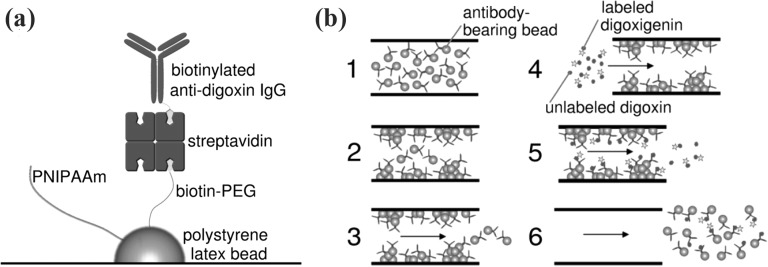
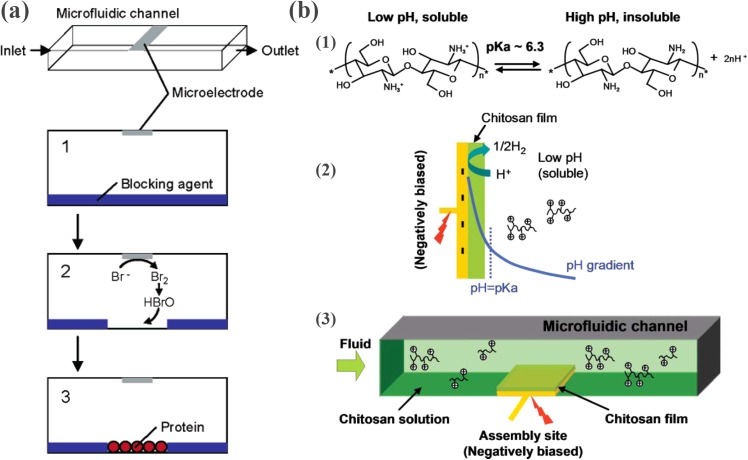
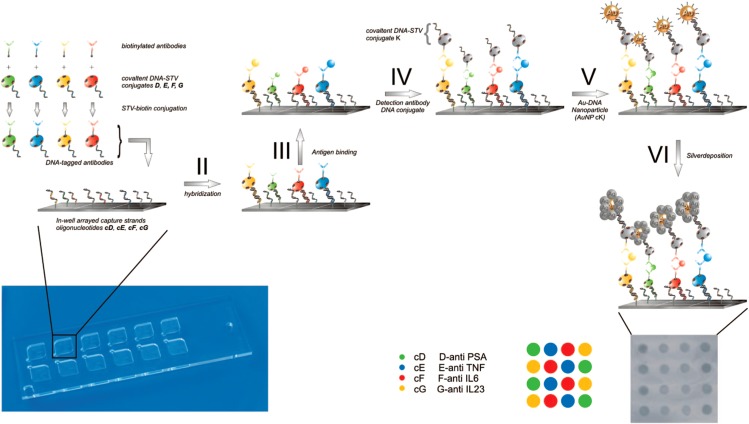
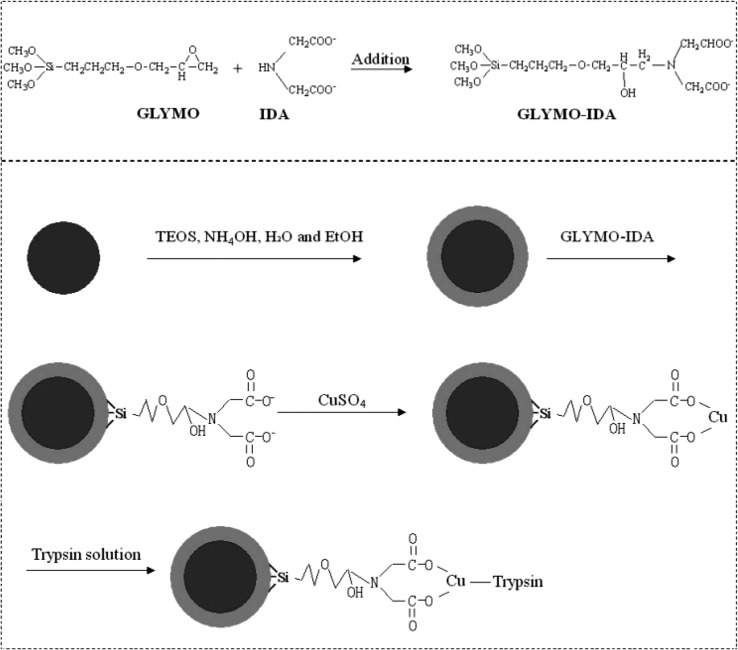
Abstract
Microfluidic systems have shown unequivocal performance improvements over conventional bench-top assays across a range of performance metrics. For example, specific advances have been made in reagent consumption, throughput, integration of multiple assay steps, assay automation, and multiplexing capability. For heterogeneous systems, controlled immobilization of reactants is essential for reliable, sensitive detection of analytes. In most cases, protein immobilization densities are maximized, while native activity and conformation are maintained. Immobilization methods and chemistries vary significantly depending on immobilization surface, protein properties, and specific assay goals. In this review, we present trade-offs considerations for common immobilization surface materials. We overview immobilization methods and chemistries, and discuss studies exemplar of key approaches—here with a specific emphasis on immunoassays and enzymatic reactors. Recent “smart immobilization” methods including the use of light, electrochemical, thermal, and chemical stimuli to attach and detach proteins on demand with precise spatial control are highlighted. Spatially encoded protein immobilization using DNA hybridization for multiplexed assays and reversible protein immobilization surfaces for repeatable assay are introduced as immobilization methods. We also describe multifunctional surface coatings that can perform tasks that were, until recently, relegated to multiple functional coatings. We consider the microfluidics literature from 1997 to present and close with a perspective on future approaches to protein immobilization.
INTRODUCTION
Proteins are biomacromolecules that play essential roles in life processes spanning from metabolic process regulation, cellular information exchange, cell-cycle control, and molecular transport to protection from the environment.1 In biomedicine, for example, proteins are of great interest as disease biomarkers. In biotechnology, as another example, the role of enzymes as biocatalysts is a topic of much study. Owing to functional involvement in physiological processes, protein state (expression levels and modifications) may be effective indicators of a disease state and/or response to therapeutic treatment.2 Biomarker detection using immunoassays has been a widely used disease diagnostic tool.3 Promising protein biomarkers benefit from further characterization by immunoassays and similar analytical tools.4 Immunoassays exploiting specific recognition of protein biomarkers by cognate antibodies have been optimized for high analytical performance (e.g., rapid assays, label-free detection, improved limits of detection, and multiplexing capability). Enzymes are a specific class of proteins that catalyze biochemical reactions. Enzymes display selectivity, accelerate reactions, provide environmentally friendly means to organic synthesis, and effectively synthesize complicated biomolecules such as DNA and RNA.5 As enzymes are selective and effective proteinaceous biocatalysts that convert substrates into products, the enzyme is actively used across agricultural feeds, polymer synthesis, biofuels production, food processing, and the paper industry.6 Enzymes are also used widely in biosciences and biotechnology such as genetic engineering (e.g., oligonucleotide manipulation) and the pharmaceutical industry (e.g., production of pharmaceutical ingredients).6, 7 In addition, enzyme-mediated fluorescence or colorimetric detection of proteins, i.e., ELISA (Enzyme-Linked Immunosorbent Assay), is a standard immunoassay technique.
In analysis of proteins and enzymes, microfluidic design has proven to be a powerful technological tool to improve performance of immunoassays,8, 9, 10 enzymatic reactors,11, 12, 13 and other biological assays.14 Importantly, manipulation of liquid inside microscale fluidic networks enables reduced consumption of reagents, compared to macroscale instruments.8, 9, 10, 13, 14 Decreased liquid volume and short diffusion lengths allow facile reactions between analyte and antibody or enzyme and substrate, resulting in reduced assay times.8, 9, 11 Using design strategies pioneered by the semiconductor industry, microfluidic integration offers a “sample-in, answer-out” capability.9, 10, 12, 14, 15, 16, 17 Microfluidic technologies make possible monolithic integration of disjoint assay steps, further underpinning automation of those steps.15, 16, 17, 18, 19, 20, 21, 22, 23, 24 As discussed in depth in this review, the fine spatial control in immobilizing proteins and biomolecules inside microchannels allows multiplexed21, 22, 25 and multiparameter assays.26 The overall form factor of self-contained microfluidic devices (and automation) reduces human errors and risks of exposure to dangerous and toxic bio-/chemical reagents.
Analytical immunoassays in microfluidic formats are designed for rapid and sensitive detection of one or several targeted antigens in clinical diagnostics,27, 28, 29, 30, 31, 32, 33 as protein sensors,34, 35, 36, 37 or in environmental analysis.38, 39, 40, 41, 42 Laboratory-grade assays such as polyacrylamide gel electrophoresis (PAGE) based immunoassays,43, 44, 45 isoelectric focusing (IEF),21 and Western blotting18, 19, 20, 22, 24, 46 provide qualitative and/or quantitative information on multiple proteins, even in complex biological fluids. Questions spanning from protein-protein interactions,47, 48 and protein binding kinetics,49, 50 to post-translational modifications23, 51 have all been pursued using analytical technologies in microfluidic formats. Recent reviews by Hanares et al.,9 Bange et al.,10 and Ng et al.8 are recommended as excellent overviews of immunoassay advances. Microfluidic enzyme reactors find use in analysis and optimization of biocatalytic process. For more detailed information on microfluidic enzyme reactors, recent reviews by Křenková et al.,11 Asanomi et al.,12 and Miyazaki et al.13 are recommended. Here, before scaling up to a large-scale batch process, the throughput and appreciable assay sensitivity of a microfluidic format can expedite candidate-enzyme screening process from mutant libraries.52 Enzyme-kinetic study has been performed in microfluidic formats.53, 54, 55, 56, 57, 58 Important to proteomics, enzymatic digestion before MALDI-TOF/MS (Matrix-assisted laser desorption-ionization time-of-fly/mass spectrometry) peptide mapping of a protein has been explored in microfluidic devices.59, 60, 61, 62, 63, 64, 65, 66 Compared to conventional in-solution enzyme digestion, which is time consuming and offers limited sensitivity, microfluidic formats have shown high conversion rates, facile replacement of inactivated enzyme, and long-term stability.54, 55, 63, 67 Enzymatic production of fluorescent and colored products for protein analysis (e.g., alkaline phosphatase (ALP) production of chemiluminescence or colorimetric product) enhances detection limits of immunoassays.
Heterogeneous assay formats where one binding or reaction partner is immobilized to a surface are widely employed and are the focus of this review. Consequently, surface immobilization is a primary design and performance consideration.8, 9, 10, 11, 12, 13 In contrast to heterogeneous formats are homogeneous approaches, where reactants are reacted in solution. This review focuses on the former category of reactions. Two examples of heterogeneous assays that are popular in microfluidic formats are immunoassays8, 9, 10 and enzyme reactors.11, 12, 13 For microfluidic immunoassays, antigen or antibody is immobilized inside microchannels. Key immunoassay performance metrics include analytical sensitivity, analytical specificity, and reproducibility. The immunoassay performance depends on the quality of protein immobilization, and thus on the immobilization surface, immobilization chemistry, and surface passivation technique (i.e., antibiofouling).68 In microfluidic enzymatic reactors where enzyme is immobilized inside microchannels, the enzyme conversion rate, long-term stability, and reusability depend on similar immobilization factors.63 We will cover the design and operation of these two canonical heterogeneous formats—the immunoassay and enzyme reactors—as we detail design and operational considerations for protein immobilization in microfluidic systems.
