Abstract
Objectives
In this study, we demonstrated that TBEV-infected ticks have been distributed in the ROK, combined with our previous results. These results suggest that TBEV may exist in the ROK, and H. longicornis, H. flava, and I. nipponensis may be potential vectors of TBEV. In addition, these results emphasize the need for further epidemiological research of TBEV.
Methods
We examined for the presence of RNA of TBEV by reverse transcriptase-nested polymerase chain reaction (RT-nested PCR) using ixodid ticks captured in 25 localities of 10 provinces. Ticks were collected by the flagging and dragging method or using sentinel BG traps at forests, grass thickets, and grassland. A total of 13,053 ticks belonging to two genera and four species were collected and pooled (1292 pools), according to collection site, species of tick, and developmental stage.
Results
Among 1292 pools, the envelope (E) protein gene of TBEV was detected using RT-nested PCR in 10 pools (3 pools of the 1,331 adult ticks and 7 pools of the 11,169 nymph ticks) collected from Gangwon-do province, Jeonrabuk-do province, and Jeju Island. The minimum infection rates for TBEV of Haemaphysalis longicornis, Haemaphysalis flava, and Ixodes nipponensis were 0.06%, 0.17%, and 2.38%, respectively. Phylogenetic analysis based on the partial E protein gene was performed to identify relationships between the TBEV strains. This showed that 10 Korean strains clustered with the Western subtype.
Conclusion
In this study, we investigated the prevalence of tick-borne encephalitis virus (TBEV) in ixodid ticks from various regions of the Republic of Korea (ROK) during 2011–2012 to identify whether TBEV is circulating and to determine the endemic regions of TBEV.
Keywords: envelope gene, ixodid ticks, Republic of Korea, tick-borne encephalitis virus, Western subtype
1. Introduction
Ixodid ticks transmit a number of zoonotic pathogens, such as Borrelia burgdorferi [1], Batonella [2], Ehrlichia and Anaplasma [3,4], Rickettsia [5], and tick-borne encephalitis virus (TBEV) [6] to mammalian hosts. As the causative agent of tick-borne encephalitis (TBE), TBEV is a member of the family Flaviviridae and genus Flavivirus, and one of the most important human infections of the central nervous system [7]. TBE occurs in endemic areas of the Eurasian continents, including Europe, Russia, and Far-Eastern Asia (China and Japan), and has a significant impact on public health in these endemic regions [8]. So far, TBEV has been subdivided into three subtypes: the Far-Eastern subtype, known as Russian spring summer encephalitis virus; the Western or European subtype, known as Central European encephalitis virus; and the Siberian subtype based on phylogenetic analysis [8,9]. The main tick vector of the Western subtype is Ixodes ricinus, and Ixodes persulcatus is the main vector for the other subtypes [10,11]. TBEVis transmitted by tick bite and is maintained in the zoonotic transmission cycle between ixodid ticks and wild vertebrate hosts, such as wild and domestic mammals, birds, and reptiles.
In the Republic of Korea (ROK), although TBE infections have not been reported among humans, we have identified molecular evidence of TBEV infections in infesting ticks of the wild animals or collected ticks such as Haemaphysalis longicornis, Haemaphysalis flava, Haemaphysalis japonica, and Ixodes niponensis, which previously had not been known as TBEV vectors, and have isolated TBEV from lung tissues of the wild rodent, Apodemus agrarius [12,13]. However, unlike our expectation, Korean isolates were identified as the Western subtype of TBEV by sequence and phylogenetic analyses compared with other TBEV strains from neighboring countries, including China, Japan, and northeastern Russia that belong to the Far-Eastern subtype [12,13].
In this study, we investigated the prevalence of TBEV in ixodid ticks from various regions of the ROK using sensitive reverse transcriptase-nested polymerase chain reaction (RT-nested PCR) method to identify whether TBEV is circulating and to determine the endemic regions of TBEV.
