Abstract
A distinctive mechanism for ubiquitin (Ub) ligation has recently been proposed for the RING1-IBR-RING2 (RBR) family of E3s: an N-terminal RING1 domain recruits a thioester-linked intermediate complex between Ub and the E2 UbcH7, and a structurally unique C-terminal RING2 domain displays a catalytic cysteine required for Ub ligation. To obtain insights into RBR E3s, we determined the crystal structure of the Human Homolog of Ariadne (HHARI), which reveals the individual RING1, IBR, and RING2 domains embedded in superdomains involving sequences specific to the Ariadne RBR subfamily. The central IBR is flanked on one side by RING1, which is exposed and binds UbcH7. On the other side, a C-terminal autoinhibitory “Ariadne domain” masks the RING2 active site. Insights into RBR E3 mechanisms are provided by structure-based mutations that indicate distinct steps of relief from autoinhibition, Ub transfer from E2 to HHARI, and ligation from the HHARI cysteine to a terminal acceptor.
INTRODUCTION
Ubiquitin (Ub) is ligated to targets through the coordinated action of E1 (ubiquitin activating), E2 (ubiquitin conjugating), and E3 (ubiquitin ligase) enzymes. First, the activated Ub protein is typically transferred between E1 and E2 active site cysteines. The resulting covalent thioester-linked E2~Ub intermediate next employs an E3 enzyme. E3s utilize a variety of mechanisms to catalyze Ub ligation to targets. Many E3s function as activating scaffolds that direct transfer of Ub's C-terminus from the E2's active site to a substrate Lys. This category is exemplified by the large family of RING E3s (Deshaies and Joazeiro, 2009). RING domains typically coordinate two zinc ions in a cross-braced structure via Cys, His, and occasionally other side-chains. Notably, structural studies have revealed how RING domains bind both E2 and Ub to optimally orient and activate the E2~Ub intermediate for nucleophilic attack, and how RING E3s can bind both a substrate and E2~Ub to position a substrate's acceptor Lys adjacent to an associated E2's active site (Calabrese et al., 2011; Dou et al., 2012a; Plechanovova et al., 2012; Pruneda et al., 2012).
Another mechanism by which E3s can promote Ub ligation involves Ub transfer from the E2 to a Cys on the E3 itself, generating a thioester-linked E3~Ub intermediate. It is thought that Ub is ultimately transferred from the E3 catalytic cysteine to the target. The first thioester-forming E3s to be identified were members of the HECT family (Huibregtse et al., 1995; Scheffner et al., 1995), and a prior structural study revealed how the HECT catalytic domain binds both E2 and Ub for Ub transfer to the E3 Cys (Kamadurai et al., 2009). More recently, several thioester-forming E3 ligases have been identified from pathogenic bacteria, which are thought to hijack the host Ub system during infection processes (Anderson and Frank, 2012). These bacterial E3 ligases are structurally unrelated to HECT E3s, but nevertheless bind E2 for Ub transfer to the E3 catalytic Cys (Diao et al., 2008; Huang et al., 1999; Lin et al., 2012; Quezada et al., 2009; Singer et al., 2008; Verdecia et al., 2003; Zhu et al., 2008). Active forms of both HECT and bacterial thioester-forming E3s often undergo autoubiquitination, reflecting the high degree of reactivity of the E3~Ub thioester intermediate.
Recently, a landmark study revealed a variant mechanism for the RING1-IBR (in-between-RING)-RING2 (RBR) family of E3s (Wenzel et al., 2011). RBR E3s have a RING1 domain for E2-binding that shares many features of standard RING E3s. However, the finding that the RING1 domain of some RBRs binds the E2 UbcH7 (Ardley et al., 2001; Moynihan et al., 1999), which was reported to transfer Ub only to cysteines and not to lysines, led to the reclassification of RBRs as “RING-HECT hybrids” (Wenzel et al., 2011). A conserved unliganded Cys in a second, structurally distinctive “RING2” domain is required for RBR E3-mediated autoubiquitination or free Ub chain formation in vitro, which are readouts for Ub ligation activity (Wenzel and Klevit, 2012; Wenzel et al., 2011). The corresponding Cys is mutated in the RBR E3 Parkin in some cases of Parkinson's Disease, and is also required for Parkin to bind its substrates, and for Parkin-mediated substrate ubiquitination in cells (Lazarou et al., 2013; Nuytemans et al., 2010; Sarraf et al., 2013).
The best-understood RBR E3s are the Autosomal Recessive Juvenile Parkinson's disease associated protein Parkin, the heterotrimeric Linear Ubiquitin Chain Assembly Complex (LUBAC) that contains both the HOIP and HOIL-1L RBR proteins, and members of the Ariadne subfamily (Marin, 2009; Wenzel and Klevit, 2012). Parkin and LUBAC have been implicated in regulating mitochondrial dynamics and NF-κB signaling, respectively (Ikeda et al., 2011; Kitada et al., 1998; Tokunaga and Iwai, 2012; Tokunaga et al., 2011). The Ariadne subfamily is the largest RBR cohort, with Ariadne-type RBR E3s found in all eukaryotes including yeast. In addition to a central RBR catalytic domain, Ariadne-family RBRs are characterized by a C-terminal “Ariadne domain” (Marin and Ferrus, 2002). Two human E3s, ARIH1/HHARI and ARIH2/TRIAD, are closely related to Drosophila Ariadne. Other more divergent Ariadne subfamily members have evolved to contain additional domains, such as CUL9/PARC which has an N-terminal cullin-homology domain, and ANKIB1 which has N-terminal ankyrin repeats (Marin and Ferrus, 2002). Although specific pathways regulated by Ariadne subfamily members are relatively poorly understood, their functional importance has been shown by genetic studies. For example, Drosophila Ariadne is essential for development (Aguilera et al., 2000). Murine Arih2 is also required for embryogenesis, and reconstitution of hematopoetic lineages with Arih2 null fetal liver cells leads to lethal activation of the immune response (Lin et al., 2013). The yeast Ariadne subfamily member Hel1 (“Histone E3 Ligase 1”) was shown to be important for degrading exogenously expressed Histone H3 (Singh et al., 2012).
Although truncated forms of the proteins are active, full-length Parkin and HOIP proteins are inactive on their own (Chaugule et al., 2011; Smit et al., 2012; Stieglitz et al., 2012). These two E3s are thought to be autoinhibited through distinct mechanisms as they lack obvious sequence homology beyond their RBR domains. In the case of Parkin, an N-terminal Ub-like domain binds between the IBR and RING2 domains to lock the E3 in an inactive conformation (Chaugule et al., 2011). In vivo, Parkin is activated through recruitment to the mitochondrial membrane and phosphorylation by the PINK1 kinase (Kondapalli et al., 2012; Lazarou et al., 2012; Lazarou et al., 2013; Sha et al., 2010). By contrast, HOIP is activated within the LUBAC complex by interaction with either of its HOIL-1L and/or SHARPIN partner proteins (Ikeda et al., 2011; Kirisako et al., 2006; Smit et al., 2012; Stieglitz et al., 2012; Tokunaga et al., 2011).
Given the emerging appreciation of RBRs as a distinctive family of E3 ligases, it is important to understand the structural basis for their functions. Although there are structures of isolated E2-binding canonical RING E3s, as well as an RBR-specific IBR, and RING2 domains, to date, there is no structure of a full RING1-IBR-RING2 E3 (Beasley et al., 2007; Capili et al., 2004; Dou et al., 2012b; Plechanovova et al., 2012; Pruneda et al., 2012; Zheng et al., 2000). To address this problem, we determined the crystal structure of HHARI. Taken together with biochemical experiments, the data provide structural insights into RBR E3s, and reveal how a thioester-forming E3 ligase is autoinhibited by a conserved domain masking its active site cysteine.