IMMOBILIZATION SURFACE
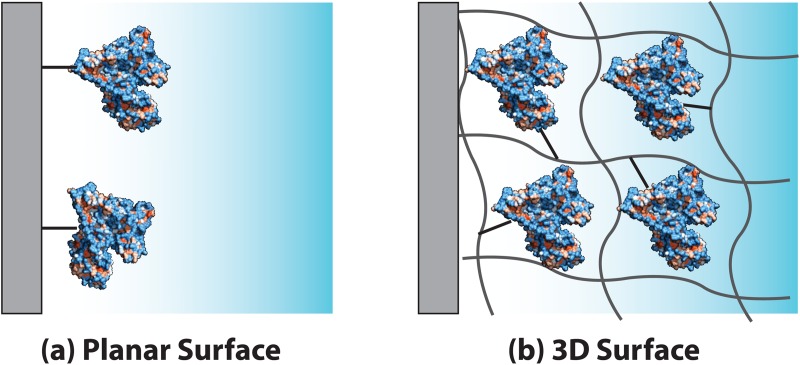
Immobilization methods vary largely with immobilization surface, protein properties, and the goals of the immunoassay or enzyme reactor. A major factor to consider is immobilization surface properties. One of the simplest surfaces on which protein is immobilized is the inner surface of microfluidic channels (Figure 1a). Traditional inorganic microfluidic device substrates are glass and silicon, which originated from the semiconductor industry and benefit from mature microfabrication techniques. For specialized detection methods such as surface plasmon resonance (SPR),8, 9, 49 Raman spectroscopy,69 and electrochemical analysis,35, 36, 70 protein is immobilized on metal films deposited on a glass or silicon surface. Silicon and glass share a similar surface chemistry, thus the route to immobilization is similar. Typically, the approach includes surface silanization followed by anchoring to a functional group of a silanizing agent. PDMS (polydimethylsiloxane), a silicon-based organic polymer, attained widespread use because of the low cost, rapid and prototype-friendly fabrication, as well as optical transparency, malleability, and gas permeability (appropriate for some applications).10, 71, 72, 73 Recently, plastic substrates such as PMMA [Poly(methyl methacrylate)], PS (polystyrene), and COC (cyclic olefin copolymer) have gained attention owing to low cost of fabrication (e.g., injection molding or hot embossing), a chemical resistance superior to PDMS, optical transparency, and low autofluorescence.74, 75, 76 PDMS and plastic surfaces are relatively inert and lack functional groups (i.e., sites for protein attachment). Thus, involved chemical surface preparation is generally required to induce surface functional groups for protein immobilization.9, 35, 77, 78, 79, 80 As immobilization on planar surfaces yields limited protein density, three dimensional (3-D) structures have been employed inside microfluidic channels for higher protein capture capacity,21, 22, 81 resulting in improved immunoassay sensitivity or enzyme conversion rates (Figure 1b). 3-D structures have been created by patterning microstructures (e.g., microposts29, 82, 83 and micropits60) or through insertion of porous membranes67, 84, 85 before assembly of microfluidic chips. In post-assembly approaches, microbeads54, 62, 86, 87, 88, 89, 90, 91, 92, 93 can be packed into enclosed channels or various polymers such as hydrogels,18, 19, 20, 21, 22, 23, 24, 57, 94, 95, 96, 97 sol-gels,64, 98, 99 polymer monoliths,61 or membranes100 can be polymerized in situ. For silica-based 3-D structures such as silica beads91 and alkoxysilane-based sol-gels,63 a similar glass/silicon surface immobilization strategy can be used. For polymer-based 3-D structures like agarose beads, hydrogels, and polymer monoliths, various immobilization methods including copolymerization of protein,18, 19, 20, 23, 94 graft polymerization,101 and oxidative activation of functional groups29, 54 can be used. Among these 3-D structures, hydrogels such as polyacrylamide gel and polyethylene glycol (PEG) gel provide hydrophilic environments conducive to good protein stability and retained protein activity.102 Paper has recently gained momentum as a 3-D substrate material for POC (point-of-care) diagnostics for low-resource settings owing to low cost, simple assay visualization, and simple reagent immobilization.103, 104, 105 In this section, we provide more detail on the most popular immobilization surfaces.
Figure 1.
Role of surface geometry in binding site density. Schematic drawing of (a) planar and (b) high surface-area-to-volume ratio three-dimensional immobilization surfaces.
Planar microchannel surface
In a large body of literature, protein is immobilized on microchannel surfaces made from silicon, glass, PDMS, plastic, or metal film deposited on channel surfaces. A protein monolayer can be formed on the surface after immobilization. The planar channel surface is a natural choice because of simplicity, a surface-to-volume ratio of microchannel surface larger than macroscopic counterparts, and the fact that fluids contact the surface. Random surface immobilization relying on multiple anchoring points can cause a protein to be denatured and lose native activity.106, 107 Also, active sites can orient towards the immobilization surface, resulting in reduced activity.106 Even though the diffusion length within microfluidic channel is short and thus the overall reaction can be faster than macroscopic counterparts, a single monolayer of protein may not provide sufficiently high analytical signal in immunoassays or a sufficiently high conversion rate in enzyme reactors. An ideal immobilization surface should have a large surface-area-to-volume ratio, a protein-friendly environment, minimal nonspecific protein adsorption, mechanical and chemical stability, and a reactive moiety for protein coupling.11
Silicon
Silicon is a most widely used and well-characterized substrate originating from integrated circuit development in the semiconductor industry. Silicon was adapted as a mechanical material with the advent of microelectromechanical systems (MEMS)108 and, subsequently, employed in the first microfluidic analytical system.109 Owing to high-resolution microfabrication technique development (feature size as low as 22 nm are attainable with mass fabrication110), fine fluidic channels and microfluidic components can be created on a silicon substrate. Naturally or artificially grown oxide on the silicon surface makes silanol-based chemistries compatible with for protein immobilization on silicon.40, 111, 112 While powerful, silicon has, however, three major drawbacks for microfluidic design: (1) opaqueness of silicon in the visible spectrum renders various optical imaging techniques irrelevant; (2) incompatibility of silicon with electrokinetic methods owing to the electrical conductivity of the silicon substrate, and (3) expense associated with the sophisticated microfabrication techniques used to micromachine silicon in a cleanroom environment. Therefore, silicon is not as widely used as initially with the exceptions of continued widespread application in electrochemical analysis113 and SPR.65
Glass
Besides silicon, glass (e.g., fused silica, soda lime, borosilicate glass), and quartz are another widely used substrate for microfluidic devices.114 Glass substrates were initially used in microfluidic electrophoresis systems, building on the earlier successes of glass capillaries as a format of choice among the electrophoresis community.115 Glass is transparent across a wide spectrum, with negligible autofluorescence.74 Therefore, glass is well-suited for fluorescence-based microfluidic assay readouts. Glass is robust, being resistant to solvents and acids and bases.10 Several commercially successful microfluidic devices use glass substrates.116 However, glass is not without disadvantages. Glass can fracture, so it must be handled with a care. Glass microfabrication can be costly, since much of the process requires a cleanroom facility. Microfabrication processes similar to those used for silicon result in patterned microchannels but acid wet etching of glass does not yield high-aspect-ratio anisotropic microchannels unlike silicon (e.g., deep reactive-ion etching process). Glass substrates benefit from immobilization chemistries including various silanol chemistries for covalent linking of proteins.117
PDMS
PDMS is currently one of the most frequently used and studied substrates in microfluidics, owing to a rapid design-fabricate-test cycle and low cost. PDMS is a rubber-like flexible polymer (i.e., elastomer) and is transparent making optical imaging possible. In contrast to rigid substrates such as silicon and glass, microfluidic actuators such as valves and pumps can be readily formed in a microdevice.118 Owing to rapid casting-based soft lithography processing, fabrication of microfluidic networks in PDMS is inexpensive, requiring low investment in infrastructure. Overall, PDMS is an excellent material for rapid prototyping of research device. After treating with oxygen plasma, PDMS can be irreversibly sealed with glass, plastic substrates, or PDMS slabs to form enclosed microchannels. Multiple layers of PDMS can be stacked yielding multifunctional microfluidic devices.118 This fabrication process stands in stark contrast to complex and time-consuming silicon or glass bonding processes.89, 93 Nevertheless, no material is well suited to every application. Drawbacks of PDMS are as follows: limited resistance to organic solvents, gas permeability, compliant characteristics and thus often inappropriate for harsh environments needing robust packaging.10 As related to protein immobilization, PDMS is hydrophobic in native form, so proteins tend to readily and nonspecifically bind to the surface. Therefore, blocking of the adsorptive surface must be done before an assay is completed. PDMS lacks functional groups for covalent derivatization. Silanol groups can be introduced after oxygen plasma treatment but these groups do not offer long-term stability.119 Therefore, immobilization methods are generally complex and require multiple steps to implement.97, 120 Because a large numbers of microfluidic devices are made by sealing microchannel-patterned PDMS slabs to glass slides, consideration of glass and PDMS surface properties is often required (e.g., care to avoid nonspecific adsorption to PDMS surfaces when protein is immobilized on glass surface).
Plastic
With a relevance to mass fabrication, cost effective, robust, and reliable substrates for microfluidics are of great interest. Microfluidic chips fabricated from plastics such as PMMA, PS, and COC are extremely cheap to mass produce when using mold-based techniques such as injection molding or hot embossing.76 Moreover, plastic is generally resistant to solvents and acids/bases, rigid but not fragile, common in the marketplace, and optically transparent.74, 75, 76 Owing to these attributes, numerous groups have investigated plastic as a material of a choice for commercial microfluidic devices.121, 122 Indeed, a few commercial microfluidic devices are made of plastic.123 Plastics share the disadvantages of PDMS, being hydrophobic in native form making hydrophobic nonspecific protein adsorption a concern. Inert plastic surfaces lack functional groups, so chemistry is employed to prepare the surface to immobilize proteins.29, 80, 124 Oxygen plasma77, 79 or strong bases/oxidizers35, 77, 78 are often used to introduce functional groups.
Metal
Metals films are sometimes deposited on silicon or glass surface. Protein is immobilized on the metal surface for detection methods other than fluorescence or colorimetric detection. Several assay readout modes are appropriate, including electrochemical sensing,35, 36, 69 Raman spectroscopy,69 and SPR.8, 9, 49 Thiol-based chemistry, although not as strong as covalent linkages, is available for protein immobilization on noble metal surfaces including gold, silver, and platinum.125
Three-dimensional materials in microchannels
In contrast to planar immobilization surfaces, three-dimensional (3-D) surfaces are often advantageous. Common formats include micro/nanostructures created using microposts, microbeads, hydrogels, sol-gels, polymer monoliths, and membranes. Fabrication approaches for these 3-D structures vary greatly, depending on the material choice and properties needed. The design rationale underpinning exploration and selection of 3-D structures stems from the increased surface-area-to-volume ratio offered (as compared to a planar surface). The increased effective surface area found in the 3-D material offers a larger number of immobilization sites, as compared to channel wall (2-D) systems.21, 22, 81 Back-of-the-envelope estimates suggest that three-dimensional gel structures provide about 100 ∼ 1000 fold increase in binding sites, as compared to immobilization sites on capillary or microchannel walls alone.21 Importantly, the diffusion length between reaction partners (e.g., antibody and antigen, or enzyme and substrate) is reduced when the immobilized partner is in a 3-D material, as opposed to patterned on a microchannel or even nanochannel wall. Therefore, high analytical sensitivity or fast conversion rates can be realized when 3-D materials are utilized in reacting systems and transport conditions are optimized.63, 70 For immobilization of proteins, structures with nanoscale features (e.g., sol-gels and nanoporous hydrogels) can physically encapsulate protein without chemically activating surfaces.64, 98 Packed bead beds can be dynamically introduced and eluted from the microchannel for quick surface regeneration.70 Clearly, 3-D structures need to be transparent for optical detection methods. For more information, the readers is referred to an excellent review on 3-D solid supports for microfluidic systems from Peterson.85
Packed bead beds
Monodisperse beads comprised of a wide-range of bead materials (i.e., PS,31, 86 silica,91 agarose,54, 126 ferromagnetic materials30, 62, 88) are a workhorse of conventional macroscale analytical chemistry, including chromatography85 and enzyme reactors.126 Bead packing inside microchannels has been accomplished using size-exclusion structures including microposts, dam, and weirs. High-sensitivity immunoassays are possible with the packed beads.93 As mentioned, the diffusion length from the solution phase to the bead surface is short in a packed bed, so enzyme conversion rates are also high.63 Magnetic beads are attractive, as these reactive 3-D surfaces can be immobilized by applying a magnetic field from outside of the microchannel (using a magnet). Regeneration after assay completion is facile, with beads flushed out of the channel after removing the magnetic field and applying a bulk flow (i.e., pressure driven).70 Protein immobilization strategies are diverse and vary with the bead material. For example, silanol chemistry can be exploited for glass or silica beads. More involved immobilization protocols are required for polymer beads such as polystyrene127 and agarose beads128 to induce functional groups on the polymeric surfaces.
Hydrogel
Hydrogels have been actively used in tissue engineering129 and protein microarray81 owing to the hydrophilic, protein-friendly microenvironment offered. Hydrogels are flexible materials with a well-ordered fibrous structure. Synthetic PEGDA (PEG diacrylate) gel57, 83 and polyacrylamide gel18, 19, 20, 21, 22, 23, 24, 94, 95, 96 are popular with natural gels like chitosan130 or agarose gel also used. Porogen is sometimes employed to further increase the surface area by inclusion of macropores.82, 83 Hydrogel is usually transparent so that sensitive fluorescence imaging is appropriate. Polyacrylamide gel can also act as a biomolecular sieve without significant nonspecific adsorption, so protein can be separated based on charge and/or size (e.g., SDS-PAGE or sodium-dodecyl-sulfate polyacrylamide gel electrophoresis) before a detection step.22, 23, 24, 131, 132 With suitable surface modification, gel regions offering different assay functions can be integrated on a single chip using fabrication via photopatterning.18, 19, 20, 23, 24, 46, 94 The swelling property of hydrogels allows integration of actuators such as valves, allowing integration of sophisticated fluid handling functions.133 A wide range of immobilization methods are available to hydrogels, including copolymerization of proteins,18, 19, 23, 94, 132 activation for covalent linking of proteins,134 or electrostatic capture on charged hydrogel.20, 24 Even with a 3-D microchannel-filling hydrogel, the microchannel surface should be functionalized for covalent anchoring of the hydrogel structure within the channel, so that the gel will not drift out of the channel under hydrodynamic pressure or applied electric field.46, 82, 83 Hydrogels provide a hydrated environment for proteins so that native activity or functionality can often be maintained.102 A disadvantage of hydrogels is the fragility of some gel structures (i.e., application of shear forces or high electric fields can damage the structure135). Once formed, a stationary hydrogel is difficult to remove from the channel if regeneration of the assay system is needed.46
Sol-gel
Sol-gels are a condensation polymerization of colloids in aqueous media. After preparation of the “sol” (i.e., metal alkoxide monomer in acid and organic/aqueous solvent), the “gel” is formed by polymerization during evaporation of the solvent. By simply adding proteins to the “sol” before gelation, proteins are encapsulated in optical transparent 3-D nanostructure. Silica sol-gels made from silicon alkoxide colloids98 are the most common, but titania64 or zeolite99 colloids are also used. Immobilization of proteins proceeds under mild conditions,64 thus proteins can retain near-native activity.