2. Materials and Methods
2.1. Collection of ixodid ticks
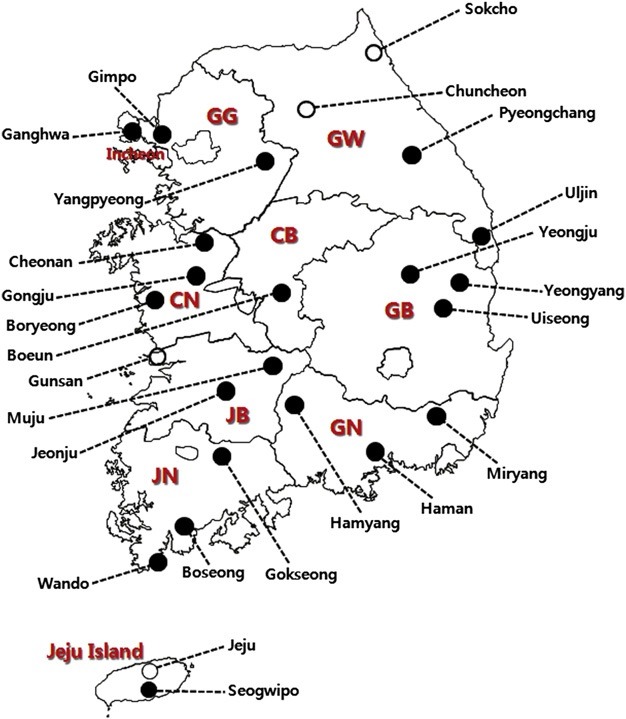
Ixodid ticks surveys were performed by the flagging and dragging method or using sentinel BG traps in various sites, including grass thicket, grassland, and broad-leaved and coniferous forests in 25 localities of 10 provinces of the ROK during 2011–2012. Figure 1 represents the geographical locations of the collection sites. After collection, ticks were placed in plastic tubes and transported to the medical entomology laboratory of Korea National Institute of Health, where they were identified according to their species and developmental stages under a dissecting microscope according to the classification method of Yamaguti et al [14]. Identified ticks were stored at 4 ℃ until further investigation.
Figure 1. Geographical location of the collection sites in the ROK. Open circles show the sites at which positive tick pools for TBEV were detected [KorVec11-215, KorVec11-224, and KorVec11-226 (N 38°00’01.5’’ E 127°44’29.3’’ , Goseong-ri, Sabuk-myeon, Chuncheon-si), KorVec11-265 (N 38°00’51.1’’ E 126°46’36.2’’, Jugok-ri, Napo-myeon, Gunsan-si), KorVec12- 1131 (N 38°12’9.37’’ E 128°30’55.32’’, Nohak-dong, Sokchosi), KorVec12-1138 (N 38°12’9.11’’ E 128°30’52.84’’, Nohakdong, Sokcho-si), KorVec12-1152 (N 38°12’12.42’’ E 128°31’2.2’’, Nohak-dong, Sokcho-si), KorVec12-1195, KorVec12-1200, and KorVec12-1217 (N 33°25’27.3’’ E 126°33’14.8’’, Ara-dong, Jeju-si)]. CB = Chungcheongbukdo province; CN = Chungcheongnam-do province; GB = Gyeongsangbuk-do province; GG = Gyeonggi-do province; GN = Gyeongsangnam-do province; GW = Gangwon-do province; JB = Jeonllabuk-do province; JN = Jeonllanam-do province.
2.2. Tick processing and RNA extraction
A total of 13,053 identified ticks were pooled according to species, developmental stage, and locality. The size of pools ranged from 1 to 50 in larvae, from 1 to 30 in nymphs, and from 1 to 5 in adult males or females (Table 1). All the pooled ticks were homogenized using Precellys 24 homogenizer (Bertin Technologies, Montigny, Bretonneux, France) with a tissue lysis buffer in RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) and 2.8 mm stainless-steel beads. The homogenate was centrifuged at 10,000 rpm for 5 minutes, and supernatant was used for RNA extraction. RNA was extracted using the RNeasy Mini Kit (Qiagen GmbH) according to the manufacturer’s instructions.
Table 1.
The number of ixodid ticks classified by species and developmental stage, and MIR of TBEV in this study
| Species | Developmental stages | Number of ticks | Number of pools | Positive pools | MIR (%)a |
|---|---|---|---|---|---|
| Haemaphysalis longicornis | Larvaeb | 350 | 22 | 0 | 0 |
| Nymphc | 10,436 | 460 | 6 | 0.06 | |
| Adult (male)d | 221 | 106 | 0 | 0 | |
| Adult (female)d | 849 | 329 | 1 | 0.12 | |
| Subtotal | 11,856 | 917 | 7 | 0.06 | |
| Haemaphysalis flava | Larvae | 197 | 11 | 0 | 0 |
| Nymph | 710 | 163 | 0 | 0 | |
| Adult (male) | 108 | 79 | 1 | 0.93 | |
| Adult (female) | 134 | 91 | 1 | 0.75 | |
| Subtotal | 1,149 | 344 | 2 | 0.17 | |
| Ixodes nipponensis | Larvae | 0 | 0 | 0 | 0 |
| Nymph | 23 | 9 | 1 | 4.35 | |
| Adult (male) | 8 | 8 | 0 | 0 | |
| Adult (female) | 11 | 11 | 0 | 0 | |
| Subtotal | 42 | 28 | 1 | 2.38 | |
| Ixodes persulcatus | Larvae | 6 | 3 | 0 | 0 |
| Nymph | 0 | 0 | 0 | 0 | |
| Adult (male) | 0 | 0 | 0 | 0 | |
| Adult (female) | 0 | 0 | 0 | 0 | |
| Subtotal | 6 | 3 | 0 | 0 | |
| Total | Larvae | 553 | 36 | 0 | 0 |
| Nymph | 11,169 | 632 | 7 | 0.06 | |
| Adult (male) | 337 | 193 | 1 | 0.30 | |
| Adult (female) | 994 | 431 | 2 | 0.20 | |
| Total | 13,053 | 1,292 | 10 | 0.08 | |
aMIR: number of positive pools/total number of ticks assayed; b1–50 larvae/pool; c1–30 nymphs/pool; d1–5 adults/pool. MIR = minimum infection rate; TBEV = tick-borne encephalitis virus.
