than the other AAV8-H1 vectors at week 1 and an increase of 2-fold at week 6, the different AAV8-H1 vectors produced barely detectable shRNA levels in the liver at weeks 1 and 6 after AAV transduction. In contrast, at week 1 after AAV transduction, the different AAV8-U6 vectors produced at least fivefold more shRNA than the corresponding AAV8-H1 vectors; however, the U6-promoter-driven HBV-S1 and sAg19 shRNAs were not stable, declining by week 6 to 25% and 11%, respectively, of their week 1 levels. Levels of U6-promoter-driven GL2 and sAg25 were only slightly decreased or unaffected over the 6-week observation period. Interestingly, in Figure 3a top panel, there was an extra band over the predicated size of mature RNAi transcripts, which might be caused by the inefficiently processed shRNA transcripts as reported in other studies (Bridge et al., 2003; Grimm et al., 2006; Boudreau et al., 2009).
FIG. 3.
Expression of shRNA and the copy number of AAV vectors in the liver. ICR/HBV mice were injected intravenously with 1012 vg per mouse of the different AAV8-H1 or AAV8-U6 vectors as in Figure 2 and then were euthanized after 1 or 6 weeks, two mice at each time point. (a) Northern blot analysis of shRNA and miR-122. Total liver RNA was analyzed by Northern blot using radiolabeled probes specific for the antisense strand of each shRNA, endogenous miR-122, or 5S rRNA. (b) Southern blot analysis of AAV vectors. Total liver DNA was digested with HindIII and XbaI and hybridized with a radiolabeled DNA probe against the nonencoded green fluorescent protein sequence present in all AAV8 vectors. Each lane represents an individual mouse sample. The figure shows one representative set of data from two independent experiments. (c-e) The density of the signals in each lane was quantitated using ImageQuant software. The amount of shRNAs (c) or miR-122 (e) in each group is presented as the ratio of the density of the band to that of 5S rRNA. (d) The AAV genome copy number per cell in each group was calculated using the reference standard generated from the pAAVEMBL plasmid. Data are the summarized results of four mice from two independent experiments and are presented as the mean±SD. AAV, adeno-associated virus.
To determine whether there was a correlation between shRNA levels and the amount of injected AAV vector DNA, total cellular DNA was isolated from the AAV-transduced livers and subjected to Southern blot analysis using a probe corresponding to the green fluorescent protein sequence, which was present in all the AAV vectors (Fig. 1a). At week 1 after AAV transduction, similar levels of vector DNA, ranging from 63±12 to 76±3 copies per cell for the different AAV8-H1 vectors and 59±10 to 74±1 copies per cell for the different AAV8-U6 vectors, were present in the livers of the mice in the different groups, irrespective of shRNA target sequence (Fig. 3b and top panel of Fig. 3d). This result showed that these different AAV vectors were roughly equivalent in their infectious titers. The amount of vector DNA then decreased with time. At week 6, the amounts of vector DNA in mice transduced with the different AAV8-H1 vectors decreased to 40±10 to 50±10 copies per cell (about 60% to 67% of their week 1 level), whereas those in mice transduced with AAV8-U6/HBV-S1 or AAV8-U6/sAg19 decreased much more rapidly to 10±1 and 7±5 copies per cell, respectively (14% and 11% of their week 1 levels). Vector DNA levels in the AAV8-U6/GL2 and AAV8-U6/sAg25 groups were more stable, being 43% and 71%, respectively, of their week 1 levels at week 6.
