Abstract
Background
Apomixis is an alternative route of plant reproduction that produces individuals genetically identical to the mother plant through seeds. Apomixis is desirable in agriculture, because it guarantees the perpetuation of superior genotypes (i.e. heterotic hybrid seeds) by self-seeding without loss of hybrid vigour. The Paspalum genus, an archetypal model system for mining apomixis gene(s), is composed of about 370 species that have extremely diverse reproductive systems, including self-incompatibility, self-fertility, full sexual reproduction, and facultative or obligate apomixis. Barriers to interspecific hybridization are relaxed in this genus, allowing the production of new hybrids from many different parental combinations. Paspalum is also tolerant to various parental genome contributions to the endosperm, allowing analyses of how sexually reproducing crop species might escape from dosage effects in the endosperm.
Scope
In this article, the available literature characterizing apomixis in Paspalum spp. and its use in breeding is critically reviewed. In particular, a comparison is made across species of the structure and function of the genomic region controlling apomixis in order to identify a common core region shared by all apomictic Paspalum species and where apomixis genes are likely to be localized. Candidate genes are discussed, either as possible genetic determinants (including homologs to signal transduction and RNA methylation genes) or as downstream factors (such as cell-to-cell signalling and auxin response genes) depending, respectively, on their co-segregation with apomixis or less. Strategies to validate the role of candidate genes in apomictic process are also discussed, with special emphasis on plant transformation in natural apomictic species.
Keywords: Apomixis, comparative mapping, molecular markers, Paspalum, transcriptomic analysis
INTRODUCTION
Modern agriculture is continuously creating new and highly productive cultivars. Traditional plant breeding methods and, more recently, genetic engineering have succeeded in steadily increasing crop production over the years. Despite these achievements, and with a global community calculated to stabilize at 8·1–11·9 billion people in the middle of the 21st century (Lutz et al., 1997), the challenges for agriculture remain overwhelming. Among promising approaches for significantly increasing crop productivity, the transfer of clonal reproduction through seeds or apomixis would represent an enormous benefit for agriculture (Vielle-Calzada et al., 1996). The genus Paspalum is an attractive biological system for studying apomixis, because it is both a model system for mining candidate gene(s) and an important target crop. Over the past five decades, a wealth of information has been produced regarding the biology, genetic and reproductive modes of many Paspalum species including: (1) cytoembryological aspects of apomixis; (2) detailed molecular maps of apomixis loci; (3) isolation of the first candidate genes; and (4) development of transformation systems for gene delivery. Moreover, P. notatum represents an unprecedented occurrence: the existence of freely crossable apomictic and sexually reproducing races of an agronomically important species.
In sexual reproduction, a single cell within the ovule typically becomes the megaspore mother cell (MMC). The MMC undergoes meiosis to form four reduced megaspores, one of which develops into an embryo sac (ES). In most plants, the ES includes the egg cell and two synergids at one pole, a large binucleated central cell, and three antipodals at the opposite pole. This is known as Polygonum-type ES. Double fertilization by two sperm released from the pollen tube results in a diploid zygote (n egg + n sperm) and a triploid central cell (2n central cell + n sperm) that develops into endosperm (Grossniklaus, 2001). Conversely, gametophytic apomixis in angiosperms relies on the formation of an ES from an unreduced ES initial (Nogler, 1984). Whether unreduced ESs arise from an MMC or from a somatic, usually nucellar cell, distinguishes diplospory from apospory, respectively. Fertilization of polar nuclei is usually required for apomictic seed formation. Both pathways have been broadly referred to as ‘apomeiosis’ or ‘apomeiotic pathways’ and encompass a variety of developmental schemes (Nogler, 1984; Asker and Jerling, 1992; Crane, 2001; Pupilli and Barcaccia, 2012). Apomeiosis was originally defined as the ‘loss of meiotic reduction’ (Renner, 1916). In our opinion, the term is not exactly applicable to apospory, because it is derived from the word ‘meiosis’ and the Greek prefix απο (apo), meaning away from, without or lacking. The term apomeiosis applies to diplospory, which involves a loss of meiosis. However, in apospory, somatic cells acquire a novel ability to develop ESs, a function normally reserved for the megaspores, whereas meiosis usually occurs in the MMC. The functional megaspore aborts or develops into a meiotic ES that usually loses functionality in competition with the aposporous sacs developing in the same ovule. Nowadays, these pathways are viewed as heterochronic traits resulting from the ectopic expression of sexual reproduction sub-programmes either temporally or spatially (Grimanelli et al., 2003; Bradley et al., 2007; Sharbel et al., 2010).
PASPALUM AGAMIC COMPLEXES
Botany, phylogeny and evolution
Paspalum (Linnaeus, 1759) is one of the ten largest genera within Poaceae. Recent systematic works have expanded the genus, including species of the genus Thrasya (Denham, 2005), and new taxa have been described (Oliveira and Rua, 2005; Rua et al., 2008; Oliveira and Valls, 2009; Sánchez-Ken, 2010; Ramos et al., 2011). In addition, several species have been transferred to Paspalum from the polyphyletic genus Panicum (Morrone et al., 2007, 2008; Zuloaga et al., 2007, 2010, 2011; Sede et al., 2008, 2009). With a wide range of morphological and ecological adaptations, the approx. 370 species of Paspalum have been classified into four subgenera.
Anachyris (Nees) Chase, a well-delimited monophyletic group of six closely related species sharing specific morphological and embryological features (Morrone et al., 2000; Urbani et al., 2002; Hojsgaard et al., 2008; Rua et al., 2010).
Ceresia (Pers.) Rchb., 25 species mainly characterized by bearing a winged rachis (Denham et al., 2002).
Harpostachis (Trin.) S. Denham (formerly genus Thrasya), 40 species distributed in Central America and northern South America (Denham, 2005; Sánchez-Ken, 2010).
Paspalum sensu stricto, which contains most of the species (approx. 300), and shows the greatest diversity (Zuloaga and Morrone, 2005).
The two most comprehensive taxonomic reviews recognized 40 infrageneric groups among these subgenera (Chase, 1939; Zuloaga and Morrone, 2005). Considerable taxonomic efforts have been devoted to the genus Paspalum, especially regarding infrageneric classification (Rua et al., 2010). Current phylogenetic analyses using morphological and/or genetic data have better clarified relationships within the genus (Denham et al., 2002; Souza-Chies et al., 2006; Denham and Zuloaga, 2007; Essi and Souza-Chies, 2007). However, the molecular relationships did not reflect morphological data as only the subgenus Anachyris formed a well-supported clade (Denham and Zuloaga, 2007; Rua et al., 2010). Moreover the informal groups in Paspalum subgenus Paspalum have not yet been delimited by molecular tools, and most of them cannot be diagnosed exclusively by morphological synapomorphies (Souza-Chies et al., 2006). Recent phylogenetic analyses using molecular data have confronted their composition, and no uniform criteria of classification have been achieved to date (Giussani et al., 2009; Rua et al., 2010). Further studies based on extensive taxon sampling and an adequate number of informative molecular markers are therefore needed to resolve relationships among both subgenera and informal groups.
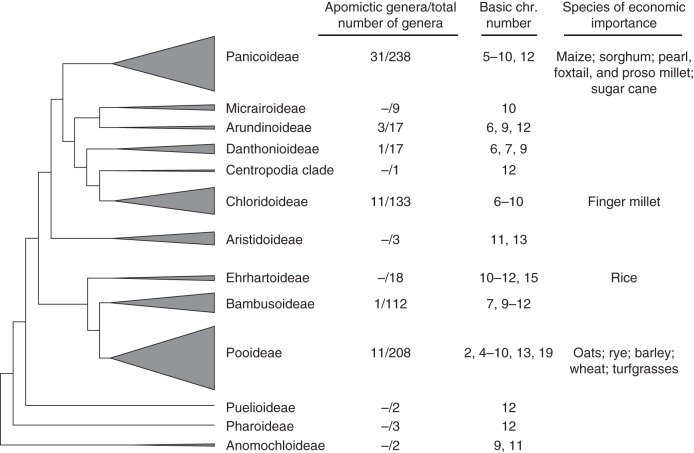
Paspalum species occupy diverse habitats in North and South America, while a few species are native to Africa, Asia and Oceania, and only three or four are cosmopolitan. The centre of origin for the genus is tropical South America (Chase, 1939; Nicora and Rúgolo de Agrasar, 1987; Judziewicz, 1990), but secondary centres of diversity have been recognized in the Brazilian cerrados and the campos of Argentina, Uruguay and Southern Brazil (Zuloaga and Morrone, 2005). The wide range of ecological adaptations found in the genus (Chase, 1939; Parodi, 1969; Zuloaga and Morrone, 2005; L. R. Parodi and E. G. Nicora, unpubl. res.) is probably related to the various reproductive strategies (i.e. sexual reproduction, auto- and allogamy, clonal reproduction through apomixis and vegetative propagation) and ploidy levels found within and across the species (Quarin, 1992). This has undoubtedly affected the evolutionary success of the genus (Bashaw et al., 1970). The observation that the three most diverse subfamilies of Poaceae have the highest proportions of genera that combine apomictic and sexual reproductive modes (i.e. 13·03 % in Panicoideae, 8·27 % in Chloridoideae and 5·29 % in Pooideae; Fig. 1) suggests that apomixis in Paspalum species was probably a key factor in their diversification through the formation of both intra- and interspecific agamic complexes with special evolutionary properties. Contrary to the classical view of the agamic complexes being closed systems (Stebbins, 1950), Carman (1997) pointed out that apomixis is a transitional evolutionary state of polyploid complexes (transition theory). In Paspalum, these complexes usually form spatially restricted diploid populations that represent the main source of variability and that co-habit and hybridize with polyploids (Urbani et al., 2002; Daurelio et al., 2004; Speranza, 2009). Polyploids exploit the advantages of apomixis, i.e. uniparental reproduction and clonality, to expand their geographical and ecological ranges (Kearney, 2005; Hörandl, 2006). Recently, Hörandl and Hojsgaard (2012) extended the transition theory to include the possibility of reversals from facultative apomixis to obligate sexuality. In Paspalum, a few species exhibit the features of a typical genetic system, but with higher ploidy levels (i.e. P. durifolium and P. ionanthum; see Table 1) and may represent cases of reversals forming new agamic complexes after second rounds of polyploidization. Under these assumptions, the morphologically diverse and species-rich genus Paspalum can be considered – according to Hörandl and Hojsgaard (2012) – as a typical evolutionary outcome of sexual–apomictic multiploid complexes within the main clades of Poaceae.
Fig. 1.
Phylogenetic tree of Poaceae subfamilies (modified after Grass Phylogeny Working Group II, 2012); the sizes of subfamilial clades are relative to the species richness of each clade. The number of apomictic genera is after Carman (1997) and Hörandl and Hojsgaard (2012). The total number of genera is according the World-Wide Phylogenetic Classification of Poaceae database (Stevens, 2001 onwards). Chromosome (chr.) numbers were collected from the Grass Genera of the World database (http://www.biologie.uni-hamburg.de/b-online/delta/grass/index.htm). Economically important species inside each subfamily are indicated.
Table 1.
Cytology and reproductive modes in Paspalum spp.
| Specie | Ploidy level | Meiosis* | Reproductive mode† | References | Referred to as |
|---|---|---|---|---|---|
| P. almum Chase | 2x | R | S ss | Quarin and Hanna (1980b) | P. hexastachyum Parodi |
| 4x | mca | Ap | Burson (1975) | ||
| P. arundinellum Mez | 4x | mca | Ap | Bashaw et al. (1970) | |
| P. atratum Swallen | 4x | mca | Ap | Quarin et al. (1997) | |
| P. bertonii Hack. | 2x | R | S ss | Quarin and Burson (1991) | |
| P. buckleyanum Vasey | 2x | R | S sf | Burson (1997) | P. alcalinum Mez |
| 4x | mca | Ap | Burson (1997) | ||
| 5x | mca | Ap | Burson (1997); Sartor et al. (2011) | P. alcalinum Mez | |
| 6x | Ap | Sartor et al. (2011) | |||
| P. chaseanum Parodi | 2x | R | S ss | Espinoza and Quarin (1997) | |
| P. chacoense Parodi | 2x | R | S | Burson (1985) | |
| P. compressifolium Swallen | 2x | R | S ss | Quarin et al. (1996) | |
| 4x | mca | Ap | Quarin et al. (1996) | ||
| 6x | mca | Ap | Quarin et al. (1996) | ||
| P. conjugatum P.J. Bergius | 4x | asy | Dp | Chao (1980); Ma et al. (2009) | |
| P. conspersum Schrad. | 4x | mca | S | Burson and Bennett (1976) | P. platyphyllum Schult. |
| 6x | R | S | Bashaw et al. (1970) | ||
| – | R | S | Quarin and Hanna (1980a) | ||
| P. coryphaeum Trin. | 2x | R | S | Quarin and Urbani (1990) | |
| 4x | mca | Ap | Burson (1975); Quarin and Urbani (1990) | ||
| P. cromyorrhizon Trin. | 2x | R | S ss + App | Quarin et al. (1982) | P. guaraniticum Parodi |
| 4x | mca | Ap | Bashaw et al. (1970); Burson and Bennett (1971); Quarin et al. (1982); Martínez et al. (1999) | ||
| P. dasypleurum Kunze ex Desv. | 4x | R | S sf | Quarin and Caponio (1995) | |
| P. dedeccae Quarin | 4x | mca | Ap | Quarin and Burson (1991) | |
| P. densum Poir. | 2x | R | S ss + App | Caponio and Quarin (1993) | |
| P.denticulatum Trin. | 2x | – | S | Sartor et al. (2011) | |
| 3x | – | Ap | Sartor et al. (2011) | ||
| 4x | mca | Ap | Sartor et al. (2011); Quarin and Burson (1991) | ||
| P. dilatatum Poir | 4x | R | S | Bashaw and Holt (1958) | |
| 5x | I | Ap | Bashaw and Holt (1958) | ||
| 6x | R | Ap | Burson et al. (1991) | ||
| 6x | I | Ap | Burson et al. (1991) | ||
| 6x | asy | Ap | Burson et al. (1991) | ||
| – | – | Ap | Brown and Emery (1958) | ||
| P. distichum L. | 4x | R | Ap | Quarin and Burson (1991); Ma et al. (2009) | |
| 6x | mca | Ap | Bashaw et al. (1970); Quarin and Burson (1991) | ||
| P. durifolium Mez | 4x | R | S ss + App | Quarin (1994) | |
| 6x | R | Ap | Burson (1985) | ||
| P. equitans Mez | 2x | R | S ss + App | Quarin and Norrmann (1987) | |
| P. exaltatum J.Presl. | 4x | R | Ap | Burson and Bennett (1971) | P. arechavaletae Hack. ex Arechav. |
| P. falcatum Ness ex Steud. | 4x | mca | Ap | Burson (1997) | |
| P. fasciculatum Willd. ex Flüggé | 2x | R | S ss | Urbani (1996) | |
| P. glaucescens Hack. | 2x | R | S ss | Pritchard (1962) | P. yaguaronense Henrard |
| P. guenoarum Arechav. | 4x | mca | Ap | Bashaw et al. (1970); Pritchard (1970); Espinoza et al. (2001) | P. rojasii Hack. |
| Burson and Bennett (1971) | |||||
