Abstract
Hepatitis B virus (HBV) can evolve by mutations in the major hydrophilic region (MHR) of the HBV surface antigen (HBsAg) to permit its escape from neutralization by antibodies such as HBV surface antibody (anti-HBs) and from host immune responses. This study investigated the prevalence and pattern of MHR mutations in North China and the clinical correlations of these mutations. The MHRs of 161 HBsAg-positive patients were amplified using nested PCR, and directly sequenced to identify MHR mutations. It was showed that among the 161 patients infected with HBV genotype C in North China, the overall frequency of MHR mutation was 46.6%. Furthermore, MHR mutations were associated with high white blood cell counts (P = 0.025), high bilirubin levels (P = 0.048), and cirrhosis (P = 0.010). The most frequent mutations in patients with both HBsAg-positive and anti-HBs-positive were located in subregion 1 and 3 of MHR, specifically, residue Q101 (29.9%) and I126 (70.6%), which was different from the mutation pattern found in Western Europe and the United States. Taken together, these data indicated important virological and clinical aspects of HBV evolution in terms of the surface gene of genotype C, which might be important for its response to the currently available HBV vaccine.
Keywords: HBV, major hydrophilic region, mutation
INTRODUCTION
Hepatitis B virus (HBV) infection is a worldwide health problem, and is responsible for one million deaths annually from HBV-related cirrhosis, liver failure, and hepatocellular carcinoma [Dienstag, 2008]. HBV infection is highly endemic in China with a prevalence rate of 8–20% in the general population [Chinese Society of Hepatology, 2007]. The genetic variability of HBV is associated with the natural history of infection and the response of HBV to antiviral therapy. The mutations tend to cluster into four regions of the HBV gene: The basal core promoter (BCP), the region encoding the pre-core region, the polymerase gene, and the area encoding the major hydrophilic region (MHR) of HBsAg [Gunther et al., 1999].
The MHR spans residues 99–169 of HBsAg, and is probably composed of several conformational epitopes. Mutations within MHR, especially in the “a” determinant region (residue 124–147), have been described in vaccinated children and liver transplant recipients who received HBIG prophylaxis [Protzer-Knolle et al., 1998]. Some specific mutations within MHR affect the antigenicity of HBsAg, result in the loss of recognition by antibodies, and lead to the escape of the virus from neutralizing antibody response [Ghany et al., 1998]. Understanding the prevalence and diversity of MHR is fundamental to assay design and the planning of vaccination programs. Genotypes B and C of HBV have been identified as the most common strains and account for approximately 95% of infections among Chinese patients [Wang et al., 2007]. Compared with HBV genotype B infections, HBV genotype C infections have been associated with lower rates of spontaneous clearance of HBsAg in serum, higher levels of virus replication, more advanced liver disease [Pujol et al., 2009], and a lower rate of response to α-interferon therapy [Kao et al., 2000]. All these reveal a poor prognosis.
In this study, the prevalence and pattern of MHR mutations in patients with chronic hepatitis B genotype C in North China, and the clinical correlations of the observed mutations were examined.
METHODS
Patients
From July 2010 to January 2011, 161 patients with chronic HBV infections from Beijing Youan Hospital (Beijing, China) were included in the study. All patients provided written and informed consent prior to entrance into this study. All samples were obtained with appropriate approval of the local ethical committee. Diagnosis of chronic HBV infection was based on a positive result for HBsAg (HBsAg+) for at least 6 months and a positive result for HBV DNA at >500 copies/ml as described below.
Serum HBsAg Quantitation and HBV DNA Viral Loads
Serum markers for HBV infection were measured using the Elecsys HBsAg II assay (Roche Diagnostics, Mannheim, Germany), and results were given in COI. Samples with HBsAg levels less than 0.90 COI were determined as HBsAg negative. Serum HBV DNA was measured using Applied Biosystems 7500/7500 Fast Real-Time PCR Systems (Applied Biosystems, Foster City, CA), and HBV DNA levels were expressed in log IU/ml.