RESULTS
Full-length HHARI binds the E2, UbcH7, but is inactive with generic readouts for E3 ligase function
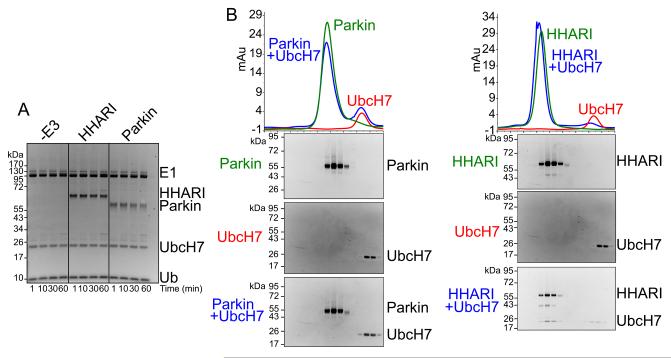
To date, in vitro activities of Parkin and HOIP have been best-characterized through either autoubiquitination, which is a common read-out for E3 ligase activity, or free Ub chain formation (Chaugule et al., 2011; Smit et al., 2012; Stieglitz et al., 2012; Wenzel et al., 2011). Whereas their RBR domains are active in these assays, both Parkin and HOIP proteins are autoinhibited in their full-length forms (reviewed in Dove and Klevit, 2012). Thus, we wondered whether a common feature of RBR E3s is their maintenance in an inactive state. We examined intrinsic, substrate-independent E3 ligase activity of the Human Homolog of Ariadne (HHARI), which shares no apparent homology to Parkin and HOIP beyond the RBR domain, and obtained a similar negative result (Fig. 1A).
Figure 1. HHARI is inactive for autoubiquitination but unlike Parkin binds UbcH7.
a. SYPRO-Ruby-stained reducing SDS-PAGE gel after time-course incubating full-length HHARI or Parkin with E1 (Uba1), UbcH7, Ub, and MgATP.
b. Gel filtration chromatography profiles of Parkin or HHARI alone (green), UbcH7 (red), and of Parkin or HHARI mixed with UbcH7 (blue). SYPRO-Ruby-stained SDS-PAGE gels of fractions from the gel filtration peaks are shown below chromatograms. For reference to relative migration during chromatography, the same data with UbcH7 are shown below both chromatograms.
A fundamental requirement for E3 ligase function is binding to the E2. We therefore examined binding of full-length Parkin and HHARI to their reported cognate E2, UbcH7, by comigration over gel filtration chromatography. Unlike Parkin, HHARI and UbcH7 copurify by this method (Fig. 1B). Taken together, the results raised the possibility that HHARI is maintained in an architecture that allows E2 binding but prevents E3 ligase activity.
Overall structure of HHARI – an elongated architecture assembled from multiple domains
To gain insights into HHARI architecture, we crystallized the full-length HHARI protein and determined the structure from crystals growing in the space group P41 to a resolution of 3.3 Å (Table 1, Fig. S1, Fig. S2). Initial phases were obtained by MAD based on the six tightly bound zinc atoms in the native protein. Anomalous data on crystals of selenomethionine-labeled HHARI, as well as data collected from 8 individual site-specific mutants, incorporating one additional selenomethionine residue at a time, guided building the structure (Fig. S1). Because the P41 crystal form contained two molecules per asymmetric unit, we also determined the structure from a second crystal form in the space group P63, with a single molecule of HHARI per asymmetric unit. Despite great effort at optimization, anisotropy of data collected from the P63 form limited the resolution and refinement. Nonetheless, the high quality electron density confirmed the overall features of the structure in the P63 form, and suggested that HHARI is a monomer (Fig. S1, S2). Indeed, the molecular weight of HHARI measured by sedimentation velocity analytical ultracentrifugation of 66.9 kDa and by size exclusion chromatography-multiangle light scattering of 62.2 kDa agree well with the calculated molecular weight of 64.2 kDa for a monomer (Fig. S2). Thus, in general a single molecule of HHARI from the P41 crystal form is described here.
Table 1.
Crystallographic Data and Refinement Statistics (supported by Supplementary Figure 2)
| Data Collection | Crystal Form 1 | Crystal Form 2 |
|---|---|---|
| Wavelength (λ) | 0.9798 | 0.9798 |
| Space Group | P41 | p63 |
| Unit cell parameters | ||
|---|---|---|
| a,b,c(Å) | 147.6, 147.6, 87.34 | 96.1, 96.1, 151.3 |
| α,β,γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 46.67 - 3.30 | 36.47 - 3.60 |
| No. of measured reflections | 84,920 | 95,159 |
| Original | Truncated* | ||
|---|---|---|---|
| No. of unique reflections | 27,938 | 9,276 | 8,348 |
| Overall Rsym (%) | 4.9 (84.3) | 9.2 (49.2) | 8.8 (15.0) |
| Completeness (%) | 98.1 (96.4) | 99.9 (99.9) | 89.9 (17.6) |
| Overall I/σI | 14.8 (1.3) | 25.8 (3.0) | 27.8 (15.9) |
| Mean Redundancy | 3.0 | 10.2 (9.4) | 10.4 (11.1) |
| Refinement | ||
|---|---|---|
| Rwork/Rfree | 0.231/0.283 | 0.270/0.299 |
| rmsd bond lengths (Å) | 0.008 | 0.011 |
| rmsd bond angles (°) | 1.4 | 1.2 |
| Subunits in asymmetric unit | 2 | 1 |
| No. of atoms | ||
|---|---|---|
| Protein | 6569 | 3133 |
| Zinc | 12 | 6 |
| Ramachandran statistics | ||
|---|---|---|
| Residues in favored regions (%) | 95.8 | 94.1 |
| Residues in disallowed regions (%) | 0.1 | 0.0 |
Truncated values correspond to statistics after elliptical truncation prompted by severe anisotropy
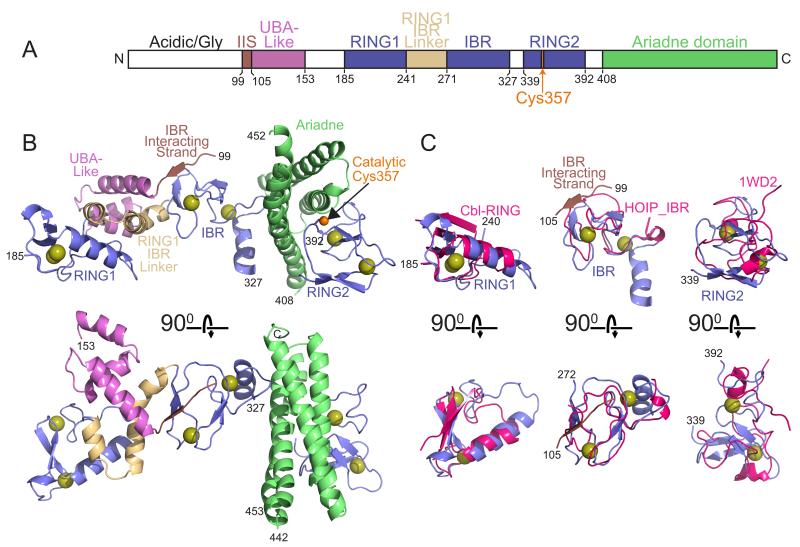
HHARI adopts an extended, roughly 90 Å-long assembly of multiple domains: a β-strand, a 3-helix bundle that unexpectedly adopts a UBA domain-like fold, RING1, a 2-helix RING1-to-IBR linker, the IBR, RING2, and the Ariadne domain. Several of these domains pack with each other to form superdomains (Fig. 2). The HHARI N-terminal 98 residues were required for expression and crystallization, although this region, which contains 30 glycines and 36 acidic residues, was not visible in the electron density from either crystal form. Residues 154–184 linking the UBA-like domain and RING1, and 328–338 linking the IBR and RING2 were likewise not visible and are not modeled.