| P. hartwegianum E. Fourn. | – | – | Ap | Brown and Emery (1958) | |
| P. haumanii Parodi | 2x | R | S ss + App | Norrmann et al. (1989) | |
| 4x | mca | Ap | Burson (1975); Norrmann et al. (1989) | ||
| P. inaequivalve Raddi | 6x | R | S sf | Quarin and Burson (1991) | |
| P. indecorum Mez | 2x | R | S | Quarin and Burson (1983) | |
| P. intermedium | 2x | R | S ss + App | Burson and Bennett (1970); Norrmann et al. (1989) | |
| Munro ex Morong & Britton | 4x | mca | Ap | Norrmann et al. (1989) | |
| P. ionanthum Chase | 4x | R | S + App | Burson and Bennett (1970) | P. guaraniticum Parodi |
| – | R | S or S + App | Martínez et al. (1999) | P. guaraniticum Parodi | |
| 8x | mca | Ap | Burson and Bennett (1970) | ||
| P. jurgensii Hack. | 2x | R | S | Bashaw et al. (1970); Burson and Bennett (1971) | |
| P. langei (E. Fourn.) Nash | – | – | S | Brown and Emery (1958) | |
| P. laxum Lam. | 6x | R | S sf | Quarin et al. (1982) | |
| P. lenticolare Kunth | 2x | R | S ss | Espinoza et al. (2001) | P. limbatum Henrard |
| 4x | mca | Ap | Espinoza et al. (2001) | ||
| P. lividum Trin. | 4x | mca | Ap | Burson and Bennett (1971); Sartor et al. (2011) | |
| P. longifolium Roxb. | 4x | desy | Dp | Chao (1974) | |
| P. maculosum Trin. | 2x | R | S ss | Norrmann et al. (1989) | |
| 4x | mca | Ap | Norrmann et al. (1989) | ||
| P. malacophyllum Trin. | 2x | R | S ss + App | Hojsgaard et al. (2008) | |
| 4x | mca | S | Bennett and Bashaw (1966) | ||
| – | mca | Ap | Burson and Hussey (1998); Hojsgaard et al. (2008) | ||
| – | – | Ap | Brown and Emery (1958) | ||
| P. mandiocanum Trin. | 6x | R | Ap | Burson and Bennett (1971) | |
| P. minus E. Fourn. | 5x | asy | Dp + Ap | Bonilla and Quarin (1997) | |
| P. modestum Mez | 2x | R | S ss | Burson (1997); Quarin and Hanna (1980a) | |
| P. monostachyum Vasey | 2x | R | S ss | Burson (1997) | |
| – | – | S | Brown and Emery (1958) | ||
| P. nicorae Parodi | 4x | mca | Ap | Bashaw et al. (1970); Burson and Bennett (1970); Sartor et al. (2011) | |
| P. notatum Flüggé | 2x | R | S ss | Burton (1948); Burton (1955); Bashaw et al. (1970) | Pensacola bahiagrass |
| – | – | S + App | Quarin et al. (2001) | ||
| 3x | – | Ap | Quarin et al. (1989) | ||
| 4x | mca | Ap | Burton (1948); Bashaw et al. (1970) | ||
| P. palustre Mez | 2x | R | S ss | Quarin and Burson (1991) | |
| P. paniculatum L. | 2x | R | S | Burson and Bennett (1971) | |
| P. pauciciliatum (Parodi) Herter | 4x | I | Ap | Bennett and Bashaw (1966); Bashaw et al. (1970) | P. dilatatum var. pauciciliatum Parodi |
| P. paucifolium Swallen | 4x | mca | Ap | Burson (1997) | |
| P. plicatulum Michx. | 2x | R | S ss | Espinoza and Quarin (1997) | |
| 4x | mca | Ap | Bashaw et al. (1970); Burson and Bennett (1971); Norrmann et al. (1989) | ||
| P. polyphyllum Nees ex Trin. | 4x | mca | Ap | Burson (1997) | |
| P. procurrens Quarin | 2x | R | S ss | Quarin (1993) | |
| 4x | mca | Ap | Hojsgaard et al. (2008) | ||
| P. proliferum Arechav. | 4x | mca | Ap | Quarin et al. (1982) | |
| 6x | I | Ap | Burson (1975) | ||
| P. pubiflorum Rupr.ex E. Fourn. | 6x | R | S | Bashaw et al. (1970); Actkinson and Burson (1999) | P. pubiflorum var. Glabrum Vasey ex Scribn. |
| – | – | S | Brown and Emery (1958) | ||
| P. pumilum Nees | 2x | R | S sf | Burson and Bennett (1971) | |
| P. quadrifarium Lam. | 2x | R | S ss + App | Norrmann et al. (1989) | |
| 3x | mca or I | Ap | Bashaw et al. (1970); Norrmann et al. (1989) | ||
| 4x | mca | Ap | Quarin and Burson (1983); Norrmann et al. (1989) | ||
| P. quarinii Morrone & Zuloaga | 2x | R | Sss + App | Norrmann et al. (1989) | P. brunneum Mez |
| 4x | mca | Ap | Burson (1975); Norrmann et al. (1989) | P. brunneum Mez | |
| P. ramboi Barreto | 6x | R | Ap | Quarin and Burson (1991) | |
| P. regnelli Mez | 4x | R | S sf | Norrmann (1981) | |
| P. repens P.J. Bergius | 2x | R | S sf | (Burson, 1997) | |
| P. rufum Nees | 2x | R | S ss + App | Norrmann et al. (1989); Siena et al. (2008) | |
| 4x | mca | Ap | Burson (1975); Norrmann et al. (1989) | ||
| P. scrobiculatum L. | 4x | R | S | Bashaw et al. (1970); Pritchard (1970) | P. commersonii Lam. |
| – | R | S sf | Quarin and Hanna (1980a) | P. boscianum Flüggé | |
| 6x | asy | Dp + App | Chao (1974) | P. commersonii Lam. | |
| 10x | – | Dp | Ma et al. (2009) | P. commersonii Lam. | |
| 12x | R | S + App | Chao (1974) | P. commersonii Lam. | |
| P. secans Itchc. & Chase | 4x | asy | Ap | Snyder (1957) | |
| I | Ap | Bashaw et al. (1970) | |||
| P. setaceum Michx. | 2x | R | S sf | Banks (1964, 1966) | P. debile Michx. |
| P. ciliatifolium Michx. | |||||
| P. longepedunculatum LeConte | |||||
| P. propinquum Nash | |||||
| P. psammophilum Nash | |||||
| P. pubescens Muhl. | |||||
| – | – | S | Brown and Emery (1958) | P. rigidifolium Nash | |
| P. simplex Morong ex Britton | 2x | R | S ss | Espinoza and Quarin (1997) | |
| 3x | – | S | Urbani et al. (2002) | ||
| 3x | – | Ap | Urbani et al. (2002) | ||
| 4x | mca | Ap | Caponio and Quarin (1987) | ||
| 6x | – | – | Urbani et al. (2002) | ||
| P. thunbergii Kunth ex Steud. | 4x | I | Ap | Ma et al. (2004) | |
| P. umbrosum Trin. | 2x | R | S | Bashaw et al. (1970) | |
| P. unispicatum (Scribn. & Merr.) Nash | 4x | mca | Ap | Burson (1997) | |
| P. urvillei Steud. | 4x | R | S | Brown and Emery (1958); Bashaw et al. (1970) | |
| P.vaginatum Sw. | 2x | R | S | Bashaw et al. (1970) | |
| P. virgatum L. | 4x | R | S | Burson and Quarin (1982) | |
| P. wrightii Hitchc. & Chase | 2x | R | S ss | Martínez et al. (1999) | P. hydrophilum Henrard |
| 4x | mca | Ap | Norrmann (1981) | P. hydrophilum Henrard |
For practical reasons, the species of the newly recognized subgenus Harpostachys (formerly genus Thrasya) are not included in this review.
*R, regular (mainly bivalent pairing); I, irregular (bivalents plus one or two unpaired genomes), mca, multivalent chromosome associations (mainly presence of quadrivalents), asy or desy, asynapsis or desynapsis (majority of chromosomes unpaired).
bS, sexual; Dp, diplosporous apomictic; Ap, aposporous apomictic; App, aposporous potential (occasional ovules with an aposporous sac beside the sexual sac); ss, self-sterile; sf, self-fertile.
The two main Paspalum species used as models for apomixis research, P. notatum and P. simplex, form agamic complexes made up of diploid sexual and autopolyploid apomictic individuals. Interestingly for molecular approaches toward gene isolation, DNA content values for these two species are among the smallest within the Poaceae tribe: 1C = 0·58 pg for diploid P. notatum (Jarret et al., 1995) and 1C = 0·75 pg for diploid P. simplex (Cáceres et al., 1999).
Cytology and reproduction
As highlighted above, the grass genus Paspalum is characterized by an extremely versatile genetic system due to extensive variation in chromosome number, meiotic chromosome behaviour and reproductive mode. Based on taxonomical reviews, the main components of the genetic system have been examined for approx. 20 % of all Paspalum species (e.g. Chase, 1939; Zuloaga and Morrone, 2005; Williams et al., 2011). From the compilation data presented in Table 1, most species are polyploids (75 %), out of which some form multiploid complexes.
Sexuality in Paspalum species is typical for that of most angiosperms and is characterized by the double fertilization of a reduced ES of the Polygonum type, typically composed of the egg apparatus (egg cell and two synergids), a large two-nucleated central cell and a mass of proliferated antipodals at the chalazal end (Quarin, 1992). Apomictic reproduction is mainly of the aposporous type, according to which unreduced ESs differentiate from particular nucellar cells, so-called aposporous initials (AIs). Typically, three or the four spores of the legitimate MMC degenerate and several nucellar cells change their fate and differentiate into AIs (Fig. 2A, B). Aposporous ES development is achieved through an unstable pattern of cell division and cell differentiation. A first mitosis of the AI nucleus produces a binucleate ES whose further development varies according to the number of both divisions and nuclei involved. Consequently, mature ESs always contains an egg cell and a binucleate central cell, though one or two synergids may often be observed beside the egg cell; antipodal cells are always absent in this so-called Paspalum type of aposporous ES (Fig. 2C, D; Burson and Bennett, 1970a, 1971; Quarin et al., 1996; Espinoza et al., 2001; Ma et al., 2004).
Fig. 2.
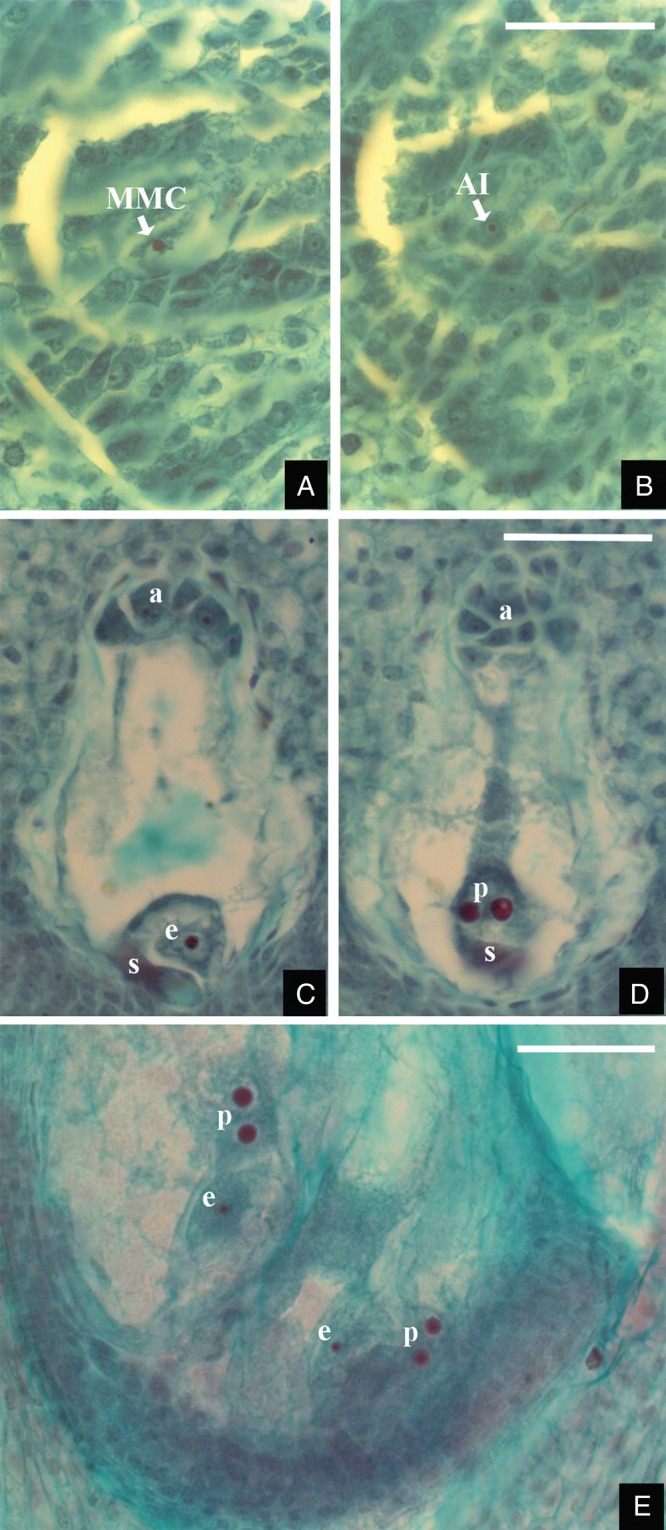
Photomicrographs of sectioned ovaries of sexual and apomictic Paspalum species. (A, B) Apomictic tetraploid cytotype of P. notatum. Two consecutive sections of a young ovary showing the megaspore mother cell (MMC) and one nucellar aposporous initial cell (AI). (C, D) Sexual diploid cytotype of P. cromyorrhizon. Two consecutive sections of a mature ovary showing a well developed sexual embryo sac bearing the egg cell (e), two synergids (s), a large central cell with two polar nuclei (p) and a mass of proliferated antipodal cells (a) at the chalazal end of the embryo sac. (E) Apomictic tetraploid cytotype of P. cromyorrhizon. Section of a mature ovule showing two aposporous embryo sacs, each containing an egg cell (e) and two polar nuclei (p). Scale bars: (A, B) = 20 µm, (C–E) = 50 µm.
Apart from the aposporous Paspalum type, the Taraxacum type of diplospory was reported for two Asian species, P. commersonii Lam. (=P. scrobiculatum L.; 2n = 6x) and P. longifolium Roxb. (2n = 4x), and for the pantropical species P. conjugatum Berg. (2n = 4x) (Chao, 1974, 1980). Hieracium-type ES development has been described in only two species of Paspalum, P. secans Hitchc. & Chase (Snyder, 1957) and P. simplex Morong (Caponio and Quarin, 1987; Cáceres et al., 2001), while both aposporous and diplosporous types were detected in P. minus E. Fourn. (Bonilla and Quarin, 1997) and P. scrobiculatum L. (Chao, 1974).
Microsporogenesis consists of the meiotic division of the male archespore that gives rise to an array of four cells (tetrad) each containing a nucleus with a reduced chromosome number. Then the nucleus of each microspore moves to the side wall before mitosis I starts, giving rise to the microgametogenesis process. This mitosis involves an unequal cell division, producing a large vegetative cell and a small generative cell having a nucleus with condensed chromatin structure. The generative cell divides into two sperm cells via mitosis II. The mature male gametophyte consists of a tricellular pollen with two sperm cells, plus a vegetative cell. It is now widely accepted that apomixis in Paspalum is always associated with irregular male meiosis in the forms of multivalent chromosome associations (mainly the presence of quadrivalents), asynapsis or desynapsis, the whole genome lasting unpaired, or appearance of chromosome bridges and micronuclei (Quarin, 1992; Table 1). Comparative cytogenetic examinations during microsporogenesis revealed meiotic abnormalities at anaphase I in P. notatum apomicts, which were attributed to genetic rearrangements, such as an inversion or a translocation in one chromosome (Stein et al., 2004). Conversely, male meiosis in natural sexual cytotypes is always regular (formation of bivalent chromosome associations and balanced gametes), with some rare cases of quadrivalent formation in sexual P. malacophyllum and P. conspersum (Table1). Colchicine-induced sexual tetraploids of P. notatum and their derivatives showed high rates of meiotic abnormalitites, though their proportions were significantly lower when compared with those detected in natural conspecific apomictic genotypes (Podio et al., 2012b). Frequent formation of 2n pollen as a consequence of abnormal cytokinesis followed by nuclear fusion of multinucleate microspores has been reported in polyploid Brazilian accessions of Paspalum by Pagliarini et al. (1999). Although there is no evidence of correlation between abnormal cytokinesis and apomixis, the occurrence of restitution nuclei as a consequence of irregular or arrested meiosis has been reported in connection with apomixis in P. secans (Snyder, 1961), P. conjugatum (Chao, 1980) and P. minus (Bonilla and Quarin, 1997). To sum up, experimental evidence supports a correlation between the occurrence of meiotic abnormalities and apomixis in Paspalum. These abnormalities might be related to the rearranged nature of the chromosome bearing the apomixis locus.
Endosperm development in angiosperms requires an exact maternal-to-paternal (2m:1p) genomic balance, and any deviation from it usually results in seed abortion, although this is not inevitable and depends on the genetic context (Schatlowski and Köhler, 2012, and references therein). All Paspalum apomicts are pseudogamous, meaning that the endosperm develops after fertilization of the polar nuclei by a reduced male gamete. In P. notatum, seed development in apomicts is insensitive to dosage effects in the endosperm in spite of the strong, maternal genomic excess, whereas in the sexual biotypes the 2m:1p balance is strictly required for normal endosperm development and seed production (Quarin, 1999).
The number and fertilization of the polar nuclei in aposporous ESs is also of practical importance in apomixis research, particularly when the method of reproduction must be determined for a large number of individuals. The development of the flow cytometric seed screen (FCSS) method (Matzk et al., 2000) has facilitated identification of the reproductive mode for large sample numbers because in Paspalum the relative ratio of embryo:endosperm DNA content distinguishes seeds of apomictic origin from those formed sexually. A sexually produced seed is formed by an embryo (n + n) which has arisen from fertilization of the reduced egg cell (n) by a reduced sperm nucleus (n), and endosperm derived from fertilization of two reduced polar nuclei by a reduced sperm nucleus [(n + n) + n]. Therefore, this seed has a 2:3 embryo:endosperm ratio of DNA content. On the other hand, a seed formed through apospory, parthenogenesis (embryo from 2n + 0) and pseudogamy [endosperm from (2n + 2n) + n] has a 2:5 embryo:endosperm DNA ratio. The FCSS technique applied to individual or bulked seeds has greatly facilitated the analyses of the reproductive mode in Paspalum species in the last decade (Cáceres et al., 2001; Siena et al., 2008; Sartor et al., 2009, 2011; Aguilera et al., 2011; Rebozzio et al., 2011; Hojsgaard et al., 2013).