2.3. Reverse transcriptase-nested polymerase chain reaction
To examine for the presence of TBEV envelope (E) gene, the one-step RT-PCR was carried out using a Maxime RT-PCR PreMix kit (iNtRON Biotechnology, Gyeonggi, Korea) with previously described primers, TBE-913F (5’-TGCACACAYYTGGAAAA CAGGGA-3’) and TBE-1738R (5’-TGGCCACTTTT CAGGTGGTACTTG-3’) [15]. The one-step RT-PCR reaction was performed in a PCR thermal cycler (GeneAmp PCR system 9700, Applied Biosystems, Foster City, CA, USA) under the following conditions: 30 minutes at 45 ℃ for reverse transcription and 5 minutes at 94 ℃ for denaturation as the initial step, followed by 25 cycles of 30 seconds at 94 ℃, 30 seconds at 52 ℃, and 1 minute at 72 ℃, and a final extension step of 5 minutes at 72 ℃. Nested PCR was carried out using an i-StarMaster Mix PCR kit (iNtRON Biotechnology) with previously designed primers, TBE- 1192F (5’-CAGAGTGATCGAGGCTGGGGYAA-3’) and TBE-1669R (5’-AACACTCCAGTCTGGTCTC CRAGGTTGTA-3’) [15]. The nested PCR reaction consisted of an initial denaturation step of 2 minutes at 94 ℃, followed by 30 cycles of 20 seconds at 94 ℃, 10 seconds at 62 ℃, and 20 seconds at 68 ℃, and a final extension step of 5 minutes at 72 ℃. To confirm the PCR products, we analyzed them by agarose gel electrophoresis, stained with SYBR safe DNA gel stain (Invitrogen, Carlsbad, CA, USA).
2.4. Sequencing and phylogenetic analysis
The TBEV-positive products of RT-nested PCR were purified using a QIAquick gel extraction kit (Qiagen GmbH) according to the manufacturer’s instructions and sequenced using ABI Prism BigDye terminator cycle sequencing kits and ABI 3730xl sequencer (Applied Biosystems) at Solgent Inc. (Daejeon, Korea). Results of sequencing were assembled using the SeqMan program implemented in DNASTAR software (version 5.0.6; DNASTAR Inc., Madison, WI, USA) to determine the consensus sequences. The TBEV strains used for phylogenetic analysis are listed in Table 2; multiple sequence alignments were performed using Clustal W implemented in MEGA software version 5 [16]. Phylogenetic analysis was performed using MEGA software version 5 by the maximum likelihood method. The sequences obtained from TBEV-positive products were submitted to GenBank (accession numbers JX282515–JX282524).
Table 2.
TBEV strains used in the phylogenetic analysis
| Strain | Subtype | Source of virus | Geographical origin | Year of isolation | GenBank accession no. |
|---|---|---|---|---|---|
| Hypr | Western | Human blood | Czech Republic | 1953 | X75286 |
| Neudoerfl | Western | Ixodes ricinus | Austria | 1971 | U27495 |
| K23 | Western | I. ricinus | Germany | 1975 | AF091010 |
| Abesettarov | Western | Human blood | Russia | 1951 | AF091005 |
| Kem I | Western | I. ricinus | Hungary | 1952 | AF091011 |
| KrM 93 | Western | A. agrarius | Republic of Korea | 2006 | EU276109 |
| KrM 213 | Western | A. agrarius | Republic of Korea | 2006 | EU276110 |
| KrM 215 | Western | A. agrarius | Republic of Korea | 2006 | EU276111 |
| KrM 216 | Western | A. agrarius | Republic of Korea | 2006 | EU276112 |
| KrM 219 | Western | A. agrarius | Republic of Korea | 2006 | EU276113 |