Porous polymer monolith
Rigid polymer monoliths are a porous polymer frit formed inside microfluidic chips (or capillaries) using polymerization of monomers such as ethylene dimethacrylate, acrylamide, or 2-hydroxyethyl methacrylate in the presence of porogens, such as dodecanol, decanol, and cyclohenxanol.61, 85 Whereas packed beads require an immobilizing structure like a weir or micropost array, polymer monoliths can be photopolymerized at any location in the channel without said retaining structures.61 Owing to pores throughout the monolith, a large surface area is available for protein immobilization, and a low back pressure allows pressure driven flow for fluidic control.61 A disadvantage of polymer monoliths lies in the difficulty of reproducible operation owing to batch-to-batch variability.85
Membrane
Membranes are porous (planar) sheets that can provide a large surface area for protein immobilization. Commercial membrane comprised of polycarbonate (PC), nitrocellulose (NC), and PVDF (polyvinylidene fluoride) have been inserted and physically clamped between a microfluidic chip patterned with channels and a blank cover chip.67, 84, 85 However, resolving fluid leakage issues from the clamped membrane can be challenging.84 Membranes can also be formed in situ by condensation100 or electrospinning.33, 136 Membranes have been extensively used in biochemistry or analytical chemistry to adsorb proteins. Such protein immobilization strategies rely on intermolecular forces137 (i.e., hydrophobic, electrostatic, and van der Waals) and simple adsorption without complicated chemical activation of the solid supports. Membranes also have additional useful properties such as ion selectivity and selected transport of small molecules such as enzyme substrate (i.e., size exclusion)100 and ions.
Paper
Paper, a cellulose membrane, offers a versatile, low-cost material for immobilization.103, 104, 105 Originated from disposable lateral-flow immunoassays, paper microfluidic devices have the potential to be more cost-effective than plastics or PDMS.138 Fluid and material transport in and through paper can be accomplished passively (without power consumption) using capillary action.103, 138 Even though paper is macroscopically planar, the material has microscopic 3-D pores. Immobilization strategies on paper are rather simple. Much like NC membrane, paper adsorbs proteins via a combination of intermolecular forces making simple reagent spotting effective for protein immobilization. Paper is optically opaque, so that sensitive detection using fluorescence imaging could be difficult. Therefore, the limit of detection is rather poor compared to immunoassays based on transparent substrates.138
Porous silicon
Porous silicon is produced by anodic electrochemical or photochemical etching of silicon in hydrofluoric acid (HF).139 Nanopores in porous silicon are usually straight (unidirectional), and perpendicular to the silicon surface. Porous silicon offers the advantage that that porosity and pore size are exquisitely controllable. Owing to sensitive refractive index change, porous silicon offers exceptional performance an optical biosensor.139 Proteins can be immobilized on the porous silicon using covalent chemistries similar to those used for planar silicon substrates.40, 60, 68
IMMOBILIZATION STRATEGY
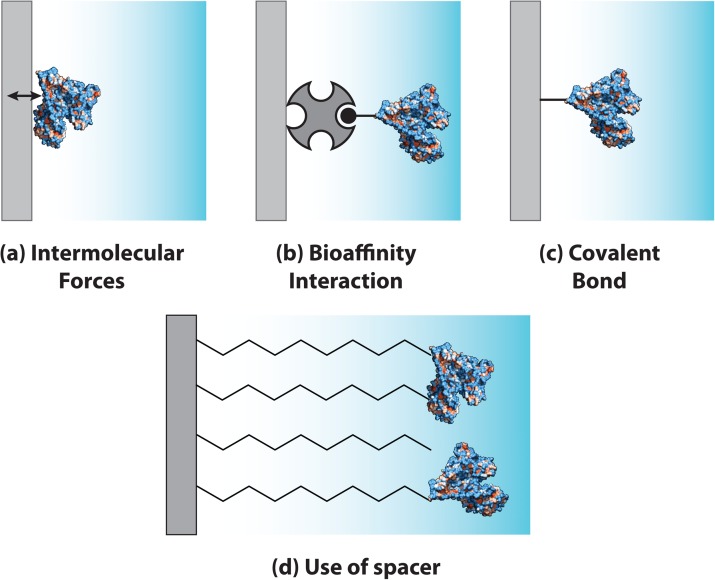
A wide variety of immobilization methods are employed to attach proteins to the immobilization surface. A specific immobilization method relies on a variety of factors including immobilization surface, sample matrix, protein property, buffer constituents, and assay performance metrics (e.g., sensitivity, reusability, selectivity, and reproducibility). Ideally, active sites for antibody binding or enzymatic conversion should be accessible to reaction partners (i.e., proteins face away from the immobilization surface to mitigate steric hindrance and are not sterically blocked by neighboring immobilized proteins). After immobilization, protein conformation should be intact so that protein functions are retained for a high-performance, reproducible assay.106 An excellent review on immobilization strategies for protein microarrays is provided by Rusmini et al.106 Useful information on enzyme immobilization inside microfluidic chips can be found in reviews by Křenková and Foret,11 Asanomi et al.,12 Miyazaki and Maeda.13 For protein immobilization in microfluidic immunoassays, readers are directed to reviews by Bange et al.,10 Ng et al.,8 and Křenková and Foret.11 Molecular mechanism of protein immobilization methods can be categorized into physisorption, bioaffinity interaction, covalent bond (Figures 2a, 2b, 2c), and combinations of the three mechanisms. The following sections detail specific aspects of the most widely used immobilization methods.
Figure 2.
Common surface immobilization methods for heterogeneous assays. Schematic of immobilization mechanisms: (a) physisorption, (b) bioaffinity interaction, and (c) covalent bond. The surface immobilization methods are often used in conjunction with (d) spacer for improved protein activity.
Physisorption
The simplest approach to immobilizing a protein on a surface is physisorption (i.e., physical adsorption). Protein can be conveniently adsorbed to various surfaces via intermolecular forces such as electrostatic, hydrophobic, van der Waals, hydrogen bonding interactions, or combination of those (Figure 2a).140 Incubation of protein in solution contacting the immobilization surface or continuous flow of solution will achieve attachment of protein without complicated chemistry or reagents. Physisorption is generally weak, and thus an adsorbed layer of proteins is not as stable as one formed by covalent or bioaffinity binding. The intermolecular forces are highly dependent on environmental condition such as pH, ionic strength, temperature, and surface condition.107 Therefore, immobilization of proteins—in a reproducible manner—can be difficult using physisorption. As protein immobilization is randomly oriented on the surface, some fraction of the binding sites within a population of immobilized proteins are likely not accessible.106 Further, protein can be immobilized to the surface via multiple binding sites, which may result in conformational change and reduction of protein activity.106, 107 Immobilization density depends on protein size, as well as physicochemical surface properties. If the immobilization density is too high, active sites could be sterically blocked.106 Therefore, the use of a spacer (e.g., PEG) with a surface-attaching head group and a protein-binding tail group has been widely adopted to reduce steric hindrance (Figure 2d).40 Blocking of uncoated surface should be performed after protein immobilization (e.g., BSA or bovine serum albumin) in order to minimize nonspecific adsorption of off-target biomolecules. Even given the low degree of control, many studies have employed physisorption of protein to a microfluidic-device surface. Popularity of the approach stems from several advantages including simple assay procedure, no toxic reagents and no sophisticated chemical protocols.125 For plastic and PDMS, proteins are often adsorbed onto the bare surface owing to the hydrophobic nature of these substrates. In such systems, intermediate molecules are frequently used in order to attach to surface by covalent linkage on one end and provide hydrophobic or charged functional group on the other end for protein physisorption. Surfaces are modified to have stronger charge or hydrophobicity to adsorb protein better than the native surface, with reports of such physisorption as practically irreversible.92 In some cases, physisorption happens instantaneously (i.e., high kon) compared to covalent or bioaffinity bonds that usually requires a substantial incubation time, and thus physisorption can be used as an intermediate immobilization step of a multistep assay sequence (e.g., Western blot).20, 24 Physisorption has a wider choice of buffer system, compared to widely used covalent bonding through primary amines where popular amine-based buffers (e.g., Tris, glycine) cannot be used.20
Electrostatic interaction
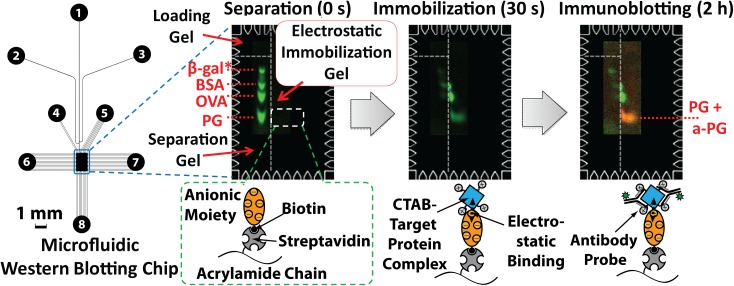
Electrostatic or ionic interaction is fundamental to biomolecular attraction (e.g., protein-protein interaction141 and DNA hybridization142). Thus, electrostatic interactions are exploited frequently in biochemical assays, for example, cell adherence to positively charged poly-L-lysine (PLL)-treated PS surfaces143 or protein blot to positively charge nylon membrane.144 Typical positively charged functional groups encountered in biochemistry are protonated amine (NH3+) and quaternary ammonium cations (NR4+). Negatively charged functional groups are carboxylic acid (—COO−) and sulfonic acid (—RSO3−). These functional groups are involved in the electrostatic interaction between protein and a surface. Complete isolation of contributions from other intermolecular forces may be difficult or impossible. However, electrostatic interactions can be studied by measuring the binding isotherm while changing the buffer ionic strength.141, 145 Electrostatic interactions find application in the protein immobilization in microfluidic assays. Protein friendly, gel-like hydrophilic environment have been created by electrostatic layering of polyelectrolytes such as PEI [poly(ethylene amine)], PDADMAC [poly(diallyldimethylammonium chloride)], PAA [poly(acrylic acid)], PAH [poly(allylamine hydrochloride)], and PSS [poly(styrene sulfonate)] on protein-unfriendly hydrophobic polymeric (e.g., PDMS, PMMA, and PS) surfaces35, 77, 78, 79, 124 or silicon surfaces.35, 40, 59, 68, 77, 79, 124 Proteins are immobilized on the composite layer directly by electrostatic interaction,146 covalent chemistry,40, 68 or bioaffinity interaction.35, 40, 68, 77, 79, 124 Electrostatic interaction has also been used to pack microbeads (with bead-surface immobilized proteins) into beds in microfluidic channels.92 Protein has also been directly immobilized inside hydrogels via electrostatic interaction. Kim et al. created negatively charge polyacrylamide (PA) gels to immobilize proteins after separation by electrophoresis to yield microfluidic Western blotting (Figure 3).20 Because of a high surface charge (−120 e) in electrophoresis buffer (pH 8.3), the enzyme β-gal (β-galactosidase) was copolymerized in PA gel to introduce a negative charge to the gel. CTAB (cetyltrimethylammonium bromide, cationic detergent) treated proteins were first separated in photopolymerized PA gel via electrophoresis then transferred to and instantaneously immobilized on the β-gal-conjugated PA gel. Immobilization was owing to strong electrostatic interaction between the positively charged protein-CTAB complex and the negatively charged PA gel. After the BSA blocking step, the immobilized target protein was detected by an antibody. In that study, the charge interaction was strong enough for immobilized antigens to sustain electrophoretic wash, blocking, and antibody introduction via electrophoresis. The role of electrostatic interaction was studied by the systematic changing of ionic strength and the associated characterizing of binding strength of the interaction.147, 148 As a follow up to this research, the same research group created a positively charged PA gel by copolymerizing PLL, thus allowing electrostatic immobilization of negatively charged SDS-treated proteins.24 Therefore, electrostatic protein immobilization after protein separation was used as a basis for automated microfluidic format SDS Western blotting.
Figure 3.
Charged PA gel allows electrostatic immobilization of CTAB-coated proteins in a microfluidic Western blotting. Negatively charged PA gel immobilizes separated proteins, followed by fluorescent detection (immunoblotting). Reprinted with permission from D. Kim et al., Anal. Chem. 84, 2533 (2012). Copyright 2012 American Chemical Society.
Hydrophobic interaction
As some designers of novel microfluidic devices aim for disposable point-of-care diagnostics applications, immobilization of biomolecules on polymeric hydrophobic materials has gained attention. COC, PMMA, and PS—transparent, hydrophobic thermoplastics—have gained attention recently for the POC application.121, 122 Bhattacharyya and Klapperich introduced a hot-embossed COC microfluidic chip for an immunoassay of CRP (C-reactive protein).28 Human CRP was introduced in the microfluidic channel and physisorbed by hydrophobic interaction, followed by BSA blocking, and chemiluminescence detection by horse radish peroxidase (HRP) conjugated antibody.