The genetic systems observed in Paspalum have been categorized into eight different groups (Table 2). Most species (approx. 70 %) belong to groups 2, 3 and 4. Group 2 comprises sexual self-sterile (outbreeder) diploids. Group 3 consists of multiploids, whose chromosome races include both diploid outbreeders and apomictic polyploids. Group 4 is formed by aposporous apomictic polyploids with usually multivalent chromosome associations at meiosis, pseudogamy and self-fertility. Because most studies have considered only one or a few individuals for each species, the published data suggest that most species in groups 2 and 4 might actually be members of group 3. Once the genetic system is screened for more individuals per natural population, diploid cytotypes might be discovered among apomictic polyploids (group 4), and apomictic polyploids might be found among sexual self-sterile diploids (group 2).
Table 2.
Summarized genetic system for 72 species of Paspalum
| Group no. | Genetic system* | No. of species |
|---|---|---|
| 1 | Diploid, regular meiosis, sexual and self-fertile. | 6 |
| 2 | Diploid, regular meiosis, sexual and self-sterile. Apospory potential observed in two species. | 12 |
| 3 | Multiploid (diploid and polyploid cytotypes): diploids with regular meiosis, sexual and self-sterile, though apospory potential was observed in eight diploid cytotypes; polyploids (mainly 4x) with usually multivalent chromosome associations at meiosis, aposporous apomictic, pseudogamous and self-fertile. | 19 |
| 4 | Polyploid (mainly 4x) usually with multivalent chromosome associations at meiosis suggesting autoploidy, exceptionally with unpaired chromosomes (alloploidy), aposporous apomictic, pseudogamous and self-fertile. | 19 |
| 5 | Polyploid (mainly 4x and some 6x), bivalent chromosome associations at meiosis indicating alloploidy, sexual reproduction, self-fertile. | 9 |
| 6 | Multiploid of alloploid origin (sexual 4x plus higher polyploid aposporous cytotypes: 5x, 6x or 8x), tetraploids with regular chromosome pairing, higher polyploids with regular or irregular meiosis. P. dilatatum, P. durifolium, P. ionanthum. | 3 |
| 7 | Polyploid, asynaptic or desynaptic chromosome behaviour at meiosis, restitution nucleus, diplosporous apomictic. P. conjugatum, P. longifolium, P. minus (diplospory + apospory). | 3 |
| 8 | Sexual tetraploid and higher polyploid cytotypes with diplosporous apomixis and some potential for apospory. P. scrobiculatum. | 1 |
| Total | 72 |
*Genetic systems are reported as described in the available literature. When information on breeding system (self-fertility or self-sterility) was missing in the references, personal data (C. L. Quarin, unpubl.) were added.
Variability in natural populations
Apomixis was first considered as a blind alley for evolution (Darlington, 1939), suggesting that clonal seed production would result in genetically uniform populations. However, studies in natural populations of apomictic Paspalum spp. revealed a high level of variation in ploidy and genetic structure (Urbani et al., 2002; Daurelio et al., 2004; Sartor et al., 2011). Chromosome counts and ploidy level estimates by flow cytometry from 32 populations of P. simplex showed that most individuals were tetraploid and that diploid populations were confined to a small area (Urbani et al., 2002). On the other hand, Sartor et al. (2011) analysed the ploidy levels and reproductive mode of 19 populations from five species and found that diploid populations reproduced sexually, while polyploids (2n = 3x, 4x, 5x and 6x) reproduced by apomixis. Interestingly, apomixis in 4x individuals was facultative (i.e. they produced some of their progeny by sexual means), while other polyploid individuals were obligate apomicts. Finally, analysis of facultative apomicts revealed variations in the degree of facultativeness (2–30 %) as well as in the origin of non-maternal progeny (via sexual reproduction or fertilization of unreduced egg cells) (Cáceres et al., 2001; Urbani et al., 2002). Daurelio et al. (2004) showed that variability in tetraploid P. notatum was significantly higher in sympatric diploid–tetraploid populations than in those tetraploid populations isolated from diploids. Similarly, Sartor et al. (2011) observed higher levels of genetic variability in mixed ploidy populations of P. rufum, P. denticulatum and P. unispicatum than in pure populations. These results support the hypothesis of recurrent polyploidization for the majority of Paspalum species proposed by Quarin (1992) (see ‘Apomixis, hybridity and polyploidy’ below for details) according to which new apomictic tetraploid genotypes would be continuously generated, thereby increasing the genetic variability of apomictic populations.
Agronomy and genetic improvement
From a plant breeding perspective, apomixis provides a unique mechanism for developing superior cultivars and preserving those genotypes indefinitely. Generally speaking, the three fundamental prerequisites of any successful plant breeding programme are: (1) availability of a diverse germplasm collection; (2) adequate knowledge of the biology, cytology and reproductive system of the available material; and (3) explicit and achievable objectives. In other sections of this review, we clearly demonstrate that Paspalum meets the first two prerequisites; here we discuss objectives and breeding strategies related to apomictic reproduction.
Among the species of Paspalum, P. notatum and P. dilatatum are the most widely cultivated forage grasses. Specific objectives for Paspalum breeding consist of the enhancement of: (1) cold tolerance and cool-season growth; (2) seed yield; (3) grazing resistance; (4) nutritive value; and (5) resistance to biotic stresses. Because most Paspalum species reproduce asexually by apomixis, specific breeding techniques must be used to enhance their genetics. These techniques are all based on fixing superior genotypes via apomixis. Ecotype selection is the oldest and the most productive breeding approach. This technique involves germplasm collection evaluation, selection, multiplication of the best ecotypes, and release of superior genotypes as new apomictic cultivars, i.e. seed-propagated clones. The success of ecotype selection depends on the number of polymorphic ecotypes within a species (Vogel and Burson, 2004). As an example, the tetraploid ‘Argentine’ (PI 148996) cultivar, released in the mid-20th century as the result of evaluating approx. 80 accessions of P. notatum, is still sown for pasture and utility turf (Blount and Acuña, 2009). Cultivars of P. dilatatum, which is better adapted to temperate areas than P. notatum, have been selected as natural variants from the apomictic pentaploid and hexaploid cytotypes (Evers and Burson, 2004; Burson et al., 2008). Of particular interest is the Plicatula group, a diverse assemblage that includes many forage types. Several cultivars have been released from this group belonging to P. atratum, P. guenoarum and P. plicatulum (Evers and Burson, 2004).
Hybridization has been used more recently to enrich cultivars for specific traits of interest. The success of hybridization in Paspalum breeding depends on the availability of sexual polyploid cytotypes. Improving apomictic Paspalum via hybridization began with the pioneering work of Dr G. W. Burton (USDA-ARS, Tifton, GA, USA) and his colleagues who generated colchicine-induced sexual tetraploid plants (Burton and Forbes, 1960) and hybridized these induced autotetraploids with naturally occurring tetraploid ecotypes to obtain many apomictic hybrids that were never released as cultivars (Burton, 1992). Twenty out of several hundreds induced hybrids created by tissue culture were selected for a breeding programme for resilience to clipping (Quesenberry et al., 2010), and a few others, characterized as highly sexual and self-incompatible, were crossed with highly productive apomictic genotypes (Acuña et al., 2007). After two cycles of hybridization, a high degree of variability and heterosis was observed (Acuña et al., 2007, 2009, 2011). Superior apomictic hybrids are currently being evaluated in different environments and are expected to result in new apomictic forage cultivars. One of the original sexual tetraploid hybrids was hybridized with an Argentinean local ecotype to yield several hybrids, from which the cultivar named ‘Boyero UNNE’ was selected and released in 2012 as the first apomictic cultivar of Paspalum developed by hybridization. In addition, two sexual clones (i.e. they generate their progeny exclusively sexually) have been released and are currently available (Quarin et al., 2003). Completely sexual tetraploid genotypes of P. simplex (Cáceres et al., 1999) and P. plicatulum Michx. (Sartor et al., 2009) have also been generated. These plants are used as female parents to enhance variability through both intra- and interspecific hybridization. Indeed, fertile hybrids can be generated by crossing the induced sexual tetraploid of P. simplex with natural tetraploid ecotypes of the same species, as well as with P. malacophyllum and P. procurrens (Pupilli et al., 2004; Hojsgaard et al., 2011, respectively). Hybrid genotypes characterized by high forage production have been obtained using this approach. Fertile hybrids have also been obtained by crossing a sexual tetraploid genotype of P. plicatulum with conspecific apomicts or with apomicts of P. guenoarum Arechav. (Aguilera et al., 2011). Increasing the low number of apomictic hybrids produced in crosses, predicting the occurrence of heterosis and selecting for highly self-incompatible hybrids would dramatically enhance the efficiency and usefulness of this breeding approach. In particular, the loss of self-incompatibility in hybrids could induce inbreeding depression and limit the yield of hybrids in crosses. Although most diploid and induced tetraploid plants of P. notatum are self-incompatible (Burton, 1955; Acuña et al., 2007), this characteristic is absent in apomictic plants (Acuña et al., 2007). Thus, modulation of self-incompatibility in breeding programmes should be further investigated.
Ionizing radiation was used to breed early introductions of P. dilatatum in the USA when breeders failed to generate variability through hybridization (Evers and Burson, 2004). Most of the resulting plants were aberrant; only a few exhibited good agronomic characteristics and none was ever released. Genetic transformation has also been used to improve Paspalum species as forage or turf. The biolistic method was used to obtain transgenic plants of diploid and tetraploid P. notatum (Altpeter and James, 2005; Gondo et al., 2005). Glufosinate-resistant plants of P. notatum were obtained by transforming plants of the cultivar ‘Argentine’ with the bar gene (Sandhu et al., 2007). These genetically modified plants proved to be highly resistant to this herbicide under field conditions, enhancing their competitive ability against weeds during pasture establishment. The transcription factor gene Hs-DREB1A from xeric Hordeum spontaneum was also introduced into ‘Argentine bahiagrass’ to enhance water stress tolerance (James et al., 2008). In addition, to increase turf quality, the endogenous gibberellin-catabolizing gene At-GA2ox1 was inserted into the genome of ‘Argentine bahiagrass’ (Agharkar et al., 2007). Transformed plants exhibited higher turf density, shorter tillers and delayed flowering. However, none of these new genetically modified plants has been released. Although Sandhu et al. (2010) showed that both apomixis and polyploidy are major barriers to pollen-mediated transgene flow from transformed bahiagrass to wild types, the public remains concerned about the safety of transgenic plants.
In summary, the coexistence of apomixis and sexuality in Paspalum species is a great advantage for breeding. A wide variety of traits of interest occur in existing germplasm collections or can be induced by traditional or biotechnological tools. These traits can be rapidly fixed in superior hybrid strains by apomixis.
INHERITANCE OF APOMIXIS IN PASPALUM SPECIES
Genetic analysis
Apomixis is a heritable reproductive system thought to have evolved through a rearrangement of the developmental programmes that constitute the normal sexual pathway (Grimanelli et al., 2001). Nogler (1984) suggested that the basic determinants of apomixis could have originated by mutation and that most of the genes involved in the process would probably be similar to those implicated in sexual reproduction. Genetic studies on the inheritance of apomixis in Paspalum spp. were performed mainly on P. notatum Flüggé and P. simplex Morong, two genetically distant species comprising sexual self-incompatible diploids and nearly obligate tetraploid apomicts. Genetic analysis of apomixis in these species is impossible unless sexual tetraploid germplasm is available. The production of artificial sexual tetraploid individuals (Quarin et al., 1984, 2001, 2003; Cáceres et al., 1999; Quesenberry et al., 2010) allowed the generation of populations segregating for the mode of reproduction, without the need for interspecific or interploid crosses.
A pioneering study on the genetic control of apomixis in Paspalum was carried out by Burton and Forbes (1960). Using progeny testing for morphological traits in P. notatum segregating populations derived from an experimentally obtained sexual female progenitor and a natural apomictic pollen donor, they proposed that apomixis was controlled by a few recessive genes. Several subsequent studies have concentrated on the inheritance of apospory i.e. the capacity to develop non-reduced ESs. Segregation analysis of apospory in F1 progeny from an interspecific cross P. ionanthum (tetraploid sexual) × P. cromyorrhizon (facultative tetraploid apomict) revealed a 3:1 aposporous:non-aposporous ratio (Martínez et al., 1999). Two models for the genetic control of the trait were proposed, but these could not be corroborated because most hybrids were male sterile.
The use of intraspecific crosses between a completely sexual tetraploid female genotype and natural apomictic progenitors in P. notatum allowed investigation of the reproductive modes across several generations (Martínez et al., 2001). Segregation ratios of 1:2.8 to 1:3 aposporous vs. sexual progeny led the authors to propose that a single tetrasomically inherited dominant allele with a pleiotropic lethal effect and incomplete penetrance controls apospory development. An excess of sexual progeny, which deviated from the expected Mendelian ratios of 1:1 or 13:15 (assuming random assortment of chromosomes or chromatids, respectively), was repeatedly observed in segregating populations of P. notatum (Stein et al., 2004; Acuña et al., 2009, 2011), P. simplex (Pupilli et al., 2001), P. malacophyllum (Pupilli et al., 2004), P. procurrens (Hojsgaard et al., 2011) and P. plicatulum (Aguilera et al., 2011). Similar distortions in favour of sexual individuals have been observed in segregating populations of several apomictic grasses (Ozias-Akins and van Dijk, 2007). The most common hypothesis to explain the low transmission rate of apomixis in segregating populations is the presence of a lethal allele linked to the apomixis locus acting at either the gametophytic or sporophytic level. Nogler (1982) postulated the existence of a dominant apospory factor that acts as a recessive lethal allele. This apospory factor could not be transmitted through monoploid gametes, explaining the absence of natural diploid apomictic plants. This hypothesis was partially confirmed in P. notatum and P. simplex where, in intraspecific crosses involving sexual diploids and tetraploids as pistillate parents and apomict triploids as pollen donors, apospory could only be transmitted by pollen through diploid or hypodiploid gametes (Martínez et al., 2007). Several pieces of experimental evidence confirmed that genetic rearrangements and meiotic abnormalities were associated with apospory in P. notatum (Pupilli et al., 2004; Stein et al., 2004; Podio et al., 2012b). The presence of an inversion or translocation at the apomixis locus could explain both the distorted segregation ratio of apospory, via differential survival of meiocytes carrying the rearranged locus, and the observed suppression of recombination near that locus (see below). In addition, meiotic drive, a mechanism that allows one of the allelic alternatives to be transmitted in excess to the progeny (Lyttle, 1991), was proposed as a cause of apomixis segregation distortion in maize–Tripsacum dactyloides hybrids (Grimanelli et al., 1998) and pearl millet–Pennisetum squamulatum hybrids (Roche et al., 2001). An explanation for the preferential transmission of sexuality in segregating populations of Hieracium invoked the presence of post-meiotic factors favouring the development of sexual embryos (Bicknell et al., 2000). Polegri et al. (2010) reported that, of nearly 200 genes differentially expressed between apomictic and sexual lines of P. simplex, only 10 % were genetically associated with apomixis (discussed below). Consequently, transferring the apomixis locus from an apomictic ‘donor’ to a sexual ‘receiver’ genotype (the parental lines of a segregating population) would reprogramme the expression of a group of genes that presumably act downstream of apomixis-linked factors. This reprogramming could affect a delicate network of gene–gene communication probably based on homology. If even a few of these interactions do not work properly, the apomictic zygotes could be lethal or disadvantaged relative to sexual zygotes. These interactions would fail more frequently as genetic distance between the parents increased, because the sexual ‘receiver’ genotype might be unable to adapt to a new apomictic condition. Therefore, zygotic lethality (or its coexistence with male gametophytic lethality) could explain the low transmission rate of apomixis in Paspalum and the striking differences in its segregation distortion between interspecific and intraspecific crosses in P. simplex.