MHR Mutant Detection
HBV DNA was extracted from 200 μl of serum using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer's protocol. MHR was amplified from each sample using nested PCR with primary primers HBVS1F (5′-ttcct gctggtggctccagttc-3′) and HBS1R (5′-tgctaggagttccgcagtatg-3′), and then with nested primers HBS2F (5′-ct cgtggtggacttctctc-3′) and HBVS2R (5′-gcaaagcccaaaagacccac-3′). Primary PCR was performed for 33 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, and elongation at 72°C for 90 sec. Secondary PCR was carried out for 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 60 sec, and elongation at 72°C for 90 sec. The predicted size of the PCR product was 766 bp. Nucleotide sequences of the PCR fragments were determined with the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). All sequences were initially aligned using the ClustalW program incorporated in Bioedit v7.0 sequence alignment software. HBV reference sequences were obtained from Genbank (AB246345.1) and used to define MHR mutations.
Statistical Analysis
Statistical analysis was performed using the SPSS software v10.1.3.
RESULTS
The study cohort consisted of 161 HBsAg positive Chinese patients, and the majority were males in their late 40s (Table I). Among the 161 patients, 75 (46.6%) had MHR mutations. Based on these results, patients were then evaluated based on the presence of MHR mutations (Group I) and the absence of MHR mutations (Group II). There was no significant difference between the two groups with regard to age, sex, platelet count, albumin, serum alanine aminotransferase level, serum HBV DNA level, and serum HBsAg level. Univariant analysis showed that the white blood cell count and bilirubin were significantly higher in Group I than in Group II (P < 0.05). Additionally, 16 patients in Group I and seven patients in Group II had cirrhosis detected by ultrasound (P = 0.010), and 17 patients in Group I (29.3%) but no patient in Group II (0%) were anti-HBs positive (P < 0.001).
TABLE I.
Clinical Characteristics of Chronic HBV-Infection Patients With (Group I) and Without (Group II) MHR Mutations
| With MHR mutations (Group I, n = 75) | Without MHR mutations (Group II, n = 86) | P | |
|---|---|---|---|
| Age (years)a | 49.13 ± 10.24 | 47.98 ± 12.55 | 0.526 |
| Gender (M/F)b | 59/16 | 67/19 | 0.907 |
| White blood cell count (×109/L)a | 4.58 ± 1.99 | 5.39 ± 2.51 | 0.025* |
| Platelet count (×109/L)a | 125.87 ± 65.49 | 138.94 ± 64.82 | 0.206 |
| Albumin (g/L)a | 41.08 ± 7.62 | 41.20 ± 5.88 | 0.907 |
| Bilirubin (mmol/L)a | 21.9 (5.8–627.4) | 16.9 (4.1–364.1) | 0.048* |
| ALT (IU/L)a | 47.0 (8.8–1001.6) | 47.3 (3.2–1109.5) | 0.740 |
| Cirrhosis by ultrasound (%)b | 16 (21.3) | 7 (8.1) | 0.010* |
| HBV DNA (log IU/ml)a | 4.13 | 4.36 | 0.584 |
| HBsAg (COI)a | 4907.3 ± 1904.4 | 5517.9 ± 2144.4 | 0.255 |
| HBsAb ( +/− )a | 17/58 | 0/86 | 0.000* |
MHR, major hydrophilic region; M, male; F, female; ALT, alanine aminotransferase.
Statistical significance was calculated using two independent samples t-test.
Gender and cirrhosis by ultrasound were calculated using Chi-square tests.
P < 0.05.
