Abstract
Most eukaryotic telomeres contain a repeating motif with stretches of guanine residues that form a 3′-terminal overhang extending beyond the telomeric duplex region. The telomeric repeat of hypotrichous ciliates, d(T4G4), forms a 16-nucleotide 3′-overhang. Such sequences can adopt parallel-stranded as well as antiparallel-stranded quadruplex conformations in vitro. Although it has been proposed that guanine-quadruplex conformations may have important cellular roles including telomere function, recombination, and transcription, evidence for the existence of this DNA structure in vivo has been elusive to date. We have generated high-affinity single-chain antibody fragment (scFv) probes for the guanine-quadruplex formed by the Stylonychia telomeric repeat, by ribosome display from the Human Combinatorial Antibody Library. Of the scFvs selected, one (Sty3) had an affinity of Kd = 125 pM for the parallel-stranded guanine-quadruplex and could discriminate with at least 1,000-fold specificity between parallel or antiparallel quadruplex conformations formed by the same sequence motif. A second scFv (Sty49) bound both the parallel and antiparallel quadruplex with similar (Kd = 3–5 nM) affinity. Indirect immunofluorescence studies show that Sty49 reacts specifically with the macronucleus but not the micronucleus of Stylonychia lemnae. The replication band, the region where replication and telomere elongation take place, was also not stained, suggesting that the guanine-quadruplex is resolved during replication. Our results provide experimental evidence that the telomeres of Stylonychia macronuclei adopt in vivo a guanine-quadruplex structure, indicating that this structure may have an important role for telomere functioning.
Telomeres are specialized DNA–protein complexes at the end of eukaryotic chromosomes. They protect the chromosome ends from recombination, from fusion, and from being mistaken as broken ends (1–3). Telomeric DNA consists of simple repetitive DNA sequences ranging in size from 36 nucleotides in hypotrichous ciliates up to 50 kb in mammals. The 3′-strand is usually G-rich and extends over the complementary strand. It has been shown that the G-rich overhang can adopt a variety of unusual DNA structures (4), of which guanine-quadruplex DNA and t-loops are stable in vitro under physiological conditions (5–8). Parallel-stranded as well as antiparallel-stranded guanine-quadruplex structures have been biophysically and structurally analyzed in detail with synthetic oligonucleotides (6–11). It has been suggested that this structure is also involved in numerous cellular processes, including transcription and recombination, in addition to telomere function (12). However, direct evidence for this DNA structure in vivo has been lacking to date.
Ciliated protozoa are a well-suited biological system to study telomere structure and function, and many important processes including telomere sequences and telomerase were first identified in these cells (1–3). Hypotrichous ciliates, such as Oxytricha, Euplotes, or Stylonychia, have two morphologically and functionally different nuclei in one cell, the DNA-rich transcriptionally active macronucleus and the diploid micronucleus. Micronuclear DNA is organized as conventional eukaryotic chromosomes. After sexual reproduction of the cells, a new macronucleus is formed from the micronucleus. In the course of macronuclear differentiation, extensive DNA reorganization and DNA elimination processes take place, leading to a macronuclear genome that consists of several millions of gene-sized chromosomes ranging in size from about 500 bp to 20 kbp (13). The ends of these DNA molecules are capped by short telomeres that are homogeneous in length and have the sequence G4(T4G4)4 in Stylonychia and Oxytricha, of which the last 16 nucleotides form a 3′-overhang of two unpaired d(T4G4) repeats (14). When macronuclei were gently lysed, the chromosomes were found to form long coherent fibers (15, 16), suggesting that the DNA is organized in a higher-order architecture in the nucleus. Purified macronuclear chromosomes can also assemble in vitro in the absence of proteins, and the 3′-telomeric overhang is involved in this end-to-end aggregation (7, 17, 18). Studies of oligonucleotides based on the telomeric sequence led to the proposal that two overhangs could mediate end-to-end association by forming four-stranded conformations of two fold-back hairpins involving quadruplex stems, stabilized by stacked layers of cyclically hydrogen-bonded guanine quartets (Fig. 1). It is now well established that DNA sequences with multiple guanine stretches can form inter- and intramolecular, parallel or antiparallel quadruplex assemblies in vitro (6–11).
Figure 1.
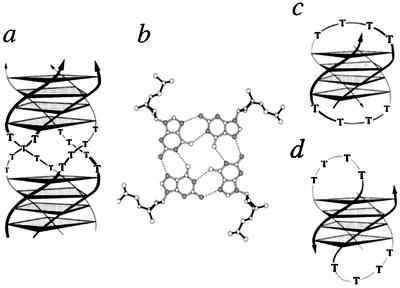
Telomeric guanine-quadruplex molecules. (a) Schematic drawing of a parallel guanine-quadruplex from Stylonychia telomeric DNA d(G4T4G4). (b) The quadruplex is stabilized by guanine quartet layers of four cyclically hydrogen-bonded guanine residues (nitrogen atoms are shaded). The Stylonychia telomeric DNA can also adopt fold-back structures as deduced in the presence of Na+ (c) and in the presence of K+ (d). The thymine residues are arranged in four-nucleotide loops spanning either the diagonal (c) or the edges (d) of the guanine-quadruplex stem (reviewed in ref. 4). Arrows indicate strand polarity.