| Oshima 5-10 | Far-Eastern | Dog blood | Japan | 1995 | AB001026 |
| Sofjin-HO | Far-Eastern | Human brain | Russia | 1937 | AB022703 |
| Senzhang | Far-Eastern | Human brain | China | 1953 | AY174188 |
| Oshima I-1 | Far-Eastern | Ixodes ovatus | Japan | 1996 | AB022292 |
| Oshima 5-11 | Far-Eastern | Dog blood | Japan | 1995 | AB022290 |
| KH98-2 | Far-Eastern | Ixodes persulcatus | Russia | 1998 | AB022295 |
| KH98-5 | Far-Eastern | I. persulcatus | Russia | 1998 | AB022296 |
| KH98-10 | Far-Eastern | I. persulcatus | Russia | 1998 | AB022297 |
| Crimea | Far-Eastern | I. persulcatus | Ukraine | 1987 | AF091008 |
| D1283 | Far-Eastern | Human brain | Russia | 1998 | AB049347 |
| RK1424 | Far-Eastern | I. persulcatus | Latvia | 1977 | AF091015 |
| T-blood | Far-Eastern | Human blood | Russia | 1939 | AF091019 |
| VL99-m11 | Far-Eastern | I. persulcatus | Russia | 1999 | AB049345 |
| N132 | Far-Eastern | I. persulcatus | Russia | 1979 | AF091013 |
| Vasilchenko | Siberian | Human blood | Russia | 1969 | M97659 |
| Aina | Siberian | Human blood | Russia | 1963 | AF091006 |
| IR99-1m1 | Siberian | I. persulcatus | Russia | 1999 | AB049348 |
| IR99-1m4 | Siberian | I. persulcatus | Russia | 1999 | AB049349 |
| IR99-2m3 | Siberian | I. persulcatus | Russia | 1999 | AB049350 |
| IR99-2m7 | Siberian | I. persulcatus | Russia | 1999 | AB049351 |
| IR99-2f7 | Siberian | I. persulcatus | Russia | 1999 | AB049352 |
| IR99-2f13 | Siberian | I. persulcatus | Russia | 1999 | AB049353 |
TBEV = tick-borne encephalitis virus.
3. Results
3.1. Numbers and identification of ixodid ticks
A total of 13,053 ticks (553 larvae, 11,169 nymphs, 337 males, and 994 females) were collected from 2011 to 2012 in 25 localities of the ROK (Table 1 and Figure 1). The ixodid tick samples were identified to belong to four species in two genera: H. longicornis, H. flava, I. niponensis, and I. persulcatus. Of the identified ticks, H. longicornis (90.8%, 11,856/ 13,053) was the most abundant species in this study, followed by H. flava (8.8%, 1149/13,053), I. niponensis (0.3%, 42/13,053), and I. persulcatus (0.05%, 6/13,053).
3.2. Prevalence of TBEV in ixodid ticks
Of the 13,053 ixodid tick samples, 10 pools were positive for TBEV according to RT-nested PCR method. Among the 10 positive pools, three pools of the adult ticks and seven of the nymph ticks were collected from Gunsan in Jeollabuk-do province, Chuncheon or Sokcho in Gangwon-do province, and Jeju in Jeju Island. Regional prevalence of TBEV for each species was shown in Table 3. The minimum infection rate (MIR, calculated with the assumption that a positive pool contains one infected tick) of TBEV in H. longicornis, H. flava, and Ixodes nipponensis was 0.06%, 0.17%, and 2.38%, respectively (Table 1). Although H. longicornis was the most common species collected in this study, its positive rates of TBEV was lower than that of other species.
Table 3.
Regional prevalence of TBEV in ixodid ticks (minimum infection rate) collected in the Republic of Korea
