Abstract
Caenorhabditis elegans lives in compost and decaying fruit, eats bacteria and is exposed to pathogenic microbes. We show that C. elegans is able to modify diverse microbial small-molecule toxins via both O-and N-glucosylation as well as unusual 3′-O-phosphorylation of the resulting glucosides. The resulting glucosylated derivatives have significantly reduced toxicity to C. elegans, suggesting that these chemical modifications represent a general mechanism for worms to detoxify their environments.
Nematodes live in complex microbial environments that present many potential challenges to their health and viability. Caenorhabditis elegans, a commonly used model nematode, has an innate immune system that shares many characteristics with mammalian defenses1, 2 and is susceptible to many mammalian pathogens.3 Its immune system is highly specific: worms can distinguish and mount distinct responses to different pathogens.4–7 C. elegans possesses an array of immune effectors and detoxification enzymes known to be involved in microbial defense and xenobiotic detoxification. Distinct sets of immune effectors, including lysozymes, lipases, and antibacterial peptides, as well as detoxification enzymes such as UDP-glucuronosyl/UDP-glucosyltransferases (UDPGT), cytochrome P450s (CYP), and glutathione S-transferases (GST) are modulated during these responses.4–8 Having a large arsenal of detoxification and defense genes is not surprising for an organism that lives in environments rich in decomposing organic material, areas in which potential pathogens are also likely to thrive. In this study we examined two unrelated bacterial toxins, 1-hydroxyphenazine (1-HP, 1) and indole (6), released by Pseudomonas aeruginosa and Escherichia coli, respectively.9 Both toxins can kill C. elegans, and we found that worms glycosylate both toxins, a modification that significantly lowers their toxicity.
P. aeruginosa is a gram-negative opportunistic human pathogen which is commonly found in soil and water, although it can survive in a number of environments and is a frequent cause of infection in immunocompromised patients.10 Several different strains have been used extensively as model pathogens to identify virulence and host defense factors, including PA14, PAK, and PAO1.3 P. aeruginosa PA14 kills C. elegans by two distinct modes of action: when grown on low-osmolarity or minimal media agar plates, the pathogen kills over the course of several days,5 while PA14 grown on high-osmolarity media is able to kill L4 stage C. elegans in as little as 6 hours through the production of diffusible toxins.11, 12 At least some of these toxins have been identified as phenazines (Brent Cezairliyan, personal communication),11, 13 which are redox-active small molecules that are thought to cause damage to cells by producing reactive oxygen species and disrupting normal redox reactions.14, 15 E. coli is typically used as a food source for C. elegans in the laboratory, but E. coli produces indole, a compound that is toxic to many animals, and has nematicidal properties.16 As described in detail below, we have found that E. coli bacterial pellets used for feeding C. elegans contain indole at concentrations of 3.3 mM, which is sufficient to kill C. elegans.
We asked whether C. elegans employs specific metabolic transformations to eliminate bacterial toxins such as phenazines and indole. After challenging young adult worms in large-scale liquid culture for 24 h with 200 μM of 1-HP (1), HPLC-UV analysis of worm media samples revealed at least three new compounds (2,4,5) present in samples from worms exposed to 1-HP but not present in control samples (Supplementary Figure 1a). In addition, we analyzed whole-body extracts of 1-HP-challenged worms and found at least one novel UV peak (3) that was absent in control extracts (Supplementary Figure 1b). These putative 1-HP metabolites were isolated via preparative HPLC and subsequently identified using MS and NMR spectroscopy (Figure 1, Supplementary Figure 2–5, Supplementary Table 1). We identified 1-O-(β-D-glucopyranosyl)-phenazine (2) and 1-O-(β-D-gentiobiosyl)-phenazine (4) as the two most abundant metabolites in the worm media samples, which were accompanied by two compounds (5) whose mass and NMR spectra suggested phenazine trisaccharides; however, strong overlap in the NMR spectra of these two compounds prevented their full characterization. In addition, we identified the major phenazine derivative detected in the worm body extracts as 1-O-(3′-O-phospho-β-D-glucopyranosyl)- phenazine (3).