To test for the actual presence of guanine-quadruplexes in the nucleus, taking into account the low percentage of telomeric sequences present in the genome, it would be very useful to have high-affinity binders specific for the guanine-quadruplex structure as a probe. Starting from the Human Combinatorial Antibody Library (19), we selected in vitro by ribosome display (20, 21) single-chain antibody fragments (scFvs) specific for Stylonychia telomeric guanine-quadruplex DNA. The characterization of two scFvs, Sty3 and Sty49, by radioimmunoassay (RIA) using a wide range of competitors demonstrates the specificity of the antibody probes. Kd measurements were performed in solution with different concentrations of guanine-quadruplex as soluble competitor either in the parallel or antiparallel conformation. We defined the epitope specifically recognized by the scFvs by competition BIAcore using parallel guanine-quadruplexes formed by sequences with shorter T- or G-stretches (Table 1). These results indicate that the optimal epitope encompasses the entire Stylonychia telomeric sequence d(T4G4) arrayed in a four-stranded assembly.
Table 1.
Dissociation constants of anti-guanine-quadruplex scFvs determined by inhibition BIAcore and definition of the minimal DNA epitope bound
| DNA sequence |
Kd, nM
|
|
|---|---|---|
| scFv Sty3 | scFv Sty49 | |
| 4⋅d(TG4T) parallel | 654 ± 56 | 249 ± 55 |
| 4⋅d(T2G4T) parallel | 117 ± 17 | 104 ± 32 |
| 4⋅d(T3G4T) parallel | 1.38 ± 0.16 | 26.1 ± 5.3 |
| 4⋅d(T4G4T) parallel | 0.126 ± 0.011 | 2.96 ± 0.53 |
| 4⋅d(T4G3T) parallel | 7.0 ± 0.9 | 61.7 ± 7.7 |
| d(T4G2T)* | —† | —† |
| d(G4T4G4T4G4T4G4) antiparallel | 171 ± 14 | 5.08 ± 1.69 |
d(T4G2T) does not adopt a quadruplex structure under the conditions studied (33).
No binding detected.
We probed Stylonychia macronuclei and micronuclei with these antibodies and show that the macronucleus but not the micronucleus reacts with the quadruplex-specific antibody Sty49. Moreover, our results show that the guanine-quadruplex structure is resolved for replication and telomere elongation in the macronuclear replication band.
Materials and Methods
Preparation of the Guanine-Quadruplexes.
For selection, we used the biotinylated Stylonychia telomeric sequence, denoted b-sty (5′-TTTTTGGGGTTTTGGGGTTTTGGGGTTTTGGGG-3′), in the parallel quadruplex conformation. Parallel and antiparallel quadruplex structures of the sequences examined were prepared as described (11, 22) and further purified to conformational homogeneity by gel filtration with a Superdex-75 column (Amersham Pharmacia). All DNA conformations were confirmed by CD spectroscopy using a Jasco 715 spectropolarimeter (11, 23).
In Vitro Selection by Ribosome Display.
The Human Combinatorial Antibody Library (19) and the C-terminal spacer derived from TonB were assembled in vitro as described (21). Ribosome display was carried out using the protocol developed in our laboratory (20, 21). Six cycles of ribosome display were performed using 75 nM oligonucleotide b-sty as the antigen.
Test of the Pools and Single scFvs by RIA.
Neutravidin-coated microtiter plates were incubated with 20 pmol of b-sty for 30 min with gentle shaking at 25°C. After washing with PBS (20 mM sodium phosphate/150 mM NaCl, pH 7.4), the wells were blocked for 2 h with 4% milk powder in PBS. For RIA, 1 μg of DNA (plasmid or PCR product with T7 promoter) was transcribed, and the mRNA was purified and in vitro translated for 30 min at 37°C in the presence of 0.3 μM [35S]methionine (50 μCi/ml; 1 μCi = 37 kBq) and then stopped with PBST (PBS with 0.05% Tween 20); subsequently, sterilized milk powder (2% final concentration) was added (20). The mixture was used either directly for binding to the coated microtiter wells or first incubated for 1 h at 25°C with antigen or control substances as competitors. Sonicated salmon sperm DNA, poly[d(GC)], poly(dT) (all from Amersham Pharmacia), Escherichia coli tRNA (Sigma), a T4-hairpin (5′-CGCGCGCGTTTTCGCGCGCG-3′), and the sequence d(T4G4) in a double-strand (5′-CGCGAATCGCTTTTGGGGTACCCCAAAAGCGATTCGCG-3′) (all from Microsynth; Balgach, Switzerland) were dissolved in 50 mM Tris/50 mM NaCl, pH 7.4. Triplex DNA competitor poly(dT)⋅poly(dA)⋅poly(dT) was prepared as described (24). Phosphatidylcholine (100 mg/ml, Sigma) was dissolved in hexane. Competitor DNA conformations were confirmed by CD spectroscopy. DNA concentrations were determined by absorbance at 260 nm (see legend of Fig. 2). The radioactive scFvs were allowed to bind 1 h at 25°C to immobilized b-sty. After washing five times with PBS, the radioactive antibodies were eluted with 0.1 M triethylamine and quantified in a scintillation counter.