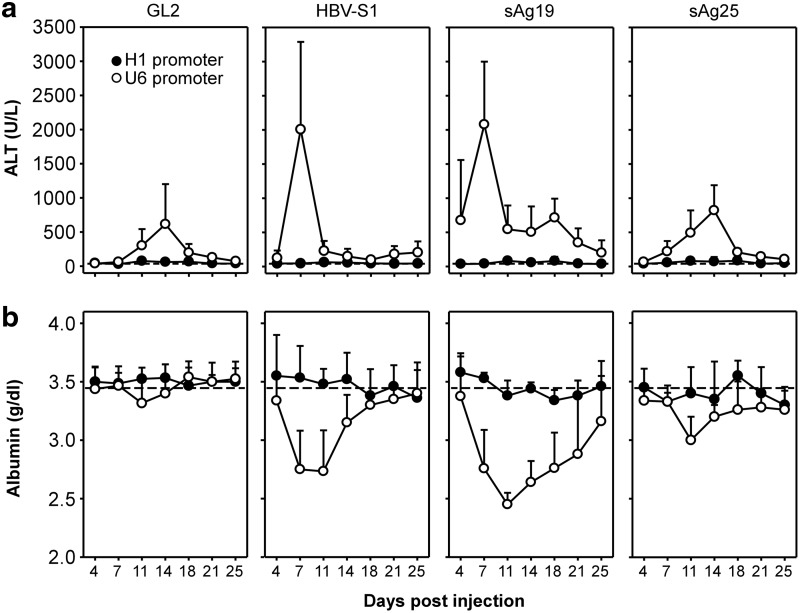
The rapid loss of AAV genomes and shRNA in hepatocytes transduced with AAV8-U6/HBV-S1 or AAV8-U6/sAg19 might have resulted from liver injury and subsequently accelerated liver regeneration, leading to dilution of the vg in hepatocytes. To test this, ICR/HBV mice (n=8) were injected intravenously with 1012 vg per mouse of various AAV8-U6 or AAV8-H1 vectors and serum samples collected at different times to measure serum ALT activity and albumin levels as indicators of liver injury. As shown in Figure 4a, serum ALT activity in mice that received the different AAV8-H1 vectors was stable and normal, ranging from 40 to 60 U/L, throughout the 25-day observation period. In marked contrast, injection of all four AAV8-U6 vectors induced a significant increase in serum ALT activity, particularly in the mice that received AAV8-U6/sAg19 and AAV8-U6/HBV-S1, with levels peaking at 2,078±921 U/L and 2,007±1,351 U/L, respectively, on day 7 after AAV transduction and then declining, but remaining higher than basal levels up to day 25. Injection of AAV8-U6/sAg25 or AAV8-U6/GL2 induced lower and delayed peak levels of ALT activity of 820±369 U/L and 614±446 U/L on day 14 after AAV transduction. The level of circulating albumin in mice treated with these different AAV vectors also reflected a similar trend in their ability to cause liver injury (Fig. 4b). A marked reduction in serum albumin was observed in mice injected with AAV8-U6/sAg19 and AAV8-U6/HBV-S1, followed by AAV8-U6/GL2 and AAV8-U6/sAg25, whereas those injected with the different AAV8-H1 vectors had relatively normal serum albumin levels throughout the observation period. Liver injury caused by these AAV vectors was also shown by histopathological examination. Severe hepatocellular damage was seen in the livers of ICR/HBV mice transduced with either AAV8-U6/HBV-S1 (Supplementary Fig. S1f) or AAV8-U6/sAg19 (Supplementary Fig. S1g), as shown by an irregular hepatocellular arrangement, spotty necrosis, and aggregation of mononuclear cells in the hepatic parenchyma and sinusoids. AAV8-U6/sAg25 (Supplementary Fig. S1h) or AAV8-U6/GL2 (Supplementary Fig. S1e) caused much milder pathological changes and less inflammatory cell aggregation in the liver, whereas no apparent pathological changes were observed in mice that received any of the AAV8-H1 vectors (Supplementary Fig. S1a–d). These results demonstrate a strong correlation between the amounts of shRNA produced by the different AAV vectors and the severity of hepatotoxicity in vivo, with AAV8-U6/sAg19 and AAV8-U6/HBV-S1 being the most toxic AAV vectors.
FIG. 4.
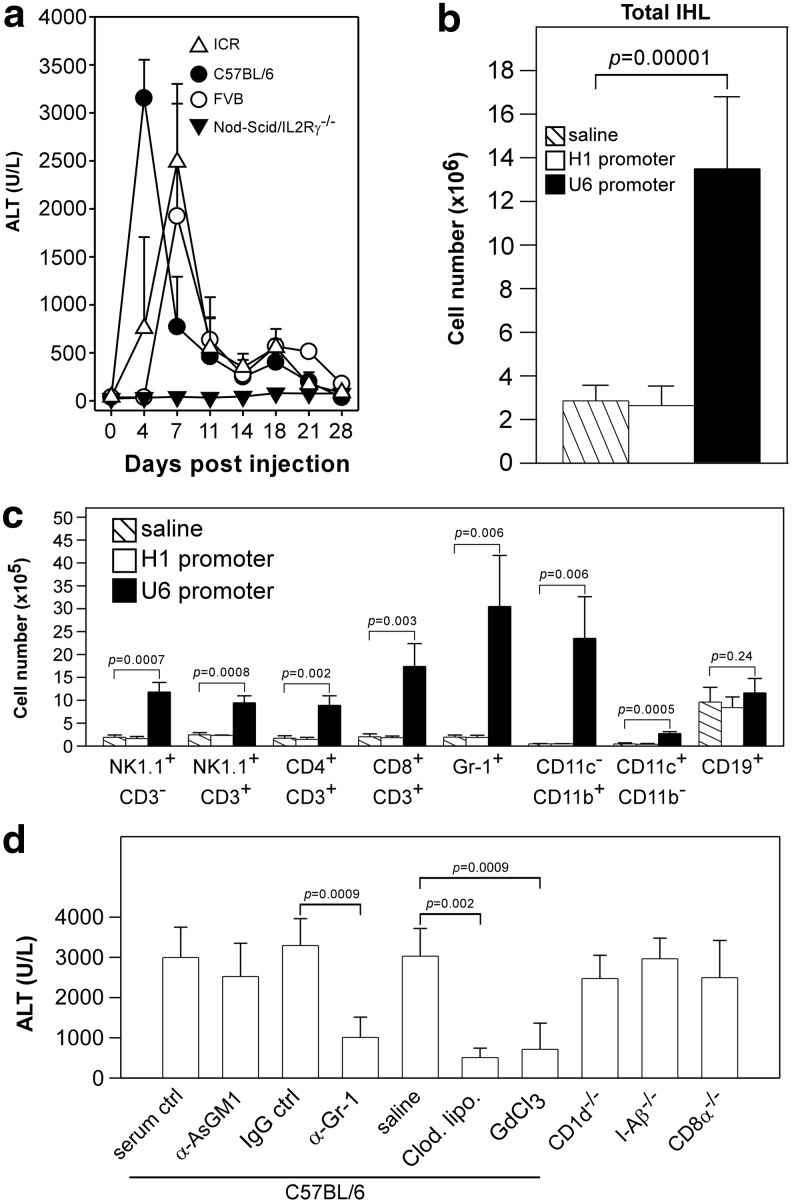
Liver injury induced by AAV8-H1- and AAV8-U6-encoded shRNAs in ICR/HBV mice. Groups of ICR/HBV mice (n=8) were injected intravenously with 1012 vg per mouse of different AAV8-H1 or AAV8-U6 vectors as in Figure 2, and serum samples collected at the indicated times for measurement of ALT activity (a) and albumin levels (b). The data (mean±SD) are for one representative result from two independent experiments. The dashed lines represent the mean value for ALT activity (a) or albumin levels (b) in untreated ICR/HBV mice (n=6). ALT, alanine aminotransferase.