Figure 1.
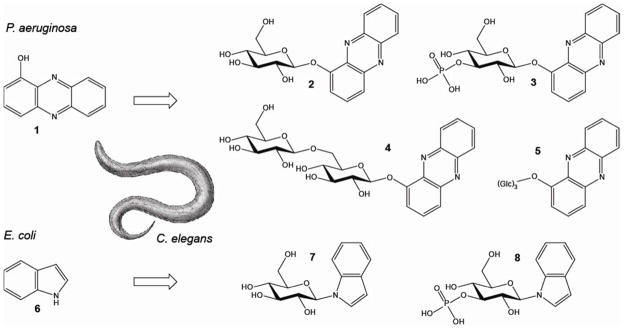
Chemical conversion of 1-hydroxyphenazine (1-HP) (1) and indole (6) by C. elegans. 1-HP is converted into 1-O-(β-D-glucopyranosyl)-phenazine (2), 1-O-(β-D-gentiobiosyl)-phenazine (4), and two phenazine-trisaccharides (5) which are found in the media and worm pellet. 1-O-(3′-O-phospho-β-D-glucopyranosyl)- phenazine (3) is found only in the worm pellet. Indole (6) is converted into N-(β-D-glucopyranosyl)- indole (7) and N-(3′-O-phospho-β-D-glucopyranosyl)-indole (8) which are found in the media and pellet.
These results show that C. elegans converts 1-HP into a series of glycosylated derivatives, including a compound featuring an unusual phosphate substitution. Next we asked whether C. elegans also modifies E. coli-derived indole (6). Notably, HPLC-analysis of C. elegans large-scale liquid culture extracts revealed only trace amounts of indole 24 h after feeding with indole-rich E. coli bacteria. Instead, HPLC-analysis revealed two prominent peaks with UV and mass spectra indicative of highly polar indole derivatives. NMR-spectroscopic analysis (Figure 1, Supplementary Figure 6, Supplementary Table 1 & 2), of extract fractions containing these compounds revealed N-(β-D-glucopyranosyl)-indole (7), of which we prepared an authentic sample via synthesis. In addition, we identified N-(3′-O-phospho-β-D-glucopyranosyl)- indole (8), in direct analogy to the phosphorylated 1-HP derivative (3). We quantified the indole glucoside by HPLC-UV and found that more than 80% of indole provided with the OP50 diet appears to be converted into 7 (Supplementary Figure 7a). Analysis of worm body and supernatant extracts further revealed that indole glucoside is predominantly released by C. elegans and accumulates in the media (Supplementary Figure 7b). We also studied the time course of the N-glucosylation reaction by feeding of [U-D7]-indole to C. elegans liquid cultures and found that the majority of exogenous indole is converted into the glucosides within 6 h (Figure 2c).
Figure 2.