Figure 2.
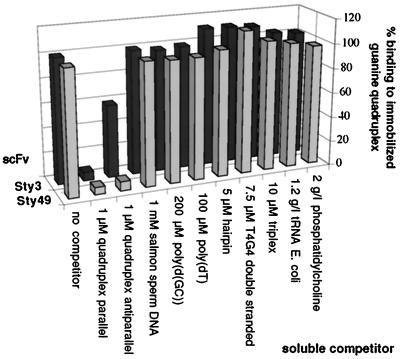
Analysis of two representative scFv proteins by RIA. mRNA of Sty3 and Sty49 was translated in vitro in the presence of radioactive methionine. After incubation with the listed substrates as competitors, the translation mixture was applied to microtiter plates with immobilized parallel guanine-quadruplex. Each bar represents the average of four measurements. Competitor concentrations are given as follows: per nucleotide for salmon sperm DNA, poly[d(GC)], and poly(dT); per oligonucleotide for hairpin and d(T4G4) double strand; and per base triplet for triplex DNA. The background signal (binding to neutravidin) corresponding to 3–5% of the total signal was subtracted. Nonselected clones did not give a signal above the background. Soluble quadruplex in the parallel conformation abrogated binding of both scFvs Sty3 and Sty49 to the immobilized DNA. Binding was also efficiently inhibited by adding the antiparallel quadruplex conformer to Sty49, but only to a much lower extent in the case of Sty3. None of the other soluble competitors inhibited binding to a detectable degree.
Cloning, Expression, and Purification of Individual scFvs.
The scFvs were cloned in the vector pTFT74, expressed as inclusion bodies, and refolded (21). Refolded His-tagged scFvs of antibodies were purified by immobilized metal ion affinity chromatography using Ni-NTA Superflow (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. The eluate was dialyzed against HBS (20 mM Hepes/150 mM NaCl, pH 7.3) and further purified by gel filtration. Gel filtration runs of scFv antibody probes (5 μM), DNA samples (5 μM), and protein–DNA complexes (4–5 μM) were performed in HBS using a Superdex-75 column and the SMART system (Amersham Pharmacia). Protein concentration was determined by OD at 280 nm [extinction coefficients: ɛ(Sty49) = 53,010 M−1⋅cm−1; ɛ(Sty3) = 49,170 M−1⋅cm−1].
Determination of Dissociation Constants in Solution by Competition BIAcore.
Competition BIAcore analysis was performed under conditions of mass-transport limitation using an SA sensor chip (Amersham Pharmacia) coated with 2,900 resonance units of b-sty, and a streptavidin surface without DNA was used as a control. The antibodies were preincubated with soluble antigen (different guanine-quadruplexes) at different concentrations for at least 1 h at 4°C before injection, and the inhibition of the mass-transport limited on-rate was measured as a function of DNA concentration. Each binding–regeneration cycle was performed at 20°C with a constant flow rate of 20 μl/min using HBST (HBS with 0.005% Tween 20), and 20 μl of 3 M guanidinium chloride were used for regeneration. Data were evaluated using the BIAEVALUATION software (Amersham Pharmacia). Slopes of linear sensorgrams during injection were plotted against the corresponding total antigen concentrations to calculate the dissociation constant including error values (21). For competition with parallel guanine-quadruplex, the sequence d(T4G4T) was chosen with a 3′-terminal thymidine to maintain sufficient solubility of the sample under the conditions used (I.B., unpublished results). For competition with the antiparallel quadruplex, the intramolecular fold-back structure formed by d(G4T4G4T4G4T4G4) was used. Thus, both the parallel and antiparallel quadruplexes used in the competition experiments offered one 4-layer guanine quartet stem per molecule for binding.
Growth of Stylonychia lemnae, synchronization of replicating cells, and isolation of macronuclei were performed as described (25, 26).
In Situ Immunofluorescence Experiments.