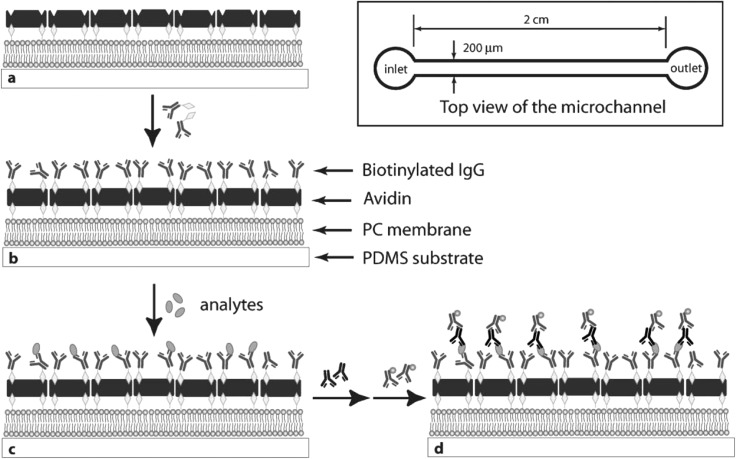
Tsougeni et al. recently presented an approach to increase protein adsorption on hydrophobic PMMA surfaces.149 Using directional O2 plasma etch and mask-based lithography, the researchers not only patterned microchannels but also roughened the channel surface. The roughened PMMA surface yielded stronger adsorption of protein (biotinylated BSA or IgG) compared to a smooth, hot-embossed PMMA surface (i.e., 120× poorer detection limit). Sia et al. reported a microfluidic device consisting of a PS lid mated to a PDMS substrate patterned with microchannels.150 HIV Env antigen (gp41) was adsorbed to the PS surface to assay antibodies in HIV-infected sera, with catalytic silver deposition using gold nanoparticle conjugated secondary antibodies. Xiang et al. designed an “H”-channel glass-covered PDMS chip.34 An Escherichia coli antigen was physisorbed to a PDMS surface and then later detected by primary and secondary antibodies. The authors used electrokinetic fluidic control.
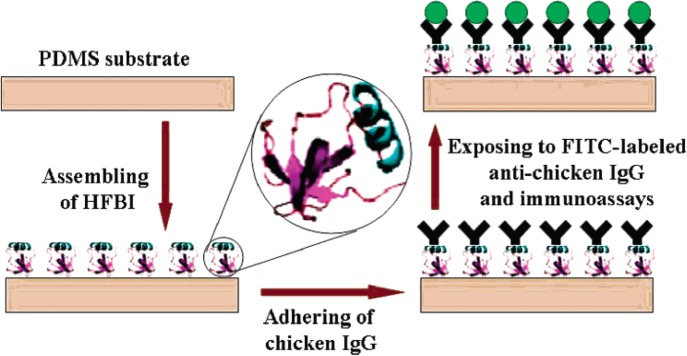
Kitamori's group published work on proteins absorbed to PS microbeads in a bead-based immunoassay.89, 93 Three glass substrates were patterned with channels using a CO2 laser and fast atom beam. Then, all the layers were thermally bonded to form a glass microfluidic device. Using a dam structure, analyte-adsorbed beads were pseudo-immobilized. Then, gold nanoparticle functionalized detection antibody was introduced for TLM (thermal lens microscopy). In a first study, secretory IgA was immobilized then analyzed by detection antibody.93 In a subsequent study, anti IFN-γ (interferon-r) capture antibody was immobilized, and IFN-γ was captured. Then biotinylated detection antibody was introduced, followed by injection of streptavidin conjugated with gold nanoparticle for improved detection limit.89 Feng's group published on a novel proteinaceous monolayer for antibody immobilization on a PDMS surface (Figure 4).151, 152 The authors used a copper TEM grid (i.e., stencil) to pattern hydrophobin, allowing conversion of the hydrophobic surface to a hydrophilic surface. Hydrophobin is a cysteine-rich small protein (∼100 amino acids) extracted from filamentous fungi. Hydrophobins can form self-assembled monolayer on hydrophilic-hydrophobic interface (e.g., air-water interface) owing to amphiphilic nature. Hydrophobic patches faced towards the PDMS while hydrophilic patches faced away. Chicken IgG was physisorbed via polar interaction to the hydrophobin SAM (self-assembled monolayer) creating a heterogeneous immunoassay format. Jo et al. reported a mass spectrometric (MS) imaging (spatially resolved MS information of attached polypeptides) modality using a PDMS microfluidic device.65 An Aplysia bag cell was attached to a PLL coated silicon surface through electrostatic interaction. Released neuropeptides were delivered through a PDMS microfluidic channel and physisorbed to another silicon surface rendered hydrophobic using octadecyltrichlorosilane (OTS) treatment. The peptides bradykinin, angiotensin II, substance P, renin substrate, and egg laying hormone were imaged by MALDI-TOF/MS.
Figure 4.
Immobilization process of chicken IgG on hydrophobin coated PDMS surface and immunoassay. Reprinted with permission from R. Wang et al., Chem. Mater. 19, 3227 (2007). Copyright 2007 American Chemical Society.
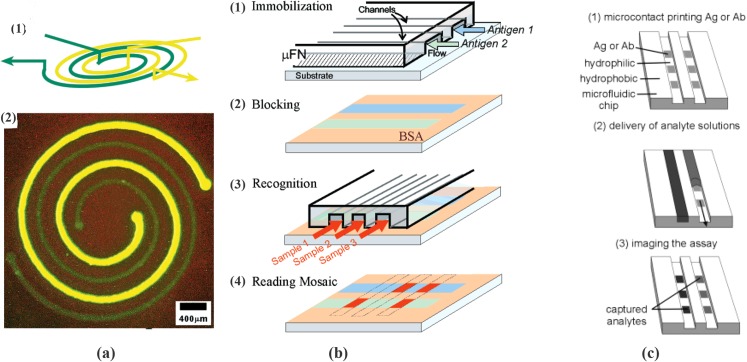
As described earlier, the simplicity of physisorption makes it a preferred method for immobilizing proteins in early, proof-of-concept experiments—a prototyping immobilization strategy. The Whitesides' group created a 3-D microfluidic stamp in PDMS as the basis for their technique called “3-D micromolding in capillaries (MIMIC),” which overcame limitations of conventional soft lithography.153 The micro contact printing can pattern complex protein patterns but requires multiple inking and stamping steps to have discrete pattern of multiple protein species.154 2-D MIMIC technique can deposit a discrete pattern of multiple protein species at a single “stamping,” but the pattern has to be continuous because the technique uses microfluidic channels that does not cross over.155 On the contrary, the 3-D MIMIC can put a discrete pattern of multiple proteins in a single stamping.153 BSA and fibrinogen were physisorbed to a PS substrate, and a complex protein pattern was created (Figure 5a). Following up on early work on “microfluidic networks (μFN),”156 Delamarche created a simple protein-microarray-like multiplexed immunoassay.157 The immunoassay was enabled by reversible sealing of silicon μFN to a PDMS slab, with simple physisorption of proteins resulting in a striped pattern on the PDMS. After patterning, the cover (housing a series of trenches) was rotated 90° and another reversible sealing of the μFN on the PDMS slab created enclosed microchannels for introduction of detection antibody and a multiplexed immunoassay was completed (Figure 5b). In later work, capillary action on wetting tissue was used to generate flow, and the silicon μFN was hydrophilized by gold-layer deposition, followed by PEG coating (HS-PEG) to reduce background signal resulting from nonspecific protein binding.27 CRP and cardiac markers (i.e., myoglobin and cardiac troponin I) were physisorbed on a PDMS slab and detected using the same sandwich immunoassay format. Nevertheless, preventing leakage of reagents through neighboring channels (i.e., cross-talk) has been a challenge for such reversible sealing. Finally, instead of a PDMS slab, Delamarche and colleagues used a gold-coated silicon μFN treated with HS-PEG for protein patterning (Figure 5c).158 The top surface of the μFN was coated with hexadecanethiol (HDT) to prevent nonspecific protein adsorption. Then, using deformable PDMS stamps, protein was microcontact-printed on the bottom surface, allowing subsequent antibody based detection. The authors observed that the HS-PEG coating promoted protein transfer from the PDMS stamp, as well as reduced nonspecific protein binding from solution. Stability of the transferred protein pattern on the hydrophilic HS-PEG surface could be an issue.
Figure 5.
Novel protein patterning methods using simple physisorption: (a) nested spirals of BSA (bright green) and fibrinogen (light green) on a PS surface using 3-D MIMIC technique. Reprinted with permission from D. T. Chiu et al., Proc. Natl. Acad. Sci. U.S.A. 97, 2408 (2000). Copyright 2000 National Academy of Science of USA. (b) Multiplexed immunoassay using μFN and reversible PDMS-to-PDMS sealing. Reprinted with permission from A. Bernard et al., Anal. Chem. 73, 8 (2001). Copyright 2001 American Chemical Society. (c) Protocol for an immunoassay in which the protein capture sites are patterned using microcontact printing and μFN. Reprinted with permission from J. Foley et al., Langmuir 21, 11296 (2005). Copyright 2005 American Chemical Society.
Unspecified combinations of intermolecular forces
Commercial NC and PVDF membranes are popular polymeric supports in molecular biology, being frequently used in Western blotting and dot blotting. These membranes now find use in microfluidic assays. While the exact immobilization mechanisms are not clear, protein immobilization on NC membranes is attributed primarily to hydrophobic interactions, hydrogen bonding, and electrostatic forces.137 For PVDF membranes, hydrophobic interaction is considered to play a major role.159 Gao et al. created an on-line protein digestion module in a PDMS microfluidic device.67 In this study, a commercial PVDF membrane (0.45 μm pore size) was clamped between two patterned PDMS substrates. Bovine pancreatic trypsin was adsorbed on a PVDF membrane by on-line injection. Denatured horse heart cytochrome C and ribonuclease A were passed through the trypsin-immobilized PVDF membrane, and then digested peptide was analyzed by ESI-MS (electrospray-ionization mass spectrometry). The authors observed that the microscopic surface area of the microporous PVDF membrane available for protein adsorption is 200 times larger than the macroscopic surface area of the membrane. Compared to solution-based trypsin digestion, the membrane reactor was 500–1000 times faster. The authors also reported that trypsin was active for more than 2 weeks. Lu et al. used wax-patterned NC membrane for their paper microfluidic device instead of pure cellulose (i.e., paper) owing to a higher binding capacity and more uniform binding patterns.160 Using a printer, a hydrophobic wax pattern was created on the NC membrane to confine antibody spots. Sandwich immunoassays using catalytic silver precipitation were demonstrated in the wax-patterned device.
Alternatively, porous membranes are fabricated in situ. These membranes benefit from tailored porosity and morphology, as well as localization in specific regions of a microfluidic channel. An NC membrane was created in situ on the glass surface by Park et al.66 After silanizing the glass surface with OTS to form a hydrophobic SAM, an NC membrane (dissolved in organic solvent) was spot-dried on the glass surface. Then, the glass was bonded to a patterned PDMS chip. The enzyme β-gal was physisorbed inside the membrane. The enzyme substrate di-β-D-galactopyranoside (FDG) was hydrolyzed to a fluorescent product and analyzed by electrophoresis. Jiang's group used an electrospinning (ES) technique to create a highly fibrous membrane as a protein-adsorption substrate in the PDMS microfluidic devices.33, 136 First, the researchers created a nanofibrous membrane using electrospinning of PC, and sandwiched the membrane between a glass substrate and a patterned PDMS chip.33 Then, HIV Env protein was physisorbed and detected by a primary antibody and a fluorescein isothiocyanate (FITC) conjugated secondary antibody. Compared with a track-etched polycarbonate (TEPC) membrane having a uniform pore size, the nanofiber membrane showed higher binding capacity. Similarly, a PVDF nanofibrous membrane was created for a similar PDMS-glass slide device.136 After adsorbing antibodies in the PVDF membrane, a multiplexed immunoassay was performed. The study reported that the protein adsorption capacity of the PVDF membrane was 8 times larger than that of TEPC membrane owing to an increased surface area.
Recently, paper (e.g., cellulose membrane) has drawn attention in the microfluidics community103, 104, 105, 138 owing to low chip material and manufacturing costs. Intermolecular forces including electrostatic and hydrophobic interactions are involved in protein adsorption to paper.161 Tan et al. reported a paper-PDMS glucose sensor.36 Enzyme GOx (glucose oxidase) was adsorbed to Whatman filter paper, then a glucose solution was flowed through a PDMS microfluidic channel to be converted into hydrogen peroxidase, which was later detected electrochemically. The stability of GOx observed in paper was excellent showing a 2.7% RSD (relative standard deviation) in repeatability and a one-month shelf life.