| Collection site | Larvae | Nymph | Male | Female | Total | Larvae | Nymph | Male | Female | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Haemaphysalis flava: no. infected/no. examined (%) | Haemaphysalis flava: no. infected/no. examined (%) | |||||||||
| Gyeonggi-do | ||||||||||
| Yangpyeong | – | 0/12(0) | – | 0/3(0) | 0/15(0) | – | 0/238(0) | 0/4(0) | 0/9(0) | 0/251(0) |
| Gimpo | – | – | 0/2(0) | 0/1(0) | 0/3(0) | – | 0/4(0) | – | 0/1(0) | 0/5(0) |
| Incheon | ||||||||||
| Ganghwa | – | 0/7(0) | – | 0/1(0) | 0/8(0) | – | 0/523(0) | 0/47(0) | 0/43(0) | 0/613(0) |
| Gangwon-do | ||||||||||
| Chuncheon | – | 0/64(0) | 1/6(16.67) | 0/5(0) | 1/75(1.33) | – | 2/354(0.56) | – | 0/4(0) | 2/358(0.56) |
| Pyeongchang | 0/17(0) | 0/6(0) | 0/11(0) | 0/11(0) | 0/45(0) | 0/147(0) | 0/1(0) | – | – | 0/148(0) |
| Sokcho | – | 0/125(0) | 0/3(0) | 1/6(16.67) | 1/134(0.75) | – | 1/122(0.82) | 0/1(0) | 0/5(0) | 1/128(0.78) |
| Chungcheongbuk-do | ||||||||||
| Boeun | 0/17(0) | 0/217(0) | 0/52(0) | 0/59(0) | 0/506(0) | 0/190(0) | 0/3598(0) | 0/25(0) | 0/353(0) | 0/4166(0) |
| Chungcheongnam-do | ||||||||||
| Boryeong | – | 0/18(0) | 0/3(0) | 0/5(0) | 0/26(0) | – | 0/436(0) | 0/14(0) | 0/27(0) | 0/477(0) |
| Cheonan | – | 0/1(0) | – | 0/1(0) | 0/2(0) | – | 0/232(0) | – | 0/41(0) | 0/273(0) |
| Gongju | – | 0/23(0) | 0/2(0) | 0/4(0) | 0/29(0) | 0/1(0) | 0/359(0) | 0/12(0) | 0/19(0) | 0/391(0) |
| Jeollabuk-do | ||||||||||
| Muju | – | 0/14(0) | 0/1(0) | 0/3(0) | 0/18(0) | – | 0/29(0) | – | – | 0/29(0) |
| Gunsan | – | 0/42(0) | 0/10(0) | 0/7(0) | 0/59(0) | – | 1/516(0.19) | – | 0/18(0) | 1/534(0.19) |
| Jeonju | – | 0/3(0) | – | – | 0/3(0) | – | 0/9(0) | – | 0/2(0) | 0/11(0) |
| Jeollanam-do | ||||||||||
| Boseong | – | 0/11(0) | 0/1(0) | 0/2(0) | 0/14(0) | – | 0/269(0) | 0/3(0) | 0/18(0) | 0/290(0) |
| Gokseong | – | 0/5(0) | – | 0/2(0) | 0/7(0) | – | 0/109(0) | 0/1(0) | 0/32(0) | 0/142(0) |
| Wando | – | 0/3(0) | 0/2(0) | 0/1(0) | 0/6(0) | – | 0/38(0) | 0/28(0) | 0/47(0) | 0/113(0) |
| Gyeongsangbuk-do | ||||||||||
| Uiseong | – | 0/21(0) | 0/2(0) | 0/2(0) | 0/25(0) | – | 0/76(0) | 0/1(0) | 0/2(0) | 0/79(0) |
| Yeongju | – | 0/57(0) | 0/1(0) | 0/5(0) | 0/63(0) | – | 0/83(0) | – | – | 0/83(0) |
| Yeongyang | – | 0/12(0) | – | – | 0/12(0) | – | 0/422(0) | 0/14(0) | 0/81(0) | 0/517(0) |
| Uljin | – | 0/15(0) | 0/1(0) | 0/3(0) | 0/19(0) | – | 0/574(0) | 0/22(0) | 0/69(0) | 0/665(0) |
| Gyeongsangnam-do | ||||||||||
| Haman | – | 0/1(0) | – | – | 0/1(0) | – | 0/52(0) | 0/9(0) | 0/33(0) | 0/94(0) |
| Hamyang | – | 0/3(0) | 0/3(0) | 0/2(0) | 0/8(0) | – | 0/1(0) | 0/1(0) | – | 0/2(0) |
| Miryang | – | 0/6(0) | 0/6(0) | 0/7(0) | 0/19(0) | – | 0/80(0) | – | 0/1(0) | 0/81(0) |
| Jeju-do | ||||||||||
| Jeju | – | 0/35(0) | – | – | 0/35(0) | 0/6(0) | 2/1,418(0.14) | 0/25(0) | 1/26(3.85) | 3/1,475(0.20) |
| Seogwipo | 0/2(0) | 0/9(0) | 0/2(0) | 0/4(0) | 0/17(0) | 0/6(0) | 0/893(0) | 0/14(0) | 0/18(0) | 0/931(0) |
| Total | 0/197(0) | 0/710(0) | 0/710(0) 1/108(0.93) | 1/134(0.75) | 2/1,149(0.17) | 0/350(0) | 6/10,436(0.06) | 0/221(0) | 1/849(0.12) | 7/11,856(0.06) |
| Gyeonggi-do | ||||||||||
| Yangpyeong | – | – | – | – | – | – | – | – | – | – |
| Gimpo | – | – | – | – | – | – | – | – | – | – |