We further investigated whether the dose of AAV-U6 vectors and the resulting shRNA expression levels can influence hepatotoxicity, and if so is there a window within which sustained HBV suppression can be achieved without inducing nonspecific toxicity. A dose titration study was carried out by injecting ICR/HBV mice with different doses of AAV8-U6/HBV-S1, ranging from 2×1011 to 8×109 vg per mouse. Injection of AAV8-U6/HBV-S1 at the dose of 6.6×1010 vg per mouse resulted in significant HBV suppression, with a peak reduction of 3,300-fold at week 3 (Supplementary Fig. S2b), and more importantly, did not induce elevation of serum ALT during the 6-week observation period (Supplementary Fig. S2a). The higher 2×1011 dose induced a mild increase in serum ALT activity and transient HBV suppression, whereas the lower 8×109 dose had very little effect on either the HBV titer or the serum ALT level.
Hepatotoxicity induced by U6-promoter-driven shRNAs is caused by inflammatory responses
It has been previously reported that hepatotoxicity caused by shRNA-expressing AAV vectors is because of disruption of endogenous microRNA biogenesis (up to 80% reduction) and function by the overexpressed shRNAs (Grimm et al., 2006). To determine whether endogenous microRNA expression was affected by shRNAs produced by our AAV8-U6 and AAV8-H1 vectors, the Northern blot membrane described in Figure 3a was stripped and rehybridized to detect miR-122, a highly expressed liver microRNA. Our previous data showed that endogenous miR-122 expression is not affected by i.v. injection with the AAV8-H1/HBV-S1 vector, the most effective and nontoxic AAV vector against HBV (Chen et al., 2007). As in this previous study, we did not detect any significant difference in miR-122 expression in mice injected with AAV8-H1/HBV-S1 or the other three AAV8-H1 vectors (Fig. 3a and e). Surprisingly, even in mice treated with the AAV8-U6 vectors, which caused mild to severe hepatotoxicity, at week 1 or week 6 after AAV transduction, there was no significant reduction in miR-122 levels compared with those in mice treated with the nontoxic AAV8-H1 vectors (Fig. 3a and e). We also used real-time reverse transcrptase PCR (see Supplementary Methods) to analyze miR-122 and other two less abundant microRNAs, let-7a and miR-26b, and showed that there was no significant differences of these microRNAs in the liver tissues of mice treated with the toxic and nontoxic AAV8 vectors (Supplementary Fig. S3). These results suggest that other host factors might play the major role in AAV8-U6 shRNA-mediated liver injury.
Since AAV8-U6/sAg19 induced the most severe hepatotoxicity, we used this vector and its nontoxic counterpart AAV8-H1/sAg19 in the following experiments to investigate the underlying mechanisms responsible for the in vivo hepatotoxicity. We first demonstrated that the presence of target (HBV) genes was not required for the AAV8-U6/sAg19-induced toxicity, as injection of this vector at the dose of 1012 vg per mouse in transgene-negative ICR mice or wild-type C57BL/6 or FVB mice still induced an early and profound increase in serum ALT activity (Fig. 5a), with peak ALT activity of 2,488±814 U/L, 3,150±400 U/L, and 1,923±1,173 U/L, respectively, being seen in these mice, similar to the results in ICR/HBV mice (Fig. 4a). As expected, the AAV8-H1/sAg19 vector did not induce an increase in ALT activity in any of the mouse strains (data not shown). Since significant infiltration of inflammatory cells was observed in the liver of mice transduced with AAV8-U6/sAg19 (Supplementary Fig. S1g), we then used Nod-scid/IL2Rγ−/− mice, which lack mature T cells, B cells, and natural killer cells and show impaired macrophage function (Shultz et al., 2005), to determine whether immune-mediated inflammation contributed to the liver injury. Injection of the AAV8-U6/sAg19 vector into this immunodeficient mouse strain did not induce an increase in serum ALT activity at any time in the entire 28-day observation period, and the amount of intrahepatic AAV DNA remained relatively stable for at least 10 weeks (Supplementary Fig. S4). These results suggest that immune-mediated inflammation was the most important host factor involved in the toxicity caused by the overproduction of shRNAs from the AAV-U6 vectors.
FIG. 5.
Liver injury induced by AAV8-U6/sAg19 in different mouse strains and analysis of intrahepatic leukocytes and their contribution to liver injury caused by toxic shRNA. (a) ICR mice, C57BL/6 mice, FVB mice, or Nod-scid/IL2Rγ−/− mice (n=5–7 per group) were injected intravenously with 1012 vg of AAV8-U6/sAg19 and serum samples collected at the indicated times for analysis of ALT activity (mean±SD). (b and c) Groups of C57BL/6 mice (n=6) were injected with saline, AAV8-H1/sAg19, or AAV8-U6/sAg19, and liver leukocytes were isolated on day 4 postinjection. The number of total intrahepatic leukocytes (b) and different cell subsets (c) per liver from 3 mice per group are shown (mean±SD).  , Saline; □, AAV8-H1/sAg19; ■, AAV8-U6/sAg19. The number of cells per liver for individual cell types was calculated by multiplying the percentage of the individual cell type by the total number of isolated liver leukocytes per liver. The experiment was performed twice with similar results. (d) Groups of wild-type C57BL/6 mice treated with anti-asialo-GM1 antiserum, anti-Gr-1 monoclonal antibody, clodronate liposome or GdCl3, CD1d−/− mice, I-Aβ−/− mice, or CD8−/− mice (n=5–8) were injected with AAV8-U6/sAg19 on day 0, and then serum samples were collected at day 4 for analysis of ALT activity (mean±SD). Wild-type C57BL/6 mice treated with control rabbit antiserum, control rat immunoglobulin G, or saline were included as controls.