To determine factors associated with anti-HBs the patients' groups were stratified into HBsAg-positive/anti-HBs-positive (Group III) and HBsAg-positive/anti-HBs-negative (Group IV) (Fig. 1). Clinical characteristics and the virological and biological features of Group III are shown in Table II. Patients in Group III had a median age of 48 years (ranging from 16 to 78 years), and 15 (88.2%) were male. Group III patients were more likely to have HBV DNA levels over 5.0 log IU/ml than Group IV (35.3% vs. 12.5%; P = 0.02), and HBsAg and anti-HBs both persisted for more than two months in all the 17 patients in Group III. The alanine aminotransferase levels were elevated in 10 (58.8%) patients in Group III. Six patients in Group III had never received anti-HBV therapy at the time of detection of the co-existence of HBsAg and anti-HBs, while the other 11 patients had received anti-HBV therapies (Table II). The most frequent mutations observed in the MHR sequences of Group I were Q101 (11.5%), I126 (23.7%), Y100 (3.6%), T131 (3.6%), G145 (4.3%), R160 (10.1%), and E164 (7.2%), which could alter immune escape [Seddigh-Tonekaboni et al., 2000]. Mutations in the MHR sequences of Group III clustered at six amino acid residues including Q101 (32%), P105 (4%), G102 (4%), S117 (4%), I126 (48%), and Y100 (8%). Overall, Group III had a higher mutation frequency than Group IV (P < 0.001) (Fig. 1). All patients in Group III and 52 patients (70%) in Group IV carried at least two MHR mutations, and three patients in Group III had more than eight mutations. Additionally, in Group III, MHR mutations only occurred in subregion 1 and subregion 3, whereas in Group IV MHR mutations occurred in five subregions (Table III). For HBsAg+/anti-HBs+ patients, only one mutation (I126) arose at the “a” determinant region.
Fig. 1.
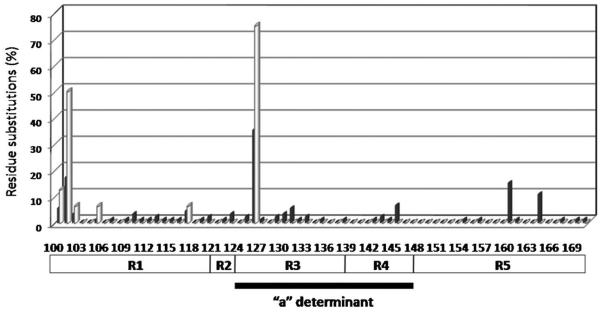
Frequencies of residue substitutions within MHR in isolates from HBsAg positive/anti-HBs positive patients (gray bars, n = 17) and HBsAg positive/anti-HBs negative patients (black bars, n = 75). Each bar represents the percentage of mutated residues per group. Amino acid variability was considered the percentage of sequences that harbored at a given position an amino acid other than the one found in HBV reference sequences of HBV genotype C (found in the genotyping reference set of HBV sequences available on the NCBI Web site; http://www.ncbi.nih.gov/projects/genotyping/view.cgi?db=2).
TABLE II.
Factors Related to MHR Mutations (Multivariate Logistic-Regression Analysis)
| β | S.E | Wald | P | OR | 95%CI | |
|---|---|---|---|---|---|---|
| Cirrhosis by ultrasound | −1.266 | 0.500 | 6.412 | 0.011 | 0.282 | 0.106 ~ 0.751 |
| HBsAb | −21.657 | 9603.767 | 0.000 | 0.998 | 0.000 | 0.000 |
| Constant | 0.573 | 0.189 | 9.221 | 0.002 | 1.773 |
TABLE III.