Figure 2. Structure of HHARI (supported by Supplementary Figure 1).
a. Schematic view of domain structure of HHARI. Colored boxes correspond to the IBR-interacting strand (IIS), UBA-like, RING1, RING1-to-IBR linker, IBR, RING2, and Ariadne domains observed in the crystal structure. White boxes correspond to regions not visible in the electron density. The location of the catalytic Cys357 is indicated in orange.
b. Two views, rotated 90° around the x axis, of one molecule in the asymmetric unit for HHARI determined from the P41 crystal form, with the IBR-interacting strand in maroon, the UBA-like domain in violet, the RING1, IBR, and RING2 domains in slate, the RING1-to-IBR linker in tan, and the Ariadne domain in green. Zinc atoms are shown as yellow spheres.
c. Individual domains from the RBR superimposed on related structures: HHARI RING1 (slate) superimposed on a canonical RING domain as shown from c-Cbl (1FBV.PDB, pink); HHARI IBR domain (slate) superimposed on the IBR domain from HOIP (2CT7.PDB, pink); HHARI RING2 manually docked via its C-terminal zinc-binding site on the NMR structure of the isolated HHARI RING2 bound to only a single zinc atom (1WD2.PDB, pink).
At the center of the HHARI structure is the IBR, which superimposes on the prior NMR structure of the IBR domain from HOIP with 3.2 Å RMSD (Fig. 2C). Embedded within the IBR structure is the most N-terminal region visible in the structure (residues 99–105), which we term “IBR-interacting strand” due to its forming part of a 3-stranded β-sheet with two strands from the IBR. On one side of the IBR, a discontinuous helical domain containing the UBA-like domain and the RING1-to-IBR linker connect the IBR and RING1. RING1 superimposes well onto structures of canonical RING domains, such as of that from c-Cbl with 2.4 Å RMSD (Fig. 2C).
C-terminal of the IBR, RING2 adopts a different zinc-binding fold. Of the eight cysteines in RING2, seven are involved in coordinating two zinc atoms (Fig. S3). One zinc is coordinated by four cysteines from two two-stranded antiparallel β-sheets. The C-terminal cysteine in the terminal β-strand connects to a loop-like structure in which two other cysteines and a histidine face inward to coordinate a second zinc atom. This latter zinc-binding site can be manually docked partially onto the corresponding region from a prior NMR structure (Fig. 2C) (Capili et al., 2004). The RING2 domain also contains a single unliganded cysteine, Cys357, in agreement with the prior finding suggesting this as the only unliganded cysteine in RING2: treatment of a purified RING2 domain with the thiol reactive 4-(2-iodoacetamido)-TEMPO results in a single modification, only to Cys357 (Wenzel et al., 2011). Cys357 is the conserved RBR catalytic cysteine shown previously to be required for autoubiquitination of a GST-fusion of the RBR domain from HHARI (Wenzel et al., 2011).
The C-terminal Ariadne domain adopts an elongated four-helix bundle consisting of an antiparallel arrangement of 7-10-turn long helices. The Ariadne domain is straddled on one side by the IBR domain, and on the other by the RING2 domain. Notably, the Ariadne domain embraces the surface of RING2 containing the catalytic Cys357 (Fig. 2B).
Ariadne domain masks the HHARI catalytic Cys357 and inhibits intrinsic E3 activity
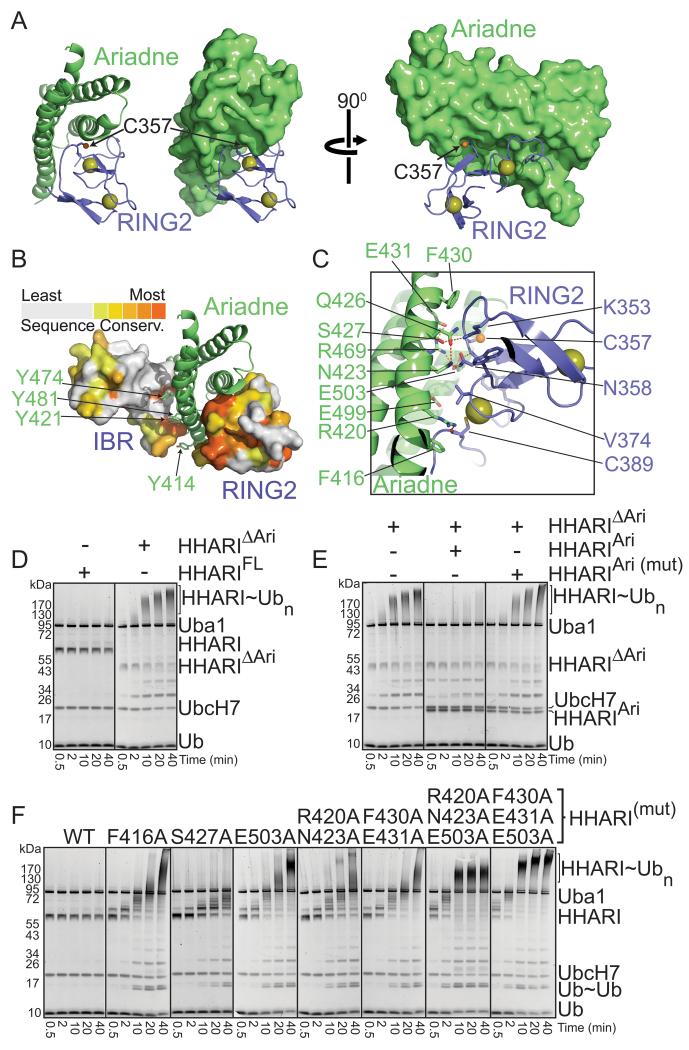
An extensive, highly conserved surface from RING2 packs in a broad 25 Å by 35 Å cleft formed by three of the four helices in the Ariadne domain (Fig. 3A, 3B, S3). This interaction buries a total of 1000 Å2 from both proteins, including more than 30 percent of the total surface from RING2 and 122 contacts between the domains (Table S1). This intramolecular interaction is secured by extensive contacts from a surface including Asn423, Ser427, Phe430, Glu431, and Glu503 from the Ariadne domain cradling the RING2 loop spanning from Lys353 to Asn358, which includes the catalytic Cys357 (Fig. 3C). The contacts extend to the C-terminus of RING2, anchored by Arg420 from the Ariadne domain making a hydrogen bond to the carbonyl from the most C-terminal zinc ligand (Cys389) from RING2, and Phe416 from the Ariadne domain making van der Waals contacts to the backbone extending from Cys389 to the terminus of the RING2 domain.