Nuclei were isolated, resuspended in PBS/2% formaldehyde for 60 min, transferred to slides, and dried overnight under vacuum (25, 26). The slides were washed four times for 15 min at 25°C with HBS, followed by blocking for 30 min in HBS/2% blocking reagent (DIG DNA labeling and detection kit, Roche Diagnostics) and incubation with the respective scFv probes (70 μg/ml in HBS) overnight. They were then washed twice for 15 min with HBS, blocked for 30 min in HBS/2% blocking reagent, and incubated with the tetra-His antibody (mouse IgG1, Qiagen) at a dilution of 1:20 for 2 h. Finally, the slides were incubated twice for 15 min with HBS, blocked for 30 min in HBS/2% blocking reagent, and incubated with a rabbit anti-mouse FITC antibody (Dako) at a dilution of 1:20 for 2 h. After washing the slides several times with HBS, antibody binding was visualized with a Leitz DM RB microscope. Identical stainings of macronuclei were done without formaldehyde to rule out any influence of fixing reagents.
In some experiments, macronuclei were digested with nucleases S1 and Bal31 or exposed to 0.2 N NaOH before antibody binding. For S1 and Bal31 digestion, macronuclei were isolated, resuspended in PBS/2% formaldehyde, followed by several washes with PBS. For S1 digestion, about 104 macronuclei (corresponding to about 8 μg of DNA) were resuspended in S1 buffer (MBI Fermentas). S1 nuclease (500 units) was added and kept at room temperature for 30 min. Nuclei were then washed several times with HBS before antibody binding. For Bal31 digestion, the same number of nuclei was resuspended in Bal31 buffer (MBI Fermentas) and digested for 30 min at 25°C with 25 units of Bal31 nuclease followed by several washes with HBS. Under these conditions, about 100 bp are digested off the ends (25). Macronuclear DNA was denatured by the addition of 0.2 N NaOH for 20 min to immobilized macronuclei on slides followed by four washes in HBS at 25°C.
Results
Selection and Characterization of Guanine-Quadruplex-Specific scFvs.
Six rounds of ribosome display were performed using synthetic Stylonychia telomeric DNA d(T4G4) in the parallel conformation as the antigen. After six selection cycles, the scFv pool was analyzed by RIA and was found to bind parallel guanine-quadruplex DNA, but not neutravidin as a control. Subsequently, single clones from the pool were isolated and characterized in detail by RIA. The ability of a range of soluble nucleic acid competitors to inhibit the binding of the radioactive scFvs to the immobilized parallel guanine-quadruplex DNA was tested. The RIA of two selected scFvs, Sty3 and Sty49, is depicted in Fig. 2. No inhibition was observed with up to 1 mM salmon sperm DNA, 200 μM of the guanine-rich DNA poly[d(GC)], or 7.5 μM Stylonychia telomeric sequence in a double-stranded B-DNA conformation. Also, no binding was observed to 100 μM poly(dT) or a B-DNA hairpin duplex with a four-thymine loop. Furthermore, 10 μM of a triplex, 1.2 g/liter tRNA, or up to 2 g/liter phospholipid did not compete with binding. On binding to telomeric guanine-quadruplex DNA, however, the two scFvs did show a difference. Sty3 bound only weakly to the intramolecular antiparallel quadruplex conformer, indicating a strong preference for the parallel conformation, whereas Sty49 bound both parallel and antiparallel conformations with similar affinity.
The different affinity of the two scFvs for the antiparallel guanine-quadruplex structure was further confirmed by competition BIAcore analysis, which provides affinities in solution (Table 1) (21). Sty3 bound the parallel guanine-quadruplex adopted by the sequence d(T4G4T) with a Kd of 126 pM and showed a 1,000-fold specificity for the parallel conformation as measured by competition with the antiparallel guanine-quadruplex structure adopted by d(G4T4G4T4G4T4G4). In contrast, Sty49 bound both conformations with a Kd of 3–5 nM. Furthermore, we defined the epitope specifically recognized by the scFvs. Shortening of either the T- or the G-stretch of the telomeric guanine-quadruplex motif resulted in progressively reduced binding (Table 1). This result indicates that the optimal epitope recognized by both scFvs with high affinity encompasses the entire Stylonychia telomeric sequence d(T4G4).
Evidence for Guanine-Quadruplex DNA in the scFv–DNA Complexes.