| Incheon | ||||||||||
| Ganghwa | – | – | – | – | – | – | – | – | – | – |
| Gangwon-do | ||||||||||
| Chuncheon | – | 0/1(0) | – | 0/1(0) | 0/2(0) | – | – | – | – | – |
| Pyeongchang | – | – | – | – | – | 0/6(0) | – | – | – | 0/6(0) |
| Sokcho | – | 1/18(5.56) | 0/1(0) | – | 1/19(5.26) | – | – | – | – | – |
| Chungcheongbuk-do | ||||||||||
| Boeun | 0/2(0) | 0/4(0) | 0/8(0) | 0/14(0) | – | – | – | – | – | |
| Chungcheongnam-do | ||||||||||
| Boryeong | – | – | 0/1(0) | – | 0/1(0) | – | – | – | – | – |
| Cheonan | – | – | – | – | – | – | – | – | – | – |
| Gongju | – | 0/1(0) | - | – | 0/1(0) | – | – | – | – | – |
| Jeollabuk-do | ||||||||||
| Muju | – | – | 0/1(0) | 0/1(0) | – | – | – | – | – | |
| Gunsan | 0/1(0) | – | – | 0/1(0) | – | – | – | – | – | |
| Jeonju | – | – | – | – | – | – | – | – | – | |
| Jeollanam-do | ||||||||||
| Boseong | – | – | – | – | – | – | – | – | – | – |
| Gokseong | – | – | – | – | – | – | – | – | – | – |
| Wando | – | – | – | – | – | – | – | – | – | – |
| Gyeongsangbuk-do | ||||||||||
| Uiseong | – | – | 0/1(0) | - | 0/1(0) | – | – | – | – | – |
| Yeongju | – | – | 0/1(0) | 0/1(0) | 0/2(0) | – | – | – | – | – |
| Yeongyang | – | – | – | – | – | – | – | – | – | – |
| Uljin | – | – | – | – | – | – | – | – | – | – |
| Gyeongsangnam-do | ||||||||||
| Haman | – | – | – | – | – | – | – | – | – | – |
| Hamyang | – | – | – | – | – | – | – | – | – | – |
| Miryang | – | – | – | – | – | – | – | – | – | – |
| Jeju-do | ||||||||||
| Jeju | – | – | – | – | – | – | – | – | – | – |
| Seogwipo | – | – | – | – | – | – | – | – | – | – |
| Total | - | 1/23(4.35) | 0/8(0) | 0/11(0) | 1/42(2.38) | 0/6(0) | – | – | – | 0/6(0) |
ROK = Republic of Korea; TBEV = tick-borne encephalitis virus.
The highest MIR of TBEV infection was identified in I. nipponensis collected from Sokcho in Gangwon-do province (5.26%). In the other localities, the MIR of infected ticks varied from 0% to 1.33%. The highest rate of TBEV infection was identified in I. nipponensis nymph (4.35%). The infection rates in males (0.30%) were higher than those in females (0.20%) and nymph (0.06%). The mean TBEV prevalence in 25 collection sites was about 0.08%.
3.3. Sequence and phylogenetic analyses
Nucleotide sequence identities between the 10 Korean strains and the other 32 TBEV strains represented that the 10 Korean strains had high identity with the Western subtype strains with 97.2–99.6%, compared with the Far-Eastern subtype with 81.1–84.3% or the Siberian subtype with 83.9–85.7%.
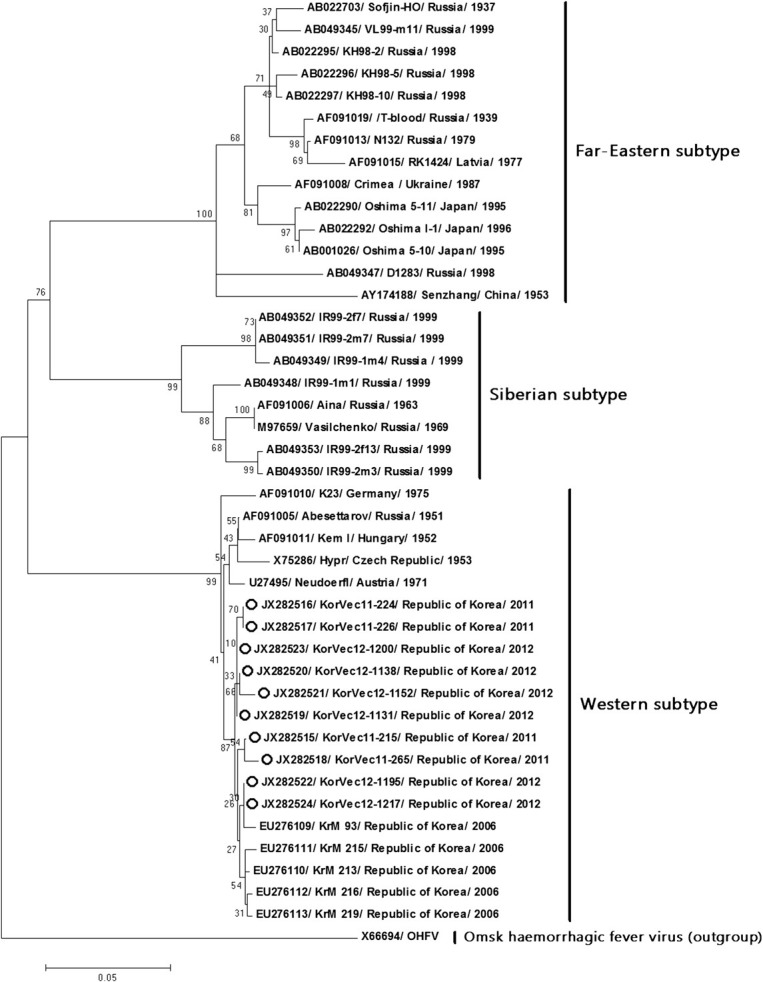
Phylogenetic trees derived from nucleotide sequences of the E gene showed that the 10 Korean strains (Kor- Vec11-215, KorVec11-224, KorVec11-226, KorVec11-265, KorVec12-1131, KorVec12-1138, KorVec12-1152, KorVec12-1195, KorVec12-1200, and KorVec12-1217) identified in this study belong to the Western subtype. An envelope gene-based phylogenetic tree was divided into three distinct groups such as Far-Eastern, Siberian, and Western subtypes and it was also shown that the 10 Korean strains belonged to the same subgroup as the Western subtype, as shown in Figure 2.