Figure 3. The Ariadne domain binds RING2 to mediate autoinhibition (supported by Supplementary Figure 3).
a. The Ariadne domain (green) masks the RING2 (slate) surface containing the catalytic Cys357 (sulfhydryl shown as orange sphere). Zinc atoms are shown as yellow spheres.
b. The IBR and RING2 domains are shown in surface representation, colored by conservation among Ariadne family members, with the Ariadne domain shown in green cartoon.
c. Close-up view of intramolecular contacts between the Ariadne domain (green) and RING2 domain (slate, with catalytic Cys357 sulfhydryl represented as a small orange sphere, and zinc atoms as yellow spheres).
d. SYPRO-Ruby stained reducing SDS-PAGE gels showing time-courses of autoubiquitination of the full-length or mutant form of HHARI with the Ariadne domain deleted (HHARIΔAri) in the presence of E1 (Uba1), E2 (UbcH7), Ub, and MgATP.
e. SYPRO-Ruby stained reducing SDS-PAGE gels showing time-courses of autoubiquitination of HHARIΔAri in the presence or absence of the isolated Ariadne domain (HHARIAri) or a mutant (HHARIAri (mut)) in the structurally-observed RING2-binding surface (F430A, E431A, E503A).
f. SYPRO-Ruby stained reducing SDS-PAGE gels showing time-courses of autoubiquitination of wild-type full-length HHARI or indicated mutants with alanine substitutions in the RING2-binding surface of the Ariadne domain.
The structure suggests that the lack of intrinsic E3 ligase activity may result from the Ariadne domain blocking RING2. We performed several assays to test this idea. First, we found that a deletion mutant of HHARI lacking the Ariadne domain (HHARIΔAriadne) is extensively autoubiquitinated (Fig. 3D). Second, the isolated Ariadne domain restores inhibition when added in trans to HHARIΔAriadne (Fig. 3E). Importantly, the negative control mutant harboring Ala substitutions in Phe430, Glu431, and Glu503 from the structurally-observed RING2-binding surface fails to block E3 ligase activity, consistent with the notion that the Ariadne domain binds to RING2 to mediate inhibition (Fig. 3E). Third, Ala mutations in the structurally-observed RING2-binding surface of the Ariadne domain are sufficient to activate both autoubiquitination of and free Ub-chain formation by full-length HHARI (Fig. 3F).
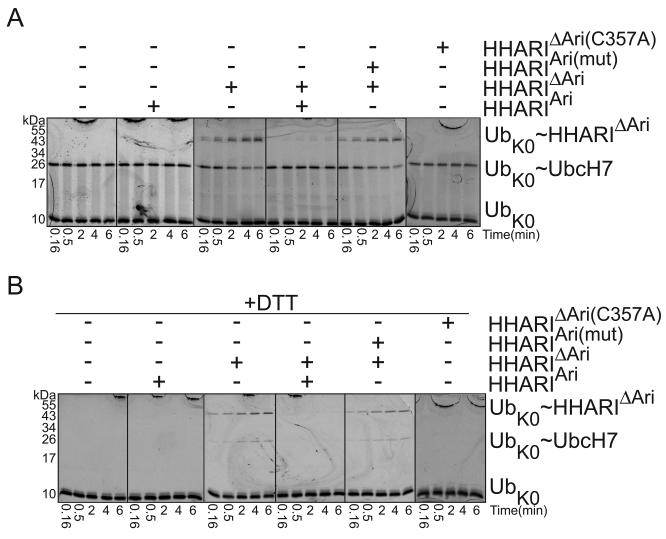
In order to determine whether the Ariadne domain blocks the initial Ub transfer from UbcH7, we developed a pulse-chase assay. First, we enzymatically generated a fluorescent Ub~UbcH7 thioester intermediate, and quenched E1-mediated loading of UbcH7 by adding apyrase to eliminate ATP and prevent recharging. The Ub~UbcH7 intermediate is stable in the presence of a mutant version of HHARIΔAriadne with the catalytic Cys357 substituted with alanine (Fig. 4A). However, addition of wild-type HHARIΔAriadne results in loss of the thioester-linked UbcH7~Ub intermediate, and concomitant formation of nonreducible complexes with the fluorescent Ub linked to both HHARIΔAriadne and UbcH7, both being sources of available lysines in the reactions (Fig. 4A, 4B). Notably, upon addition of the isolated Ariadne domain, but not the version mutated in the RING2-binding surface, the thioester-linked UbcH7~Ub intermediate persists and Ub ligation is inhibited (Fig. 4A). The results are consistent with the Ariadne domain blocking the catalytic cysteine from RING2 to prevent formation of a HHARI~Ub intermediate.
Figure 4. The Ariadne domain inhibits Ub transfer from UbcH7 to HHARI (supported by Supplementary Table 1).
a. Fluorescence scans of nonreducing SDS-PAGE gels showing time-courses of the “chase” in which the thioester-linked fluorescent lysinelesss UbK0~UbcH7 intermediate was incubated with HHARIΔAri in the presence or absence of the isolated Ariadne domain (HHARIAri) or the mutant (HHARIAri(mut)) in the structurally-observed RING2-binding surface (F430A, E431A, E503A).
b. Same experiment as panel a, but after samples were reduced with DTT.
The canonical RING1 packs against the RING1-to-IBR linker helices for recruitment but not activation of E2
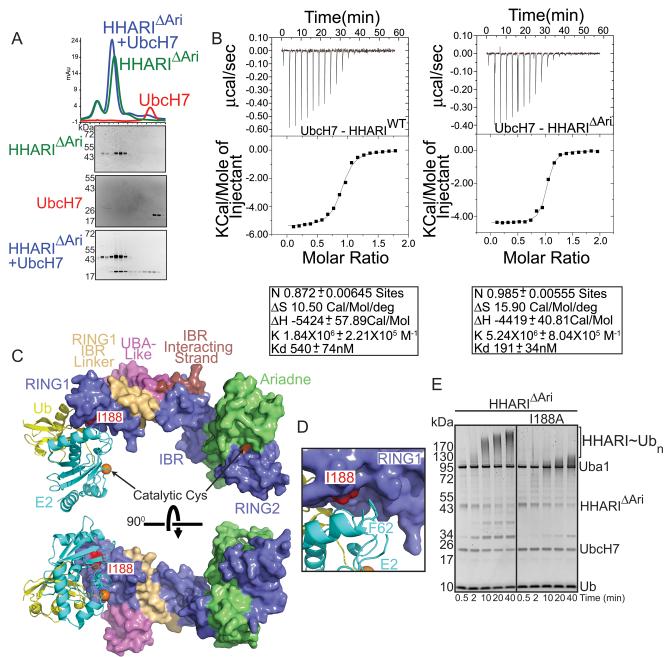
Our identification of the Ariadne-RING2 domain interaction as responsible for autoinhibition enabled interrogation of other functional regions of HHARI through biochemical assays with the active HHARIΔAriadne mutant. The HHARI structure provides a framework for understanding previous mutagenesis data defining requirements for interacting with the E2, UbcH7: coimmunoprecipitation of HHARI and UbcH7 from mammalian cells required both HHARI's RING1 and residues 238–273 (Moynihan et al., 1999) (Ardley et al., 2001). Residues 238–273 form the RING1-to-IBR linker, which packs tightly against RING1 to form an integrated RING1/RING1-to-IBR1 structural unit (Fig. 2). The RING1-to-IBR1 linker also packs against the N-terminal UBA-like domain. Although it is tempting to speculate a role for the UBA-like domain, we see no effect of making aspartate substitutions in place of surface hydrophobic residues that might be predicted to bind Ub's hydrophobic Ile44 patch (Fig. S4). Thus, future studies will be required to ascertain functional roles of the UBA-like domain from HHARI. In a related vein, in previous studies, deleting the Ariadne domain had no effect on association of HHARI and UbcH7 in mammalian cells (Moynihan et al., 1999). Accordingly, the Ariadne domain is structurally distal and separated from RING1 and the RING1-to-IBR linker, and is dispensable for their copurification by gel filtration in vitro (Fig. 5A). Interestingly, the activated version of HHARI lacking the autoinhibitory Ariadne domain binds slightly tighter to UbcH7 (roughly 190 versus 540 nM Kd measured by isothermal titration calorimetry), although future studies will be required to determine the basis for this enhanced affinity (Fig. 5B).