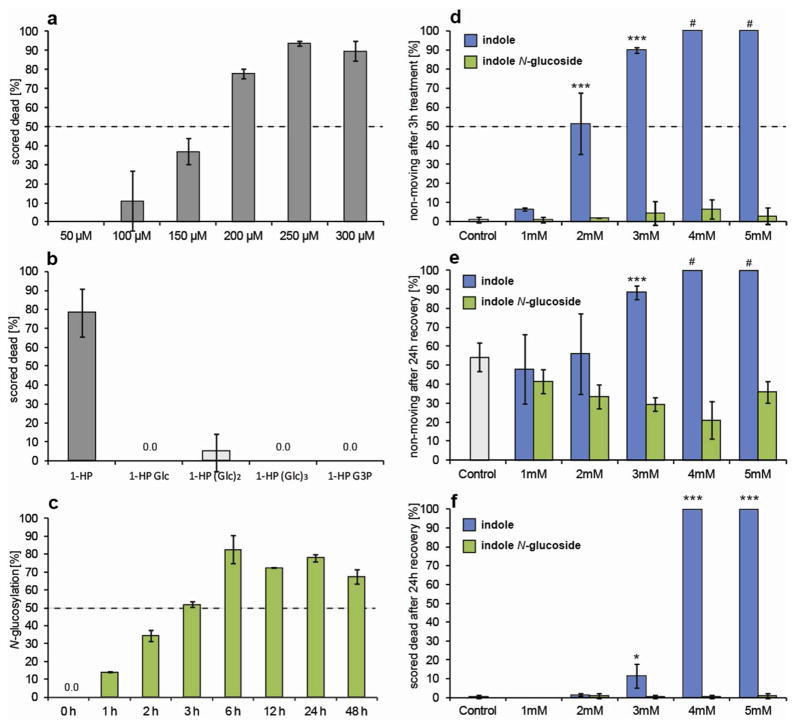
Toxicity Assays. Error bars represent mean ± s.d. (a) Killing of L4 stage worms on M9 agar plates containing increasing concentrations of 1-HP after 6 hours (N = 2 or 3 with 20–40 worms per trial). (b) Killing of L4 stage worms on M9 agar plates containing 200 μM of each phenazine glucoside after 6 hours (N = 3 with 40–50 worms). (c) Time course for N-glucosylation of indole by C. elegans determined by application of 0.5 mg/100 ml [U-D6]-indole. (d) Paralysis of L4 worms after treatment with increasing amounts of indole or indole-N-glucoside in M9 media for 3 h (# worms scored dead after recovery: see f). (e) Paralysis of L4 worms after treatment with indole or indole-N-glucoside for 3 h and 24 h recovery in M9 (# worms scored dead after recovery: see f). (f) Killing of L4 worms after treatment with indole or indole-N-glucoside for 3 h and 24 h recovery in M9 (N = 4 with 50 worms per trial).
Our results demonstrate that C. elegans modifies two chemically distinct bacterial toxins, 1-HP and indole, by glycosylation and additional 3′-O-phosphorylation, suggesting that these modifications may be part of a general detoxification pathway in the nematode. Therefore, we tested the modified glycosides for their toxicity to C. elegans. For this purpose we used synthetic N-(β-D-glucopyranosyl)-indole (6) and isolated samples of the 1-HP glucosides (2–5). Using a plate-based assay, we found that whereas 1-HP at concentrations of 200 μM kills about 80% of L4 worms after 6 h, exposure to the same concentrations of 1-HP glycosides results in less than 5% mortality (Figure 2a–b). Next we compared the toxicity of indole and N-(β-D-glucopyranosyl)-indole in plate and liquid-culture assays. We found that indole is more toxic in liquid media assays in comparison to plate-based assays (not shown). In liquid media indole kills C. elegans at concentrations of 4 mM, whereas lower concentrations of 2 and 3 mM result in reversible paralysis. In contrast, indole glucoside did not kill or elicit paralysis at concentrations as high as 5 mM (Figure 2d–f). Although our toxicity assays differ significantly from the large-scale liquid culture conditions employed for isolation of glucosylation products, our results demonstrate that the bacterial toxins, 1-HP and indole, kill C. elegans, whereas their glucosylation products are much less toxic.
C. elegans possesses at least 60 ABC transporter genes, 15 of which are p-glycoproteins (pgp), a subfamily of ABC transporters that is presumed, by homology to mammalian genes, to be involved in drug resistance.17 Several C. elegans pgp’s have been shown to be involved in xenobiotic transport.18 A previous study showed that a C. elegans pgp-1;pgp-3 double mutant was much more susceptible to fast-killing by PA14 than wild type worms (N2), and that this susceptibility was dependent on production of phenazines by PA14.11 Although we confirmed increased susceptibility of the pgp-1;pgp-3 mutants to PA14, there was no increase in lethality of the double mutant compared to wild-type when exposed to 1-HP (Supplementary Figure 8). We tested a selection of pgp knockout strains and identified a second strain (a pgp-12 knockout, VC26) that is more susceptible than N2 worms to PA14. Furthermore, VC26 worms are more susceptible to 1-HP than wild type, suggesting that pgp-12 is involved in detoxification of 1-HP (Supplementary Figure 8). However, pgp-12 mutant worms are able to produce and excrete 1-HP glycosides (Supplementary Figure 9), suggesting that other transporters contribute to detoxification in addition to pgp-12. These results show that the C. elegans detoxification machinery is based, at least in part, on components that are homologues of genes involved in mammalian small-molecule detoxification. Because transporters and glycosyltransferases often have broad specificity and thus their substrate ranges may overlap, more detailed characterization of this pathway will require systematic study of deletion mutants of different combinations of transporter and glycosyltransferase genes.