, Saline; □, AAV8-H1/sAg19; ■, AAV8-U6/sAg19. The number of cells per liver for individual cell types was calculated by multiplying the percentage of the individual cell type by the total number of isolated liver leukocytes per liver. The experiment was performed twice with similar results. (d) Groups of wild-type C57BL/6 mice treated with anti-asialo-GM1 antiserum, anti-Gr-1 monoclonal antibody, clodronate liposome or GdCl3, CD1d−/− mice, I-Aβ−/− mice, or CD8−/− mice (n=5–8) were injected with AAV8-U6/sAg19 on day 0, and then serum samples were collected at day 4 for analysis of ALT activity (mean±SD). Wild-type C57BL/6 mice treated with control rabbit antiserum, control rat immunoglobulin G, or saline were included as controls.
We next investigated the cellular mechanisms involved in the hepatotoxicity caused by U6-promoter-driven shRNA. Groups of C57BL/6 mice (n=6) were injected intravenously with 1012 vg per mouse of AAV8-U6/sAg19 or AAV8-H1/sAg19 or with saline, and then were examined 4 days later for total cell number and cellular composition of hepatic mononuclear cells. As shown in Figure 5b, treatment with AAV8-U6/sAg19 significantly increased the number of total hepatic mononuclear cells by 4.7-fold (p=0.00001) compared with mice treated with saline. Flow cytometric analysis (Fig. 5c and Supplementary Fig. S5) revealed a pronounced increase in the numbers of Kupffer cells (macrophages) (CD11c−CD11b+, 49.3-fold, p=0.006), granulocytes (Gr-1+, 15.7-fold, p=0.006), NK cells (NK1.1+CD3−, 6.2-fold, p=0.0007), and CD8+ T cells (CD8+CD3+, 8.6-fold, p=0.003) in mice treated with AAV8-U6/sAg19 compared with those in the saline group, and the number of NKT cells (NK1.1+CD3+) and CD4+ T cells (CD4+CD3+) was also increased, but to a lesser extent (3.9-fold, p=0.0008 and 5.2-fold, p=0.002, respectively, vs. the saline group). Treatment with AAV8-H1/sAg19 had no effect on the number of total mononuclear cells or that of any of the leukocyte subpopulations in the liver.
We then used a panel of gene knockout mice and mice depleted of a specific leukocyte subpopulation by antibodies or chemicals to further clarify the cellular mechanisms in the liver toxicity caused by U6-promoter-driven shRNA. C57BL/6 mice with targeted disruption of CD1d (CD1d−/−), H2-I-A beta chain (I-Aβ−/−), or CD8 alpha chain (CD8α−/−) were used to assess, respectively, the relative contribution of NKT cells, CD4+ T cells, and CD8+ T cells. The role of NK cells and granulocytes in shRNA-mediated liver toxicity was evaluated by depletion of these cell types in wild-type C57BL/6 mice using, respectively, rabbit anti-asialo GM1 antiserum or rat anti-Gr-1 mAb and controls treated with normal rabbit serum or an irrelevant rat IgG at the same dose and schedule. Kupffer cell depletion was achieved using two well-established approaches involving i.v. injection of clodronate liposomes or GdCl3 and saline-injected mice as controls. All mice (n=5–8 in each group) were injected intravenously with 1012 vg per mouse of AAV8-U6/sAg19, and some of the animals were injected with the indicated antibodies or chemicals according to the schedule described in the Methods section, and serum from each mouse was collected 4 days later for measurement of ALT activity. As shown in Figure 5d, AAV8-U6/sAg19 treatment induced an increase in ALT activity in CD1d−/−, I-Aβ−/−, and CD8α−/−, and NK-cell-depleted mice, with titers ranging between 2,474±577 U/L and 2,964±513 U/L, comparable to those in the control mice (wild-type C57BL/6 mice treated with saline, irrelevant IgG, or normal rabbit serum), in which titers ranged between 2,996±754 U/L and 3249±669 U/L. In contrast, mice treated with clodronate liposomes, GdCl3, or anti-Gr-1 mAb showed a significant reduced increase in ALT activity to, respectively, 509±233 U/L (p=0.002 vs. saline group), 714±649 U/L (p=0.0009 vs. saline group), or 1,011±502 U/L (p=0.0009 vs. irrelevant IgG group). These results strongly suggest that Kupffer cells and granulocytes are the major cell types responsible for AAV8-U6 shRNA-mediated liver toxicity.