Apomixis, hybridity and polyploidy
Hybridization and polyploidization represent two important processes in the evolution of angiosperms. Both mechanisms were investigated with regard to their role in the emergence of apomixis from sexuality (Carman, 1997; Ozias-Akins and van Dijk 2007; Pupilli and Barcaccia, 2012). In Paspalum, as in many other agamic complexes, the sexual–diploid/apomictic–polyploid conditions seem to constitute a common genetic system for a large number of species (Quarin, 1992). However, the relative contributions of these processes as yet remain unclear. Ernst (1918) maintained that all apomicts are of hybrid origin, and Stebbins (1941) added that the great majority of apomicts are probably allopolyploids of hybrid origin. Nogler (1984) in his comprehensive review of apomixis in plants also considered alloploidy and hybridity essential for the occurrence of apomixis. More recently, these concepts have been reformulated by Carman (1997) as the ‘hybridization theory’. This theory suggests that hybridization of species with dissimilar ecological affinities and reproductive developmental programme timing contributes to the induction of apomixis. Thus, asynchronous expression of the two parental gene sets could lead to the aberrant initiation of embryological processes during ovule development, causing a shift from sexual to apomictic reproduction. Cytogenetic studies and breeding behaviour analyses of several species and of interspecific hybrids, as well as the segregation analysis in apomictic tetraploid species and induction of artificial apomictic tetraploids from sexual diploids, partially support these views. On one hand, polyploidy seems to be a prerequisite for the expression of apomixis (Quarin and Hanna, 1980a; Quarin et al., 1998, 2001). On the other hand, cytogenetic studies in apomictic polyploids have suggested an autoploid rather than an alloploid origin of most Paspalum apomicts (Bennett and Bashaw, 1966; Norrmann et al., 1989; Pupilli et al., 1997; Stein et al., 2004; Hojsgaard et al., 2008). In addition, artificial autopolyploidization of sexual diploids in several Paspalum species triggered and maintained apomictic reproduction (Quarin and Hanna, 1980a; Quarin et al., 1998, 2001). Nevertheless, there are a few examples of apomictic allopolyploid species: (1) 5x and 6x dallisgrass, P. dilatatum Poir., derived from hybridization between 4x dallisgrass apomicts and 2x P. urvillei (Bashaw and Forbes, 1958; Bashaw and Holt, 1958; Burson et al., 1991); and (2) the tetraploid P. dasypleurum Kunze & Desv. (the Paspalum species with the southernmost distribution in South America), another P. dilatatum sexual relative (Quarin and Caponio, 1995). These apomictic 5x and 6x cytotypes of P. dilatatum and their tetraploid relatives constitute the Dilatata group of Paspalum. They share two basic genomes: the genome I of sexual self-sterile diploid P. intermedium Munro ex Morong & Britton and the genome J, which belongs to P. jurgensii Hack, a sexual self-fertile diploid species (Burson, 1991, 1992). An allopolyploid origin of the apomictic pentaploid dallisgrass (the common biotype) was proposed by both Burson (1992) and Speranza (2009), but their interpretations of the evolutionary patterns are different. Burson (1991, 1992) proposed that the common biotype (IIJJX) originated by natural hybridization between a sexual tetraploid cytotype (genome formula IIJJ) and an apomictic hexaploid form (genome formula IIJJXX). Speranza (2009) considered that the pentaploid cytotype was probably the first apomictic form of the group that can produce new IIJJX pentaploids through the formation of euploid IJX male gametes and their fusion with egg cells derived from sexual IIJJ tetraploids. Regardless of how the common apomictic pentaploid cytotype of P. dilatatum evolved, it clearly had an allopolyploid origin, and the control of apomixis is in the non-recombining X genome. The apomictic common type of dallisgrass is one of the first and most widely investigated species of the genus and could be regarded erroneously as a paradigm for apomixis research in Paspalum. The fact is that most apomictic Paspalum entities belong to multiploid species of autoploid origin. Each multiploid contains a sexual self-sterile diploid cytotype and a series of aposporous apomictic autopolyploid cytotypes, usually from 3x to 6x, with tetraploids as the most common cytotype (Table 2). Autopolyploidy may evolve stepwise through fertilization of occasional aposporous ESs which have arisen in diploids beside the normal meiotic sac (Quarin et al., 1982, 2001; Norrmann et al., 1989). In this way, diploids could give rise to triploids by 2n + n fertilization (2x + x = 3x). New tetraploids could be established in the same way from rare apomictic triploids and sympatric diploids (2n + n; 3x + x = 4x) or via fertilization of unreduced gametes from diploids by reduced gametes of naturally occurring tetraploids (n = 2x), i.e. 2x + 2x = 4x (Quarin, 1992; Siena et al., 2008). Whether the tetraploid cytotype or the entire series of polyploid cytotypes in multiploid species have autoploid origins may be questionable. Segmental allopolyploidy has been proposed for several species (groups 3 and 4 in Table 2), but the classification as autopolyploids or segmental allopolyploids is uncertain for several species (e.g. Burson and Bennett, 1970b, 1971; Quarin and Burson, 1991).
Although there is a strong link between apomixis and polyploidy, a few cases of gametophytic apomixis have been described at the diploid level. Ovules bearing both an aposporous and a meiotic ES were sporadically observed in several diploid species of Paspalum (Quarin and Norrmann, 1987; Norrmann et al., 1989; Quarin et al., 2001; Hojsgaard et al., 2008). These observations suggested the potential for apomictic reproduction at the diploid level, although evidence of parthenogenesis from those rare aposporous sacs was lacking. Recently, Siena et al. (2008) showed that a diploid plant of P. rufum, when exposed simultaneously to its own reduced haploid pollen (n = x) and mentor reduced pollen (n = 2x) from a P. urvillei tetraploid strain, produced some diploids and polyploid descendants by apomixis. These data indicate that the factor(s) responsible for apomixis are effectively expressed in diploid plants, but at very low rates. In addition, polyploidization might also lead to the normal expression of apomixis, as occurred when new tetraploids were induced by colchicine from sexual diploid plants in P. hexastachyum (= P. almum), P. rufum and P. notatum (Quarin and Hanna, 1980b; Quarin et al., 1998, 2001). Why apomixis is poorly expressed or silent in diploids is still unclear. This is a critical issue regarding the use of apomixis in diploid crops.
MOLECULAR DISSECTION OF THE APOMIXIS-CONTROLLING REGION
Molecular markers linked to apomixis and comparative mapping analyses
Apomictic reproduction in Paspalum is controlled by a single dominant locus that, when present, confers nearly 100 % apospory, a variable degree of parthenogenesis and full capacity to form endosperm with 4:1 maternal:paternal genome ratios (Cáceres et al., 2001; Martínez et al., 2001, 2003; Pupilli et al., 2004; Stein et al., 2004). These three apomixis components are probably inherited as a linkage block, because no recombination event has been documented to date. Molecular mapping of apomixis in Paspalum led to three main findings: (1) validation of segregation distortion and lack of genetic recombination around the apomixis locus; (2) establishment of syntenic relationships between apomixis-related markers and the rice map; and (3) narrowing of the chromosome region containing the apomixis locus by interspecific comparative mapping.
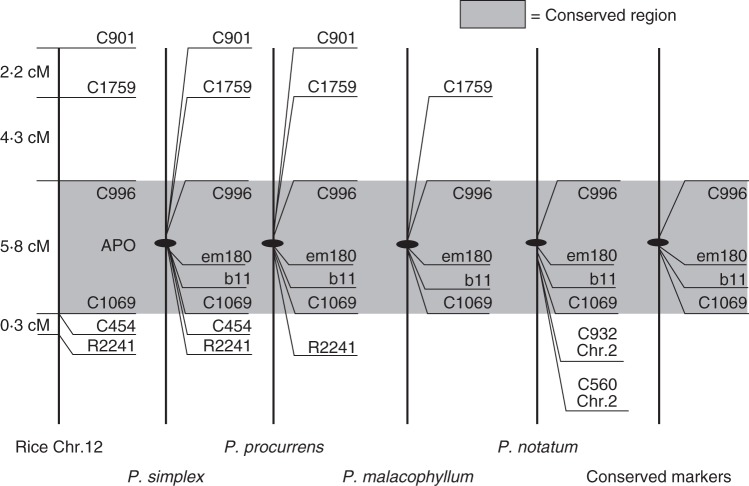
Using heterologous probes, Pupilli et al. (2001) identified a set of markers that, although they spanned 15 cM apart in a distal region of the long arm of rice chromosome 12, strictly co-segregated with apomixis in P. simplex. Similar results were obtained in P. notatum, but synteny was detected for rice chromosomes 12 and 2 as well as for maize chromosomes 3 and 5 (Martínez et al., 2003; Pupilli et al., 2004; Podio et al., 2012a; Fig. 3). Moreover, various random molecular markers [randomly amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP)] completely linked to the apospory locus were detected in P. simplex (Labombarda et al., 2002) and P. notatum (Martínez et al., 2003; Pupilli et al., 2004; Stein et al., 2004, 2007). Overall, comparative molecular analyses and cytological studies revealed that the apomixis-controlling region (ACR) in both P. simplex and P. notatum species appears to be located in a chromosome region where genetic recombination is suppressed. This structure seems to be highly conserved across apomictic races of P. notatum (Rebozzio et al., 2012).
Fig. 3.
Conservation of apomixis-linked markers in four Paspalum species in relation to their position on the homoeologous rice chromosome counterpart. The markers included in the highlighted area are conservatively linked to apomixis in the four Paspalum species considered.
Lack of recombination at the apomixis locus has been observed in several apomictically reproducing plants (Ozias-Akins and van Dijk, 2007), and this fact was attributed to its hypothetical location in a heterochromatic pericentromeric position (Ozias-Akins et al., 1998). However, syntenic relationships with rice (Pupilli et al., 2001) and fluorescence in situ hybridization (FISH) analysis in P. simplex (Calderini et al., 2006) suggested that the ACR is located in a non-pericentromeric and heterochromatin-poor region where genes are transcriptionally active (Polegri et al., 2010). An alternative hypothesis which posits that recombination suppression at the ACR is caused by a DNA rearrangement immediately after (or as a consequence of) polyploidization was supported by evidence obtained from P. simplex and P. notatum (Pupilli et al., 2001; Urbani et al., 2002; Stein et al., 2004; Podio et al., 2012b). Moreover, loss of pairing after local chromosome rearrangement was also reported in other apomictic species such as Pennisetum squamulatum (Ozias-Akins et al., 1998) and Cenchrus ciliaris (Goel et al., 2003). However, the non-recombining ACR of Paspalum appears relatively modest in size if compared with other models (Roche et al., 2002) as single apomixis-linked bacterial artificial chromosome (BAC) clones were identified by multiple markers that were independently developed (Calderini et al., 2011).
Fine structure of the ACR inferred from sequencing of an apomixis-related BAC and of molecular markers completely linked to the trait
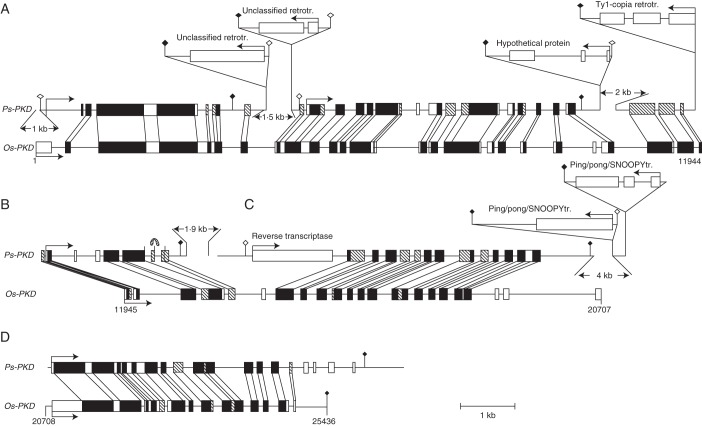
The apomixis-linked BAC clone 346H10, isolated from a genomic BAC library of apomictic P. simplex, was shotgun-sequenced at 10× coverage (Calderini et al., 2006, 2011). Annotation of the 129 046 bp revealed approx. 10 % non-coding sequences, 13 sequences related to transposons (i.e. ping/pong/SNOOPY, En/Spm and mariner sub-classes) and retrotransposons (i.e. ty3-gypsy and ty1-copia sub-classes) and four putative genes matching at high similarity (e-values ≤−34) with known genes. Of these genes, two that co-segregated with apomixis in several Paspalum species were considered as good candidates for apomixis (Calderini et al., 2006). Functional annotation showed that they encode a protein with significant homology with a protein kinase domain (Ps-PKD) and a protein of the ERD1/XPR1/SYG1 family (Ps-EXS). Detailed comparative analysis of Ps-PKD and its rice homologue is shown in Fig. 4. Both large- and small-scale rearrangements occurred in the structures of Ps-PKD compared with its rice homologue, which was assumed to represent the sexual counterpart of the apomixis-linked alleles of Paspalum. Large-scale rearrangements mainly due to insertions of transposable elements (TEs), probably resulting in aberrant transcription patterns (i.e. multiple independent transcriptional units or long chimeric mRNAs), were observed. Small-scale rearrangements included a 110 bp duplication, frequent small deletions, and occasional point mutations creating premature stop codons (Fig. 4). The mRNA resulting from transcription of this gene is probably unable to be translated into a protein. A similar gene structure including deletion of some exons compared with the homologous rice sequence and loss of coding capacity was detected in the second candidate sequence Ps-EXS (Calderini et al., 2006).
Fig. 4.
Microcollinearity of the Ps-PKD gene of P. simplex with its rice homologue Os-PKD. (A–D) Four non-overlapping contigs. Homologous areas are connected by vertical lines, arrows indicate the gene orientation and position of the first exon, black and white boxes indicate homologous coding and non-homologous coding regions, respectively, while dashed boxes indicate homologous non-coding regions. Open and filled rhombuses indicate putative initiations of transcription and polyadenylation sites, respectively. The curved arrow in (B) indicates a 110 bp duplication (the figure is reproduced from Calderini et al., 2006, with permission).
In P. notatum, characterization of the ACR by sequencing of a group of molecular markers completely linked to apospory revealed the presence of both low and high copy number sequences including Ty1-copia retroelements (Podio et al., 2012a). Interestingly, one sequence (Pn-GSA3) obtained by chromosome walking from one marker mapped in the ACR showed high similarity with maize and rice loci encoding MT-A70-like (mRNA N6-adenosine-methyltransferase) family proteins. Functional roles of these candidates in apomictic reproduction are discussed in the section ‘Candidate and downstream genes identified by large-scale sequencing analysis’.
COMPARATIVE TRANSCRIPTOMICS
Transcriptomic landscapes: towards the identification of candidates
The characterization of the Paspalum ACR in P. notatum and P. simplex revealed a strong repression of recombination and, probably as a consequence, accumulation of repetitive elements and non-coding DNA disrupting map collinearity with the homologous region on rice chromosome 12 (Calderini et al., 2006). Although several candidates were identified, the difficulties in the assembly of a considerable amount of sequences composed mainly of repetitive elements prompted groups researching apomixis in Paspalum to adopt a two-step approach to enhance the probabilities of identifying the genetic determinants of the trait. This is based on: (1) the identification of differentially expressed transcripts in reproductive organs of apomictic and sexual plants; and (2) mapping the differentially expressed transcripts, to restrict the number of candidates to the ACR-linked genes. Comparative transcriptomic analysis of apomixis in Paspalum faced several major drawbacks common in apomictic systems: (a) the lack of genuine near-isogenic apomictic and sexual lines; (b) the fact that apomictic species are highly heterozygous, thus complicating data interpretation; and (c) the lack of microarray reference systems allowing high-throughput analyses of gene expression. However, mRNA profiling assays based on differential display and cDNA-AFLP were carried out to overcome these difficulties and generate new data for describing transcriptomic landscapes in reproductive tissues of sexual and apomictic Paspalum spp.