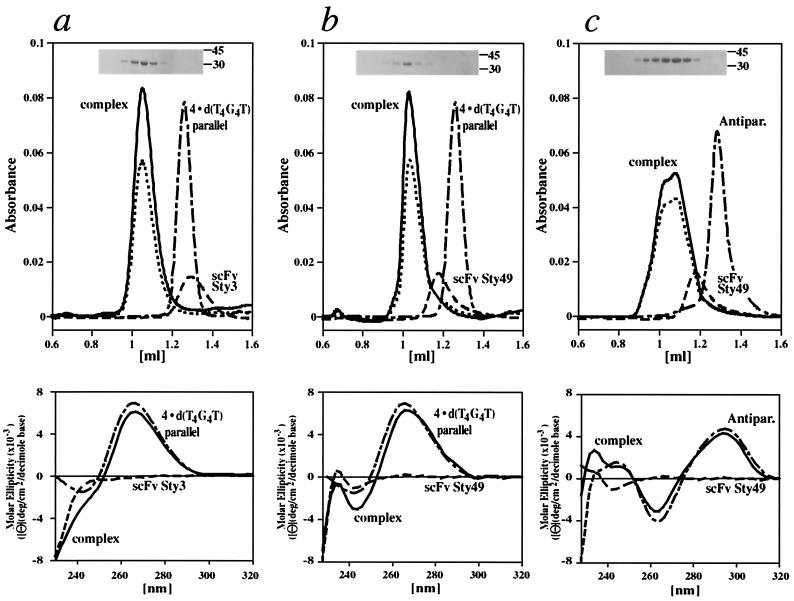
We then isolated the complex of scFv Sty3 bound to the parallel guanine-quadruplex formed by d(T4G4T) using gel filtration and analyzed the purified protein–DNA complex by SDS/PAGE and CD spectroscopy (Fig. 3a). The DNA peak was quantitatively shifted to the higher molecular weight peak in the elution profile by adding equimolar amounts of scFv Sty3 to the parallel guanine-quadruplex. The CD spectrum identified the parallel-stranded conformer in the scFv–DNA complex (Fig. 3a). We carried out analogous experiments with a mixture of scFv Sty49 and either the parallel d(T4G4T) quadruplex or the antiparallel fold-back conformer adopted by d(G4T4G4T4G4T4G4) (Fig. 3 b and c). Gel filtration of the mixture of Sty49 and the antiparallel guanine-quadruplex gave rise to a doublet peak for the complex, detected at 260 nm and 280 nm, possibly indicating two different binding modes of the protein to its DNA target. In gel-filtration control experiments with single-stranded and duplex forms of Stylonychia telomeric DNA mixed with excess of antibody, the scFvs and DNA eluted separately (data not shown), indicating again the specificity of recognition.
Figure 3.
Evidence for guanine-quadruplex in the scFv–DNA complexes. Elution profiles in all gel-filtration experiments were recorded at 280 nm for proteins (Sty3, Sty49; dashed line), 260 nm for DNA (broken line), and at both wavelengths for the complexes (solid line, 260 nm; dotted line, 280 nm). (a) The complex formed by scFv Sty3 and the parallel quadruplex adopted by four strands of d(T4G4T) eluted at higher apparent molecular weight than the individual components in isolation (Upper) with an equimolar mixture of scFv and guanine-quadruplex DNA. Fractions from the higher molecular weight peak were loaded on an SDS gel and stained by Coomassie blue (Inset), confirming the presence of the scFv. The eluate was analyzed by CD spectroscopy (Lower). In the region from 250 nm to 320 nm, the spectrum of the complex (solid line) is virtually superimposable on the spectrum (broken line) of four-stranded d(T4G4T), exhibiting a maximum at 264 nm characteristic for the parallel quadruplex conformer (23). In this region, the protein alone (dashed line) does not have a significant CD signal. (b) The analogous experiment performed with scFv Sty49 also resulted in a quantitative shift toward higher molecular weight of the DNA when mixed with equimolar amounts of protein (line coding as for scFv Sty3). SDS/PAGE (Inset) and CD spectroscopy confirmed the presence of Sty49 and parallel guanine-quadruplex DNA in this peak, with the complex showing again the characteristic CD signal of the parallel conformer (Lower). (c) scFv Sty49 was also mixed with d(G4T4G4T4G4T4G4) DNA, forming an antiparallel fold-back quadruplex (labeled Antipar.). The mixture eluted in a doublet at higher apparent molecular weight than either DNA or Sty49 in isolation (Upper). The CD spectrum of the complex peaks showed a maximum at 295 nm and a minimum at 265 nm, characteristic of the antiparallel quadruplex conformation of this DNA (23).
In Situ Analyses.
We probed Stylonychia macronuclei and micronuclei for reaction with the selected telomeric guanine-quadruplex DNA-specific scFv antibody probes Sty3 and Sty49 by indirect immunofluorescence (Fig. 4). Incubation of the macronuclei with Sty3 did not produce a signal above the background level where no scFv was used (Fig. 4b). Sty49, on the other hand, produced a clear signal when applied to vegetative macronuclei (Fig. 4a). Although Sty3 and Sty49 both recognize the parallel guanine-quadruplex adopted by Stylonychia telomeric DNA with nanomolar or better affinity, only Sty49 has a high affinity for the antiparallel quadruplex (Table 1). This observation indicates that it is the antiparallel conformer that is bound in the macronuclei in our experiment, consistent with the model for the end-to-end linkage of macronuclear chromosomes mediated by a fold-back quadruplex (7).