Physical encapsulation and entrapment
Alternately, a protein immobilization strategy that relies on physically encapsulating proteins in nanoporous structures has been employed. Compared to polymer monoliths or membranes having pore sizes on the order of a few hundred to thousands nanometers,84, 162 various nanoporous structures afford pore sizes of less than a few tens of nanometers.163 Owing to the small pore sizes, protein can be effectively encapsulated. A trade-off is seen in assays where an interaction with large binding partner (e.g., antibody or enzyme) could be hindered by slow diffusion through the nanoporous structure to the immobilized protein. Hydrogel has been used for protein encapsulation by using high monomer content and a suitable crosslinker to achieve small pore sizes.57, 95 Sol-gels are another popular material that can generate nanoporous structures. Some of the benefits of using sol-gels are the excellent enzymatic activity owing to high encapsulation concentration, mild immobilization conditions,64 and optical transparency for imaging.98 Common sol-gels are silica based, made by polycondensation of alkoxysilane monomers. Sakai-Kato et al. reported a PMMA microfluidic enzyme reactor based on silica sol-gel encapsulation of trypsin.98 The sol-gel was prepared with tetramethoxysilane (TMOS) in water and HCl. TMOS was hydrolyzed to form SiOH4−n(OMe)n. After addition of trypsin, a trypsin-entrapped sol-gel was formed inside the PMMA chip. On-chip digestion was characterized with electrophoresis of digested amino acids (ArgOEt, arginine ethyl ester) and proteins (bradykinin and casein). The immobilized trypsin was active for two days, whereas in-solution trypsin lost activity within a day. The enzyme reactor was stored for one week without loss in activity.
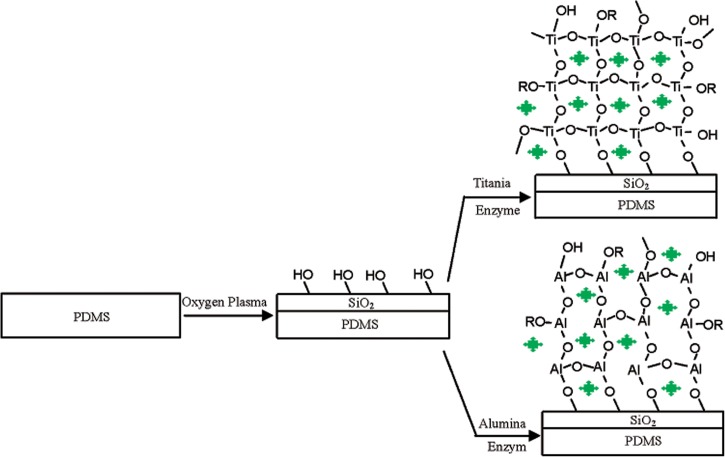
Common silica-based sol-gels are, however, fragile, experience pore shrinkage as well as pore collapse, and sometimes offer poor adhesion to the substrate.64 Consequently, new sol-gel materials have been explored. Wu et al. created titania and alumina sol-gels on a sandwiched PDMS microfluidic device (Figure 6).64 First, PDMS was treated with oxygen-plasma to generate silanol group. Then, titania sol and alumina sol were prepared by heating tetrabutyl titanate and aluminum isopropoxide in solvent. After adding trypsin to the sol, the PDMS microfluidic channel was filled with the sol. The silanol groups on the plasma-treated PDMS surface covalently anchored the hydroxyl group of the sol by condensation, such that a stable sol-gel formed on the PDMS surface with trypsin encapsulated within the sol-gel. BSA was digested, and peptides were analyzed by MS. A faster digestion time and longer enzyme lifetime were observed, as compared to those of a homogeneous (solution phase) reaction. Baohong Liu's group employed a similar strategy for enzyme encapsulation using nanozeolite.99 The Liu group's microfluidic device was fabricated using thermally laminated poly(ethylene terephthalate) (PET) sheets patterned via photoablation. Then, PSS polyelectrolyte was adsorbed to the PET surface yielding a negative surface charge. A layer-by-layer assembly technique59 was subsequently employed to build three layers of electrostatically combined polyelectrolyte PDADMAC (positively charged) and nanozeolite colloid crystal (negatively charged, 80 nm diameter). Finally, trypsin was adsorbed to the assembled nanozeolites in order to digest BSA and protein extract from mouse macrophage. After digestion, peptides were analyzed by MALDI-TOF MS. The zeolite encapsulated trypsin showed more stability and faster digestion compared to free-solution trypsin.
Figure 6.
Process of enzyme-encapsulated sol-gel inside microchannel of PDMS functionalized by oxidation in an oxygen plasma. Reprinted with permission from H. Wu et al., J. Proteome Res. 3, 1201 (2004). Copyright 2004 American Chemical Society.
Bioaffinity immobilization
The bioaffinity interaction or biospecific adsorption (Figure 2b) exploits specific binding phenomena existing in nature. The bioaffinity interaction has advantages over physisorption. A bioaffinity interaction yields relatively stronger, highly specific, and oriented protein immobilization.11, 106, 107 Therefore, protein leakage can be minimized and immobilized protein offers better accessibility to binding partners than random orientation strategies. Additionally, bioaffinity immobilization can be reversed using chemical treatment, pH change, or heat treatment.68, 77 In most of cases, bioaffinity interactions are used in conjunction with other immobilization mechanisms (i.e., physisorption and covalent bonding) with the bioaffinity reagent used as an intermediate binding molecule between the surface and proteins. Avidin-biotin, protein A/G-antibody, genetically engineered protein affinity ligands, DNA hybridization, and aptamers have been employed in microfluidic devices.
Avidin-biotin
One of the most widely employed immobilization partners is avidin (66–69 kDa tetrameric glycoprotein) and biotin (water-soluble vitamin B). Avidin binds to biotin via an exceptionally strong non-covalent interaction.164 The binding interaction is rapid and nearly insensitive to pH, temperature, proteolysis, and denaturing agents.106 Biotin is a small molecule and conjugation to proteins does not significantly affect protein functionality or conformation. One downside of using the avidin-biotin system is the high cost of the proteinaceous binding reagent. NHS (N-hydroxysuccinimide) ester is a popular commercial biotinylation reagent, which allows covalent linking of protein amine groups with biotin. Natural avidin or engineered avidin (e.g., streptavidin, neutravidin, and nitrividin) can be physisorbed or covalently linked to a surface for subsequent immobilization of biotinylated proteins. Sometimes, avidin is attached to surfaces functionalized with biotin, leaving available unoccupied biotin binding sites on the avidin for immobilization of two biotinylated proteins.38
Owing to the popularity of the avidin-biotin immobilization strategy, streptavidin coated PS, agarose, and glass beads are commercially available. These beads are used extensively in microfluidic assays. Typically, proteins are incubated with streptavidin-functionalized microbeads, then pseudo-immobilized in a microfluidic device using size-exclusion structures, such as dams, weirs, and microposts. Seong and Crooks packed enzyme functionalized PS beads on a PDMS microfluidic device with a weir structure.86 Biotinylated GOx and HRP were immobilized on the streptavidin-coated PS beads. The system was used to study the mixing efficiency of enzymatic substrate in a bead-packed microfluidic channel. Wang and Han adapted a nanofluidic pre-concentrator to concentration of the fluorescent proteins GFP (green fluorescent protein) and R-PE (R-phycoerythrin). A bead-based immunoassay was employed to detect the concentrated proteins.87 Biotinylated GFP and R-PE antibodies were attached to commercial streptavidin-coated PS beads. The bead was then pseudo-immobilized by a microfluidic weir. An impressive 50 pM to sub 100 fM detection limit was observed.
Protein A/G—antibody
Protein A and protein G are both popular antibody (IgG) immobilizing reagents, extracted from bacteria. Protein G is known to have a wider immunoreactivity to mammalian IgGs than protein A. Protein A or G specifically binds to the constant Fc region of IgG. Thus, the variable Fab region of IgG is accessible to antigen binding.11 A key consideration is the orientation of protein G or A, so that bound IgGs are away from the immobilization surface and accessible to antigen binding. Like the avidin-biotin system, a spacer that links proteins to the planar surface is frequently used in conjunction with protein A/G.106 An additional benefit of using protein A/G is that antibody can be detached by acid treatment and the surface made reusable.68
Affinity capture ligand
The C- or N-terminus of proteins can be genetically engineered to have an oligohistidine (His) segment that specifically chelates with metal ions (e.g., Ni2+).165 Ni2+ is then bound to another chelating agent such as NTA (nitriloacetic acid), which is typically covalently bound to an immobilization surface. Although the affinity of His tagged-NTA is much weaker than that of the streptavidin-biotin linkage, advantages include: (1) ready reversal of immobilization by adding completing chelating reagents such as EDTA (ethylenediaminetetraacetic acid), and (2) the controlled orientation of immobilized proteins is possible, as the His tag is on the C- or N-terminus of each protein. In a similar sense, GST (glutathione S-transferase) is tagged onto the N-terminus of proteins by genetic engineering. GST-fused target proteins are strongly attached to glutathione (GSH) functionalized surfaces,107 for example, GSH-coated agarose beads. By adding a high concentration of GST, GST-fused proteins can be released. Disadvantages of the GST tagging approach are: (1) cost and time associated with producing recombinant proteins and (2) irrelevance to endogenous proteins.
DNA hybridization
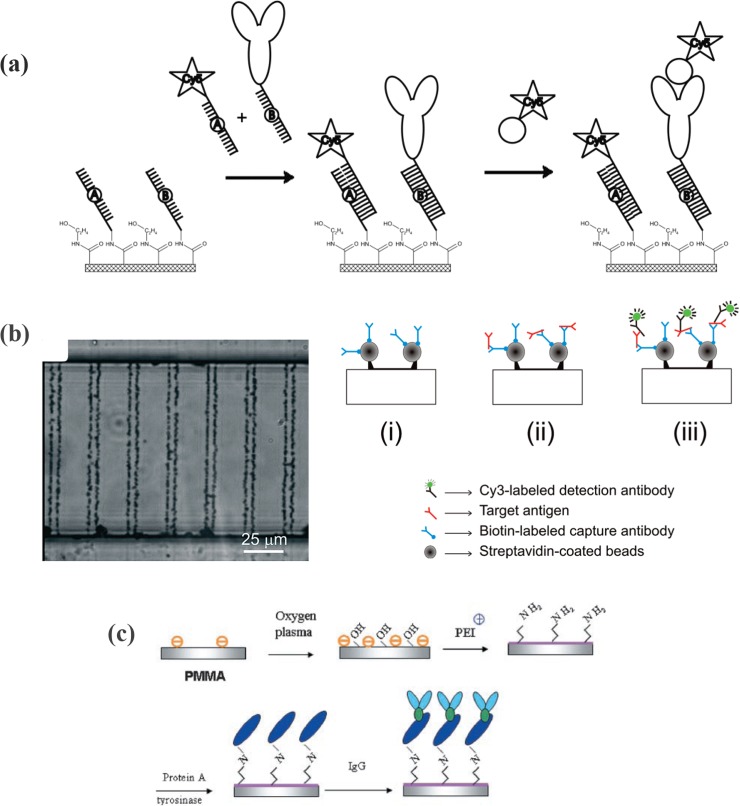
Specific hybridization of single-strand DNA (ssDNA) with the complementary DNA (cDNA) has been employed to immobilize proteins as well (i.e., DNA-directed protein immobilization).25, 26, 77 Oligonucleotide hybridization is exceptionally stable and selective.106 Protein is first coupled to ssDNA and then ssDNA that is linked to the protein is hybridized to the surface where complementary ssDNA are immobilized. An ssDNA tag may be attached to the protein via covalent linkage or a biotin-streptavidin linkage.106 The spatially encoded “DNA-directed” protein immobilization is originated from microarray technology.166, 167 Immobilization of multiple antibodies and multiplexed microfluidic immunoassays can be performed using the DNA-encoded immobilization method.25, 26 Additional advantages are (1) hybridized DNA acts as a spacer arm, and (2) immobilization is reversible by temperature control or alkaline denaturation.77
Antibody
Regardless of the high cost, variable affinity, and short shelf life, an antibody is a ubiquitous biomolecule for immobilizing proteins. The wide-spread use of antibodies arises from the exceptional specificity toward binding partner (i.e., immunoadsorption). Sandwich immunoassays in which immobilized antibody captures a target antigen are a workhorse bioanalytical assay.168 Microbeads coated with antibody having immunoreactivity toward an IgG of a specific animal species are commercially available. Shin et al. created a microfluidic SPE (solid phase extraction) device for improved immunoassay sensitivity.31 In this study, PS beads coated with goat anti-mouse IgG were incubated with mouse anti-CRP IgG. Then the beads were pseudo-immobilized onto a frit structure of a hybrid glass substrate–PDMS device. Immobilized CRP was eluted by an acidic 0.1 M glycine buffer (pH 1.8) treatment and the fluorescence signal was detected by a photodiode integrated in the microfluidic device. Competitive immunoassays were demonstrated.