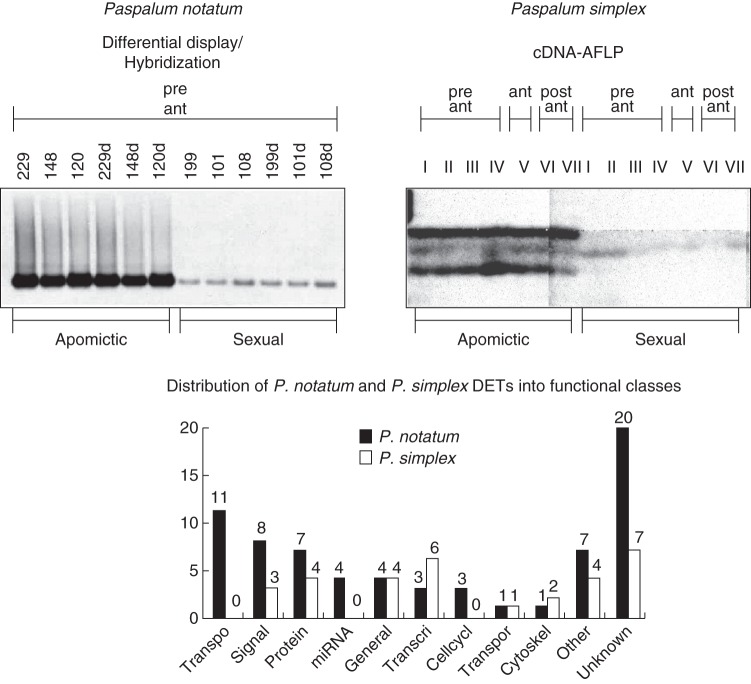
In P. notatum, differential display was first carried out to explore differences between mRNA bulks obtained from immature inflorescences of sexual and apomictic plants. This approach led to the identification of a transcript annotated as containing a KPS multiphosphorylation domain previously detected in several cdc2-regulated cytoskeletal proteins and highly expressed during early megagametophyte development in apomictic plants (Pessino et al., 2001). This approach was further completed by a more comprehensive analysis taking advantage of the reproductive calendar proposed by Laspina et al. (2008) according to which mRNA was extracted from spikelets at stage I, i.e. immediately prior to AI development. Differential display experiments allowed the identification of 65 DETs (differentially expressed tags) selected as expressed in only one of the plants (apomictic or sexual; Laspina et al., 2008). Further characterization showed that 45 DETs were protein-coding fragments, while the remainder were homologous to retroelements and putative microRNA precursors (Laspina et al., 2008; Ochogavía et al., 2011). Quantitative and/or spatial differential expression was confirmed for ten selected DETs using real-time PCR and/or in situ hybridization. In order to select candidates of interest for the molecular characterization of apomixis, the DETs were mapped in silico on the rice chromosomes (Laspina et al., 2008). Distribution was strongly biased toward chromosome 2 but not chromosome 12 (12 and four transcripts, respectively, compared with 6·8 transcripts per chromosome on average). Moreover, some of the DETs mapping in silico onto rice chromosomes 2 and/or 12 were experimentally mapped in P. notatum, but none of them co-segregated strictly with apomixis, suggesting that these could be downstream-acting genes rather than the genetic determinants of the trait (Laspina et al., 2008; Felitti et al., 2011; Ochogavía et al., 2011). Interestingly, some DETs were classified as TEs carrying transduplicated segments of genes previously associated with apomictic development (SERK, CYT, P450), suggesting that they might regulate the expression of these genes through mechanisms involving transcriptional and/or post-transcriptional gene silencing (Ochogavía et al., 2011). A total of 202 DETs were identified in P. simplex by cDNA-AFLP profiling using mRNA samples collected at several stages of development in P. simplex (Polegri et al., 2010). The majority of them were expressed exclusively at specific stages of the apomictic development or were misregulated, as temporal ectopic expression could be observed in apomictic genotypes. In contrast to DETs obtained in P. notatum, in silico mapping onto the rice genome showed no bias towards chromosomes 12 or 2. Interestingly, despite the high density of TEs in the ACR (Calderini et al., 2006), only a few DETs showed homology with these elements, suggesting that many of the apomixis-linked TEs are transcriptionally silent.
Identifying common features in transcriptomic analysis of related apomictic species is extremely useful to identify key conserved steps in apomictic development. The most evident similarity is the ontology classes to which the candidates correspond. In both species, the main classes were ‘signal transduction’, ‘nucleic acid binding’, ‘protein metabolism’, ‘transcription’ and ‘transport’ (Fig. 5). Interestingly, several DETs belong to the same annotation classes including extensins (P. simplex E1/124-6 and P. notatum N31), yoda-like MAP3Ks (P. simplex A/148-3 and P. notatum N46), LRR-like proteins (P. simplex A/124-3, P. notatum N78 and P. notatum N79) transferase proteins (P. simplex C1/121-7 and P. notatum N91) and retrotransposon proteins (P. simplex A/121-1, P. simplex E/120-1 and P. notatum N92) (Laspina et al., 2008; Polegri et al., 2010). All of these belong to P. simplex DET sub-classes that were differentially expressed between apomictic and sexual flowers at early developmental stages, because this was the only developmental stage analysed in P. notatum.
Fig. 5.
Characterization of apomixis-associated DETs identified by transcriptomic analysis in Paspalum genotypes. Top left: a representative example of DET (apo417/N114) identified in P. notatum flowers at late pre-meiotic/meiotic stage by differential display experiments. Differential display amplicons originating from three apomictic and three sexual F1 plants (in duplicate) were transferred to nylon membranes and hybridized with an N114 probe labelled with digoxigenin (Pessino et al., 2001). Top right: a representative example of a Class A DET identified in P. simplex flowers by cDNA-AFLP analysis at pre-anthesis, anthesis and post-anthesis stages (Polegri et al., 2010). Bottom: histogram representing the relative occurrence of DETs belonging to particular ontology classes in P. notatum, as identified from DD experiments and in P. simplex, as identified from cDNA-AFLP experiments. Ontology classes (from left to right): transposon/retrotransposon, signal transduction, protein metabolism, miRNA precursors, general metabolism, transcription, cell cycle, transport, cytoskeleton, other, unknown.
Finally, a remarkable correlation was observed between the identity of genes modulated during reproductive development in P. simplex/P. notatum (Laspina et al., 2008; Polegri et al., 2010) and those regulated during autopolyploidization in P. notatum (Martelotto et al., 2005). Several common annotations were detected between the P. simplex apomixis-associated candidates and the P. notatum ploidy-regulated ones: P. simplex B4/120-3 and P. notatum DDT13522x2 corresponded to glucose 6P-P translocators; P. simplex B4/122-1 and P. notatum DDT13682x to F-box proteins; P. simplex D4/153-8 and P. notatum DDT32844x1 to ubiquitin-conjugating enzymes; P. simplex B4/137-2 and P. notatum DDT43722x2 to chitinases; and P. simplex A/148-4 and P. notatum DDT43964x1 to DNAJ domain-containing proteins. Moreover, P. notatum apomixis-associated sequences N7 (unknown), N14 (ribosomal protein S12), N16 (acetolactate synthase) and N108 (transposon protein) corresponded to ploidy-regulated sequences DDT32852x1, DDT32834x2, DDT32774x2 and DDT32884x, respectively, in BLAST2seq searches (Laspina et al., 2008).
These observations imply that a considerable number of sequences involved in apomictic development are transcriptionally modulated by a change in ploidy. These sequences may represent the molecular link between apomixis and polyploidy. The presence of at least one of these sequences mapping onto the P. simplex ACR (P. simplex A/148-4 DNAJ domain-containing protein) was previously confirmed by RFLP mapping (Polegri et al., 2010). This sequence belonged to class A, whose members are constitutively expressed at low levels in apomictic individuals, indicating that gene de-regulation, polyploidization and apomixis are closely inter-related phenomena.
Possible causes of expression deregulation at the ACR
The transcription deregulation observed in the ACR could be due to its highly rearranged nature. In P. notatum, a major rearrangement, possibly an inversion or a translocation, was reported in some apomictic genotypes (Stein et al., 2004; Podio et al., 2012b). In P. simplex, an apomixis-linked BAC was characterized by the presence of large and small indels, translocations and TE insertions (Calderini et al., 2006). All of these rearrangements can deregulate transcriptional activity. Genetic rearrangements following gene migration or the inversion/translocation of large chromosomal areas can relocate genes near cis-regulatory elements that can deregulate their transcription or they can move away boundary elements, allowing heterochromatin to invade new areas.
The Paspalum ACR is plagued with abundant TEs (Calderini et al., 2006; Podio et al., 2012a). Expression deregulation at the ACR could be mediated by these repetitive elements by several mechanisms. TEs may contain sequences that bind a regulator protein or, once inserted, they may create new binding sites that act as transcriptional regulators (Lerat and Sémon, 2007). Alternately, TE insertion may induce gene inactivation and affect gene transcription via a mechanism similar to the random inactivation model for the neo-Y genes proposed in Drosophila miranda (Bachtrog, 2006). According to this model, genes located in a recombinationally repressed area are inactivated by random insertion of TEs, and these mutations induce downregulation of the Y-linked genes. In the Paspalum system, a general downregulation of apomixis-linked genes compared with their sexual homologues was noticed (Polegri et al., 2010). Recently, a central role for silencing involving retrotransposons in determining gametic fate was reported in Arabidopsis thaliana (Olmedo-Monfil et al., 2010). Inactivation of the A. thaliana gene At-AGO9 decreased the generation of retrotransposon-related small interfering RNAs (siRNAs) and induced the formation of multiple non-reduced ESs within the nucellus. This phenotype strongly resembled apospory (Olmedo-Monfil et al., 2010). At-AGO9 predominant TE targets were located in the pericentromeric regions of all five arabidopsis chromosomes, suggesting a link between the At-AGO9-dependent siRNA pathway and heterochromatin formation (Durán-Figueroa and Vielle-Calzada, 2010). A similar apospory-like phenotype was induced in maize by inactivating the DNA methyltransferases Zm-DMT102 and Zm-DMT103 (Garcia-Aguilar et al., 2010). Taken together, the mutant phenotypes ago9 in arabidopsis and dmt103 in maize suggest that retrotransposon-mediated siRNA generation and DNA methylation pathways control the switch between apomictic and sexual reproduction. Considering the possible role of retrotransposon-mediated silencing in the regulation of sexual reproduction revealed for the model species, the functional analysis of ACR-linked candidates in natural apomictic plants should be redirected. Future work in the genus Paspalum should be oriented not only to phenotypic analysis of null and gain-of-function mutants of protein-coding candidates, but also to determine how altered retroelement activity within the ACR could influence gene expression and condition the fate of the female gametes in natural apomictic species.
Studies of synthetic allopolyploids have revealed that the genomic response to allopolyploidy usually involves (retro)transposon mobility, sequence rearrangements and losses, DNA methylation changes and chromatin remodelling. All of these features have a significant effect on gene silencing and up- or downregulation of the duplicated genes (Adams and Wendel, 2005).
Martelotto et al. (2007) showed that polymorphisms between diploids and their polyploid counterparts were mainly related to band loss and retrotransposon mobilization, and that sequence modifications associated with polyploidization occurred at cytosine-methylated regions, although the genetically modified regions remained identically methylated after polyploidization. Similarly, Rodriguez et al. (2012) reported no differences in the average proportions of methylated CCGG sites between diploid and tetraploid cytotypes of P. notatum, but methylation patterns were significantly more variable in the tetraploids, and sequence analysis of new epialleles which emerged after polyploidization revealed homology with TEs.
To determine which genes were regulated by ploidy, a comparative transcriptome analysis was conducted in flowers of sexual diploid and tetraploid P. notatum genotypes (Martelotto et al., 2005). The 64 validated clones showing differential expression between diploid and tetraploid sexual cytotypes belonged to the following ontology classes: (1) chromatin remodelling; (2) protein trafficking, folding and degradation; (3) carbohydrate and lipid metabolism; (4) cell cycle regulation; (5) transcription; and (6) signal transduction. Interestingly, several of the genes regulated by ploidy had identical annotations regarding other differentially modulated genes during apomictic development (see the previous section ‘Transcriptomic landscape: towards the identification of candidates’ for details).
To sum up, the genome of Paspalum is genetically modified at regions encoding retrotransposons probably as a consequence of polyploidization; this modification in turn modulates the representation of protein-coding transcripts through an unknown mechanism (Martelotto et al., 2005, 2007; Rodriguez et al., 2012). Interestingly, several families of retroelements carrying transduplicated segments of genes, some of which are associated with apomixis, were described in P. notatum, suggesting that the molecular pathways involved in reproduction can be affected by polyploidization at specific levels (Ochogavía et al., 2011).
FURTHER STRATEGIES FOR MINING APOMIXIS GENES IN PASPALUM SPP.
Candidates genes identified by large-scale sequence analyses
Comparative molecular genetic analysis of the ACR in Paspalum has shown that a restricted region homologous to a specific chromosome area of rice was conservatively linked to apomixis in several Paspalum spp. (Pupilli et al., 2004; Hojsgaard et al., 2011; Fig. 3). Large-scale sequencing of this conserved region and of DETs mapped in the same area should disclose genes that on the basis of their homology with other genes of known function are worth considering as candidate genetic determinants of apomictic reproduction in Paspalum. Here we report and discuss some of these genes.
As a first example, consider the case of Ps-EXS (Calderini et al., 2006). Proteins containing the EXS motif include: (1) SYG1, a signal transduction protein that in Saccharomyces cerevisiae was associated with the G-protein; (2) sequences thought to be murine leukaemia virus receptors (XPR1) (Battini et al., 1999); and (3) ERD1 proteins, involved in the localization of endogenous endoplasmic reticulum proteins in S. cerevisiae (Hardwick et al., 1990). Deletion mutants of SYG1 protein can suppress cell cycle arrest and differentiation in yeast, and the suppression capacity is related to the loss of specific portions of the gene (Spain et al., 1995). The same authors hypothesized that one of these deletion mutants (Syg1Δ340p) can promote cell division in otherwise arrested (differentiated) cells. Compared with the Os-EXS gene structure, Ps-EXS lacks some rice exons entirely and coding capacity in others, suggesting a possible mechanism in which deleted (or rearranged) supernumerary copies of genes involved in sexual development can reprogramme differentiated cells into the apomictic developmental process.
The alignment of the apomixis-linked clone Pn-MAI3 of P. notatum, extended by chromosome walking, showed significant homology with a maize cDNA related to an N6-adenosine-methyltransferase (MT-A70; Podio et al., 2012a). This enzyme catalyses the N6-adenosine methylation in nascent mRNA and was shown to play key roles in cell fate decision in multiple eukaryote systems (Jia et al., 2012). MTA-70 expression in A. thaliana is associated with dividing tissues, as inactivation of this enzyme leads to failure of the developing embryo to progress past the globular stage (Zhong et al., 2008). MT-A70 in Paspalum might play a role in one or more aspects of apomictic development, i.e. the parthenogenenetic development of the unreduced egg cell.
To date, transcriptional surveys have identified several candidates which can play a role either as genetic determinants of or as downstream players in (depending on their co-segregation or less with apomixis, respectively) apomictic reproduction in Paspalum. An interesting candidate is represented by the Pn-N46 clone which showed homology to a gene belonging to the mitogen-activated protein 3kinase (MAP3K) gene family and was identified as differentially expressed in apomictic and sexual flowers of P. notatum. The arabidopsis predicted orthologue to n46, At1g53570, belong to a gene family formed by 12 members (Ichimura et al., 2002). Of these, YDA, which was reported to be more closely related to At1g53570, promotes the zygote elongation and suspensor development in arabidopsis (Lukowitz et al., 2004). In flowers of P. simplex, an apomixis-specific allele homologous to Pn-n46, constitutively expressed across all the developmental stages taken into account, was also identified (Polegri et al., 2010). It could be that n46 may have a role in the parthenogenetic development of the embryo in both species.
Another remarkable candidate as a downstream acting gene is the P. notatum clone N20, homologous to a LORELEI-family member, which was overexpressed in apomictic plants relative to sexual plants at pre-meiosis, post-meiosis and anthesis (maximal expression) but downregulated at meiosis (Laspina et al., 2008; Felitti et al., 2011). Curiously, arabidopsis null mutants for the At-LORELEI (At4g26466) gene showed impaired sperm cell release into the egg cell (Capron et al., 2008). The fact that egg cell fertilization is generally absent in P. notatum apomictic plants (embryos are formed through parthenogenesis) but the central cell is fertilized to produce the endosperm, suggests an active mechanism involving LORELEI-family members, which might operate to prevent fertilization in apomictic plants (Felitti et al., 2011).
Another likely downstream-acting gene, showing homology to the member 1 of the AUXIN-RESPONSE FACTOR family proteins (ARF-1), was identified in P. simplex by Polegri et al. (2010). These proteins are transcription factors that regulate the expression of auxin-responsive genes in both activation and repression modes (Guilfoyle and Hagen, 2007). The developmental fate of non-reproductive cells has been switched in female gametophytes of arabidopsis by manipulating auxin response genes (Pagnussat et al., 2009). A homologue of arabidopsis At-ARF1 was expressed early in P. simplex apomictic ovule formation, suggesting that the auxin response may affect the differentiation of AIs from nucellar cells. Ps-ARF1 may repress a class of auxin-responsive genes that maintain the undifferentiated state of nucellar cells once the MMC is formed.
An example of a structural gene whose expression might be influenced by the upstream genetic determinants of apomixis are α-zeins, which are seed-storage proteins whose accumulation in the endosperm is developmentally regulated at the transcriptional level (Sabelli and Larkins, 2009). Apomictic flowers of P. simplex began to accumulate zein-homologue transcripts about 5 d after pollination, while those transcripts were absent in sexual flowers (Polegri et al., 2010). In particular, the transcription of zein homologues in P. simplex may begin earlier in apomictic endosperm than in sexual endosperm. Because parthenogenetic embryos do not need fertilization, embryo development in apomictic plants could be accelerated compared with their sexual counterparts, and fast-developing embryos could send positive signals to the central cell of the ES, which is then ‘primed’ for faster cell cycle activity. These signals might include those committing to earlier storage protein accumulation.