Figure 2. Phylogenetic analysis of TBEV strains based on the envelope region. Phylogenetic trees constructed using the ML method based on the Tamura-Nei model in MEGA version 5 (1000 bootstrap replicates). The ML tree were rooted with the sequences of the OHFV (Genbank accession no. X66694). The scale bar indicates the nucleotide substitutions per position. The 10 Korean strains identified in this study are marked with open circles. ML = maximum likelihood; OHFV = Omsk hemorrhagic fever virus.
4. Discussion
So far, TBEV vectors have been reported in 10 species of three genera (I. ricinus, I. persulcatus, Ixodes hexagonus, Ixodes arboricola, Ixodes ovatus, Haemaphysalis punctata, Haemaphysalis concinna, Haemaphysalis inermis, Dermacentor marginatus, and Dermacentor reticulatus) [17,18]. Among the known vectors, the total distribution of five species of TBEV vector (I. persulcatus, I. ovatus, Haemaphysalis concinna, D. marginatus, and D. reticulatus) in the ROK have been reported [19]. In our previous study, we suggested that TBE might exist in the ROK based on the following evidence. First, there was molecular evidence of TBEV infection in mammalian hosts and in potential vector ticks (H. longicornis, H. flava, H. japonica, and I. nipponensis) collected from the ROK [12,13,20], although these ticks had not been previously reported as TBEV vectors. Second, we reported on the isolation and identification of five TBEV strains from lung tissue homogenates of wild rodents captured in the ROK using in vitro and in vivo experiments [12]. Contrary to our expectations, the sequence comparisons and phylogenetic analyses based on the complete E gene or fullgenome of TBEV Korean isolates compared with other TBEV strains indicated that all the five Korean strains belonged to the Western subtype [21,22].
Based on this evidence, we carried out more extensive survey throughout the ROK for the prevalence of TBEV in ixodid ticks from various localities to identify whether TBEV is circulating and to determine the endemic areas of TBEV. A total of 13,053 ticks were divided into 1292 pools to identify for TBEV by RTnested PCR method, which detected 10 TBEV-positive pools. Sequence and phylogenetic analyses based on the envelope gene sequence revealed that the 10 Korean strains from infected ticks in this study belonged to the Western subtype, in accordance with our previous results.
The principal tick vector of the Western subtype of TBEV is I. ricinus [8], but in this study, it is reasonable to suppose that H. longicornis, H. flava, and I. nipponensis serve as the potential vectors of TBEV in the ROK. Until recently, the molecular evidence of TBEV infection was identified in ixodid ticks collected from Yangpyeong or Dongducheon in Gyeonggi-do province, Pyeongchang or Jeongseon in Gangwon-do province, and Jeju Island [13,20]. In this study, ixodid ticks such as H. longicornis, H. flava, and I. nipponensis collected from Gunsan in Jeollabuk-do province, Chuncheon or Sokcho in Gangwon-do province, and Jeju in Jeju Island were identified according to the molecular evidence of TBEV infection. These results indicated that TBEV may be endemic in these localities of theROKand H. longicornis, H. flava, and I. nipponensis may be potential vectors of the TBEV Western subtype, combined with previous findings. However, in order to prove these points, isolation of TBEV from infected vectors and characterization of TBEV isolates will be required.
In summary, we found that TBEV-infected ticks have been distributed in some localities of the ROK. These results emphasize the need for further epidemiological research of TBEV and preventive measures against the occurrence of TBE in the ROK.
Acknowledgments
This research was funded by a grant from the Korea National Institute of Health.