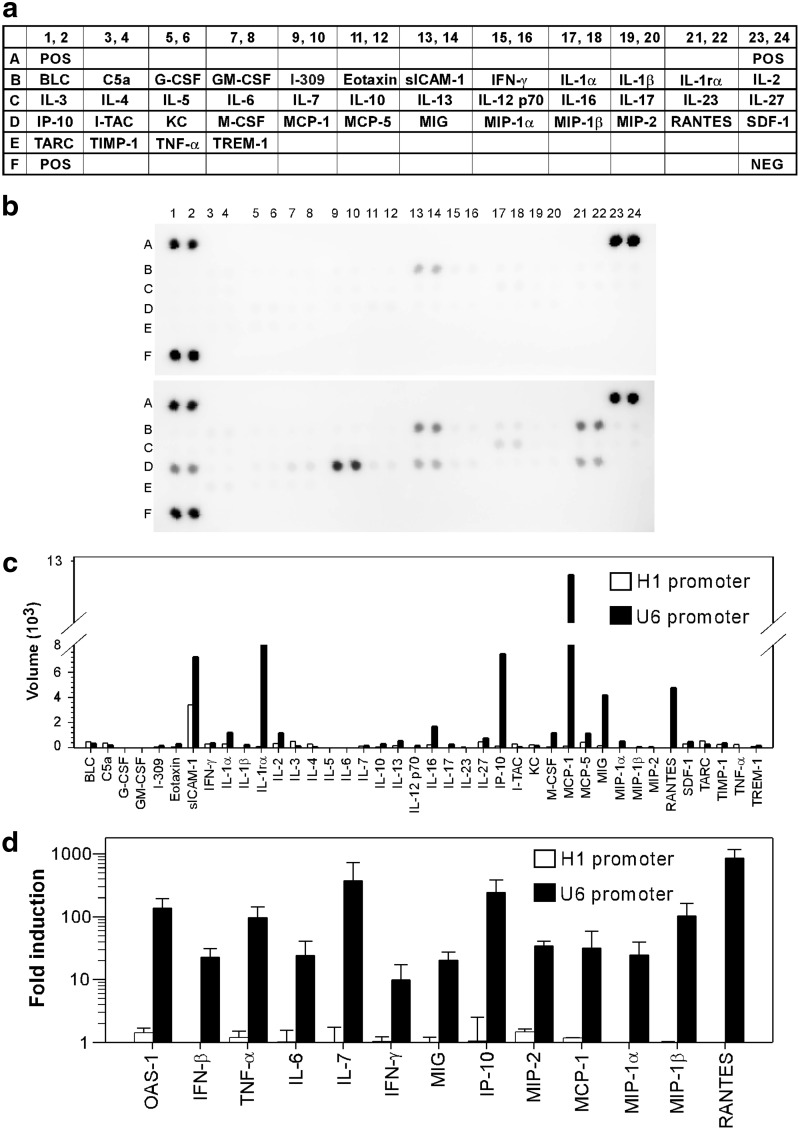
Next we determined which cytokine/chemokines were associated with AAV8-U6 shRNA-induced liver toxicity. C57BL/6 mice (n=3 in each group) were injected intravenously with 1012 vg per mouse of AAV8-U6/sAg19 or AAV8-H1/sAg19, and then, 4 days later, liver tissue lysates were prepared from the individual mice in each group and pooled and analyzed using a cytokine array membrane coated with antibodies against 40 different inflammatory proteins (Fig. 6a). In the AAV8-H1/sAg19-transduced mice, the only change in inflammatory proteins found was a moderate increase in sICAM-1, an inflammatory biomarker (Fig. 6b top panel and 6c). In contrast, AAV8-U6/sAg19 treatment induced significant expression of several proinflammatory chemokines, including IP-10, MIG, MCP-1, and RANTES, and of inflammatory biomarkers, including sICAM-1 and IL-1RA (Fig. 6b bottom panel and 6c). These results were confirmed using a more sensitive quantitative PCR assay to detect mRNAs for several selected cytokines and chemokines, including IFN-β and its target protein, OAS-1, which were not included in the cytokine array. As shown in Figure 6d, AAV8-U6/sAg19 treatment generated high levels of mRNAs coding for IFN-β, OAS-1, TNF-α, IL-6, IL-7, IFN-γ, MIG, IP-10, MIP-2, MCP-1, MIP-1α, MIP-1β, and RANTES, whereas negligible amounts of mRNAs for these inflammatory proteins were present in liver samples from mice treated with AAV8-H1/sAg19.
FIG. 6.
Analysis of hepatic cytokine profiles in C57BL/6 mice treated with shRNA-encoding AAV vectors. Total liver protein (a–c) or RNA (d) from C57BL/6 mice (n=3 per group) collected 4 days after injection of 1012 vg of AAV8-H1/sAg19 or AAV8-U6/sAg19 was assayed by the mouse cytokine array (a–c) or real-time reverse transcriptase polymerase chain reaction (d). (a) List of antibodies on the cytokine array membrane. Each antibody is represented by duplicate spots. (b) A representative blot for two independent experiments. (c) Average net optical intensity for each pair of cytokine spots. (d) Cytokine and chemokine mRNAs in AAV8-vector-treated animals measured by real-time reverse transcrptase polymerase chain reaction. mRNAs are expressed as a fold-change compared with the control (mean±SD).