Gene transfer as a definitive tool to validate candidate apomixis genes
Functional analysis of the selected candidate genes would require the establishment of an efficient transformation system in Paspalum. In the last few years, several biolistic transformation protocols were reported for P. notatum (Grando et al., 2002; Smith et al., 2002; Gondo et al., 2005). More recently P. notatum has also been used as a model system to assess the risk of pollen-mediated transgene flow in the field (Sandhu et al., 2009, 2010). Our research group adapted these protocols for use on sexual and apomictic tetraploid genotypes (unpublished results). In the next few years, we expect to produce a number of transformants with modified expression of several candidate genes related to apospory. The introduction of apomixis by de novo engineering will require the identification of candidate genes, the isolation of promoters that control gene expression spatially and temporally, and the development of technologies to introduce the transgenes (Grossniklaus, 2001). Although some species have evolved complex mechanisms to control apomixis, this trait could be engineered through a synthetic approach targeting the key regulatory steps: non-reduction, parthenogenesis and seed development. Genes controlling each step should be identified and regulated to produce a synthetic apomict. Endosperm development is a crucial challenge (Savidan, 2001). A balanced maternal:paternal genome ratio (2m:1p) is an absolute requirement for endosperm development in cereals, due to specific imprinting of gametic nuclei. Deviation from this ratio leads to embryo abortion or seeds with diminished fertility. Therefore, detailed knowledge on the behaviour of genes controlling autonomous endosperm development might be necessary to introduce apomixis into sexual crops. Promising results have already been obtained in arabidopsis; a combination of maternal hypomethylation with loss of fie function led to the development of endosperm without fertilization (Vinkenoog et al., 2000). To date, several promoters have been described that are active in the ovule or the gametophyte (Yu et al., 2005; Nain et al., 2008; Dwivedi et al., 2010). However, the choice of the promoter for transformation strategies to produce an artificial apomict would depend on the temporal and spatial expression patterns of the particular candidate gene targeted, and therefore should be selected on an individual basis. All these aspects should be taken into account when attempting to develop an apomixis system for crops without closely related apomictic relatives. Wild apomictic systems with sexual and asexual cytotypes could serve as models to understand the complex functional network of gene–gene communications that could transform a sexual plant into an asexual one after it has received an apomixis-controlling locus.
CONCLUDING REMARKS
Transferring apomixis to economically important crops could have enormous benefits for agriculture, making understanding the molecular mechanisms of this trait of outstanding importance in agricultural biotechnology. The manipulation of the trait is currently having a direct impact on the breeding of natural apomictic forage grasses, allowing a potential increase in cattle production in tropical, sub-tropical and temperate areas. Moreover, the possibility to clone superior genotypes and hybrids of crops such as maize, rice, wheat and soybean could greatly benefit farmers, allowing them to sustain high yields over many years by planting a sub-set of their harvested seed, without losses due to recombination and/or segregation. New interspecific and intergeneric hybrids could be obtained and propagated, allowing the development of genotypes better adapted to the different environments. Apomixis would also facilitate the use of transformants, considering that an apomictic transgenic plant would immediately fix the new trait and become a cultivar after multiplication. Thus far, introgression of apomixis into crops from wild relatives has failed. Introducing the trait through genetic engineering is not yet viable, mainly because the apomixis determinants remain unknown. Loci controlling apomixis in natural apomicts are often large, complex and, in some cases, recalcitrant to recombination-based mapping approaches, all of which hinder candidate identification and, therefore, transgenesis-based breeding programmes. Impressive progress has been made in identifying genes mimicking some elements of apomixis in arabidopsis (Ravi et al., 2008; Olmedo-Monfil et al., 2010) and maize (García-Aguilar et al., 2010; Singh et al., 2010). Arabidopsis artificial plants producing partial clonal progeny have been obtained (Marimuthu et al., 2011). Although these plants cannot be defined as genuine apomictic genotypes because they still rely on crossing to express full maternal inheritance, these constitute the first proof of principle of the possibility to develop a synthetic apomictic system in a diploid sexual species. In our view, unravelling the apomixis enigma will require the implementation of three inter-related approaches that should be combined to develop an apomixis system suitable for each target crop. First, forward genetics should be carried out in the closest-relative apomictic model to target the crop to look for promising mutations. Secondly, related genes should be validated in natural apomicts and model sexual species by reverse engineering; and, thirdly, the new information and tools should be combined to introgress validated genes into target crops. Genetic affinity between target and model is crucial, as gene networks controlling complex traits are not always conserved.
The genetic variability of the genus Paspalum is currently being exploited via sexual hybridization followed by apomictic fixation of successful polyploid genotypes. If this potential could be harnessed in full, considerable advances could be made not only in Paspalum breeding but also in other species with both sexual and asexual modes of reproduction. However, in our view, the major impact of research on Paspalum is and will be related to its role as a model system to mine apomixis genes and to develop an apomixis system for major crops. Paspalum provides a unique opportunity to identify the apomixis determinants, because its ACR is smaller than that of other apomictic systems. Moreover, its relatively small genome avoids the problems related to gene redundancy when performing functional analysis of candidate genes. In addition, the availability of multiple apomixis-segregating populations derived from different parent combinations narrows the portion of the ACR that carries the genetic determinant(s) for apomixis, enabling the genes that have been screened out by speciation to be discarded as apomixis candidates. The functional constraint of the 2:1 female:male parental genome contribution to the endosperm and the need to introgress apomixis in diploid backgrounds are major drawbacks to introducing apomixis in crop species. In both cases, Paspalum species show promising features (i.e. a tendency to yield viable hybrid seeds from parental lines of various ploidy levels and the presence of some elements of apomixis in diploids) that should be further studied. Further research is needed to define the relationships between structure, position and function of the known apomixis-linked genes. Many of these genes are pseudogenes characterized by a deregulated constitutive expression, but questions remain. (1) Are all of the genes in the ACR pseudogenes? (2) Is their constitutive expression related to their pseudogene nature? Although pseudogene expression might be related to silencing of their sex-related homologues, the presence of some genes that act as positive activators of apomixis cannot be ruled out. Other relevant issues are the epigenetic landscape and the proliferation of retrotransposon sequences in the ACR: (3) are these repetitive sequences simply accumulating in a heterochromatic chromosomal sector with impaired coding and functional capacity without any selective constraints, or (4) could these sequences be controlling transcription of neighbouring genes/pseudogenes and therefore influencing expression of sequence-related functional gene members? There is mounting evidence in model species that at least some elements of apomixis are under epigenetic control. Analysis of TE activity, DNA methylation and chromatin remodelling in the ACR will enhance knowledge on epigenetic regulation of apomixis. Whole-genome epigenetic analyses of DNA methylation and chromatin remodelling will clarify the complex network of gene interactions, perhaps mediated by TEs that silence sex-related genes and trigger the positive effectors of apomixis.
Major constrains for the identification of genes involved in apomixis in Paspalum as well as in other natural apomictic systems are due to the small size, rare abundance and inaccessibility of the cells within the ovule and the substantial amount of sequencing efforts needed at both the genomic and transcriptomic levels. We believe that modern tools of molecular and cell biology should be used to restrict the number of cells to be analysed to eliminate unspecific background on the one hand and to give a comprehensive global view of the gene action across all stages of apomictic development on the other. In this perspective, the newly developed laser-assisted microdissection procedures coupled with linear amplification of mRNA can provide useful information on genes acting on decision fate at the level of a single or a few cells. High-throughput sequencing procedures of linearly amplified mRNA adapted to organisms for which a reference genome is not yet available should then be used to obtain a global transcriptomic platform of developing tissues.
Apomixis is a fascinating research field. Many of its features are simultaneously intriguing and complex to elucidate. The benefits to be derived from controlling apomixis are promising and the trait has been described as ‘the holy grail of agriculture’, ‘the plant breeders’ dream' and ‘the second green revolution’. The evidence summarized here indicates that Paspalum constitutes a very interesting system to study the physiological, genetic, evolutionary, ecological and agronomical aspects of apomixis and that research on this model has and will contribute significantly to understanding and harnessing this important trait.
ACKNOWLEDGEMENTS
We thank Florencia Galdeano of the Instituto de Botánica del Nordeste (CONICET-UNNE) for technical assistance; Dr Thomas Giesecke for helping us with the German translation of Renner's original work, and Dr Gabriel Rua for valuable suggestions on the systematics of the genus. The authors would like to thank the anonymous reviewers for their constructive criticism that helped to improve the manuscript. This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Argentina [PICT 2007 no. 00476]; Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina [PIP 112-200801-01378]; Facultad de Ciencias Agrarias-UNNE, Universidad Nacional de Rosario, Argentina [AGR189]; and by the Italian Ministero degli Affari Esteri, Direzione Generale per la Promozione e la Cooperazione Culturale (L401/1990) for the project ‘Isolation of genetic determinants of apomixis in Paspalum simplex’ (Italy–Argentina bilateral agreement, 2006–2007).
LITERATURE CITED
- Actkinson JM, Burson BL. Cytogenetic relationships between Paspalum pubiflorum and three South American Paspalum species. International Journal of Plant Sciences. 1999;160:775–781. [Google Scholar]
- Acuña CA, Blount AR, Quesenberry KH, Hanna WW, Kenworthy KE. Reproductive characterization of bahiagrass germplasm. Crop Science. 2007;47:1711–1717. [Google Scholar]
- Acuña CA, Blount AR, Quesenberry KH, Kenworthy KE, Hanna WW. Bahiagrass tetraploid germplasm: reproductive and agronomic characterization of segregating progeny. Crop Science. 2009;49:581–588. [Google Scholar]
- Acuña CA, Blount AR, Quesenberry KH, Kenworthy KE, Hanna WW. Tetraploid bahiagrass hybrids: breeding technique, genetic variability and proportion of heterotic hybrids. Euphytica. 2011;179:227–235. [Google Scholar]
- Adams KL, Wendel JW. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Agharkar M, Lomba P, Altpeter F, Zhang H, Kenworthy K, Lange T. Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flüggé) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnology Journal. 2007;5:791–801. doi: 10.1111/j.1467-7652.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- Aguilera PM, Sartor ME, Galdeano F, Espinoza F, Quarin CL. Interspecific tetraploid hybrids between two forage grass species: sexual Paspalum plicatulum and apomictic P. guenoarum. Crop Science. 2011;51:1544–1550. [Google Scholar]
- Altpeter F, James VA. Genetic transformation of turf-type bahiagrass (Paspalum notatum Flüggé) by biolistic gene transfer. International Turfgrass Society Research Journal. 2005;10:1–5. [Google Scholar]
- Asker SE, Jerling L. Apomixis in plants. Boca Raton, FL: CRC Press; 1992. [Google Scholar]
- Bachtrog D. Expression profile of a degenerating neo-y chromosome in Drosophila. Current Biology. 2006;16:1694–1699. doi: 10.1016/j.cub.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Banks DJ. Cytological studies in Paspalum, group Setacea (Gramineae) Sida. 1964;1:306–312. [Google Scholar]
- Banks DJ. Taxonomy of Paspalum setaceum (Gramineae) Sida. 1966;2:269–284. [Google Scholar]
- Bashaw EC, Forbes I. Chromosome numbers and microsporogenesis in Dallisgrass: Paspalum dilatatum Poir. Agronomy Journal. 1958;50:441–445. [Google Scholar]
- Bashaw EC, Holt EC. Megasporogenesis, embryo sac development and embryogenesis in Dallisgrass, Paspalum ditatatum Poir. Agronomy Journal. 1958;50:753–756. [Google Scholar]
- Bashaw EC, Hovin AW, Holt EC. Apomixis, its evolutionary significance and utilization in plant breeding. In: Norman MJT, editor. Proceedings of the 11th International Grasslands Congress. Surfers Paradise: Australia University of Queensland Press; 1970. pp. 245–248. [Google Scholar]
- Battini JL, Rasko JA, Miller AD. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proceedings of the National Academy of Sciences, USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett HW, Bashaw EC. Interspecific hybridization with Paspalum spp. Crop Science. 1966;6:52–58. [Google Scholar]
- Bicknell RA, Borst NK, Koltunow AM. Monogenic inheritance of apomixis in two Hieracium species with distinct developmental mechanisms. Heredity. 2000;84:228–237. doi: 10.1046/j.1365-2540.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- Blount AR, Acuña CA. Bahiagrass. In: Singh RJ, editor. Genetic resources, chromosome engineering, and crop improvement series: forage crops. Boca Raton, FL: CRC Press; 2009. pp. 81–102. [Google Scholar]
- Bonilla JR, Quarin CL. Diplosporous and aposporous apomixis in pentaploid Paspalum minus. Plant Science. 1997;127:97–104. [Google Scholar]
- Bradley JE, Carman GC, Jamison MS, Naumova TM. Heterochronic features of the female germline among several sexual diploid Tripsacum L. (Andropogoneae, Poaceae) Sexual Plant Reproduction. 2007;20:9–17. [Google Scholar]
- Brown WV, Emery HP. Apomixis in the Gramineae: Panicoideae. American Journal of Botany. 1958;45:253–263. [Google Scholar]
- Burson BL. Cytology of some apomictic Paspalum species. Crop Science. 1975;15:229–232. [Google Scholar]
- Burson BL. Cytology of Paspalum chacoense and P. durifolium and their relationship to P. dilatatum. Botanical Gazette. 1985;146:124–129. [Google Scholar]
- Burson BL. Genome relationships between tetraploid and hexaploid biotypes of Dallisgrass. Paspalum dilatatum. Botanical Gazette. 1991;152:219–223. [Google Scholar]
- Burson BL. Cytology and reproductive behaviour of hybrids between Paspalum urvillei and two hexaploid P. dilatatum biotypes. Genome. 1992;35:1002–1006. [Google Scholar]
- Burson BL. Apomixis and sexuality in some Paspalum species. Crop Science. 1997;37:1347–1351. [Google Scholar]
- Burson BL, Bennett HW. Cytology, method of reproduction and fertility of Brunswickgrass, Paspalum nicorae Parodi. Crop Science. 1970a;10:184–187. [Google Scholar]
- Burson BL, Bennett HW. Cytology and reproduction of three Paspalum species. Journal of Heredity. 1970b;61:129–132. [Google Scholar]
- Burson BL, Bennett HW. Chromosome numbers, microsporogenesis, and mode of reproduction of seven Paspalum species. Crop Science. 1971;11:292–294. [Google Scholar]