Figure 5. Recruitment of UbcH7 to HHARI RING1 is required for intrinsic autoubiquitination activity (supported by Supplementary Figure 4).
a. Gel filtration chromatography profiles of HHARIΔAriadne (HHARIΔAri) alone (green), UbcH7 alone (red) or a mixture of HHARIΔAri and UbcH7 (blue). SYPRO-Ruby-stained SDS-PAGE gels of fractions from the gel filtration peaks are shown below chromatograms.
b. Isothermal Titration Calorimetry data for interactions between UbcH7 and wild-type (WT) HHARI or HHARIΔAri. Upper panels show raw power data recorded during the titration experiments, and lower panels show fits of standard binding equations after integration of the raw data, using Origin (v. 7.0) software provided by MicroCal.
c. Surface representation of HHARI with UbcH5a~Ubiquitin from the RNF4~UbcH5a~Ubiquitin complex (4AP4.pdb) modeled onto RING1. The positions of the catalytic cysteines are marked by orange spheres. Ile188, a mutant defective in E2 binding, is shown in red.
d. Close-up view of the predicted E2-RING1 interaction showing the positions of UbcH7 Phe62 and HHARI Ile188.
e. SYPRO-Ruby-stained SDS-PAGE gel showing a time course for autoubiquitination of HHARIΔAri or HHARIΔAri I188A
Docking models of a HHARI-E2~Ub complex, generated by superimposing RING domains from HHARI and RNF4 or BIRC7 in complexes with E2~Ub (Dou et al., 2012b; Plechanovova et al., 2012), shows Ile188 at the center of the interaction with E2 (Fig. 5C, D). Previously, mutation of Ile188, or zinc-binding ligands required for formation of the RING1 fold, abolished HHARI-UbcH7 complex formation (Ardley et al., 2001; Wenzel et al., 2011). We find that an Ile188Ala mutation substantially decreases intrinsic E3 ligase activity for the activated HHARIΔAriadne construct (Fig. 5E).
RING2 active site loop residues contribute to Ub ligation
Although HHARI's Cys357 is required for Ub ligation, we inferred that an intermediate must be highly reactive because we were unable to unambiguously trap a substantial fraction of HHARIΔAriadne thioester-linked to Ub. This agrees with previous studies of truncated, active forms of Parkin, HOIP, and HHARI that either likewise failed to observe an intermediate or reported only an extremely small proportion of total E3 as forming a reducible thioester-linked E3~Ub complex (Smit et al., 2012; Stieglitz et al., 2012; Wenzel et al., 2011). Thus, to gain insights into RBR-mediated catalysis, we examined the structure around the HHARI active site (Fig. 6A) to identify residues potentially involved in ligation. If residues exist that specifically contribute to ligation, then in theory their mutation would allow stabilization of the thioester-linked E3~Ub intermediate.
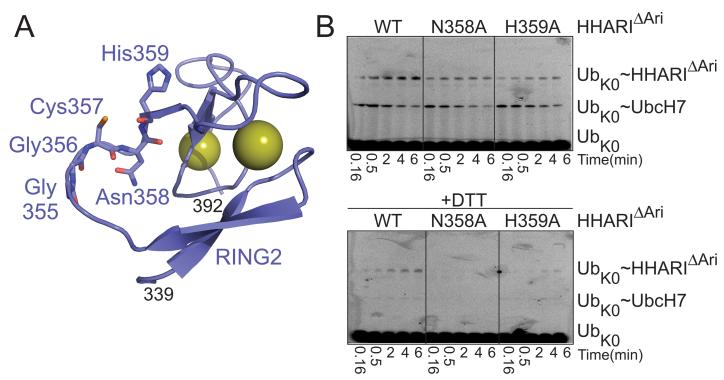
Figure 6. RING2 mutations that stabilize E3 ubiquitin-thioester intermediate reveal residues required for ligation reaction.
a. Cartoon representation of RING2 (slate) with showing residues from the catalytic loop. RING2 zinc ions are shown as yellow spheres.
b. Fluorescence scans of SDS-PAGE gels showing time-courses of the “chase” in which the thioester-linked fluorescent Ub~UbcH7 intermediate was incubated with HHARIΔAri or HHARIΔARI catalytic loop mutants N358A or H359A. Top panel is nonreducing. Bottom panel is reducing.
Immediately C-terminal of the catalytic Cys357 are two polar residues, Asn358 and His359, conserved among Ariadne subfamily members. Although Asn358 does not directly contact Cys357, and the Cys357 sulfhydryl makes a hydrogen bond to the backbone oxygen from His359, we considered that the present structure represents an inactive conformation, and it also lacks a covalently-linked Ub. Thus, these side-chains are poised to potentially contribute to E3 ligase activity in an activated but presently unknown conformation whereby HHARI is thioester-bonded to Ub. In our pulse-chase assay, the UbcH7~Ub thioester intermediate decreased in the presence of the activated HHARIΔAriadne construct, and for Asn358Ala and His359Ala mutants. However, under the conditions of our assay, the majority of the HHARIΔAriadne~Ub product formed by the two mutants was the DTT-reducible thioester intermediate (Fig. 6B). We conclude that both Asn358 and His359 are important for the reaction mediating Ub ligation to a terminal acceptor, at least in the context of the intrinsic reactivity reflected by autoubiquitination.
DISCUSSION
The structure of HHARI represents one of few for a full-length E3 ligase, and the first structure of any RBR E3. Taken together with previous data, the structure provides insights into the how the RING1, IBR, and RING2 domains common among RBR E3s are assembled, reveals that HHARI and most likely other Ariadne subfamily members share in common with Parkin and HOIP the property of being autoinhibited, and offers molecular details into the basis of autoinhibition and the two step Ub ligation mechanism. It is of great interest to understand mechanisms for both activating and attenuating RBR E3 activity for therapeutic purposes in Parkinson's Disease and immune hyperactivation (Kitada et al., 1998; Lin et al., 2013; Tokunaga and Iwai, 2012). Analysis of the Parkin sequence in light of the HHARI structure reveals several Parkinson's Disease mutations poised to disrupt the IBR structure, RING1 recruitment or positioning of E2~Ub, or catalytic activities of the RING2 domain (Fig. S3).
Surprisingly, RING1, IBR, and RING2, which are the only regions conserved among all RBR E3s, are widely separated from each other but form integral interactions with sequences that are found only in Ariadne subfamily members. For RING1, this involves the region leading to the IBR domain that likely structurally stabilizes the RING1 domain, explaining requirement of RING1-to-IBR linker helices for binding to HHARI's partner E2 UbcH7 (Ardley et al., 2001). Despite displaying the overall fold of a RING E3, HHARI's RING1 lacks an arginine utilized by some RING domains to structurally intercalate between E2 and Ub to support “folding back” of Ub against E2 and allosterically activate discharge of some thioester-linked E2~Ub intermediates (Dou et al., 2012b; Hamilton et al., 2001; Plechanovova et al., 2012; Pruneda et al., 2012; Reverter and Lima, 2005; Saha et al., 2011; Wickliffe et al., 2011). This is consistent with our finding that the UbcH7~Ub is stable in the presence of the HHARIΔAriadne catalytic Cys357Ala mutant (Fig. 4) and the previous finding that a construct encompassing the HHARI RING1 and IBR domains was inactive at stimulating UbcH5C~Ub discharge to lysine (Wenzel et al., 2011). Thus, taken together, the data suggest that the RING1/RING1-to-IBR module recruits UbcH7 much like other RING domains bind to E2s, but like HECT E3s may proceed through catalysis with E2~Ub in different conformation than that utilized by canonical RING E3s (Kamadurai et al., 2009).