Summary of Clinical Characteristics, Drug Resistance Resistance, and the Virological and Biological Features of Patients With Both HBsAg and HBsAb (Group III)
| Patient no. | Gender/age | HBsAg (COI) | HBsAb (IU/L) | HBV DNA (log IU/ml) | HBeAg/HBeAb | ALT (IU/L) | HBV treatment | Drug resistant mutation |
|---|---|---|---|---|---|---|---|---|
| 1 | M/76 | 5509 | 196.4 | 3.47 | +−/+− | 13.4 | None | L180LMV, M204FILMV |
| 2 | M/63 | 5139 | 273.8 | 6.55 | +/− | 38.5 | None | A181APST |
| 3 | F/58 | 7715 | 23.02 | 7.52 | +/− | 262.1 | None | None |
| 4 | M/36 | 3430 | 13.97 | 5.23 | +/− | 74.2 | ADV | None |
| 5 | M/48 | 1114 | 19.08 | 4.91 | −/+ | 821.6 | IFN + LAM + ETV | None |
| 6 | M/25 | 1481 | 35.38 | 7.56 | +/− | 75.9 | None | None |
| 7 | M/35 | 6095 | 64.59 | 2.72 | −/+ | 47 | ADV | M204IM |
| 8 | M/54 | 633.7 | 127.2 | 3.32 | +/− | 117.8 | LAM + ADV | L180LMV, V173LMV, M204I |
| 9 | M/78 | 4136 | 99.33 | 3.77 | +/− | 27.2 | LAM | L180M, M204V |
| 10 | M/43 | 3447 | 30.91 | 2.94 | +/− | 35.5 | ADV + ETV + LdT | M204IM, A181APST |
| 11 | M/48 | 4634 | 262.5 | 2.70 | +/− | 19.2 | None | L180LMV, M204FILMV |
| 12 | M/52 | 6192 | 100.2 | 7.41 | −/+ | 266.2 | LAM | L180M, M204I |
| 13 | M/53 | 3295 | 50.74 | 2.70 | −/− | 21.5 | IFN + LAM + ADV | None |
| 14 | M/50 | 23.88 | 12.94 | 3.13 | +/− | 74.6 | LAM | None |
| 15a | M/31 | 5698 | 14.35 | 4.34 | +/− | 64.6 | LAM + ETV | A181V |
| 16a | M/16 | 5476 | 43.27 | 4.13 | +/− | 31.8 | None | M204IM, L180LMV |
| 17 | F/59 | 7038 | 38.64 | 5.11 | +/− | 306.1 | ADV | None |
M, male; F, female; ALT, alanine aminotransferase; ADV, adefovir dipivoxil; IFN, peginterferon; LAM, lamivudine; ETV, entecavir; LdT, telbivudine.
Patients who had received HBV vaccine.
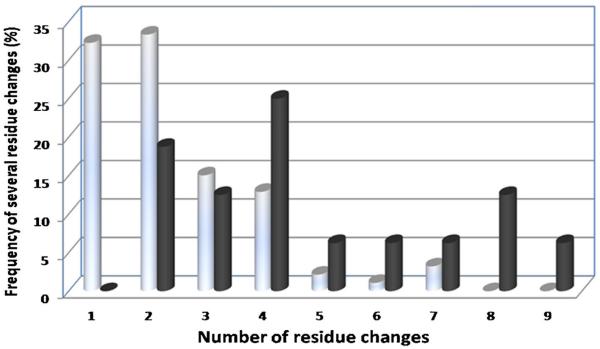
The frequencies of several residue changes for group III patients with HBsAg-positive/anti-HBs-positive were also much higher than those for group IV patients with HBsAg-positive/anti-HBs-negative (Fig. 2). Indeed, 81.25% patients from group III and only 34.4% patients from group IV carrying at least two residue changes had altered immunogenicity of the protein. Additionally, in both Group III and Group IV Amino Acid substitutions occurred mostly in `a' determinant region, Subregion 1 and Subregion 3, and but significant difference was found between the two groups (Table IV).
Fig. 2.
Frequencies of several residue changes in MHR of HBsAg+/anti-HBsAb+ patients (Group III) and HBsAg+/anti-HBsAb− patients (Group IV). The percentages of different residue change for Group III (black bars) and Group IV (gray bars) are shown. Statistical significance was calculated using Mann-Whitney test.
TABLE IV.