Figure 4.
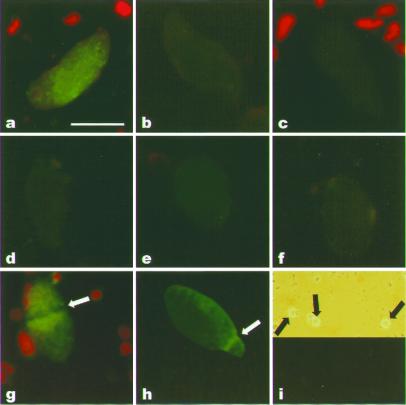
Indirect immunofluorescence staining of S. lemnae nuclei. Isolated macronuclei were incubated with scFvs Sty49 or Sty3 and analyzed by fluorescence microscopy. Synchronization of Stylonychia cells (g and h) was done as described (25). Sty49 was used in a, d–g, and i; Sty3 was used in b. (Bar in a represents 10 μm.) (a) Binding of Sty49 to a macronucleus. (b) Binding of Sty3 to a macronucleus. (c) Antibody staining in the absence of scFv probe. (d) Macronuclei were digested with S1 nuclease before Sty49 binding. (e) Macronuclei were digested with Bal31 nuclease before Sty49 binding. (f) Macronuclei were incubated with 0.2 M NaOH before Sty49 binding. (g) Binding of Sty49 to a replicating macronucleus; arrow points to the replication band. (h) Hybridization of an FITC-labeled telomeric probe to a replicating macronucleus; arrow points to the replication band. (i) Binding of Sty49 to micronuclei (Upper, phase contrast; Lower, fluorescent microscopic analysis; arrows point to micronuclei). As an internal control, in some experiments a few algae were added to the nuclei (red fluorescence in a, c, and g).
In comparison with the macronuclei, S. lemnae micronuclei comprise few (≈120) chromosomes, containing on average 18⋅106 bp (13). The telomeres of micronuclear chromosomes are organized in paired duplex d(T4G4) repeats ranging from 3 to 20 kbp in length, but their 3′-overhangs have not yet been characterized in detail. Even though the total DNA concentration in micronuclei is at least as high as in macronuclei (26), the telomeric sequences of micronuclear chromosomes, although substantially larger, are therefore far less abundant. When applied to micronuclei, Sty49 did not produce a detectable signal (Fig. 4i), thereby providing further evidence that the antibody does not bind unspecifically to DNA.
Replication and telomere elongation occurs in the macronucleus in a morphologically distinct region, the replication band (13). No binding of Sty49 to this nuclear region was observed (Fig. 4g), even though the DNA concentration, and thus the concentration of telomere sequences, is higher there than elsewhere in the macronucleus, as evidenced by in situ hybridization with an FITC-labeled telomeric probe (Fig. 4h).
We performed a number of control experiments (Fig. 4 d–f). Digestion of the macronuclei with excess amounts of S1 or Bal31 nucleases abrogated the antibody reaction. S1 nuclease treatment was shown to abolish the ability of macronuclear DNA to form end-to-end aggregates when used at concentrations above 5 units/μg DNA (18), and Bal31 exonuclease has a similar effect at the concentrations applied (25). Sty49 did not react with macronuclei that were pretreated with high pH to denature the DNA (Fig. 4f). This is in agreement with the previous observation that end-to-end aggregation of macronuclear chromosomes can be resolved by treatment with NaOH (16).
Discussion
Telomeric guanine-quadruplex DNA has been shown to form in vitro under physiological conditions (6–8). It has been suggested that this structure plays an important role in vivo in the protection of telomeric ends, and attempts have been made to use the guanine-quadruplex structure as a target for cancer therapy (12). However, until now, to the best of our knowledge, no experimental data exist that this structure actually occurs in the eukaryotic cell. We therefore decided to generate in vitro specific antibodies against the Oxytricha/Stylonychia telomeric guanine-quadruplex structure in the parallel conformation, using a synthetic antibody library and a cell-free selection and evolution system. The selected antibodies were applied for in situ localization of quadruplex DNA in the nuclei of the ciliated protozoan S. lemnae.
We selected our scFvs in vitro from a large synthetic library by ribosome display in six selection cycles. In RIA control experiments with a variety of competitors, we demonstrate that none of the control deoxynucleic acids, nor excess RNA or phospholipids, inhibited the binding of these scFvs to their specific target DNA to a detectable degree. In our experiments, considerable amounts of RNA are present in the in vitro translation mix. Furthermore, we supplemented the mixture with heparin as an RNase inhibitor and to reduce protein aggregation. Heparin mimics the sugar phosphate backbone of nucleic acids. Thus, both RNA and heparin could compete with the guanine-quadruplex target for less specific binders in the library and thereby prevent enrichment of cross-reactive antibodies.