- Burson BL, Bennett HW. Cytogenetics of Paspalum conspersum and its genomic relationship with yellow-anthered P. dilatatum and P. malacophyllum. Canadian Journal of Genetics and Cytology. 1976;18:701–708. [Google Scholar]
- Burson BL, Hussey MA. Cytology of Paspalum malacophyllum and its relationship to P. jurgensii and P. dilatatum. International Journal of Plant Sciences. 1998;159:153–159. [Google Scholar]
- Burson BL, Quarin CL. Cytology of Paspalum virgatum and its relationship with P. intermedium and P. jurgensii. Canadian Journal of Genetics and Cytology. 1982;24:219–226. [Google Scholar]
- Burson BL, Voigt PW, Evers GW. Cytology, reproductive behavior and forage potential of hexaploid dallisgrass biotypes. Crop Science. 1991;31:636–641. [Google Scholar]
- Burson BL, Venuto BC, Hussey MA. Registration of ‘Sabine’ Dallisgrass. Journal of Plant Registration. 2008;3:132–137. [Google Scholar]
- Burton GW. The method of reproduction in common Bahia grass. Paspalum notatum. Journal of the American Society of Agronomy. 1948;40:443–452. [Google Scholar]
- Burton GW. Breeding Pensacola bahiagrass, Paspalum notatum: I. Method of reproduction. Agronomy Journal. 1955;47:311–314. [Google Scholar]
- Burton GW. Manipulating apomixis in Paspalum. In: Elgin JH, Miksche JP, editors. Proceedings of the Apomixis Workshop. ARS Research Bulletin 104. Washingon, DC: USDA-ARS; 1992. pp. 16–19. [Google Scholar]
- Burton GWM, Forbes I. The genetics and manipulation of obligate apomixis in common Bahia grass (Paspalum notatum Flugge) Proceedings of the 8th International Grassland Congress. Reading. 1960:66–71. [Google Scholar]
- Cáceres ME, Pupilli F, Quarin CL, Arcioni S. Feulgen-DNA densitometry of embryo sacs permits discrimination between sexual and apomictic plants in Paspalum simplex. Euphytica. 1999;110:161–167. [Google Scholar]
- Cáceres ME, Matzk F, Busti A, Pupilli F, Arcioni S. Apomixis and sexuality in Paspalum simplex: characterization of the mode of reproduction in segregating progenies by different methods. Sexual Plant Reproduction. 2001;14:201–206. doi: 10.1007/s00497-001-0109-1. [DOI] [PubMed] [Google Scholar]
- Calderini O, Chang SB, de Jong H, et al. Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcollinearity with rice. Theoretical and Applied Genetics. 2006;112:1179–1191. doi: 10.1007/s00122-006-0220-7. [DOI] [PubMed] [Google Scholar]
- Calderini O, Donnison I, Polegri L, et al. Partial isolation of the genomic region linked with apomixis in Paspalum simplex. Molecular Breeding. 2011;28:265–276. [Google Scholar]
- Caponio I, Quarin CL. El sistema genético de Paspalum simplex y de un híbrido interespecífico con P. dilatatum. Kurtziana. 1987;19:35–45. [Google Scholar]
- Caponio I, Quarin CL. Cytology and reproduction of P. densum and its genomic relationship with P. intermedium and P. urvillei. Journal of Heredity. 1993;84:220–222. [Google Scholar]
- Capron A, Gourgues M, Neiva LS, et al. Maternal control of male-gamete delivery in Arabidopsis involves a putative GPI-anchored protein encoded by the LORELEI gene. The Plant Cell. 2008;20:3038–3049. doi: 10.1105/tpc.108.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman JG. Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biological Journal of the Linnean Society. 1997;61:51–94. [Google Scholar]
- Chao CY. Megasporogenesis and megagametogenesis in Paspalum commersonii and P. longifolium at two polyploid levels. Botaniska Notiser. 1974;127:267–275. [Google Scholar]
- Chao CY. Autonomous development of embryo in Paspalum conjugatum Berg. Botaniska Notiser. 1980;133:215–222. [Google Scholar]
- Chase A. Washington DC: Botany Department Smithsonian Institution; 1939. Paspalum of South America. Unpublished manuscript. Hitchock and Chase Library. [Google Scholar]
- Crane CF. Classification of apomictic mechanisms. In: Savidan Y, Carman JG, Dresselhaus T, editors. The flowering of apomixis: from mechanisms to genetic engineering. Mexico City: CIMMYT, IRD, European Commission DG VI (FAIR); 2001. pp. 24–43. [Google Scholar]
- Darlington CD. Evolution of genetic systems. Cambridge: Cambridge University Press; 1939. [Google Scholar]
- Daurelio LD, Espinoza F, Quarin CL, Pessino SC. Genetic diversity in sexual diploid and apomictic tetraploid populations of Paspalum notatum situated in sympatry or allopatry. Plant Systematics and Evolution. 2004;244:189–199. [Google Scholar]
- Denham SS. Revisión sistemática del subgenero Harpostachys de Paspalum (Poaceae: Panicoideae: Paniceae) Annals of the Missouri Botanical Garden. 2005;92:463–532. [Google Scholar]
- Denham SS, Zuloaga FO. Phylogenetic relationships of the Decumbentes group of Paspalum, Thrasya, and Thrasyopsis (Poaceae: Panicoideae: Paniceae) Aliso. 2007;23:545–562. [Google Scholar]
- Denham SS, Zuloaga FO, Morrone O. Systematic revision and phylogeny of Paspalum subgenus Ceresia (Poaceae: Panicoideae: Paniceae) Annals of the Missouri Botanical Garden. 2002;89:227–399. [Google Scholar]
- Durán-Figueroa N, Vielle-Calzada JP. ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signaling and Behavior. 2010;5:1476–1479. doi: 10.4161/psb.5.11.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi SL, Perotti E, Upadhyaya HD, Ortiz R. Sexual and apomictic plant reproduction in the genomics era: exploring the mechanisms potentially useful in crop plants. Sexual Plant Reproduction. 2010;23:265–279. doi: 10.1007/s00497-010-0144-x. [DOI] [PubMed] [Google Scholar]
- Ernst A. Bastardierung als Ursache der Apogamie im Pflanzenreich. Jena: Fischer; 1918. [Google Scholar]
- Espinoza F, Quarin CL. Cytoembyology of Paspalum chaseanum and sexual diploid biotypes of two apomictic Paspalum species. Australian Journal of Botany. 1997;45:871–877. [Google Scholar]
- Espinoza F, Urbani MH, Martínez EJ, Quarin CL. The breeding system of three Paspalum species. Tropical Grassland. 2001;35:211–217. [Google Scholar]
- Essi L, Souza-Chies TT. Phylogeny of Linearia and Notata groups of Paspalum L. (Poaceae, Panicoideae, Paniceae) and related species. Genetic Resources and Crop Evolution. 2007;54:779–791. [Google Scholar]
- Evers GW, Burson BL. Dallisgrass and other Paspalum species. In: Moser LE, editor. Warm-season (C4) grasses. Agronomy Monography 45. Madison: ASA, CSSA, and SSSA; 2004. pp. 681–713. [Google Scholar]
- Felitti SA, Seijo JG, Gonzáles AM, et al. Expression of lorelei-like genes in aposporous and sexual Paspalum notatum plants. Plant Molecular Biology. 2011;77:337–354. doi: 10.1007/s11103-011-9814-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. The Plant Cell. 2010;22:3249–3267. doi: 10.1105/tpc.109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giussani LM, Zuloaga FO, Quarin CL, Cota-Sánchez H, Ubayasena K, Morrone O. Phylogenetic relationships in the genus Paspalum (Poaceae: Panicoideae: Paniceae): an assessment of the Quadrifaria and Virgata informal groups. Systematic Botany. 2009;34:32–43. [Google Scholar]
- Goel S, Chen Z, Conner JA, Akiyama Y, Hanna WW, Ozias Akins P. Delineation by fluorescence in situ hybridization of a single hemizygous chromosomal region associated with aposporous embryo sacs formation in Pennisetum squamulatum and Cenchrus ciliaris. Genetics. 2003;163:1069–1082. doi: 10.1093/genetics/163.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondo T, Tsuruta S, Akashi R, Kawamura O, Hoffmann F. Green, herbicide-resistant plants by particle inflow gun-mediated gene transfer to diploid bahiagrass (Paspalum notatum) Journal of Plant Physiology. 2005;162:1367–1375. doi: 10.1016/j.jplph.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Grando MF, Franklin CI, Shatters RG. Optimizing embryogenic callus production and plant regeneration from ‘Tifton 9’ bahiagrass seed explants for genetic manipulation. Plant Cell, Tissue and Organ Culture. 2002;71:213–222. [Google Scholar]
- Grass Phylogeny Working Group II. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytologist. 2012;193:304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- Grimanelli D, Leblanc O, Espinosa E, Perotti E, Gonzalez de Leon D, Savidan Y. Non-Mendelian transmission of apomixis in maize–Tripsacum hybrids caused by a transmission ratio distortion. Heredity. 1998;80:40–47. doi: 10.1046/j.1365-2540.1998.00264.x. [DOI] [PubMed] [Google Scholar]
- Grimanelli D, Leblanc O, Perotti E, Grossniklaus U. Developmental genetics of gametophytic apomixis. Trends in Genetics. 2001;17:597–604. doi: 10.1016/s0168-9525(01)02454-4. [DOI] [PubMed] [Google Scholar]
- Grimanelli D, García M, Kaszas E, Perotti E, Leblanc O. Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics. 2003;165:1521–1531. doi: 10.1093/genetics/165.3.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U. From sexuality to apomixis: molecular and genetic approaches. In: Savidan Y, Carman JG, Dresselhaus T, editors. The flowering of apomixis: from mechanisms to genetic engineering. Mexico City: CIMMYT, IRD, European Commission DG VI (FAIR); 2001. pp. 168–211. [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Current Opinion in Plant Biology. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Lewis MJ, Semenza J, Dean N, Pelham HR. ERD1 a yeast gene required for the retention of luminal endoplasmic reticulum proteins affects glycoprotein processing in the Golgi apparatus. EMBO Journal. 1990;9:623–630. doi: 10.1002/j.1460-2075.1990.tb08154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojsgaard DH, Schegg E, Valls JFM, Martínez EJ, Quarin CL. Sexuality, apomixis, ploidy levels, and genomic relationships among four Paspalum species of the subgenus Anachyris (Poaceae) Flora. 2008;203:535–547. [Google Scholar]
- Hojsgaard DH, Martínez EJ, Acuña CA, Quarin CL, Pupilli F. A molecular map of the apomixis-control locus in Paspalum procurrens and its comparative analysis with other species of Paspalum. Theoretical and Applied Genetics. 2011;123:959–971. doi: 10.1007/s00122-011-1639-z. [DOI] [PubMed] [Google Scholar]
- Hojsgaard DH, Martínez EJ, Quarin CL. Competition between meiotic and apomictic pathways during ovule and seed development results in clonality. New Phytologist. 2013;197:336–347. doi: 10.1111/j.1469-8137.2012.04381.x. [DOI] [PubMed] [Google Scholar]
- Hörandl E. The complex causality of geographical parthenogenesis. New Phytologist. 2006;171:525–538. doi: 10.1111/j.1469-8137.2006.01769.x. [DOI] [PubMed] [Google Scholar]
- Hörandl E, Hojsgaard DH. The evolution of apomixis in angiosperms: a reappraisal. Plant Biosystems. 2012;146:681–693. [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- James VA, Neibaur I, Altpeter F. Stress inducible expression of the DREB1A transcription factor from xeric, Hordeum spontaneum L in turf and forage grass (Paspalum notatum Flugge) enhances abiotic stress tolerance. Transgenic Research. 2008;17:93–104. doi: 10.1007/s11248-007-9086-y. [DOI] [PubMed] [Google Scholar]
- Jarret RL, Ozias-Akins P, Phatak S, Nadimpalli R, Duncan R, Hiliard S. DNA contents in Paspalum ssp. determined by flow cytometry. Genetic Resources and Crop Evolution. 1995;42:273–242. [Google Scholar]
- Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends in Genetics. 2012;29:108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judziewicz E. 187. Poaceae. In: Görts-Van Rijn ARA, editor. Flora of the Guianas. 1990. pp. 1–127. Series A: Phanerogams, Vol. 8. Königstein: Koeltz Scientific Books. [Google Scholar]
- Kearney M. Hybridization, glaciations and geographical parthenogenesis. Trends in Ecology and Evolution. 2005;20:495–502. doi: 10.1016/j.tree.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Labombarda P, Busti A, Cáceres ME, Quarin CL, Pupilli F, Arcioni S. An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome. 2002;45:513–519. doi: 10.1139/g02-014. [DOI] [PubMed] [Google Scholar]
- Laspina NV, Vega T, Seijo JG, et al. Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Molecular Biology. 2008;67:615–628. doi: 10.1007/s11103-008-9341-5. [DOI] [PubMed] [Google Scholar]
- Lerat E, Sémon M. Influence of the transposable elements neighbourhood on human gene expression in normal and tumor tissues. Gene. 2007;396:303–311. doi: 10.1016/j.gene.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. Systema naturae. 10th edn. 1759. Stockholm. [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;116:109–119. doi: 10.1016/s0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- Lutz W, Sanderson W, Scherbov S. Doubling of world population unlikely. Nature. 1997;387:803–805. doi: 10.1038/42935. [DOI] [PubMed] [Google Scholar]
- Lyttle TW. Segregation distorters. Annual Review of Genetics. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- Ma G, Huang X, Zhao N, Xu Q. Apospory in Paspalum thunbergii. Australian Journal of Botany. 2004;52:81–86. [Google Scholar]
- Ma G, Huang XL, Xu QS, Bunn E. Multiporate pollen and apomixis in Panicoideae. Pakistan Journal of Botany. 2009;41:2073–2082. [Google Scholar]
- Marimuthu MP, Jolivet S, Ravi M, et al. Synthetic clonal reproduction through seeds. Science. 2011;331:876. doi: 10.1126/science.1199682. [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Ortiz JPA, Stein J, Espinoza F, Quarin CL, Pessino SC. A comprehensive analysis of gene expression alterations in a newly synthesized Paspalum notatum autotetraploid. Plant Science. 2005;169:211–220. [Google Scholar]
- Martelotto LG, Ortiz JPA, Stein J, Espinoza F, Quarin CL, Pessino SC. Genome rearrangements derived from autopolyploidization in Paspalum sp. Plant Science. 2007;172:970–977. [Google Scholar]
- Martínez EJ, Quarin CL, Hayward MD. Genetic control of apospory in apomictic Paspalum species. Cytologia. 1999;64:425–433. [Google Scholar]
- Martínez EJ, Urbani MH, Quarin CL, Ortiz JPA. Inheritance of apospory in bahiagrass, Paspalum notatum. Hereditas. 2001;135:19–25. doi: 10.1111/j.1601-5223.2001.00019.x. [DOI] [PubMed] [Google Scholar]
- Martínez EJ, Hopp HE, Stein J, Ortiz JPA, Quarin CL. Genetic characterization of apospory in tetraploid Paspalum notatum based on the identification of linked molecular markers. Molecular Breeding. 2003;12:312–327. [Google Scholar]
- Martínez EJ, Acuña CA, Hojsgaard DH, Tcach MA, Quarin CL. Segregation for asexual seed production in Paspalum achieved by male gametes of apomictic triploid plants. Annals of Botany. 2007;100:1239–1247. doi: 10.1093/aob/mcm201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzk F, Meister A, Schubert I. An efficient screen for reproductive pathways using mature seeds of monocots and dicots. The Plant Journal. 2000;21:97–108. doi: 10.1046/j.1365-313x.2000.00647.x. [DOI] [PubMed] [Google Scholar]
- Morrone O, Denham SS, Aliscioni SS, Zuloaga FO. Revisión de las especies de Paspalum (Panicoideae: Paniceae), subgénero Anachyris. Candollea. 2000;55:105–135. [Google Scholar]
- Morrone O, Scataglini MA, Zuloaga O. Cyphonanthus, a new genus segregated from Panicum (Poaceae: Panicoideae: Paniceae) based on morphological, anatomical and molecular data. Taxon. 2007;56:521–532. [Google Scholar]
- Morrone O, Denham SS, Aliscioni SS, Zuloaga FO. Parodiophyllochloa, a new genus segregated from Panicum (Paniceae, Poaceae) based on morphological and molecular data. Systematic Botany. 2008;33:66–76. [Google Scholar]
- Nain V, Verma A, Kumar N, Sharma P, Ramesh B, Kumar PA. Cloning of an ovule-specific promoter from Arabidopsis thaliana and expression of beta-glucuronidase. Indian Journal of Experimental Biology. 2008;46:207–211. [PubMed] [Google Scholar]
- Nicora EG, Rúgolo de Agrasar ZE. Los géneros de gramíneas de América Austral: Argentina, Chile, Uruguay y áreas limítrofes de Bolivia, Paraguay y Brasil. Buenos Aires: Editorial Hemisferio Sur; 1987. [Google Scholar]
- Nogler GA. How to obtain diploid apomictic Ranunculus auricomus plants not found in the wild state. Botanica Helvetica. 1982;92:13–22. [Google Scholar]
- Nogler GA. Gametophytic apomixis. In: Johri BM, editor. Embryology of angiosperms. Berlin: Springer; 1984. pp. 475–518. [Google Scholar]
- Norrmann GA. Citología y método de reproducción en dos especies de Paspalum (Gramineae) Bonplandia. 1981;5:149–158. [Google Scholar]
- Norrmann GA, Quarin CL, Burson BL. Cytogenetics and reproductive behaviour of different chromosome races in six Paspalum species. Journal of Heredity. 1989;80:24–28. [Google Scholar]
- Ochogavía AC, Seijo JG, González MA, et al. Characterization of retrotransposon sequences expressed in inflorescences of apomictic and sexual Paspalum notatum plants. Sexual Plant Reproduction. 2011;24:231–246. doi: 10.1007/s00497-011-0165-0. [DOI] [PubMed] [Google Scholar]