Discussion
To be beneficial for chronic HBV patients, treatment must be extremely powerful and able to reduce serum HBV DNA levels from over 108–109 genome copies/ml to less than 105, a critical threshold level for reducing risk of developing severe hepatic complications, such as cirrhosis and hepatocellular carcinoma (Chen et al., 2006; Iloeje et al., 2006). AAV-mediated RNAi represents a promising therapy for chronic HBV (Ivacik et al., 2011; Van Der Laan et al., 2011), but hepatotoxicity is a major safety concern. In this study, we used ICR/HBV transgenic mice, which maintain greater than 108 genome copies/ml of serum HBV DNA throughout their life span and thus closely mimic the heavy viral load in chronic HBV patients, as an animal model to evaluate the therapeutic efficacy of three previously validated shRNAs, HBV-S1, sAg19, and sAg25. These HBV-specific shRNAs were designed under the control of either the U6 or H1 RNA polymerase III promoter. All AAV8-U6 vectors at the dose of 1012 vg per mouse, which produced abundant shRNAs, were able to induce rapid and significant HBV suppression of between 3,900- and 8,000-fold. However, the U6 shRNA-mediated suppressive effect was not sustained, and, within a few weeks, serum HBV DNA levels were substantially increased in the U6/HBV-S1 and U6/sAg25 groups and even reached the pretreatment titer in the U6/sAg19 group (Fig. 2). Severe liver injury was observed in mice treated with these U6 shRNAs, supported by induction of hepatic necrosis (Supplementary Fig. S1e–h), elevated serum ALT activity (Fig. 4a), and decreased serum albumin levels (Fig. 4b). These results suggest that, in addition to the specific gene silencing effect of the RNAi, the reduction in HBV DNA levels in the U6 shRNA-treated mice was, at least partially, caused by RNAi-mediated hepatic cell death.
The U6 shRNA-associated hepatotoxicity was prevented by packaging the shRNA under the control of the H1 promoter. None of the H1-promoter-driven shRNAs elicited detectable liver toxicity (Fig. 4a and b and Supplementary Fig. S1a–d). However, H1/sAg19 and H1/sAg25 were much less effective than U6/sAg19 or U6/sAg25 in HBV suppression in the ICR/HBV mice, with H1-sAg19 reducing HBV DNA levels by 10- to 20-fold and H1-sAg25 by 16- to 84-fold (Fig. 2). This result is consistent with a previous report which showed that AAV8-H1/sAg19 reduces serum HBsAg levels by about 20-fold in another HBV transgenic mouse line (Grimm et al., 2010). Compared with the other HBV-targeting shRNAs, H1/HBV-S1 was unique in its ability to inhibit HBV. In ICR/HBV mice, H1/HBV-S1 induced a sustained and marked reduction in HBV DNA level of 6,450- to 14,700-fold (Fig. 2), a result consistent with that in our previous report (Chen et al., 2009). The mechanisms that make HBV-S1 such a potent shRNA are not entirely clear but may be because of its high affinity to RNA-induced silencing complex determined by the thermodynamic stability, which is thought to influence the loading process of the guide strand for incorporation into RNA-induced silencing complex (Schwarz et al., 2003; Gu et al., 2011), or cis-elements in small RNA sequences and RNA-binding proteins impacting small RNA stability (Ji and Chen, 2012). Importantly, AAV-H1/HBV-S1 does not induce detectable liver injury, even over a 1-year observation period (Chen et al., 2009, 2012). Its safety for long-term use in vivo and its high potency for HBV suppression make AAV8-H1/HBV-S1 an excellent candidate for development for clinical applications. In fact, the beneficial effect of AAV8-H1/HBV-S1 was demonstrated in our previous study, which showed that long-term HBV suppression by AAV8-H1/HBV-S1 avoided the liver damage and the occurrence and progression of liver tumors that develop spontaneously in ICR/HBV mice (Chen et al., 2012).
The association of shRNA levels and cellular toxicities has been reported in several previous studies (Grimm et al., 2006, 2010; Giering et al., 2008; McBride et al., 2008; Khodr et al., 2011). Our results also suggest that the levels of shRNA produced by AAV vectors are highly associated with the anti-HBV effect and hepatotoxicity. The U6 promoter is reported to be a much stronger promoter than the H1 promoter (An et al., 2006; Makinen et al., 2006). Consistent with these previous reports, we demonstrated that AAV8-U6 vectors produced at least fivefold more shRNAs than the corresponding AAV8-H1 vectors at 1 week after AAV transduction (Fig. 3a and c). Although the extremely high levels of shRNAs generated in the AAV8-U6-treated mice resulted in rapid and significant HBV suppression, they also caused severe hepatotoxicity, leading to the loss of the AAV vectors and subsequent loss of HBV suppression. In most cases, the weaker H1 promoter generated low levels of shRNAs, which, although nontoxic, were inefficient in reducing HBV gene expression in animals with a high viral load. The only exception was HBV-S1, which was produced in substantial amounts by the H1 promoter and these levels were sustained over the observation period of 6 weeks (Fig. 3a and c), explaining its high potency in HBV suppression. We also demonstrated that lowering the dose of AAV-U6/HBV-S1 to 6.6×1010 vg per mouse can achieve sustained HBV suppression without inducing liver toxicity (Supplementary Fig. S2a and b). These results suggest that controlling the levels of shRNA expression is important in achieving effective and stable control of chronic viral infection.