The glycosylation and release of a common anthelminthic drug, albendazole,19 and of several small molecules from a synthetic library20 have been described, but the function of glycosylation in C. elegans has not previously been explored. In this paper, we demonstrate that N- and O-glycosylation serve to detoxify two chemically very different xenobiotics, indole and 1-HP, indicating that glycosylation and subsequent phosphorylation represent general mechanisms to convert and remove environmental toxins. This pathway may involve specific glycosylation enzymes and transporter proteins, whose identification will enable comparing the detoxification systems of nematodes with those of other metazoans. Furthermore, existing treatments of parasitic nematode infections in humans, livestock, and plants could be augmented by inhibiting small molecule glycosyltransferases and/or transporters to increase drug efficacy by slowing down or preventing release into the environment.19, 21
Methods
Toxicity Assays (Phenazine)
C. elegans N2 eggs were arrested at the L1 stage for 24 hours in M9 buffer and then grown for 42–48 h at 20 °C on NGM plates to L4 stage as verified by observation with a stereoscope. Worms were washed off plates, allowed to settle, and dispensed onto test plates made with M9 buffer and 2% agar (a small lawn of OP50 was placed on the plate to discourage worms from crawling off). After 6 h, worms that failed to respond to physical touch with a platinum pick were scored as dead.
Toxicity Assays (Indole)
Toxicity was determined in 12 well plates using 50 worms per well, which were placed into 0.5 ml M9 buffer (control), or buffer containing 1, 2, 3, 4, or 5 mM indole or indole glucoside. 0.1% methanol was present in all experiments. Worms were inspected for spontaneous movement after 3 h, the solution removed, and the worms washed 2 times with 2 ml fresh M9 buffer. After 24 h recovery, worms were inspected for spontaneous movement and scored for survival by physical touch with a platinum pick.
Quantification of Indole in E. coli
E. coli OP50 from a 1 L culture was centrifuged at 5251 g for 45 min and the resulting bacterial pellet (3.1 mL) was lyophilized. The resulting material (637 mg) was sonicated in 5 mL methanol (10 min) and extracted with methanol (3 × 15 mL). The filtered extract was concentrated in vacuo and the residue taken up in 3 mL methanol and analyzed by HPLC using a DAD-detector. Indole was quantified by comparison of the UV absorbance at 230 and 260 nm with those of a synthetic standard of known concentration.
Glucoside Collection and Analysis (Phenazine)
Adult C. elegans were synchronized with a bleach solution. The eggs were hatched overnight into L1s, which were then grown at a worm density of 10,000 worms/mL at 22 °C at 250 rpm in S-complete medium supplemented with 2% E. coli (strain HB101) for 43 hours until they reached young adult stage verified by observation with a stereoscope. The worms were concentrated to 30,000 worms/mL, washed once with M9, and fed 1% E. coli HB101. 1-HP from a DMSO stock, or the appropriate amount of DMSO was added to the cultures, which were incubated at 22°C at 250 rpm for 24 hours. The supernatant was removed by centrifugation (2,000 rpm, 2 min), filtered through a 0.22 μm nitrocellulose filter, lyophilized, extracted with 5 mL methanol, filtered, and concentrated. The residue was resuspended in 1 mL of methanol and used for HPLC analysis. The worm pellet was washed 4 times in M9 buffer, resuspended in one volume of 80% methanol and homogenized with a BioSpec MiniBeadbeater-8 (3 cycles of 30 sec, with 1 min on ice). The lysate was spun at 14,000 rpm for 10 min, and the supernatant dried under N2 gas. The residue was resuspended in 1 mL of methanol and used for HPLC analysis.