- Oliveira RC, Rua GH. A new species of Paspalum (Poaceae, Paniceae) from Central Brazil. Systematic Botany. 2005;30:530–532. [Google Scholar]
- Oliveira RC, Valls JFM. Duas novas espécies de Paspalum L. (Poaceae: Paniceae) do Brasil. Revista Brasileira de Botânica. 2009;32:89–94. [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, et al. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins P, van Dijk PJ. Mendelian genetics of apomixis. Annual Review of Genetics. 2007;41:509–537. doi: 10.1146/annurev.genet.40.110405.090511. [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, Roche D, Hanna WW. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by divergent locus that may have no allelic form in sexual genotypes. Proceedings of the National Academy of Sciences, USA. 1998;95:5127–5132. doi: 10.1073/pnas.95.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini MS, Takayama SY, de Freitas PM, et al. Failure of cytokinesis and 2n gamete formation in Brazilian accessions of Paspalum. Euphytica. 1999;108:129–135. [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science. 2009;324:1684–1689. doi: 10.1126/science.1167324. [DOI] [PubMed] [Google Scholar]
- Parodi LR. Estudios sistemáticos sobre las Gramineae-Paniceae argentinas y uruguayas. Darwiniana. 1969;15:65–109. [Google Scholar]
- Pessino SC, Espinoza F, Martínez EJ, Ortiz JPA, Valle EM, Quarin CL. Isolation of cDNA clones differentially expressed in flowers of apomictic and sexual Paspalum notatum. Hereditas. 2001;134:35–42. doi: 10.1111/j.1601-5223.2001.00035.x. [DOI] [PubMed] [Google Scholar]
- Podio M, Rodríguez MP, Felitti S, et al. Sequence characterization, in silico mapping and cytosine methylation analysis of markers linked to apospory in Paspalum notatum. Genetics and Molecular Biology. 2012a;35:827–837. doi: 10.1590/S1415-47572012005000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podio M, Siena LA, Hojsgaard D, Stein J, Quarin CL, Ortiz JPA. Evaluation of meiotic abnormalities and pollen viability in aposporous and sexual tetraploid Paspalum notatum (Poaceae) Plant Systematics and Evolution. 2012b;298:1625–1633. [Google Scholar]
- Polegri L, Calderini O, Arcioni S, Pupilli F. Specific expression of apomixis-linked alleles revealed by comparative transcriptomic analysis of sexual and apomictic Paspalum simplex Morong flowers. Journal of Experimental Botany. 2010;61:1869–1883. doi: 10.1093/jxb/erq054. [DOI] [PubMed] [Google Scholar]
- Pritchard AJ. The cytology and reproduction of Paspalum yaguaronense. Australian Journal of Agricultural Research. 1962;13:206–211. [Google Scholar]
- Pritchard AJ. Meiosis and embryo sac development in Urochloa mosambicensis and three Paspalum species. Australian Journal of Agricultural Research. 1970;21:648–652. [Google Scholar]
- Pupilli F, Barcaccia G. Cloning plants by seeds: inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. Journal of Biotechonology. 2012;159:291–311. doi: 10.1016/j.jbiotec.2011.08.028. [DOI] [PubMed] [Google Scholar]
- Pupilli F, Cáceres ME, Quarin CL, Arcioni S. Segregation analysis of RFLP markers reveals a tetrasomic inheritance in apomictic Paspalum simplex. Genome. 1997;40:822–828. doi: 10.1139/g97-806. [DOI] [PubMed] [Google Scholar]
- Pupilli F, Labombarda P, Cáceres ME, Quarin CL, Arcioni S. The chromosome segment related to apomixis in Paspalum simplex is homoeologous to the telomeric region of the long arm of rice chromosome 12. Molecular Breeding. 2001;8:53–61. [Google Scholar]
- Pupilli F, Martínez EJ, Busti A, Calderini O, Quarin CL, Arcioni S. Comparative mapping reveals partial conservation of synteny at the apomixis locus in Paspalum spp. Molecular Genetics and Genomics. 2004;270:539–548. doi: 10.1007/s00438-003-0949-5. [DOI] [PubMed] [Google Scholar]
- Quarin CL. The nature of apomixis and its origin in Panicoid grasses. Apomixis Newsletter. 1992;5:8–15. [Google Scholar]
- Quarin CL. Morfologia, citologia y sistema reproductivo de una nueva especie de graminea: Paspalum procurrens. Bulletin of the Botanical Society of Argentina. 1993;29:73–76. [Google Scholar]
- Quarin CL. The tetraploid cytotype of Paspalum durifolium: cytology, reproductive behavior and its relationship to diploid P. intermedium. Hereditas. 1994;121:115–118. [Google Scholar]
- Quarin CL. Effect of pollen source and pollen ploidy on endosperm formation and seed set in pseudogamous apomictic Paspalum notatum. Sexual Plant Reproduction. 1999;11:331–335. [Google Scholar]
- Quarin CL, Burson BL. Cytogenetic relations among Paspalum notatum var. saurae, P. pumilum, P. indecorum, and P. vaginatum. Botanical Gazette. 1983;144:433–438. [Google Scholar]
- Quarin CL, Burson BL. Cytology of sexual and apomictic Paspalum species. Cytologia. 1991;56:223–228. [Google Scholar]
- Quarin CL, Caponio I. Cytogenetics and reproduction of Paspalum dasypleurum and its hybrids with P. urvillei and P. dilatatum ssp. flavescens. International Journal of Plant Sciences. 1995;156:232–235. [Google Scholar]
- Quarin CL, Hanna WW. Chromosome behavior, embryo sac development, and fertility of Paspalum modestum, P. boscianum, and P. conspersum. Journal of Heredity. 1980a;71:419–422. [Google Scholar]
- Quarin CL, Hanna WW. Effect of three ploidy levels on meiosis and mode of reproduction in Paspalum hexastachyum. Crop Science. 1980b;20:69–75. [Google Scholar]
- Quarin CL, Norrmann GA. Cytology and reproductive behavior of Paspalum equitans, P. ionanthum, and their hybrids with diploid and tetraploid cytotypes of P. cromyorrhizon. Botanical Gazette. 1987;148:386–391. [Google Scholar]
- Quarin CL, Urbani MH. Cytology and reproduction of diploid and tetraploid cytotypes of Paspalum coryphaeum. Apomixis Newsletter. 1990;2:44–46. [Google Scholar]
- Quarin CL, Hanna WW, Fernández A. Genetic studies in diploid and tetraploid Paspalum species. Embryo sac development, chromosome behavior, and fertility in P. cromyorryzon, P. laxum, and P. proliferum. Journal of Heredity. 1982;73:254–256. [Google Scholar]
- Quarin CL, Burson BL, Burton GW. Cytology of intra- and interspecific hybrids between two cytotypes of Paspalum notatum and P. cromyorrhizon. Botanical Gazette. 1984;145:420–426. [Google Scholar]
- Quarin CL, Norrmann GA, Urbani MU. Polyploidization in aposporous Paspalum species. Apomixis Newsletter. 1989;1:28–29. [Google Scholar]
- Quarin CL, Pozzobon MT, Valls JFM. Cytology and reproductive behaviour of diploid, tetraploid and hexaploid germplasm accessions of a wild forage grass. Paspalum compressifolium. Euphytica. 1996;9:345–349. [Google Scholar]
- Quarin CL, Valls JFM, Urbani MH. Cytological and reproductive behaviour of Paspalum atratum, a promising forage grass for the tropics. Tropical Grasslands. 1997;31:114–116. [Google Scholar]
- Quarin CL, Norrmann GA, Espinoza F. Evidence for autoploidy in apomictic Paspalum rufum. Hereditas. 1998;129:119–124. [Google Scholar]
- Quarin CL, Espinoza F, Martínez EJ, Pessino SC, Bovo OA. A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sexual Plant Reproduction. 2001;13:243–249. [Google Scholar]
- Quarin CL, Urbani MH, Blount AR, et al. Registration of Q4188 and Q4205, sexual tetraploid germplasm lines of bahiagrass. Crop Science. 2003;43:745–746. [Google Scholar]
- Quesenberry KH, Dampier JM, Lee YY, Smith RL, Acuña CA. Doubling the chromosome number of bahiagrass via tissue culture. Euphytica. 2010;175:43–50. [Google Scholar]
- Ramos DM, Valls JFM, Oliveira RC, Graciano-Ribeiro D. A new awned species of Paspalum (Poaceae, Panicoideae, Paniceae) from Brazil. Novon. 2011;21:368–372. [Google Scholar]
- Ravi M, Marimuthu MPA, Siddiqi I. Gamete formation without meiosis in Arabidopsis. Nature. 2008;451:1121–1124. doi: 10.1038/nature06557. [DOI] [PubMed] [Google Scholar]
- Rebozzio RN, Sartor ME, Quarin CL, Espinoza F. Residual sexuality and it seasonal variation in natural apomictic Paspalum notatum accessions. Biologia Plantarum. 2011;55:391–395. [Google Scholar]
- Rebozzio RN, Rodrıíguez MP, Stein J, Ortiz JPA, Quarin CL, Espinoza F. Validation of molecular markers linked to apospory in tetraploid races of bahiagrass, Paspalum notatum Flüggé. Molecular Breeding. 2012;29:189–198. [Google Scholar]
- Renner O. Zur Terminologie des pflanzlichen Generationswechsels. Biologishes Zentrablatt. 1916;36:337–374. [Google Scholar]
- Roche D, Chen Z, Hanna WW, Ozias-Akins P. Non-Mendelian transmission of an apospory-specific genomic region in a reciprocal cross between sexual pearl millet (Pennisetum glaucum) and an apomictic F1 (P. glaucum×P. squamulatum) Sexual Plant Reproduction. 2001;13:217–223. [Google Scholar]
- Roche D, Conner JA, Budiman MA, et al. Construction of BAC libraries from two apomictic grasses to study the microcollinearity of their apospory-specific genomic regions. Theoretical and Applied Genetics. 2002;104:804–812. doi: 10.1007/s00122-001-0795-y. [DOI] [PubMed] [Google Scholar]
- Rodriguez MP, Cervigni GDL, Quarin CL, Ortiz JPA. Frequencies and variation in cytosine methylation patterns in diploid and tetraploid cytotypes of Paspalum notatum. Biologia Plantarum. 2012;56:276–282. [Google Scholar]
- Rua GH, Valls JFM, Graciano-Ribeiro D, Oliveira RC. Four new species of Paspalum (Poaceae, Paniceae) from Central Brazil, and resurrection of an old one. Systematic Botany. 2008;33:267–276. [Google Scholar]
- Rua GH, Speranza PR, Vaio M, Arakaki M. A phylogenetic analysis of the genus Paspalum (Poaceae) based on cpDNA and morphology. Plant Systematics and Evolution. 2010;288:227–243. [Google Scholar]
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiology. 2009;149:14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Ken JG. Two new species of Paspalum (Paniceae: Panicoideae: Poaceae), a preliminary checklist of the genus in Mexico, and the identity of P. crinitum. Revista Mexicana de Biodiversidad. 2010;81:629–647. [Google Scholar]
- Sandhu S, Altpeter F, Blount AR. Apomictic bahiagrass expressing the bar gene is highly resistant to glufosinate under field conditions. Crop Science. 2007;47:1691–1697. [Google Scholar]
- Sandhu S, James VA, Quesenberry KH, Altpeter F. Risk asseessment of transgenic apomictic tetraploid bahiagrass cytogenetics, breeding behavior and performance of intra-specific hybrids. Theoretical and Applied Genetics. 2009;119:1383–1395. doi: 10.1007/s00122-009-1142-y. [DOI] [PubMed] [Google Scholar]
- Sandhu S, Blount AR, Quesenberry KH, Altpeter F. Apomixis and ploidy barrier suppress pollen-mediated gene flow in field grown transgenic turf and forage grass (Paspalum notatum Flüggé) Theoretical and Applied Genetics. 2010;121:919–929. doi: 10.1007/s00122-010-1360-3. [DOI] [PubMed] [Google Scholar]
- Sartor ME, Quarin CL, Espinoza F. Mode of reproduction of colchicine-induced Paspalum plicatulum tetraploids. Crop Science. 2009;49:1270–1276. [Google Scholar]
- Sartor ME, Quarin CL, Urbani MH, Espinoza F. Ploidy levels and reproductive behaviour in natural populations of five Paspalum species. Plant Systematics and Evolution. 2011;293:31–41. [Google Scholar]
- Savidan Y. Transfer of apomixis through wide crosses. In: Savidan Y, Carman JG, Dresselhaus T, editors. The flowering of apomixis: from mechanisms to genetic engineering. Mexico: Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT); 2001. pp. 153–167. [Google Scholar]
- Schatlowski N, Kölher C. Tearing down barriers: understanding the molecular mechanisms of interploidy by hybridization. Journal of Experimental Botany. 2012;63:6059–6067. doi: 10.1093/jxb/ers288. [DOI] [PubMed] [Google Scholar]
- Sede SM, Morrone O, Giussani LM, Zuloaga FO. Phylogenetic studies in Paniceae (Poaceae): a realignment of section Lorea of Panicum. Systematic Botany. 2008;33:284–300. [Google Scholar]
- Sede SM, Zuloaga FO, Morrone O. Phylogenetic studies in the Paniceae (Poaceae-Panicoideae); Ocellochloa, a new genus from the New World. Systematic Botany. 2009;34:684–693. [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, et al. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. The Plant Cell. 2010;22:655–671. doi: 10.1105/tpc.109.072223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siena LA, Sartor ME, Espinoza F, Quarin CL, Ortiz JPA. Genetic and embryological evidences of apomixis at the diploid level in Paspalum rufum support recurrent auto-polyploidization in the species. Sexual Plant Reproduction. 2008;21:205–215. [Google Scholar]
- Singh M, Goel S, Meeley RB, et al. Production of viable gamets without meiosis in maize deficient for an ARGONAUTE protein. The Plant Cell. 2011;23:443–458. doi: 10.1105/tpc.110.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RL, Grando MF, Li YY, Seib JC, Shatters RG. Transformation of bahiagrass (Paspalum notatum Flüggé) Plant Cell Reports. 2002;20:1017–1021. [Google Scholar]
- Snyder LA. Apomixis in Paspalum secans. American Journal of Botany. 1957;44:318–324. [Google Scholar]
- Snyder LA. Asyndesis and meiotic non-reduction in microsporogenesis of apomitic Paspalum secans. Cytologia. 1961;26:50–61. [Google Scholar]
- Souza-Chies LE, Rua GH, Valls JFM, Rogéria B, Miz RB. A preliminary approach to the phylogeny of the genus Paspalum (Poaceae) Genetica. 2006;126:1–2. doi: 10.1007/s10709-005-1428-1. [DOI] [PubMed] [Google Scholar]
- Spain BH, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J. Truncated forms of a novel yeast protein suppress the lethality of a G protein alpha subunit deficiency by interacting with the beta subunit. Journal of Biological Chemistry. 1995;270:25435–25344. doi: 10.1074/jbc.270.43.25435. [DOI] [PubMed] [Google Scholar]
- Speranza PR. Evolutionary patterns in the Dilatata group (Paspalum, Poaceae) Plant Systematics and Evolution. 2009;282:43–56. [Google Scholar]
- Stebbins GL. Apomixis in angiosperms. Botanical Review. 1941;7:507–542. [Google Scholar]
- Stebbins GL. Variation and evolution in plants. New York: Columbia University Press; 1950. [Google Scholar]
- Stein J, Quarin CL, Martínez EJ, Pessino SC, Ortiz JPA. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theoretical and Applied Genetics. 2004;109:186–191. doi: 10.1007/s00122-004-1614-z. [DOI] [PubMed] [Google Scholar]
- Stein J, Pessino SC, Martínez EJ, et al. A genetic map of tetraploid Paspalum notatum Flügge (bahiagrass) based on single-dose molecular markers. Molecular Breeding. 2007;20:153–166. [Google Scholar]
- Stevens PF. (onwards) Angiosperm Phylogeny Website. Version 12, July 2012. 2001 http://www.mobot.org/MOBOT/research/APweb/ [Google Scholar]
- Urbani MH. Estudios sobre citología, sistema reproductivo y compatibilidad polen–pistilo de Panicum dichotomiflorum y Paspalum fasciculatum (Gramineae:Paniceae) Darwiniana. 1996;34:193–198. [Google Scholar]
- Urbani MH, Quarin CL, Espinoza F, Penteado MIO, Rodrigues IF. Cytogeography and reproduction of the Paspalum simplex polyploid complex. Plant Systematics and Evolution. 2002;236:99–105. [Google Scholar]
- Vielle-Calzada JP, Crane CF, Stelly DM. Apomixis: the asexual revolution. Science. 1996;274:1332–1333. [Google Scholar]
- Vinkenoog R, Spielman M, Adams S, Fischer RL, Dickinson HG, Scott RJ. Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. The Plant Cell. 2000;12:2271–2282. doi: 10.1105/tpc.12.11.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel KP, Burson BL. Breeding and genetics. In: Moser LE, Burson BL, Sollenberger LE, editors. Warm-season (C4) grasses. Agronomy Monograph 45. Madison: ASA, CSSA, SSSA; 2004. pp. 51–94. [Google Scholar]
- Williams WM, Williamson ML, Real D. Paspalum In. In: Kole C, editor. Wild crop relatives: Genomic and breeding resources: Millets and grasses. Berlin: Springer, 197–215; 2011. [Google Scholar]
- Yu HJ, Hogan P, Sundaresan V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiology. 2005;139:1853–1869. doi: 10.1104/pp.105.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. The Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga FO, Morrone O. Revisión de las especies de Paspalum para América del Sur austral. Annals of the Missouri Botanical Garden; Monographs in Systematic Botany. 2005;102:1–297. [Google Scholar]
- Zuloaga FO, Giussani L, Morrone O. Hopia, a new monotypic genus segregated from Panicum (Poaceae) Taxon. 2007;56:145–156. [Google Scholar]
- Zuloaga FO, Scataglini MA, Morrone O. A phylogenetic evaluation of Panicum sects. Agrostoidea, Megista, Prionita and Tenera (Panicoideae, Poaceae): two new genera, Stephostachys and Sorengia. Taxon. 2010;59:1535–1546. [Google Scholar]
- Zuloaga FO, Morrone O, Scataglini MA. Monograph of Trichanthecium, a new genus segregated from Panicum (Poaceae, Paniceae) based on morphological and molecular data. Systematic Botany Monographs. 2011;94 [Google Scholar]