Grimm et al. (2006) first reported that AAV-mediated shRNA overexpression in the livers of adult mice can trigger severe side effects and that substantial hepatotoxicity and morbidity are observed in mice treated with AAV vectors expressing different shRNAs in a target-gene-independent manner (Grimm et al., 2006, 2010). They proposed a microRNA saturation model suggesting that the toxic side effect was caused by competition by the transduced shRNAs for the endogenous microRNA pathway, thereby interfering with endogenous microRNA biogenesis and functionality. Subsequently, several other groups reported that shRNA overexpression from AAV vectors, most using the strong U6 promoter, causes cytotoxicity in different cell types and organs (McBride et al., 2008; Boudreau et al., 2009; Bish et al., 2011; Martin et al., 2011). Lowering the AAV dose reduced the adverse effects (Grimm et al., 2006; Ulusoy et al., 2009; Ehlert et al., 2010), suggesting that the toxic effect is dependent upon shRNA levels, a result supported by our findings. Analysis of cellular microRNAs was carried out in several of these studies, and the results showed that some microRNAs were decreased to various levels, but others were not changed (Grimm et al., 2006, 2010; Witting et al., 2008; Ahn et al., 2011; Bish et al., 2011; Martin et al., 2011; Pan et al., 2011). We also investigated whether microRNA saturation correlated with the hepatotoxicity induced by our AAV8-U6 vectors, but did not find significant changes in levels of miR-122 (Fig. 3a and e), a liver-specific microRNA, and two less abundant liver microRNAs, let-7a and miR-26b, even in mice with severe liver injury caused by treatment with AAV8-U6/sAg19 or AAV8-U6/HBV-S1, suggesting that other host factors might be involved in the U6 shRNA-induced hepatotoxicity.
It is well known that, as a result of their recognition by RNA-sensing toll-like receptors or RIG-I-like receptors, siRNAs can activate cells of the innate immune system (Rossi, 2009; Couto and High, 2010; Olejniczak et al., 2010; Sioud, 2010). In this study, we provided evidence that shRNAs produced by AAV8-U6 vectors had potent immunostimulatory effects, which were highly associated with hepatotoxicity in mice treated with these vectors. First, treatment with AAV8-U6/sAg19 significantly increased the number of total mononuclear cells (Fig. 5b) and the number of several leukocyte subpopulations in the liver (Fig. 5c and Supplementary Fig. S5), in particular Kupffer cells and granulocytes, and hematoxylin and eosin staining clearly showed infiltration of mononuclear leukocytes in these mice (Supplementary Fig. S1g). Second, AAV8-U6/sAg19 induced a significant increase in liver levels of type I interferon and many inflammatory cytokines and chemokines (Fig. 6a–d). Third, no AAV8-U6/sAg19-associated hepatotoxicity was seen in immunodeficient mouse strains, Nod-scid/IL2Rγ−/− (Fig. 5a). Further experiments revealed that Kupffer cells and granulocytes were the major cell types mediating liver toxicity, since depletion of either of these two cell populations greatly reduced serum ALT activity in mice transduced with AAV8-U6/sAg19 (Fig. 5d). The inflammatory responses induced by AAV8-U6/sAg19 depended on the overexpressed shRNAs, but not the AAV vector itself, since the same amount of AAV8-H1/sAg19 did not increase hepatic leukocyte numbers (Fig. 5b and c) or trigger cytokine/chemokine expression (Fig. 6b–d). In addition to shRNA, it is possible that part of the excessive innate immune activation observed in toxic-AAV-treated mice might be further enhanced by liver injury. The contribution of inflammatory responses to shRNA-induced liver toxicity is further supported by a report by Witting et al. (2008), in which they demonstrated that overexpression of shRNA by helper-dependent adenoviruses activated the interferon response but did not alter the levels of cellular microRNAs.
In summary, our results show that the toxicity associated with AAV8-mediated RNAi therapy is immune-mediated, with saturation of endogenous microRNA synthesis pathways having only a little effect. The toxicity is probably caused by overexpression of shRNA by the strong U6 promoter and can be alleviated by using the weaker H1 promoter or decreasing the dose of AAV with the U6 promoter. However, a highly potent shRNA is required to achieve effective HBV therapy using AAV vectors.
Supplementary Material
Acknowledgments
We are grateful to Dr. Chyung-Ru Wang (Northwestern University Feinberg School of Medicine) for providing CD1d−/− mice and Dr. Fang Liao (Institute of Biomedical Sciences, Academia Sinica) for providing PCR primers for cytokine and chemokine gene analysis. This work was supported by Academia Sinica and Grants NSC100-3112-B-001-013 from National Research Program for Genomic Medicine and NSC99-2320-B-001-015-MY3 from National Science Council, Taiwan.
Author Disclosure Statement
The authors declare that no conflicts of interest exist.