Aptamers
Aptamers are oligonucleotide bioaffinity capture reagents that have drawn significant attention. Aptamers that show the highest affinity toward a target protein are selected from a synthetic, combinational nucleotide library called SELEX (systematic evolution of ligands by exponential enrichment).169 Aptamers are smaller than antibodies, thus a higher density of capture agents can be coated on surfaces to yield a large binding capacity for target proteins. Compared to antibodies, in particular polyclonal antibodies, aptamers are produced in vitro—eliminating the need for animals in production.170, 171 In addition, aptamers are touted to have a longer shelf life and less sensitivity to environmental change. For a detailed review of aptamers in microfluidics, readers are directed to the paper by Xu et al.170 and Mosing and Bowser.171 For aptamers for protein immobilization, readers are directed to the review by Nakanishi et al.107
Yang et al. used aptamer-functionalized magnetic beads in a multilayer PDMS microfluidic device to complete a CRP immunoassay.30 A CRP-specific aptamer sequence was screened using SELEX, followed by biotin conjugation. Aptamers were attached to commercial streptavidin-coated magnetic beads. After high-sensitivity, CRP (hs-CRP) was captured by aptamers, an acridinium-ester conjugated CRP antibody was introduced for chemiluminescence detection. Tennico et al. used a similar bead system (streptavidin-coated magnetic beads and biotinylated aptamer) to detect thrombin in a sandwich immunoassay format.90 They showed that aptamers have negligible affinity toward prothrombin and HSA, which are proteins similar to thrombin. The simple microfluidic device consisted of PMMA (or PC) top and bottom layers, which sandwiched an intermediate double-adhesive tape layer housing the microfluidic channel. Two aptamers having affinity toward different epitopes of thrombin were employed as detection and capture probes in the sandwich immunoassay, whereas antibody is commonly used as the detection probe in aptamer-based immunoassays.30
Covalent bond
Covalent bonds are a frequently used immobilization mechanism in microfluidic assays (Figure 2c).11 The immobilization surface is activated via reactive reagents. The activated surface reacts with amino acid residues on the protein exterior and forms an irreversible linkage. One tends to rely on covalent immobilization if high, stable protein coverage is required. Bifunctional spacer molecules are a common approach to forming an irreversible bond between proteins and the immobilization surface. In such an approach, one end of a spacer molecule is covalently linked to an activated surface, and then a protein is covalently linked to the other end of the spacer. Alternatively, another spacer or protein capture agent (e.g., streptavidin) is crosslinked on the other end. Unreacted active functional groups are blocked or deactivated (e.g., BSA,106 hydroxylamine,83 ethanolamine,172 or lysin40). Disadvantages of covalent linkage include reduced activity of proteins (by forming linkage on active sites),106 toxic reagents,125 and complicated chemistry.125 A covalent bond can be formed on active sites of proteins, resulting in reduced activity.11 The covalent attachment reaction is that the reaction is usually slow, so that the protein and surface require long incubation times of hours up to a full day (e.g., epoxide61). Therefore, covalent linking is usually performed as a preparatory step before performing microfluidic assays. An enormous variety of covalent conjugation chemistries are available. In this review, only a small subset of conjugation chemistries employed especially in microfluidic devices is introduced. For more information beyond the present scope, refer to a review by Feijen et al.106 and the excellent reference by Hermansson.125
Amine—glutaraldehyde—amine
Amino groups (-NH2) of lysine are the most common covalent binding sites because lysine residues are usually present on the exterior of proteins. Aldehyde is a reactive compound that forms the labile Schiff base with the amine and can be further reduced to form a stable secondary amine bond using NaCNBH3 or NaBH4. Glutaraldehyde (GA) is a bis-aldehyde compound that has two reactive ends. GA can crosslink two amine functional groups, for example, between two proteins or between a protein and a surface-immobilized polymer with amine groups (e.g., PEI). The Schiff bases formed on proteins are stable without further reduction by NaCNBH3 or NaBH4;125 and indeed, a large number of protein immobilization strategies that rely on GA in microfluidic devices are completed without further reduction. When used with a glass or a silicon surface, aminosilane compounds like APTES [(3-aminopropyl)triethoxysilane] are used because they can bind to hydroxyl functional groups of glass at one end and possess an amine functional group to facilitate covalent linkage with proteins on the other end.117 GA had been extensively used to form stable protein microarrays on glass substrates.173 Regnier et al. used APTES to coat wet-etched glass microfluidic channels with aminosilane.174 Then, GA was electroosmotically injected for 2 h and the enzyme β-gal was covalently immobilized on the glass channel walls. Porous-silicon micropits were treated with APTES and GA for trypsin immobilization by Marko-Varga's group.60 The Schiff base was further reduced by NaCNBH3 for stable bonding. Digested proteins were analyzed by MALDI-TOF/MS. Wang et al. used a similar approach in immobilizing proteins and antibodies in a device consisting of a silicon substrate and a PDMS microfluidic array.111 Ellipsometry was used for label-free microarray immunoassays. Richter et al.91 used the same chemistry to immobilize enzymes xanthine oxidase (XOD) and HRP on porous silica beads. After trapping the silica beads behind a weir structure, they successfully detected hydrogen peroxide in a glass microfluidic device.
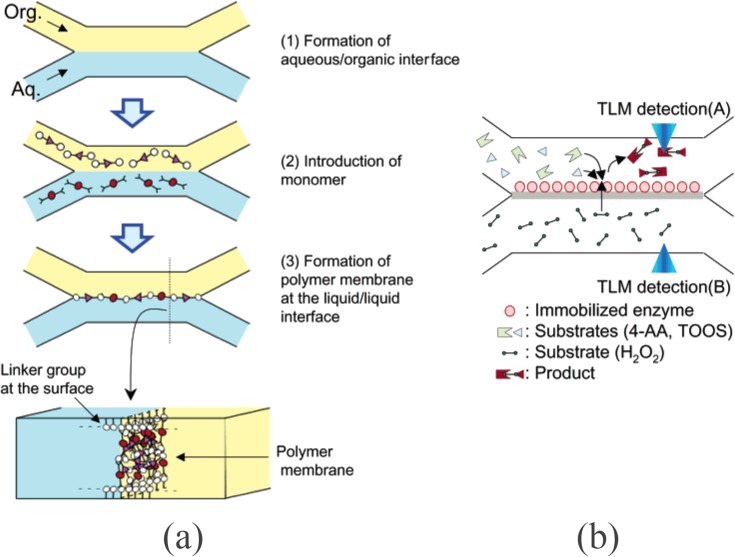
As described earlier, proteins are usually physisorbed to polymer membranes (e.g., NC, PS, and PVDF) clamped in a microfluidic device. Kitamori's group immobilized enzymes onto an in situ fabricated nylon membrane using covalent bonding (Figure 7).100 First, a glass microfluidic channel was treated with APTES and then a vertical nylon membrane was fabricated by interfacial polycondensation. Co-injecting immiscible laminar flows of adipoyl chloride in 1,2-dichloroethane and hexamethylenediamine (HMD) in 0.1 M NaOH created 10-μm thick nylon membrane. Amino groups on the membrane were activated by GA, followed by immobilization of HRP and further reduction of the Schiff base using NaBH4. The activity of HRP was tested by introducing substrate on one side and hydrogen peroxide on the other side of the membrane. Hydrogen peroxide diffused through the membrane and HRP generated a colored product.
Figure 7.
In situ polymerized nylon membrane for an enzyme assay. (a) In situ polymerization of nylon membrane using organic/aqueous two-phase flow in an X-shaped microfluidic chip, and (b) TLM (thermal-lens microscopy) was used to detect product and substrate. Reprinted with permission from H. Hisamoto et al., Anal. Chem. 75, 350 (2003). Copyright 2003 American Chemical Society.
Yu et al. used an intermediate PVA [poly(vinyl alcohol)] layer to minimize nonspecific protein adsorption to PDMS.175 The researchers silanized an oxygen-plasma-treated PDMS surface with APTES. In order to form the hydrophilic layer, the amine group on APTES was activated with GA to attach PVA (via hydroxyl group of PVA176) to PDMS. Then, GA was used once more to covalently link proteins such as IgM, BSA, and IgG to the PVA layer. A sandwich immunoassay was demonstrated. Compared to the native PDMS surface, the SNR (signal-to-noise ratio) was improved owing to low nonspecific adsorption and high antibody binding capacity. Thomsen et al. took an alternative approach to functionalizing a PDMS surface lacking functional groups. These authors applied pyrogenic silicic acid (i.e., silicon oxide powder prepared in oxyhydrogen flame) to PDMS, so that the PDMS surface exhibited hydroxyl groups.52 Using the added hydroxyl group and APTES, then activating with GA, the enzyme CelB (β-glycosidase) was immobilized to hydrolyze lactose to glucose and galactose.
Another aminosilane coupling reagent, 3-aminopropyl)trimethoxysilane (APTMS),120 is also popular because of better reactivity than APTES. Glass et al.117 provide a review of various organosilanes and deposition techniques of the organosilanes. A spacer of APTES or APTMS may be too short to properly immobilize large proteins (such as antibodies and enzymes) to planar channel surfaces in high capacity owing to steric hindrance. The scheme may also cause reduced activity owing to partial physisorption to channel surface.40 Therefore, a hydrophilic polymer matrix such as dextran (DEX),29, 68 chitosan,130 and PEG40, 120 are often used as longer spacers for improved assay sensitivity.
Amine—in situ generated aldehyde
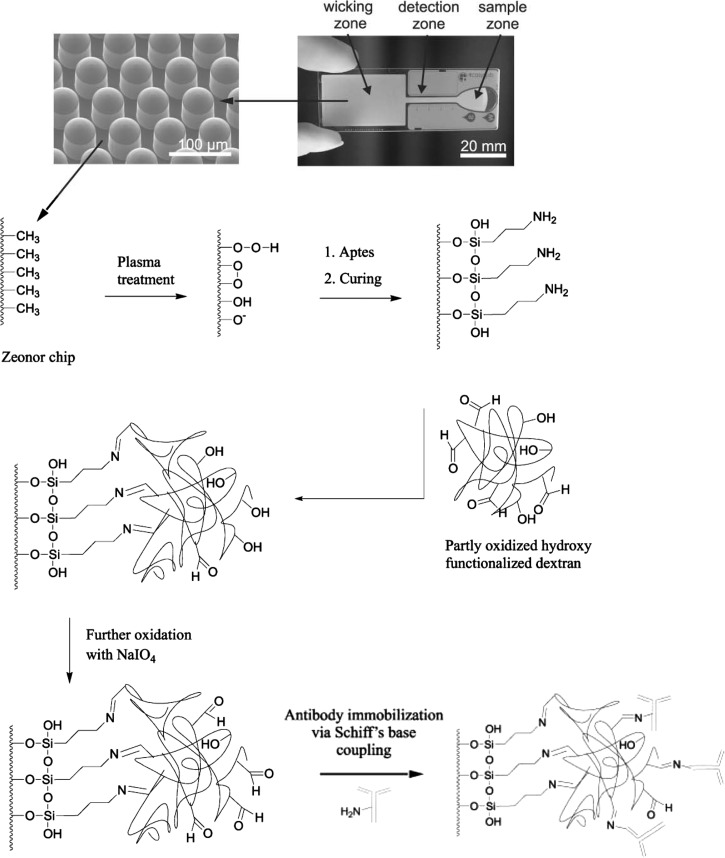
Instead of adding GA, nucleophilic aldehyde groups can be generated in situ. While paper microfluidics is generally considered a low cost approach for diagnostic devices without external fluid handling systems (e.g., lateral-flow immunoassay), Jönsson et al. fabricated such a zero-powered device using a COC substrate (Figure 8).29 Their COC microfluidic chip has a capillary-action wicking zone consisting of a micropillar array. After oxygen-plasma treatment, the chip surface was amine-functionalized with APTES. They attached dextran (40 kDa), partly oxidized to have aldehyde groups, and the aldehyde groups formed the Schiff bases with the amine groups of the APTES. After dextran immobilization, dextran was further oxidized by NaIO4 adding more aldehyde groups for capture-antibody immobilization. A sandwich immunoassay of CRP was demonstrated with an improved detection limit (two orders of magnitude), as compared to a paper-based immunoassay. The authors reasoned that dextran improved assay performance by providing a hydrophilic surface for better capillary action, a hydrogel-like protein-friendly environment, and an enlarged surface area for a high antibody binding capacity. Baeza et al. took an in situ aldehyde generation approach in order to immobilize enzyme in LTCC (low temperature cofired ceramics) microfluidic device.54 A commercial agarose bead was activated to have a glyoxal functional group (agarose-O-CH2-CHO) by etherification with 2,3-epoxypropanol and oxidation with NaIO4.128 The aldehyde functional end of glyoxal agarose was used to immobilize β-gal after the Schiff-base reduction with NaBH4. The beads were pseudo-immobilized using microcolumns in a reaction chamber. The enzyme activity was measured colorimetrically with enzyme stability observed for 6 months.
Figure 8.
Zero-powered COC immunoassay chip with patterned micropillars for covalent antibody attachment. The COC surface is oxidized in oxygen plasma and silanized with APTES. The resultant amino terminated surface is subsequently functionalized with a dextran matrix. Finally, antibody is immobilized via Schiff's base coupling to the dextran matrix. Adapted from C. Jönsson et al., Lab Chip 8, 1191 (2008) with permission from The Royal Society of Chemistry.