Distribution of Amino Acid Substitutions Within MHR Between HBsAg+/Anti-HBs+Patients and HBsAg+/Anti-HBs-Patients
| Frequency of amino acid substitutions within MHR |
|||
|---|---|---|---|
| Subregion of MHR | HBsAg+/anti-HBs+ patients Group III (n = 17) | HBsAg+/anti-HBs-patients Group IV (n = 58) | Fisher's exact test(P) |
| “a” determinant (%) | 76.5 | 77.6 | 1.000 |
| Subregion 1 (%) | 29.4 | 37.9 | 0.579 |
| Subregion 2 (%) | 0.0 | 3.4 | 1.000 |
| Subregion 3 (%) | 70.6 | 63.8 | 0.774 |
| Subregion 4 (%) | 17.6 | 20.7 | 1.000 |
| Subregion 5 (%) | 17.6 | 0.0 | 0.010 |
For analysis, MHR of HBsAg was divided into subregions corresponding to structural and/or functional domains: subregion 1 (amino acid 100 to 120), subregion 2 (amino acid 121 to 123), subregion 3 (amino acid 124 to 137), subregion 4 (amino acid 138 to 147), and subregion 5 (amino acid 148–169) [Cooreman et al., 2001]. The “a” determinant region (amino acid 124 to 147) spans HBsAg subregion 3 and 4. Statistical significance was calculated using Fisher's exact-test.
DISCUSSION
This study evaluated the variability of the HBV surface gene (S) in patients from North China who were infected chronically with HBV, and 75 of the 161 patients infected chronically with HBV had amino acid substitutions in MHR (46.6%). The frequency of MHR mutation is about 1% in all genotypes [Ma and Wang, 2012]. The frequency of MHR mutation detected in this study was far higher than those in previous reports, but was similar to the results reported by Song et al. [2005] (39.2%–54.0%). Mutations in the HBV S gene can help the virus to escape from detection and neutralization by vaccine induced antibodies [Shizuma et al., 2003]. Additionally, genetic alterations of S gene increase with the progression of the disease, and there is an accumulation of mutations in S gene in HBV-related end-stage liver disease [Rodriguez-Frias et al., 1999]. The dominant epitope cluster in HBsAg is in the MHR, and most antibodies are induced by vaccines target HBsAg at amino acid residues 139–147 [Brown et al., 1984]. Although S gene variations are known to be associated with vaccine escape and may cause false negative results in HBsAg assays [Weber, 2006], in this study, there was no difference in HBsAg positivity between patients with MHR mutations (Group I) and patients without MHR mutations (Group II) (Table I). Amino acid substitutions in MHR seemed to have little association with the reactivity by the Elecsys HBsAg II test used in this study, likely because monoclonal and polyclonal anti-HBs antibodies (mouse and sheep) were used [Gerlich, 2004].
In the study cohort, the most frequent mutations were observed at residues Q101 and I126. Interestingly, G145R mutation, the most important and best-documented mutation in Western Europe and the United States, was observed rarely in this study. On the other hand, the I126T/INST was an unique substitution to HBV genotype C and was observed frequently in this study. Residue I126 is located in the first-loop structure of the a-determinant, and amino acid changes in this region may lead to adaptive evolution of the viral gene, resulting in altered antigenicity and increased virulence [Howard, 1995, Ren et al., 2006]. Specifically, in the HBsAg positive/anti-HBspositive patients (Group III), 29.3% of mutated amino acids resided in the “a” determinant region, which was similar to the HBsAg positive/anti-HBs negative patients (Group IV) (26.6%), but all the 17 HBsAg positive/anti-HBs positive patients had more than two residue changes. This may indicate that accumulation of mutations helps the virus to escape from anti-HBs neutralization.
There were no significant clinical differences between patients with and without MHR mutations, except for the presence of anti-HBs. Seventeen patients in this study were HBsAg-positive/anti-HBs-positive, so the prevalence of HBsAg-positive/anti-HBs-positive patients was 10.56% (17/161), which was higher than the results reported previously (3.1%) [Colson et al., 2007]. This was likely because that the levels of HBV DNA of the patients selected in this study were greater than 500 copies/ml, and that all the infecting HBV were genotype C, which had been associated with a poorer prognosis [Lin and Kao, 2011]. Interestingly, all of the HBsAg-positive/anti-HBs-positive patients demonstrated mutations in MHR (Group I) (22.7%), which might suggest that although amino acid substitutions in MHR failed to cause false negative results in HBsAg assay, they could help the virus to escape from neutralization by antibodies directed toward HBsAg, i.e., anti-HBs. Of these 17 patients who were HBsAg positive and anti-HBs positive, HBV DNA levels were higher than patients who were anti-HBsAb negative, which indicated a worse clinical outcome.