References
- Ahn M. Witting S.R. Ruiz R., et al. Constitutive expression of short hairpin RNA in vivo triggers buildup of mature hairpin molecules. Hum. Gene Ther. 2011;22:1483–1497. doi: 10.1089/hum.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D.S. Qin F.X. Auyeung V.C., et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol. Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bish L.T. Sleeper M.M. Reynolds C., et al. Cardiac gene transfer of short hairpin RNA directed against phospholamban effectively knocks down gene expression but causes cellular toxicity in canines. Hum. Gene Ther. 2011;22:969–977. doi: 10.1089/hum.2011.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau R.L. Martins I. Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge A.J. Pebernard S. Ducraux A., et al. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Carmona S. Jorgensen M.R. Kolli S., et al. Controlling HBV replication in vivo by intravenous administration of triggered PEGylated siRNA-nanoparticles. Mol. Pharm. 2009;6:706–717. doi: 10.1021/mp800157x. [DOI] [PubMed] [Google Scholar]
- Chang C.M. Lo C.H. Shih Y.M., et al. Treatment of hepatocellular carcinoma with adeno-associated virus encoding interleukin-15 superagonist. Hum. Gene Ther. 2010;21:611–621. doi: 10.1089/hum.2009.187. [DOI] [PubMed] [Google Scholar]
- Chen C.J. Yang H.I. Su J., et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- Chen C.C. Ko T.M. Ma H.I., et al. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 2007;14:11–19. doi: 10.1038/sj.gt.3302846. [DOI] [PubMed] [Google Scholar]
- Chen C.C. Sun C.P. Ma H.I., et al. Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol. Ther. 2009;17:352–359. doi: 10.1038/mt.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C. Chang C.M. Sun C.P., et al. Use of RNA interference to modulate liver adenoma development in a murine model transgenic for hepatitis B virus. Gene Ther. 2012;19:25–33. doi: 10.1038/gt.2011.60. [DOI] [PubMed] [Google Scholar]
- Chou Y.C. Jeng K.S. Chen M.L., et al. Evaluation of transcriptional efficiency of hepatitis B virus covalently closed circular DNA by reverse transcription-PCR combined with the restriction enzyme digestion method. J. Virol. 2005;79:1813–1823. doi: 10.1128/JVI.79.3.1813-1823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto L.B. High K.A. Viral vector-mediated RNA interference. Curr. Opin. Pharmacol. 2010;10:534–542. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Ehlert E.M. Eggers R. Niclou S.P., et al. Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. 2010;11:20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. Prince A.M. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Giering J.C. Grimm D. Storm T.A., et al. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- Giladi H. Ketzinel-Gilad M. Rivkin L., et al. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Grimm D. Streetz K.L. Jopling C.L., et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Grimm D. Wang L. Lee J.S., et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S. Jin L. Zhang F., et al. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iloeje U.H. Yang H.I. Su J., et al. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678–686. doi: 10.1053/j.gastro.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Ivacik D. Ely A. Arbuthnot P. Countering hepatitis B virus infection using RNAi: how far are we from the clinic? Rev. Med. Virol. 2011;21:383–396. doi: 10.1002/rmv.705. [DOI] [PubMed] [Google Scholar]
- Ji L. Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodr C.E. Sapru M.K. Pedapati J., et al. An alpha-synuclein AAV gene silencing vector ameliorates a behavioral deficit in a rat model of Parkinson's disease, but displays toxicity in dopamine neurons. Brain Res. 2011;1395:94–107. doi: 10.1016/j.brainres.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I. Shin D. Choi T.H., et al. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol. Ther. 2007;15:1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- Klein C. Bock C.T. Wedemeyer H., et al. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- Kwon H. Lok A.S. Hepatitis B therapy. Nat. Rev. Gastroenterol. Hepatol. 2011;8:275–284. doi: 10.1038/nrgastro.2011.33. [DOI] [PubMed] [Google Scholar]
- Makinen P.I. Koponen J.K. Karkkainen A.M., et al. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. J. Gene Med. 2006;8:433–441. doi: 10.1002/jgm.860. [DOI] [PubMed] [Google Scholar]
- Martin J.N. Wolken N. Brown T., et al. Lethal toxicity caused by expression of shRNA in the mouse striatum: implications for therapeutic design. Gene Ther. 2011;18:666–673. doi: 10.1038/gt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride J.L. Boudreau R.L. Harper S.Q., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey A.P. Nakai H. Pandey K., et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Mingozzi F. High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Morrissey D.V. Blanchard K. Shaw L., et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005a;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- Morrissey D.V. Lockridge J.A. Shaw L., et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005b;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Olejniczak M. Galka P. Krzyzosiak W.J. Sequence-non-specific effects of RNA interference triggers and microRNA regulators. Nucleic Acids Res. 2010;38:1–16. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q. De Ruiter P.E. Von Eije K.J., et al. Disturbance of the microRNA pathway by commonly used lentiviral shRNA libraries limits the application for screening host factors involved in hepatitis C virus infection. FEBS Lett. 2011;585:1025–1030. doi: 10.1016/j.febslet.2011.02.035. [DOI] [PubMed] [Google Scholar]
- Rossi J.J. Innate immunity confounds the clinical efficacy of small interfering RNAs (siRNAs) Gene Ther. 2009;16:579–580. doi: 10.1038/gt.2009.26. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S. Hutvagner G. Du T., et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shlomai A. Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Shultz L.D. Lyons B.L. Burzenski L.M., et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- Sioud M. Recent advances in small interfering RNA sensing by the immune system. N. Biotechnol. 2010;27:236–242. doi: 10.1016/j.nbt.2010.02.015. [DOI] [PubMed] [Google Scholar]
- Ulusoy A. Sahin G. Bjorklund T., et al. Dose optimization for long-term rAAV-mediated RNA interference in the nigrostriatal projection neurons. Mol. Ther. 2009;17:1574–1584. doi: 10.1038/mt.2009.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uprichard S.L. Boyd B. Althage A., et al. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Laan L.J. Wang Y. Tilanus H.W., et al. AAV-mediated gene therapy for liver diseases: the prime candidate for clinical application? Expert Opin. Biol. Ther. 2011;11:315–327. doi: 10.1517/14712598.2011.548799. [DOI] [PubMed] [Google Scholar]
- Wang Z. Ma H.I. Li J., et al. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- Witting S.R. Brown M. Saxena R., et al. Helper-dependent adenovirus-mediated short hairpin RNA expression in the liver activates the interferon response. J. Biol. Chem. 2008;283:2120–2128. doi: 10.1074/jbc.M704178200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.