One of the antibodies, Sty3, binds the parallel guanine-quadruplex with a 1,000-fold specificity compared with the antiparallel conformation. In contrast, the other antibody, Sty49, binds both quadruplex conformations with similar affinity. The parallel guanine-quadruplex is bound with picomolar affinity by Sty3, whereas Sty49 binds both conformations with 3–5 nM affinity. By gel filtration, stable scFv DNA complexes with both guanine-quadruplex conformations could be quantitatively purified and analyzed by CD spectroscopy. The characteristic CD spectrum clearly demonstrated the presence of guanine-quadruplex DNA in the complex.
It is interesting to note that Sty49 recognizes an epitope that is present in both conformers of the quadruplex. The strand orientation in one antiparallel guanine-quadruplex conformer (10) linked over the diagonal (Fig. 1c) shows two edge stands, each aligned antiparallel and two aligned parallel in the four-stranded stem, whereas in the parallel conformer all edge strands have the same orientation. Thus, the grooves lined by coaligned sugar phosphate backbones could constitute the similar epitope in both conformers, contributing to recognition by this antibody. The exact mode of how our antibodies bind to the different guanine-quadruplex conformers will be elucidated by more detailed biophysical and structural characterization of the scFv–DNA complexes.
Hypotrichous ciliates, such as Oxytricha or Stylonychia, provide an excellent model system to study telomere behavior in vivo. The macronuclear genome occurs in small gene-sized DNA molecules, and each is terminated by telomeric sequences of homogenous length. Each macronucleus contains millions of minichromosomes, and thus the concentration of telomeric DNA is orders of magnitude higher than in any other organism. For these reasons, we probed Stylonychia macronuclei and micronuclei in situ for a reaction with the selected telomeric guanine-quadruplex DNA-specific scFvs. It has long been speculated that macronuclear telomere capping could be achieved by interaction of 3′-overhangs in a telomeric guanine-quadruplex, giving rise to the observed end-to-end association of ciliate macronuclear minichromosomes (7, 16). In fact, Oxytricha telomere-binding protein (TEBP), which is highly homologous to the Stylonychia telomere binding protein, was shown to promote guanine-quadruplex formation in vitro (27).
Our results clearly show that a specific antibody binding to the macronucleus is observed with Sty49, which recognizes both conformers, whereas no reaction was obtained with Sty3, recognizing only the parallel conformation with high affinity. Various control experiments demonstrate that the observed signal is indeed caused by specific interaction with the 3′-telomeric overhang. Staining of nuclei in the absence of the primary antibody Sty49 yielded no staining; pretreatment of the macronuclei with high pH to denature the DNA abrogated antibody binding. The same was true after digestion of the 3′-overhang with an excess of S1 nuclease and after exonucleolytic digestion with Bal31 (25).
No binding of Sty49 to the micronucleus could be detected. Two explanations are possible. The micronuclear genome is organized as conventional chromosomes, thus the number of telomeres is per se significantly lower than in the macronucleus, even though the d(T4G4) repeats are present in high amounts. Furthermore, electron microscopy studies of ciliate polytene chromosomes led to the proposal that micronuclear chromosomes may also be capped by t-loops, similar to mammalian chromosomes (28). In t-loops, the 100- to 200-bp-sized 3′ single-stranded overhang invades the duplex region of the telomeric repeats of the same chromosome and displaces the guanine-rich strand, but presumably maintains B-DNA structure (28).
Replication of macronuclear DNA and telomere elongation occurs in the replication band (13, 29), a structure easily visible in the light microscope. The DNA concentration in this region is considerably higher than in the residual macronucleus, as demonstrated by staining the nucleus with an anti-B-DNA antibody (26) and by in situ hybridization with a telomeric probe (Fig. 4h). No binding of Sty49 to the macronuclear replication band was observed. Indeed, the presence of a telomeric guanine-quadruplex structure in replicating DNA would be very unlikely, as chromosomes need to be disaggregated to provide accessible ends for telomerase function (30). This picture is reminiscent of results of the in situ localization of the telomeric DNA–TEBP complex in the macronucleus of S. lemnae (25). It was shown that this complex is associated with the nuclear matrix, but this association becomes resolved during replication, suggesting that the DNA molecules occur in the macronucleus in a higher architecture that has to be reorganized during replication.