The HBV S gene is overlapped completely by the polymerase gene [Torresi, 2002]. Therefore, mutations in the S gene may also cause changes in the overlapping polymerase gene. Several types of drug resistance mutations were detected in the HBsAg positive/anti-HBs positive patients: rtL180LMV, M204FILMV, A181APST, and V173LMV (Table II). These mutations were associated with LAM, LdT, and ADV.
Two out of the 17 HBsAg-positive/anti-HBs-positive patients had received HBV vaccine 7 or 10 years ago. Both patients had lower levels of anti-HBs (14.35 and 43.27 IU/L), less S mutations and less drug resistance mutations (Table II). The significance of infection in vaccinated donors is not clear. It has been reported that such HBsAg-mutated HBV strains may be insensitive to vaccine-induced anti-HBs. The prevalence of “a” determinant mutant is higher in vaccinated children than in unvaccinated children [Hsu et al., 2004]. Although the protective effect of the vaccine has been emphasized previously [Stramer et al., 2011], the vaccine escape mutants still infect vaccinated patients. The mutants may potentially cause infections in a vaccinated population [Fortuin et al., 1994].
In summary, patients infected with genotype C HBV were evaluated, and it was found that the prevalence of patients with both HBsAg positive and anti-HBs positive was higher than that in Western Europe and in the United States [Colson et al., 2007]. Additionally, these patients had more mutations in the MHR. The most common mutation I126 detected in our study is relatively rare in Western Europe and the United States. Since MHR is important for viral escape from anti-HBs, these results may be important in evaluating the efficacy of the HBV vaccine in the North Chinese population.
Acknowledgments
Grant sponsor: National Natural Science Foundation of China; Grant numbers: 30910103915; 81100288; Grant sponsor: CFAR Translational Virology Core; Grant number: AI36214; Grant sponsor: HNRC Neurovirology Core; Grant number: MH62512; Grant sponsor: Key Project of Beijing Natural Science Foundation; Grant number: 7101005.
REFERENCES
- Brown SE, Howard CR, Zuckerman AJ, Steward MW. Determination of the affinity of antibodies to hepatitis B surface antigen in human sera. J Immunol Methods. 1984;72:41–48. doi: 10.1016/0022-1759(84)90431-9. [DOI] [PubMed] [Google Scholar]
- Chinese Society of Hepatology CMaCSOID. Chinese Medical Association Guideline on prevention and treatment of chronic hepatitis B in China (2005) Chin Med J (Engl) 2007;120:2159–2173. [PubMed] [Google Scholar]
- Colson P, Borentain P, Motte A, Henry M, Moal V, Botta-Fridlund D, Tamalet C, Gerolami R. Clinical and virological significance of the co-existence of HBsAg and anti-HBs antibodies in hepatitis B chronic carriers. Virology. 2007;367:30–40. doi: 10.1016/j.virol.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237–247. doi: 10.1007/BF02256597. [DOI] [PubMed] [Google Scholar]
- Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- Fortuin M, Karthigesu V, Allison L, Howard C, Hoare S, Mendy M, Whittle HC. Breakthrough infections and identification of a viral variant in Gambian children immunized with hepatitis B vaccine. J Infect Dis. 1994;169:1374–1376. doi: 10.1093/infdis/169.6.1374. [DOI] [PubMed] [Google Scholar]
- Gerlich WH. Diagnostic problems caused by HBsAg mutants- a consensus report of an expert meeting. Intervirology. 2004;47:310–313. doi: 10.1159/000080873. [DOI] [PubMed] [Google Scholar]
- Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, Lok AS. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology. 1998;27:213–222. doi: 10.1002/hep.510270133. [DOI] [PubMed] [Google Scholar]