In the context of full-length HHARI, the catalytic RING2 domain differs from the previous NMR structure of the isolated domain (Capili et al., 2004), but agrees with biochemical data showing Cys357 as the sole unliganded Cys in this domain (Wenzel et al., 2011). Our finding of mutations that allow persistence of an E3~Ub thioester intermediate in a pulse-chase assay both confirms the proposed RING-HECT hybrid mechanism for RBRs (Wenzel et al., 2011), and suggests that there are distinct requirements for the two steps of transferring Ub from E2 to an RBR E3 and ligating Ub from the RBR active site cysteine to a terminal lysine (Fig. 6). It will be exciting in the future to learn if these features reflect conformational malleability of RING2 for catalysis.
Our biochemical data allows HHARI to join the growing list of E3 ligases that are autoinhibited in their full-length forms, albeit through a wide-range of mechanisms that include blocking substrate or E2-binding, masking the active site cysteine, and preventing attaining an active conformation (Dou et al., 2012a; Dove and Klevit, 2012; Du et al., 2002; Dueber et al., 2011; Gallagher et al., 2006; Kobashigawa et al., 2011; Lopez et al., 2011; Wiesner et al., 2007; Yamoah et al., 2008). Although the reasons why E3s may be autoinhibited remain largely unknown, it seems likely that this could provide a mechanism for preventing misdirected autoubiquitination, thereby steering Ub ligation to substrates in the correct biological context.
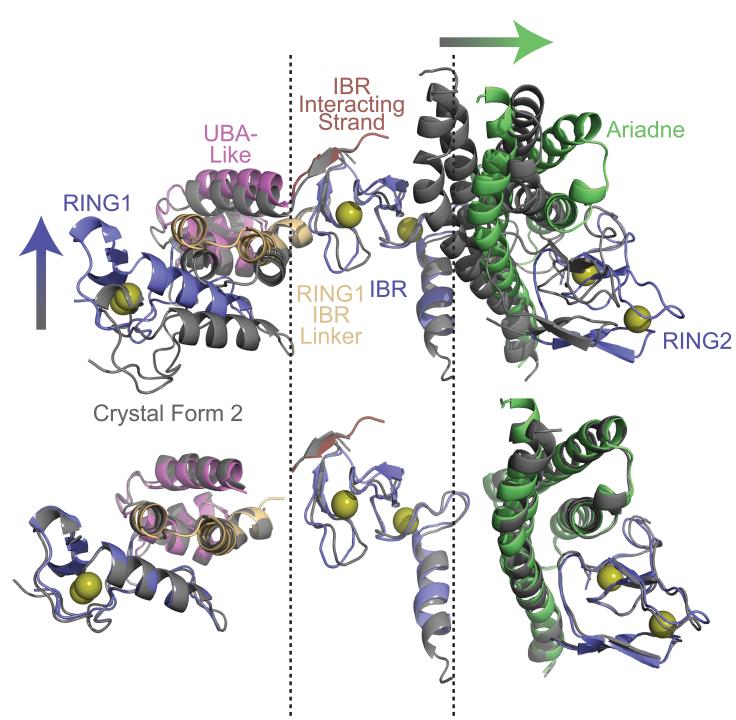
Interestingly, each family of RBR E3 seems to display unique mechanisms of autoinhibition, with the Ariadne-family-specific “Ariadne domain” blocking HHARI activity in at least two ways. First, the catalytic Cys357 is masked and thus inaccessible for forming an intermediate. Second, in the structure, the Ariadne domain is intercalated between the IBR and RING2, contributing to a wide separation between the predicted active site on a RING1-bound E2 and HHARI's catalytic Cys357 in RING2 (Fig. 5). Thus, in addition to the requirement for exposing the RING2 active site, HHARI apparently must undergo substantial structural rearrangement for Ub transfer from E2 to the catalytic cysteine. Indeed, our data do reveal structural variability even in full-length HHARI. Although overall features of HHARI are shared in the P41 and P63 crystal forms, superimposition of the two structures reveals hinges between the IBR domain and the RING/RING-to-IBR linker on one side, and the RING2 and Ariadne domain on the other (Fig. 7). These and other unknown but malleable regions of the structure may enable the conformational changes required for catalysis of E2-to-E3 Ub transfer and subsequent Ub ligation. We anticipate many exciting future studies revealing structural mechanisms of RBR E3 activities.
Figure 7. Superposition of the crystal forms shows domain variation hinging at the IBR.
Superposition of the P63 crystal form (gray) with the P41 crystal form of HHARI aligned over their IBR domains to show structural variability. Arrows indicate directions of domain rotations between the P41 and P63 crystal forms. The individual domains all superimpose well as shown below.
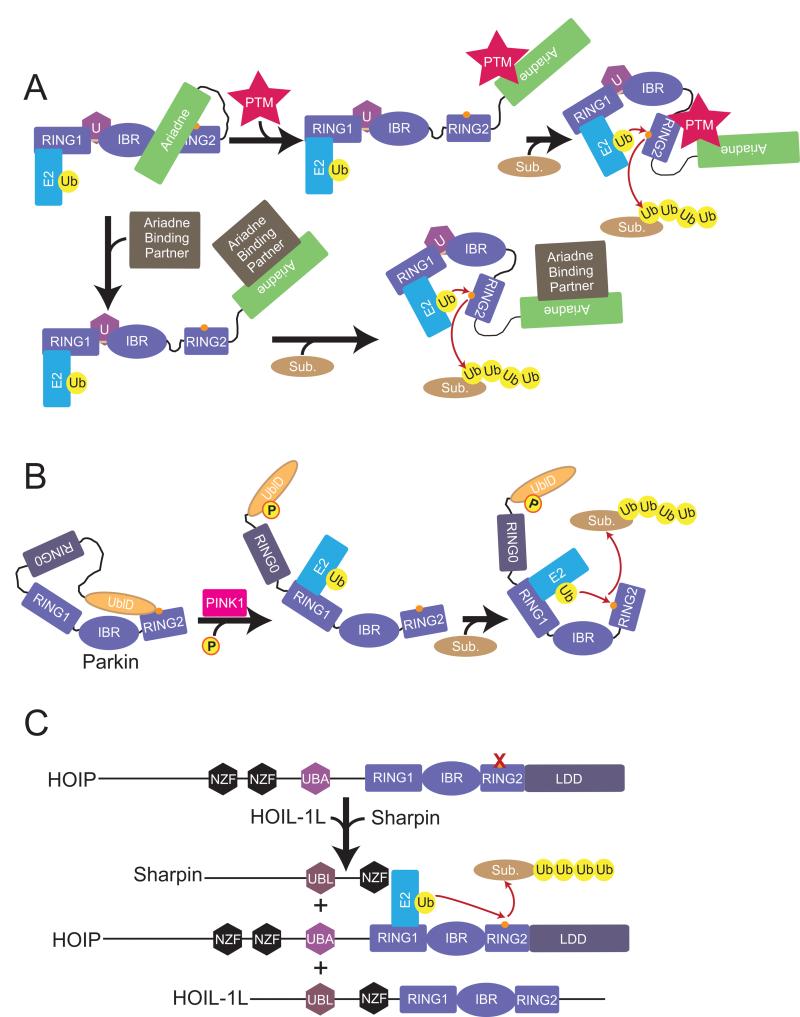
Although at this point the physiological mechanisms relieving autoinhibition and allowing UbcH7~Ub to approach HHARI's active site are unknown, it seems likely future studies will reveal activation mechanisms related to those stimulating Parkin or HOIP, such as post-translational modifications at HHARI's Ariadne-RING2 interface, or the Ariadne domain binding to other partners to liberate RING2 (Fig. 8). Interestingly, different structure-based activating HHARI mutants displayed a range of activities and substrate-independent ubiquitination patterns in our assay. Future studies may reveal gradients of endogenous mechanisms regulating autoinhibition to allow fine-tuning rather than binary control of enzyme activity for HHARI and other RBR E3s.