There are indications that telomere interaction with TEBP and guanine-quadruplex formation may be interconnected (27). TEBP is abundant in the macronucleus and was shown to bind the 3′-overhang as a single strand, burying it completely (16, 31, 32). Oxytricha TEBP α- and β-subunits form the heterodimeric TEBP only in complex with DNA. Free α- and β-subunits are likely to be present in the nuclei. Free β-subunit can promote guanine-quadruplex formation and may also facilitate its transitory unfolding (27), thus rendering the 3′-overhang accessible for TEBP or telomerase binding or dimerization to an antiparallel quadruplex. Both TEBP binding (32) and the formation of a stable antiparallel fold-back quadruplex require the entire 16 nucleotides of the overhang (7), and both could contribute to the tightly regulated telomeric length. Our results provide experimental evidence that guanine-quadruplex structures are actually present in vivo in Stylonychia macronuclei and imply that guanine-quadruplex formation can be a third mechanism for telomere capping, in addition to t-loops and TEBP binding.
Acknowledgments
We thank the members of the Plückthun Lab for assistance. MorphoSys AG (Martinsried, Germany) is acknowledged for constructive collaboration on the Human Combinatorial Antibody Library. This work was supported by Schweizerischer Nationalfonds Grant 31-46624.96 (to A.P.) and by a grant from the Deutsche Forschungsgemeinschaft (to H.J.L.). C.S. is supported by a Kékule Fellowship from the Fonds der Chemischen Industrie (Germany); I.B. is a Liebig Fellow of the Fonds der Chemischen Industrie.
Abbreviations
- scFv
single-chain fragment of an antibody
- TEBP
Oxytricha telomere-binding protein
References
- 1.Kipling D. The Telomere. Oxford: Oxford Univ. Press; 1995. [Google Scholar]
- 2.Blackburn E H, Greider C W. Telomeres. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 3.McEachern M J, Krauskopf A, Blackburn E H. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 4.Henderson E. In: Telomeres. Blackburn E H, Greider C W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 11–34. [Google Scholar]
- 5.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 6.Williamson J R, Raghuraman M K, Cech T R. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 7.Sundquist W I, Klug A. Nature (London) 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 8.Sen D, Gilbert W. Nature (London) 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 9.Kang C, Zhang X, Ratliff R, Moyzis R, Rich A. Nature (London) 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 10.Smith F W, Feigon J. Nature (London) 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 11.Jin R, Gaffney B L, Wang C, Jones R A, Breslauer K J. Proc Natl Acad Sci USA. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han H, Hurley L H. Trends Pharmacol Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 13.Prescott D M. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klobutcher L A, Swanton M T, Donini P, Prescott D M. Proc Natl Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer G F, Lipps H J. Chromosoma. 1981;82:309–314. doi: 10.1007/BF00286113. [DOI] [PubMed] [Google Scholar]
- 16.Lipps H J, Gruissem W, Prescott D M. Proc Natl Acad Sci USA. 1982;79:2495–2499. doi: 10.1073/pnas.79.8.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipps H J. Proc Natl Acad Sci USA. 1980;77:4104–4107. doi: 10.1073/pnas.77.7.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka Y, Thomas C A., Jr Nucleic Acids Res. 1987;15:8877–8898. doi: 10.1093/nar/15.21.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wölle J, Plückthun A, Virnekäs B. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 20.Hanes J, Plückthun A. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanes J, Schaffitzel C, Knappik A, Plückthun A. Nat Biotechnol. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Patel D J. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 23.Giraldo R, Suzuki M, Chapman L, Rhodes D. Proc Natl Acad Sci USA. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kan Y, Armitage B, Schuster G B. Biochemistry. 1997;36:1461–1466. doi: 10.1021/bi962335s. [DOI] [PubMed] [Google Scholar]
- 25.Postberg J, Juranek S A, Feiler S, Kortwig H, Jönsson F, Lipps H J. J Cell Sci. 2001;114:1861–1866. doi: 10.1242/jcs.114.10.1861. [DOI] [PubMed] [Google Scholar]
- 26.Lipps H J, Nordheim A, Lafer E M, Ammermann D, Stollar B D, Rich A. Cell. 1983;32:435–441. doi: 10.1016/0092-8674(83)90463-4. [DOI] [PubMed] [Google Scholar]
- 27.Fang G, Cech T R. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 28.Murti K G, Prescott D M. Proc Natl Acad Sci USA. 1999;96:14436–14439. doi: 10.1073/pnas.96.25.14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang G, Cech T R. J Cell Biol. 1995;130:243–253. doi: 10.1083/jcb.130.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahler A M, Williamson J R, Cech T R, Prescott D M. Nature (London) 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 31.Gottschling D E, Zakian V A. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 32.Horvath M P, Schweiker V L, Bevilacqua J M, Ruggles J A, Schultz S C. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 33.Sen D, Gilbert W. Biochemistry. 1992;31:65–70. doi: 10.1021/bi00116a011. [DOI] [PubMed] [Google Scholar]