- Gunther S, Fischer L, Pult I, Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res. 1999;52:25–137. doi: 10.1016/s0065-3527(08)60298-5. [DOI] [PubMed] [Google Scholar]
- Howard CR. The structure of hepatitis B envelope and molecular variants of hepatitis B virus. J Viral Hepat. 1995;2:165–170. doi: 10.1111/j.1365-2893.1995.tb00024.x. [DOI] [PubMed] [Google Scholar]
- Hsu HY, Chang MH, Ni YH, Chen HL. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut. 2004;53:1499–1503. doi: 10.1136/gut.2003.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol. 2011;26:123–130. doi: 10.1111/j.1440-1746.2010.06541.x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Wang Y. Comprehensive analysis of the prevalence of hepatitis B virus escape mutations in the major hydrophilic region of surface antigen. J Med Virol. 2012;84:198–206. doi: 10.1002/jmv.23183. [DOI] [PubMed] [Google Scholar]
- Protzer-Knolle U, Naumann U, Bartenschlager R, Berg T, Hopf U, Meyer Zum Buschenfelde KH, Neuhaus P, Gerken G. Hepatitis B virus with antigenically altered hepatitis B surface antigen is selected by high-dose hepatitis B immune globulin after liver transplantation. Hepatology. 1998;27:254–263. doi: 10.1002/hep.510270138. [DOI] [PubMed] [Google Scholar]
- Pujol FH, Navas MC, Hainaut P, Chemin I. Worldwide genetic diversity of HBV genotypes and risk of hepatocellular carcinoma. Cancer Lett. 2009;286:80–88. doi: 10.1016/j.canlet.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Ren F, Tsubota A, Hirokawa T, Kumada H, Yang Z, Tanaka H. A unique amino acid substitution, T126I, in human genotype C of hepatitis B virus S gene and its possible influence on antigenic structural change. Gene. 2006;383:43–51. doi: 10.1016/j.gene.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Frias F, Buti M, Jardi R, Vargas V, Quer J, Cotrina M, Martell M, Esteban R, Guardia J. Genetic alterations in the S gene of hepatitis B virus in patients with acute hepatitis B, chronic hepatitis B and hepatitis B liver cirrhosis before and after liver transplantation. Liver. 1999;19:177–182. doi: 10.1111/j.1478-3231.1999.tb00032.x. [DOI] [PubMed] [Google Scholar]
- Seddigh-Tonekaboni S, Waters JA, Jeffers S, Gehrke R, Ofenloch B, Horsch A, Hess G, Thomas HC, Karayiannis P. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J Med Virol. 2000;60:113–121. doi: 10.1002/(sici)1096-9071(200002)60:2<113::aid-jmv2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Shizuma T, Hasegawa K, Ishikawa K, Naritomi T, Iizuka A, Kanai N, Ogawa M, Torii N, Joh R, Hayashi N. Molecular analysis of antigenicity and immunogenicity of a vaccine-induced escape mutant of hepatitis B virus. J Gastroenterol. 2003;38:244–253. doi: 10.1007/s005350300043. [DOI] [PubMed] [Google Scholar]
- Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, Yoon JH, Lee HS, Cha CY, Kook YH, Kim BJ. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76:194–202. doi: 10.1002/jmv.20354. [DOI] [PubMed] [Google Scholar]
- Stramer SL, Wend U, Candotti D, Foster GA, Hollinger FB, Dodd RY, Allain JP, Gerlich W. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236–247. doi: 10.1056/NEJMoa1007644. [DOI] [PubMed] [Google Scholar]
- Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002;25:97–106. doi: 10.1016/s1386-6532(02)00049-5. [DOI] [PubMed] [Google Scholar]
- Wang Z, Huang Y, Wen S, Zhou B, Hou J. Hepatitis B virus genotypes and subgenotypes in China. Hepatol Res. 2007;37:S36–S41. doi: 10.1111/j.1872-034X.2007.00102.x. [DOI] [PubMed] [Google Scholar]
- Weber B. Diagnostic impact of the genetic variability of the hepatitis B virus surface antigen gene. J Med Virol. 2006;78:S59–S65. doi: 10.1002/jmv.20610. [DOI] [PubMed] [Google Scholar]