Amine—NHS (N-hydroxysuccinimide)
Probably, the most commonly used covalent linking agent is NHS ester.106 NHS ester reacts with amines on proteins and yield stable amide bonds while releasing NHS leaving groups.125 Sulfo-NHS (N-hydroxysulfosuccinimide) is preferred over NHS because sulfo-NHS is more water soluble while showing the same reactivity and specificity as NHS. Delamarche et al. provided an early report of protein immobilization in microfluidic device156 by introducing a PDMS microfluidic network (μFN) comprised of PDMS irreversibly bound to glass or silicon. The surface of the glass or silicon wafer was coated with aminosilane. Amine functional groups of the aminosilane were activated by the NHS ester for antibody immobilization. Palecek's group used PEG hydrogel micropillars copolymerized with an acryloyl-functionalized NHS ester for microfluidic kinase activity assays and immunoassays.82, 83 The microfluidic-chip fabrication process is rather interesting in that the authors fabricated a microfluidic channel using in situ polymerization of the monomer isobornyl acrylate (IBA) in an empty chamber made of a bottom glass slide, a polycarbonate top, and an intermediate adhesive rim, whereas typical microfluidic chip fabrication is done by etching bulk material.133 After the glass slide was silanized with (3-acryloxypropyl)-trimethoxysilane to provide an acryloyl group, macroporous PEG pillars were photopolymerized using PEGDA monomer. PEG porogens of various molecular masses were used to form macropores. A macroporous structure is favorable over a nanoporous gel for incorporating large proteins such as antibodies and kinase. The 6-((acryloyl)amino)hexanoic acid NHS ester was copolymerized in the hydrogel pillars for covalent immobilization of proteins such as GST-fused GFP and GST-fused CrkL (Crk-like) protein. GST antibody and kinase activity assays (phosphorylation) of K562 cell lysate were demonstrated.83 Characterization of diffusion of 250 kDa dextran and GST-GFP showed improved mass transport through the gel as compared to nanoporous PA or PEGDA hydrogels.82
Carboxylate—1-ethyl-3-(3-dimethylamonipropyl) carbodiimide (EDC)—amine
Carbodiimide is used to form amide linkage between carboxylates and amines. EDC is the most frequently used carbodiimide.125 Advantages of EDC include water solubility; EDC can be directly used with proteins in aqueous buffer unlike NHS, which is dissolved in organic solvent (e.g., DMSO). EDC reacts with carboxylic acid to form an o-acrylisourea intermediate, which subsequently reacts with primary amines to form amide bonds.119 Reaction of o-acrylisourea with amines is slow and can be hydrolyzed in aqueous solution. Thus, EDC is usually used with NHS.
Carboxylate—EDC + NHS—amine
NHS esters can be formed on-demand using carbodiimide (e.g., EDC), NHS, and carboxylates to immobilize proteins.177 Didar et al. created multiplexed protein microarrays in a microfluidic device consisting of a glass substrate and PDMS microchannels after microcontact printing of APTES spots.177 NHS and EDC were used to covalently link proteins to the amine group of APTES. Carboxylate groups of antibodies were linked to the end of APTES after releasing the NHS leaving group. Antibodies for CD34, CD31, and CD36 proteins were immobilized on the patterned APTES spots and multiplexed sandwich immunoassays were demonstrated. Hu et al. used EDC and NHS chemistries to immobilize detection antibody to aqueous quantum dots (aqQD) for microfluidic sandwich immunoassays.178 Usually, high quantum-yield QDs are synthesized in an organic phase, which requires post treatment for water solubility. In contrast, the CdTe core/CdS shell QD was prepared in an aqueous phase.179 The QD was covered with 3-mercaptopropionic acid (MPA) in order to yield a carboxylic-acid functionalized surface. The carboxylic group was later activated with the NHS ester, and then covalently linked to the amine groups of antibodies. Capture antibody was covalently attached to commercial silanized glass slides, against which the PDMS microchannels were sealed. Sandwich immunoassays of the cancer biomarkers carcinoma embryonic antigen (CEA) and α-fetoprotein (AFP) were demonstrated using this approach.
Amine/sulfhydryl—epoxide
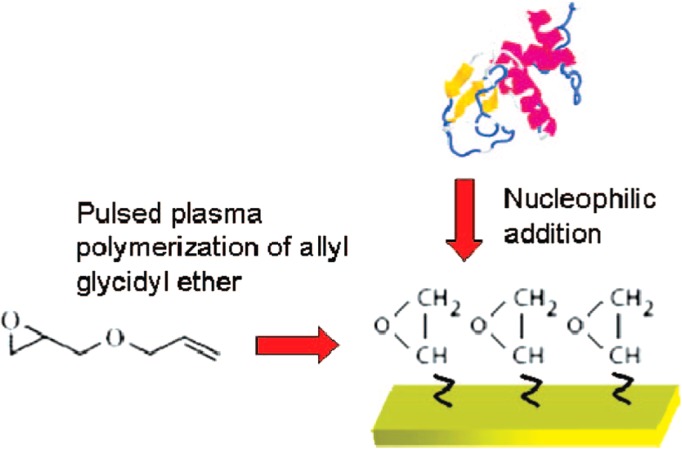
Epoxides form covalent bonds with primary amines at mild alkaline pH or with sulfhydryl groups (-SH) in the physiological pH range.125 The advantages of using epoxides include a simple protocol, a neutral pH range, and relevance to aqueous conditions. Due to slow kinetics, protocols suggest using high concentrations of epoxide and additional functional groups to promote the adsorption of proteins.106 A common epoxide functional group used in protein immobilization is glycidyl.96, 172, 180, 181 Thierry et al. reported epoxide-based capture of tyrosine-kinase human epidermal growth factor receptor (HER2) on PDMS microfluidic devices.181 They coated a PDMS microchannel and the surface of a glass substrate using a pulsed plasma polymerization of allyl glycidyl ether (AEG) monomer (Figure 9).172 The authors reasoned that a common epoxidation reagent GOPTS (3-glycidoxypropyltrimethoxysilane) was unstable owing to hydrolysis, but pulsed plasma yielded a conformal, adherent, defect-free glycidyl coatings on the surface.172 Herceptin, a commercial HER2 antibody, was attached to the coating via the epoxide group of the AEG polymer allowing capture of CTCs (circulating tumor cells). The authors also tested a multilayer coating consisting of AEG polymer and PEGDA deposited via epoxide crosslinking. The PEGDA layer was activated with NHS and EDC to immobilize Herceptin. In comparison to the AEG-only coating, reduced nonspecific protein adsorption was observed but lower Herceptin density was obtained.
Figure 9.
Pulsed plasma epoxidation of PDMS surfaces and bioconjugation of proteins. Reprinted with permission from B. Thierry et al., Langmuir 24, 10187 (2008). Copyright 2008 American Chemical Society.
Amine—isothiocyanate
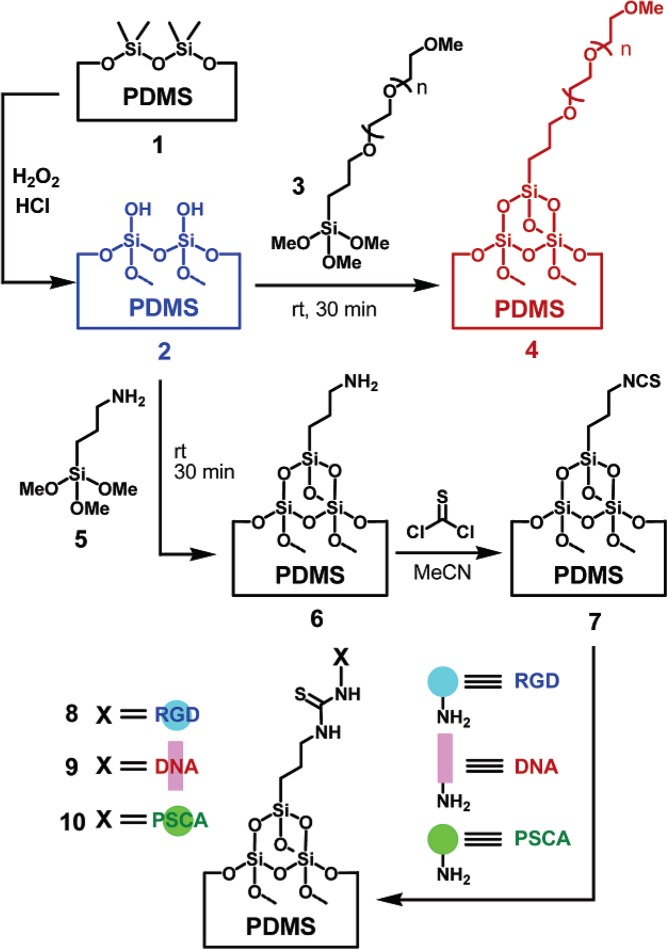
The reaction of an aromatic amine with thiophosgene (CSCl2) yields isothiocyanate (-NCS), which forms a stable bond with primary amine groups.125 Sui et al. used the isothiocyanate chemistry to immobilize proteins, peptides, and DNA on the PDMS surface of a glass-PDMS microfluidic device (Figure 10).120 The approach used solution-based oxidation of PDMS, not common plasma-based oxidation, owing to better stability of the oxidized surface. The PDMS channel was oxidized with H2O2 and HCl, then silanized with 2-[methoxy(polyethyleneoxy)propyl] trimethoxysilane to graft PEG onto the PDMS, followed by APTES treatment for amination. Subsequently, thiophosgene was introduced to convert amine groups to reactive isothiocynate groups. Later, in situ generated isothiocynate reacted with amine-terminated PSCA (prostate stem cell antigen) protein, tripeptide RGD (arginine-glycine-aspartic acid), and aminated DNA for immobilization. Sandwich immunoassays of PSCA, DNA hybridization, and RGD-mediated cell adhesion were demonstrated. The authors found that PEG was effective in repelling proteins from a surface for more than 2 months.
Figure 10.
PEG-grafted PDMS surface for biomolecule immobilization. (4) Preparation of the PEG-grafted PDMS microchannels and (6) amine-grafted PDMS microchannels. The amine-grafted microchannels can be activated by thiophosgene to obtain (7) the isothiocyanate-grafted PDMS microchannels as a precursor for (8) the RGD-grafted, (9) DNA-grafted, and (10) PSCA-grafted PDMS microchannels. Reprinted with permission from G. Sui et al., Anal. Chem. 78, 5543 (2006). Copyright 2006 American Chemical Society.
Amine—azlactone
Azlactone is formed by the cyclization of N-acyl-α-amino acids. Azlactone readily reacts with nucleophiles such as amines and thiols at room temperature to form amide bonds.182 Peterson et al. reported a microfluidic protein digestion device using a photopatterned polymer monolith where trypsin was immobilized using azlactone.61 Glass chips were silanized with 3-(trimethosysilyl)propyl methacrylate, leaving the methacryloyl group to covalently link polymer monolith to the glass substrate.183 Azlactone functionalized monomer (2-vinyl-4,4-dimethylazlactone + ethylene dimethacrylate + acrylamide or + 2-hydroxyethyl methacrylate), porogenic solvent (dodecanol, decanol, or its mixtures with cyclohexanol), and photoinitiator (2,2-dimethoxy-2-phenylacetophenone) were used for UV polymerization of the monolith. After trypsin was covalently immobilized on the azlactone-functionalized surface, unreacted azlactone was blocked by ethanolamine. Casein was digested and analyzed by MALDI-TOF/MS. The reaction time was ∼10 s, whereas in-solution digestion required ∼10 min.
Amine—p-nitrophenyl ester
p-nitrophenyl ester is reactive to amines across the slightly basic pH range spanning 7–9. The ester forms stable amide bond with proteins.125, 184 The p-nitrophenyl ester shows a similar reactivity to the NHS ester and is frequently used as one end of heterobifunctional linker (e.g., p-nitrophenyl iodoacetate and biotin-4-nitrophenyl ester).
Amine—tyrosinase (TR)—tyrosine
An biocatalyzed protein immobilization strategy employed enzyme TR. Tyrosinase is a phenol oxidase that oxidizes phenols into O-quinone (i.e., 1,2-benzoquinone), which is reactive and undergoes reaction with various nucleophiles such as primary amines. In nature, this reaction is found in the hardening of insect shells or the crosslinking of an adhesive protein-gel network.185 In an engineered system, a pro-tag (tyrosine) conjugated enzyme was immobilized on chitosan hydrogel using tyrosinase.186
Sulfhydryl—maleimide
Maleimide is a popular reagent for the crosslinking of sulfhydryl groups. Thus, maleimide is used to form covalent links with the cysteine residues of proteins. Maleimide reactions with thiols are specific at pH 6.5–7.5. However, maleimide also reacts with amines at higher pH values.106 Alkylation reaction of the maleimide double bond with sulfhydryl forms stable thioether bonds.125 Maleimide is often used as one end of a heterobifunctional cross linker such as sulfo-SMPB (sulfosuccinimidyl 4-[p-maleimidophenyl]butyrate).187 Hydrolysis in aqueous solution may present issues.
Reactive hydrogen—benzophenone
Benzophenone residues become highly-reactive triplet-state intermediates when exposed to UV light. During UV exposure, the benzophenone couples with a protein via reactive hydrogen compounds on the protein. If benzophenone residues are incorporated onto a surface, the protein can be immobilized to the surface via exposure to UV light. Such photopatterning allows highly spatially controlled immobilization.21, 22 An advantage of this strategy is that activated benzophenone does not degrade readily even if exposed to UV light and is not involved in bond formation. As such, after UV exposure, unreacted benzophenone can be photolyzed again to form a covalent bond.125 Benzophenone can be also used for free-radical polymerization of hydrogels97 and surface-grafted polymers.119, 188
Combination of two or more immobilization mechanisms
A large body of literature reports on combinations of physisorption, bioaffinity interactions, and covalent bonds for immobilizing proteins. As described earlier, each immobilization mechanism has drawbacks and advantages with relative importance depending on fabrication constraints and application requirements. Thus, attempts have been made to address the limitations of individual mechanisms, aiming for better protein activity, more control for protein orientation, less steric hindrance, more protein patterning density, and less nonspecific protein adsorption using a combined method, as compared to only a single immobilization mechanism.