Figure 8. Models for RBR E3 ligase activation.
a. Two possible models for HHARI activation. Postranslational modification (PTM) of the Ariadne domain could disrupt the interaction with RING2 and allow for RING2~Ub thioester formation and subsequent ubiquitination of substrate. Another possible activation mechanism would involve another protein binding to HHARI to release the Ariadne domain from RING2.
b. Current model for Parkin activation whereby PINK1 phosphorylates the UblD domain and releases its autoinhibitory effect.
c. Current model for HOIP activation where HOIL-1L and Sharpin UBL domains interact with HOIP to release autoinhibitory domains and promote ubiquitination of NEMO.
EXPERIMENTAL PROCEDURES
X-Ray Crystallography
Crystals of full length HHARI were grown by the hanging drop vapor diffusion method and appeared in two distinct morphologies. Crystal form 1 grew in 1.0 – 1.4M Potassium/Sodium Tartrate / 0.1M CHES pH 9.5 / 0.2M LiSO4 at room temperature, in the space group P41 with a=b=146.9, c=86.62 and α=β=γ=90°. Crystal form 2 grew in 0.5 – 0.55M Tri-Sodium Citrate / 0.1M Citric acid pH 5.2 / 0.2M Lithium Acetate at room temperature, in the space group P63 with a=b=96.1, c=151.3, α=β=90 and γ=120deg.
The structure was of Crystal Form 1 was initially determined by zinc MAD in P41212 with 6 sites found using HKL2MAP (Sheldrick and Schneider, 1997). In space group P41212 one molecule was found in the asymmetric unit corresponding to a 74% solvent content according to the Matthews coefficient (Matthews, 1968). Viewing the packing along the C-axis showed large solvent channels 48 Å across which account for the high solvent content. The resolution of the phases went to 3.5 Å, although the quality of the maps appeared much lower because of missing electron density for several sections of main-chain residues and nearly all the side-chains. The structures of several domains solved previously were docked into the electron density maps to guide manual model building, but the R and Rfree failed to drop below 0.45. To further guide structure building the method of selenomethionine scanning (SeMet) was used where eight individual leucine to methionine mutations were made, SeMet labeled protein was produced and crystallized (Huang et al., 2004). Se-SAD datasets were collected for each of these 8, which confirmed the position by the appearance of new anomalous difference density surrounding the site but failed to yield superior electron density maps using space group P41212. Overall the density was still of poor quality and difficult to build. At this point, a systematic approach was followed to determine what could be causing the refinement problem. The results indicated the presence of non-crystallographic symmetry (NCS), namely, rotational pseudo-symmetry (RPS). RPS can occur when the point-group symmetry of the lattice is higher than the crystal (Zwart et al., 2005). Similar cases of RPS have been seen before in other structures (Chacko et al., 2012; Lougheed et al., 2001; Zagotta et al., 2003). In our case, the true space group is P41 with two molecules in the asymmetric unit. The NCS operator has a kappa value of 179.94°, only 0.06° from being crystallographic in space group P41212 and the overall RMSD between the C-alpha atoms is 0.6 Å. The resulting electron density map in P41 is superior in quality showing continuous density and nearly all of the side-chains were visible allowing for unambiguous sequence assignment and the refinement proceeded to acceptable R-factor and R-free values (Table 1). Since merohedral twinning is a possibility in P41, the structure was refined in Phenix (Afonine et al., 2012) including the twin operator (h, -k, -l), but the resulting R-factor and Rfree increased by 1 and 2 %, respectively, and the electron density maps showed no visible improvement compared to refinement excluding the twin operator. Therefore, the structure was refined without a twin operator.
Initial electron density maps for Crystal Form 2 were obtained using MR-SAD with a partial molecular replacement model taken from the model built for crystal form 1 dissected into domains. Due to severe anisotropy of the data (3.9Å along a and b and 3.4Å along c), the initial electron density maps were relatively featureless and hindered building and refinement. Therefore, the data were anisotropically scaled using the UCLA Anisotropy Server (Strong et al., 2006). Following ellipsoid truncation using a reciprocal lattice origin with vertices 1/3.9, 1/3.9 and 1/3.4 Å along a, b, and c, respectively, the completeness of the highest resolution bin was only 2% (96% complete prior to truncation). The data was rescaled to 3.6Å and the 9,276 merged reflections were elliptically truncated using 3.8Å along a and b and 3.6Å along c (resolution limits determined by the server), resulting in 8,348 reflections. A negative B factor correction was applied along c of −79.20Å2. The anisotropically scaled data was used in refinement with Phenix (Afonine et al., 2012). The quality of the electron density was significantly improved and refinement proceeded as expected. Model building and refinement for both crystal forms were done in Coot and Phenix (Afonine et al., 2012; Emsley and Cowtan, 2004).
Enzyme and Binding Assays
Multiple turnover assays were done at room temperature with 150 nM Uba1, 500 nM UbcH7, 20 μM Ubiquitin and 250 nM HHARI. All reaction components were mixed in 25 mM Tris, 2.5 mM MgCl2, 2.5 mM ATP and 20 mM NaCl and reactions initiated with the addition of Uba1. Reactions were quenched at the indicated time points in SDS-PAGE sample buffer containing 50mM DTT except where indicated as nonreducing, resolved on 4–12% gradient gels (Invitrogen) and visualized with SYPRO-Ruby stain. For inhibition assays, HHARIΔAri and HHARIArI were mixed on ice for 10 minutes prior to reaction initiation.
Pulse-chase assays were done on ice. For fluorescent labeling, a cysteine residue was introduced into lysineless ubiquitin N-terminal to methionine 1. Fluoresceine-5-maleimide was used to modify this engineered cysteine. 20 μM UbcH7 was reacted with 150 nM Uba1, 20 μM fluoresceine-Ub in 25 mM Hepes, 2.5 mM MgCl2, 25 mM ATP, 50mM NaCl pH 7.0 to form the fluoresceine-Ub~UbcH7 thioester intermediate. This reaction was quenched by addition of apyrase followed by 5 fold dilution into 20mM Hepes, 100mM NaCl pH 7.0. Pulse reaction was then added to chase reactions containing 1uM of the indicated HHARI mutant in 25mM Hepes, 50mM NaCl pH 7.0. Reactions were quenched at the indicated time points in 2X SDS-PAGE running buffer, nonreducing or supplemented with DTT as indicated.