Worm water was analyzed on an Agilent 1100 Series HPLC system equipped with a diode array detector and an automated fraction collector. Absorbance was monitored at 254 nm. For worm media separation, an aqueous methanol gradient was used from 5–95% on a Zorbax SB C-18 column (4.6 cm × 150 mm, 5 μm particle diameter). For worm pellet separation, 5% methanol (A) and 95% 5mM Phosphate buffer pH 7.2 (B) was held isocratically for 4 min, increasing to 95% A and 5% B over 30 min and then held for 5 min, followed by reequilibriation of the column, at a flow rate of 2 mL min−1 in a Zorbax SB C-18 column (9.4 cm × 250 mm, 5 μm). Fractions were collected automatically by peak detection. Aliquots of each fraction were analyzed by high-resolution mass spectrometry by the University of Florida Spectroscopy Service in the Chemistry Department or in the Biomedical Mass Spectrometry Core at the Clinical and Translational Science Institute at the University of Florida.
Glucoside Collection and Analysis (Indole)
C. elegans worms from 10 cm NGM plates were washed using M9 medium into a 100 ml S-medium pre-culture where they were grown for four days at 22°C on a rotary shaker at 220 rpm. Concentrated OP50 derived from 1 L of bacterial culture (grown for 16 h in LB media) was added as food at days 1 and 3. Subsequently, the pre-culture was divided equally into four 1 L Erlenmeyer flasks containing 400 mL of S-medium for a combined volume of 425 mL of S-medium, which was then grown for an additional 10 d at 22 °C on a rotary shaker. Concentrated OP50 derived from 1 L of bacterial culture was added as food every day from days 1 to 9. Subsequently, the cultures were centrifuged and the supernatant media and worm pellet were lyophilized separately. The lyophilized materials were extracted with 95% ethanol (250 mL 2 times) at room temperature for 12 h. The resulting yellow suspensions were filtered and the filtrate evaporated in vacuo at room temperature, producing media and worm pellet metabolite extracts. The media metabolite extract from two cultures was adsorbed on 6 g of octadecyl-functionalized silica gel and dry loaded into an empty 25 g RediSep Rf sample loading cartridge. The adsorbed material was then fractionated via a reversed-phase RediSep Rf GOLD 30 g HP C18 column using a water-methanol solvent system, starting with 100% water for 4 min, followed by a linear increase of methanol content up to 100% methanol at 42 min, which was continued up until 55 min. The fractions generated from this fractionation were evaporated in vacuo and the residue was analyzed by HPLC-MS and 2D-NMR spectroscopy.
Nuclear Magnetic Resonance
Pooled fractions were dried and resuspended in 150 μL of 99.95% methanol-d4 with 0.111 mM TSP as an internal standard, and transferred into 2.5 mm NMR tubes. 1D-1H and 2D COSY spectra along with 1H-13C HSQC and 1H-1H NOESY where appropriate were collected on a Bruker Avance II 600 MHz spectrometer using a 5 mm TXI cryoprobe or an Agilent 600 MHz spectrometer using a 5 mm cryoprobe in the AMRIS facility at the University of Florida or Varian INOVA 600, INOVA 500, and INOVA 400 spectrometers at Cornell’s NMR facility. Spectra were processed and analyzed with MestReNova 7.0 (Mestrelab Research) or Varian VNMR.
Feeding experiment with [U-D7]-indole
C. elegans were cultivated in 100 mL S. complete medium by providing E. coli OP50 as a food source. After a worm density of 88,000 worms/mL was reached, 0.6 mg [U-D7]-indole in 100 μL methanol was added to the culture. Aliquots of 6 mL were taken after 0, 1, 2, 3, 6, 12, and 24 h, centrifuged at 5251 g for 10 min, and 5 mL of supernatant was lyophilized, extracted with 2 mL methanol, filtered and concentrated in vacuo. The residues were taken up in 200 μL methanol and analyzed by HPLC with UV and ESI-MS detection.