References
- 1.Burgdorfer W, Barbour AG, Hayes SF, et al. Lyme disease-a tickborne spirochetosis? Science. 1982 Jun;216(4552):1317–9. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 2.Kim CM, Kim JY, Yi YH, et al. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci. 2005 Dec;6(4):327–34. [PubMed] [Google Scholar]
- 3.Chae JS, Kim CM, Kim EH, et al. Molecular epidemiological study for tick-borne disease (Ehrlichia and Anaplasma spp.) surveillance at selected U.S. military training sites/installations in Korea. Ann N Y Acad Sci. 2003 Jun;990:118–25. doi: 10.1111/j.1749-6632.2003.tb07349.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim CM, Kim MS, Park MS, et al. Identification of Ehrlichia chaffeensis, Anaplasma phagocytophilum, and A. bovis in Haemaphysalis longicornis and Ixodes persulcatus ticks from Korea. Vector Borne Zoonotic Dis. 2003 Spring;3(1):17–26. doi: 10.1089/153036603765627424. [DOI] [PubMed] [Google Scholar]
- 5.Kim CM, Yi YH, Yu DH, et al. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. Appl Environ Microbiol. 2006 Sep;72(9):5766–76. doi: 10.1128/AEM.00431-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silber LA, Soloviev VD. Eastern tick-borne springesummer (spring) encephalitis. Am Rev Sov Med. 1946;1(80) Spec Suppl. [PubMed] [Google Scholar]
- 7.Dumpis U, Crook D, Oksi J. Tick-borne encephalitis. Clin Infect Dis. 1999 Apr;28(4):882–90. doi: 10.1086/515195. [DOI] [PubMed] [Google Scholar]
- 8.Ecker M, Allison SL, Meixner T, et al. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J Gen Virol. 1999 Jan;80(Pt. 1):179–85. doi: 10.1099/0022-1317-80-1-179. [DOI] [PubMed] [Google Scholar]
- 9.Lindquist L, Vapalahti O. Tick-borne encephalitis. Lancet. 2008 May;371(9627):1861–71. doi: 10.1016/S0140-6736(08)60800-4. [DOI] [PubMed] [Google Scholar]
- 10.Bakhvalova VN, Rar VA, Tkachev SE, et al. Tick-borne encephalitis virus strains of Western Siberia. Virus Res. 2000 Sep;70(1-2):1–12. doi: 10.1016/s0168-1702(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 11.Gritsun TS, Nuttall PA, Gould EA. Tick-borne flaviviruses. Adv Virus Res. 2003;61:317–71. doi: 10.1016/s0065-3527(03)61008-0. [DOI] [PubMed] [Google Scholar]
- 12.Kim SY, Yun SM, Han MG, et al. Isolation of tick-borne encephalitis viruses from wild rodents, South Korea. Vector Borne Zoonotic Dis. 2008 Spring;8(1):7–13. doi: 10.1089/vbz.2006.0634. [DOI] [PubMed] [Google Scholar]
- 13.Kim SY, Jeong YE, Yun SM, et al. Molecular evidence for tickborne encephalitis virus in ticks in South Korea. Med Vet Entomol. 2009 Mar;23(1):15–20. doi: 10.1111/j.1365-2915.2008.00755.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguti N, Tipton VJ, Keegan HL, et al. Ticks of Japan, Korea, and the Ryukyu Islands. Brigham Young Univ Sci Bull Biol Ser. 1971;15:1–226. [Google Scholar]
- 15.Ternovoi VA, Kurzhukov GP, Sokolov YV, et al. Tick-borne encephalitis with hemorrhagic syndrome, Novosibirsk region, Russia, 1999. Emerg Infect Dis. 2003 Jun;9(6):743–6. doi: 10.3201/eid0906.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 Oct;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suss J. Epidemiology and ecology of TBE relevant to the production of effective vaccines. Vaccine. 2003 Apr;21(Suppl. 1):S19–35. doi: 10.1016/s0264-410x(02)00812-5. [DOI] [PubMed] [Google Scholar]
- 18.Takeda T, Ito T, Chiba M, et al. Isolation of tick-borne encephalitis virus from Ixodes ovatus (Acari: Ixodidae) in Japan. J Med Entomol. 1998 May;35(3):227–31. doi: 10.1093/jmedent/35.3.227. [DOI] [PubMed] [Google Scholar]
- 19.Lee HI. Medical entomologydfamily acari. 2nd ed. Komunsa Press; Seoul: 1999. pp. 365–412. [Google Scholar]
- 20.Ko S, Kang JG, Kim SY, et al. of tick-borne encephalitis virus in ticks fromsouthern Korea. JVetSci. 2010 Sep;11(3):197–203. doi: 10.4142/jvs.2010.11.3.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun SM, Kim SY, Han MG, et al. Analysis of the envelope (E) protein gene of tick-borne encephalitis viruses isolated in South Korea. Vector Borne Zoonotic Dis. 2009 Jun;9(3):287–93. doi: 10.1089/vbz.2008.0085. [DOI] [PubMed] [Google Scholar]
- 22.Yun SM, Kim SY, Ju YR, et al. First complete genomic characterization of two tick-borne encephalitis virus isolates obtained from wild rodents in South Korea. Virus Genes. 2011 Jun;42(3):307–16. doi: 10.1007/s11262-011-0575-y. [DOI] [PubMed] [Google Scholar]