For analytical gel filtration binding assays all components were mixed at 10 μM in a 200 μl final reaction volume for 1 hour on ice prior to loading onto an SD200 column (GE Healthcare) equilibrated with 25 mM Tris pH 7.6, 100 mM NaCl, 1 mM DTT. 500 μl fractions were collected and run on 4–12% gradient gels (Invitrogen) and visualized with SYPRO-Ruby stain.
Isothermal calorimetry measurments were taken on an ITC200 (MicroCal). HHARI, HHARIΔARI and UbcH7 were buffer exchanged into phosphate buffered saline pH 7, 1 mM DTT using NAP5 desalting columns (GE Healthcare). Proteins were concentrated to 30 μM (HHARI, HHARIΔARI) and 300 μM (UbcH7). UbcH7 was titrated against either HHARI or HHARIΔARI in the sample cell at a constant temperature of 25deg. 19 titration measurements were made and the data analyzed using Origin software provided by MicroCal.
Supplementary Material
HIGHLIGHTS
Structure of HHARI shows RING1-IBR-RING2 ubiquitin E3 organization
Full-length Ariadne subfamily RBR E3 HHARI is autoinhibited
Ariadne domain masks catalytic Cys from RING2 to block E3 ligase activity
Activation, Ub transfer from E2 to E3, and ligation are distinct reaction steps
ACKNOWLEDGMENTS
This work was supported by ALSAC, PHS grants 5P30CA021765, and R01GM069530 (BAS). BAS is an Investigator of the Howard Hughes Medical Institute. This work was supported by the Scottish Institute for CeLL Signalling (SCILLS) and pharmaceutical companies supporting the Division of Signal Transduction Therapy (AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Janssen Pharmaceutica, Merck-Serono and Pfizer). We are grateful to S Bozeman, DW Miller, and J Bollinger for administrative/computational support, and to I Kelsall, W Harper, D Scott and members of the Schulman lab for helpful discussions. We thank D. King for mass spec. The Northeastern Collaborative Access Team at the Advanced Photon Source (APS) is supported by the NCRR (5P41RR015301-10) and NIH (8 P41 GM103403-10). Use of APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by U.S. DOE Contract No. DE-AC02-06CH11357.
Footnotes
ACCESSION CODES: 4KBL.PDB (P41 crystal form) and 4KC9.PDB (P63 crystal form)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera M, Oliveros M, Martinez-Padron M, Barbas JA, Ferrus A. Ariadne-1: a vital Drosophila gene is required in development and defines a new conserved family of ring-finger proteins. Genetics. 2000;155:1231–1244. doi: 10.1093/genetics/155.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Frank DW. Five mechanisms of manipulation by bacterial effectors: a ubiquitous theme. PLoS Pathog. 2012;8:e1002823. doi: 10.1371/journal.ppat.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley HC, Tan NG, Rose SA, Markham AF, Robinson PA. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. The Journal of biological chemistry. 2001;276:19640–19647. doi: 10.1074/jbc.M011028200. [DOI] [PubMed] [Google Scholar]
- Beasley SA, Hristova VA, Shaw GS. Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson's disease. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3095–3100. doi: 10.1073/pnas.0610548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese MF, Scott DC, Duda DM, Grace CR, Kurinov I, Kriwacki RW, Schulman BA. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nature structural & molecular biology. 2011;18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili AD, Edghill EL, Wu K, Borden KL. Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. Journal of molecular biology. 2004;340:1117–1129. doi: 10.1016/j.jmb.2004.05.035. [DOI] [PubMed] [Google Scholar]
- Chacko AR, Zwart PH, Read RJ, Dodson EJ, Rao CD, Suguna K. Severe diffraction anisotropy, rotational pseudosymmetry and twinning complicate the refinement of a pentameric coiled-coil structure of NSP4 of rotavirus. Acta crystallographica. 2012;68:1541–1548. doi: 10.1107/S090744491203836X. [DOI] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. The EMBO journal. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nature structural & molecular biology. 2008;15:65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nature structural & molecular biology. 2012a;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nature structural & molecular biology. 2012b;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove KK, Klevit RE. RING-between-RINGs--keeping the safety on loaded guns. The EMBO journal. 2012;31:3792–3794. doi: 10.1038/emboj.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM, Zobel K, Maurer B, Varfolomeev E, Wu P, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science (New York, NY. 2011;334:376–380. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. A unique E1–E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nature structural & molecular biology. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science (New York, NY. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Molecular cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. The EMBO journal. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20579–20584. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open biology. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Developmental cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. The Journal of cell biology. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Ebert G, Ow Y, Preston SP, Toe JG, Cooney JP, Scott HW, Sasaki M, Saibil SD, Dissanayake D, et al. ARIH2 is essential for embryogenesis, and its hematopoietic deficiency causes lethal activation of the immune system. Nature immunology. 2013;14:27–33. doi: 10.1038/ni.2478. [DOI] [PubMed] [Google Scholar]
- Lin DY, Diao J, Chen J. Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1925–1930. doi: 10.1073/pnas.1115025109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J, John SW, Tenev T, Rautureau GJ, Hinds MG, Francalanci F, Wilson R, Broemer M, Santoro MM, Day CL, et al. CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration. Molecular cell. 2011;42:569–583. doi: 10.1016/j.molcel.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Lougheed JC, Holton JM, Alber T, Bazan JF, Handel TM. Structure of melanoma inhibitory activity protein, a member of a recently identified family of secreted proteins. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5515–5520. doi: 10.1073/pnas.091601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I. RBR ubiquitin ligases: Diversification and streamlining in animal lineages. Journal of molecular evolution. 2009;69:54–64. doi: 10.1007/s00239-009-9252-3. [DOI] [PubMed] [Google Scholar]
- Marin I, Ferrus A. Comparative genomics of the RBR family, including the Parkinson's disease-related gene parkin and the genes of the ariadne subfamily. Molecular biology and evolution. 2002;19:2039–2050. doi: 10.1093/oxfordjournals.molbev.a004029. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. Journal of molecular biology. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Moynihan TP, Ardley HC, Nuber U, Rose SA, Jones PF, Markham AF, Scheffner M, Robinson PA. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. The Journal of biological chemistry. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Human mutation. 2010;31:763–780. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2 approximately Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box Ligases. Molecular cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada CM, Hicks SW, Galan JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4864–4869. doi: 10.1073/pnas.0811058106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Molecular cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013 doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Human molecular genetics. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM, Schneider TR. SHELXL: high-resolution refinement. In: Carter CW, Sweet RM, editors. Methods in enzymology. Academic Press; San Diego: 1997. pp. 319–343. [PubMed] [Google Scholar]
- Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, Chirgadze NY, Cuff ME, Joachimiak A, Tyers M, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nature structural & molecular biology. 2008;15:1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Gonzalez M, Kabbaj MH, Gunjan A. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS ONE. 2012;7:e36295. doi: 10.1371/journal.pone.0036295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. The EMBO journal. 2012;31:3833–3844. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO reports. 2012;13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Iwai K. LUBAC, a novel ubiquitin ligase for linear ubiquitination, is crucial for inflammation and immune responses. Microbes and infection / Institut Pasteur. 2012;14:563–572. doi: 10.1016/j.micinf.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Molecular cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- Wenzel DM, Klevit RE. Following Ariadne's thread: a new perspective on RBR ubiquitin ligases. BMC biology. 2012;10:24. doi: 10.1186/1741-7007-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425:200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nature structural & molecular biology. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- Zwart PH, Grosse-Kunstleve RW, Adams PD. Characterization of X-ray Data Sets. CCP4 Newsletter. 2005;42 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.