Synthesis of N-(β-D-glucopyranosyl)indole (7)
A solution of indoline (1.0 g, 8.4 mmol) in ethanol (60 mL) was treated with β-D-glucose (0.7 g, 3.9 mmol) in water (2 mL) and stirred at 90 °C. Additional water (0.8 mL) was added after 7 and 14 h. After 28 h the solution was concentrated in vacuo and the residue fractionated on silica using a gradient of 0 – 10% methanol in dichloromethane to get N-(β-D-glucopyranosyl) indoline (1.05 g, 3.7 mmol, 96% yield) as a yellowish solid. A solution of N-(β-D-glucopyranosyl) indoline (85 mg, 300 μmol) in 1,4-dioxane (15 mL) was treated with 2,3-dichloro-5,6-dicyano-1,4-benzochinone (82 mg, 360 μmol). After stirring for 14 h the mixture was concentrated. Flash column chromatography on silica gel using 20% methanol in dichloromethane afforded N-(β-D-glucopyranosyl) indole (70 mg, 251 μmol, 83% yield). Reverse phase HPLC on a C18 column afforded a pure sample for toxicity testing.
Supplementary Material
Acknowledgments
We thank F. Ausubel and B. Cezairliyan (Harvard University) for helpful discussions and information about phenazines. P. Gulig (University of Florida) provided useful advice about P. aeruginosa. This work was supported in part by the National Institutes of Health (GM008500 to Y.I., R01GM088290 to F.C.S, R01GM085285 and 3R01GM085285-01A1S1 to A.S.E and F.C.S).
Footnotes
The authors declare no competing financial interests.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nature reviews Immunology. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nature reviews Genetics. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 3.Sifri CD, Begun J, Ausubel FM. The worm has turned--microbial virulence modeled in Caenorhabditis elegans. Trends in microbiology. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Molecular and cellular biology. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS pathogens. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shivers RP, Youngman MJ, Kim DH. Transcriptional responses to pathogens in Caenorhabditis elegans. Current opinion in microbiology. 2008;11:251–256. doi: 10.1016/j.mib.2008.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. Plos Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. Journal of experimental zoology Part A, Comparative experimental biology. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. Journal of bacteriology. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes and infection/Institut Pasteur. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 11.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 12.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. P Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brent Cezairliyan NV, Grenfell-Lee Daniel, Yuen Grace J, Saghatelian Alan, Ausubel Frederick M. Identification of Pseudomonas aeruginosa Phenazines that Kill Caenorhabditis elegans. PLoS pathogens. doi: 10.1371/journal.ppat.1003101. In-Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laursen JB, Nielsen J. Phenazine natural products: Biosynthesis, synthetic analogues, and biological activity. Chemical reviews. 2004;104:1663–1685. doi: 10.1021/cr020473j. [DOI] [PubMed] [Google Scholar]
- 15.Price-Whelan A, Dietrich LE, Newman DK. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 16.Hu KJ, Li JX, Webster JM. Nematicidal metabolites produced by Photorhabdus luminescens (Enterobacteriaceae), bacterial symbiont of entomopathogenic nematodes. Nematology. 1999;1:457–469. [Google Scholar]
- 17.Kerboeuf D, Riou M, Neveu C, Issouf M. Membrane Drug Transport in Helminths. Anti-Infective Agents in Medicinal Chemistry. 2010;9:113–129. [Google Scholar]
- 18.Sheps JA, Ralph S, Zhao ZY, Baillie DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laing ST, Ivens A, Laing R, Ravikumar S, Butler V, Woods DJ, Gilleard JS. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem J. 2010;432:505–514. doi: 10.1042/BJ20101346. [DOI] [PubMed] [Google Scholar]
- 20.Burns AR, Wallace IM, Wildenhain J, Tyers M, Giaever G, Bader GD, Nislow C, Cutler SR, Roy PJ. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- 21.James CE, Davey MW. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int J Parasitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.