Physisorption—covalent bond
Yakovleva et al. studied five different polymer coatings on porous silicon surfaces40 (Figure 11). In the first system, porous silicon was treated with APTES, then with GA for antibody immobilization. Unreacted GA was blocked by L-lysine. In the second system, BPEI (branched PEI) was directly adsorbed on the porous silicon surface by electrostatic interaction between negatively charged silicon and positively charged PEI. Amine groups on BPEI were activated with GA and antibody was immobilized. In the third system, porous silicon was epoxide-functionalized with GOPTS. The amine groups of BPEI were first linked to GOPTS, then the same procedure used in the second system studied was employed to immobilize antibody. In the fourth system, positively charged LPEI (linear PEI) was physisorbed onto a silicon surface by electrostatic interaction. Then, the same procedure used in the second system studied was applied to immobilize antibody. Finally, in the fifth system, LPEI was physisorbed on a silicon surface, then antibody also physisorbed to the LPEI surface. The authors observed that the fourth system showed the best sensitivity and antibody reliability, better than direct immobilization. The authors claimed that, owing to the long chain of LPEI and BPEI, antibody was situated away from the silicon surface, and covered the silicon surface effectively. The PEI layers also prevented nonspecific protein adsorption and provided more antibody binding sites to antigens than the bare porous silicon surface.
Figure 11.
Structures of the five tested antibody functionalized porous silicon surfaces. Reprinted with permission from J. Yakovleva et al., Anal. Chem. 74, 2994 (2002). Copyright 2002 American Chemical Society.
As a native surface of PS, COC, and PMMA lacks a ligand for covalent linkage, the surfaces were modified with various methods. Besides common oxygen-plasma treatment, one approach to surface coating has been used as an intermediate polymer layer that has an attractive interaction with the surface for further protein immobilization. Another approach has been to forcibly introduce a ligand to the plastic. Bai et al. performed a study of protein adsorption on PMMA microfluidic chips.78 The PMMA chips were first treated with 1 M NaOH for hydrophilization, and then coated by physisorption of BPEI, PAH, and HMD. The amine functional group on the coatings was then activated by GA for antibody covalent immobilization. After blocking by BSA, sandwich immunoassays were performed. Among bare PMMA and BPEI-, PAH-, and HMD-treated PMMA surfaces, the signal was highest for the BPEI-treated PMMA. The authors attributed the result to the spacer effect for keeping the antibody away from the hydrophobic surface thereby minimizing steric hindrance to immunobinding.
Covalent immobilization—bioaffinity immobilization
Eteshola and Leckband compared (a) passive adsorption of a capture antibody (rabbit anti-sheep IgM) to a PDMS microchannel with (b) site-specific immobilization of the capture antibody using protein A.189 For their site-specific binding, BSA was first physisorbed to the microchannel. The adsorbed BSA was activated using GA, then protein A was covalently attached to BSA. Finally, capture antibody was immobilized on the protein A. Using sandwich immunoassays, the IgM concentration was characterized. The authors reported that the protein A and BSA coated surfaces were better than the bare PDMS surface in terms of higher antigen capture and lower nonspecific binding. Instead of using GA reagent, Yang et al. used in situ generation of aldehyde groups in macroporous agarose beads in order to covalently attach protein A.134 Macropores (average pore size of 28 μm) in the bead can improve hydrodynamic mass transport through packed agarose beads, so a high flow rate can be used in chromatography without a large backpressure. Agarose beads were hydrolyzed in 0.2 M HCl at 55 °C to form aldehyde groups.190 Protein A was immobilized to the agarose bead and the Schiff base was further reduced by NaCNBH3 to form stable bonds. Anti-goat IgG was immobilized to the beads, which later were pseudo-immobilized in a PDMS microfluidic device for sandwich immunoassays. The authors observed higher binding capacity with this approach than commercial protein-A conjugated nanoporous agarose beads.
Physisorption—bioaffinity immobilization
Schult et al. exploited the hydrophobicity of PMMA to physisorb neutravidin, then immobilized a biotinylated monoclonal capture antibody.37 hCG (pregnancy hormone chorionic) was detected by sandwich immunoassay. The approach did not use any blocking, but showed insignificant nonspecific protein adsorption. Linder et al. used three layers of biopolymer (biotinylated IgG, neutravidin, and biotin-conjugated dextran, in order of coating) to prevent nonspecific binding of protein on the PDMS surface and to introduce surface charge for cathodic electro-osmotic flow.191 For immunoassays, biotin-conjugated antibody was attached to the neutravidin layer instead of the dextran. The authors observed that nonspecific binding was significantly reduced in the composite coating. However, using costly IgGs in surface coating may not be practical. Wen et al. employed a complex multilayer approach to immobilize antibody to PMMA surfaces (Figure 12).80 First, they prepared two positively charged linkers PLL-g-PEG-biotin and PLL-g-PEG using graft copolymerization. Then, the PMMA surface was treated with oxygen plasma to become hydrophilic, and then acrylic acid was UV graft polymerized on the PMMA surface. This process resulted in a polyacrylic-acid coated PMMA surface that was negatively charged. The mixture of PLL-g-PEG-biotin and PLL-g-PEG was electrostatically attached to the surface. Neutravidin and biotin-conjugated protein A were attached to the surface successively. Finally, anti-IFN-γ was immobilized to the protein A for an IFN-γ immunoassay. The mixture of PLL-g-PEG-biotin and PLL-g-PEG has multiple roles: (1) to prevent protein A from crowding on the surface and to reduce steric hindrance of antibody bound to the protein A, (2) to prevent neutravidin from denaturing, and (3) to reduce nonspecific binding of protein on the surface. ELISA signal was greatly improved in this approach, compared to the case of simple physisorption to the PMMA surface.
Figure 12.
Schematic illustration of the stepwise process involved in the biotin-PLL-g-PEG and protein A-based antibody immobilization. Reprinted with permission from X. Wen et al., J. Immunol. Methods 350, 97 (2009). Copyright 2009 Elsevier.
A majority of immunoassays in microfluidic format employs fluorescent detection. However, highly-sensitive label-free methods such as SPR are employed as well. Lagally's group created an impressive SPRi (SPR imaging) device with 264-element multiplexing capability. Binding affinity of antibody to antigen (human α thrombin) was characterized using this device with serial dilution.49 The gold microarray was patterned on the glass slide, and the PDMS microfluidic network was irreversibly bound to the slide. Streptavidin was physisorbed to the gold microarray and biotinylated α thrombin was immobilized to the streptavidin. Then, thrombin antibody was injected to each gold patch for binding affinity characterization. However, they did not characterize how stable the physisorbed streptavidin layer was.
Mäntymaa et al. reported on laser-welded PS microfluidic chips,192 which employed a streptavidin-based protein immobilization approach adopted from Ylikotila et al.193 The authors polymerized streptavidin into a large conjugate to offer enhanced binding of biotinylated antibody and stable surface attachment of streptavidin to the PS surface. The primary amine of streptavidin was converted into a sulfhydryl group by reacting with s-acetylthioacetic acid NHS ester. The sulfhydryl group was then deacetylated using hydroxylamine and EDTA, turning the sulfhydryl group into a thiol group (-SH). Thiols were oxidized to form disulfide bonds, thus polymerizing streptavidin into large conjugates (>80 kDa). This streptavidin-conjugate coating was stable on the PS surface. A thyroid stimulation hormone (TSH) immunoassay was demonstrated.
A supported lipid bilayer (SLB) is a method for coating channel surfaces.38, 39, 55 Key benefits of using SLB are (1) biology-like (i.e., protein-friendly) environment, (2) reduced nonspecific protein adsorption, (3) ability to accommodate membrane proteins, and (4) lateral fluidity allowing 2-D rearrangement of biomolecules in the membrane, which may result in increased binding strength of receptor-ligand via multivalent interaction.38 Cremer's group explored the use of SLB in microfluidic systems.39, 55 In their first work, unilamellar vesicles were formed using egg yolk phosphatidylcholine and phosphatidylethanolamine (PE) conjugated with environmental contaminant DNP (dinitrophenyl).39 After the PDMS microfluidic channel was hydrophilized by oxygen plasma treatment, the vesicles were injected to form SLB on the channel surface via physisorption. Anti-DNP antibody was injected to conduct an immunoassay. The authors reported that nonspecific adsorption of protein was reduced by two orders of magnitude. In follow-up studies, the kinetics of enzymes immobilized to a biotinylated SLB were studied.55 The biotinylated SLB allowed immobilization of streptavidin-conjugated enzymes such as ALP to a PDMS surface and prevented the enzymes from denaturing. Biotinylated unilamellar vesicles were formed with 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) and biotinylated PE. Streptavidin-conjugated ALP was injected and immobilized, and here enzyme kinetics was studied. The authors reported that the enzymes were stable for two days. A major disadvantage of the lipid bilayer is air instability—the layer peels off when dehydrated. Inspired by Cremer's work,55 Cheng's group used a so-called reinforced supported bilayer membrane (r-SBM) for Staphylococcus enterotoxin B (SEB) immunoassays.38 The main constituent of r-SBM is egg yolk phosphatidylcholine and biotin-conjugated PE for attachment of streptavidin (Figure 13). Although not fully understood, the immobilized streptavidin layer improved air stability of the r-SBM. Also, the r-SBM showed excellent lateral fluidity and low nonspecific protein adsorption (albeit not compared with a bare PDMS surface). The same research group studied the impact of cholesterol in a SLB, made of 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) and DNP-conjugated dipalmitoylphosphatidylcholine (DP-PE), on binding of DNP antibody. Cholesterol is an important component of natural lipid bilayers such as those in cells.41 The authors found that the antibody binding improved up to 20 mol. % of cholesterol.
Figure 13.
Fabrication of an r-SBM for SEB immunoassay in a PDMS microfluidic chip. (a) Streptavidin reinforced SBM, (b) surface functionalization with biotinylated anti-SEB IgG, (c) capture of SEB, and (d) SEB-antibody binding followed by incubation with fluorescently labeled secondary antibody to generate a signal. A top view of the microchannel is also shown. Reprinted with permission from Y. Dong et al., Lab Chip 6, 675 (2006). Copyright 2006 The Royal Society of Chemistry.
Covalent immobilization—physisorption—bioaffinity immobilization
Glass is a common substrate material for microfluidic devices.9, 10 Glass is a natively hydrophilic material but can be made hydrophobic by covalently attaching silanizing reagents with hydrophobic tails. Dodge et al. used CDMODS (chlorodimethyloctadecylsilane) to form a hydrophobic monolayer on wet-etched microchannels in Pyrex7740 glass.194 Proteins tend to physisorb better on hydrophobic methylated surfaces than on hydrophilic glass surfaces.175 Protein A was first physisorbed to the modified surface, followed by attachment of fluorescently labeled rabbit IgG. Using this intermediate layer (i.e., spacer), the authors conjectured that the antibody was offset from the glass surface. The use of a physical spacer would result in an antibody that is less conformationally perturbed (i.e., denatured) and oriented, so that the antigen can be captured effectively, as compared to antibodies directly adsorbed to the glass surface. Laib and MacCraith used a combinatory approach to immobilize antibodies on inert COC plastic surfaces.77 First, the authors treated a COC surface with a hot ammonia and hydrogen peroxide solution, followed by oxygen plasma treatment to create a hydrophilic surface. The plasma-treated surface was observed to return to a hydrophobic state after 6 days. Next, the authors studied coating the surface with alternating layers of PEI (positive) and PAA (negative) via electrostatic interaction, which yielded a stable hydrophilic surface. Aminated ssDNA was covalently immobilized on carboxylic acid on PAA via EDC and sulfo-NHS chemistry. Rabbit IgG conjugated with a complementary ssDNA was hybridized to the ssDNA on the COC surface and an immunoassay was demonstrated (Figure 14a). The authors found that the polyelectrolyte-coated COC surface was stable for one month. DNA-DNA binding was stable for a week and reversible with denaturation. The polyelectrolyte layers showed a low nonspecific adsorption of DNA, but showed significant adsorption of protein, requiring BSA and ethanolamine blocking. Wang et al. reported an electrochemical thrombin sensor based on aptamers immobilized on PMMA microfluidic chip.35 The PMMA chip was hydrophilized in a hot NaOH solution. Then, three alternating layers of PDADMAC (positive) and gold nanoparticles (negative) were formed by electrostatic interaction. This composite layer improved surface-area-to-volume ratio for aptamer attachment. Then, thiolized aptamers were attached to the gold nanoparticles for thrombin capture. Thrombin was injected, and subsequently an ALP-functionalized detection aptamer was introduced. Finally, the enzyme ALP converted the substrate 4-aminophenyl phosphate to the product p-aminophenol, which was amperometrically detected. The authors reported a 1-pM detection limit. Sivagnanam et al. patterned antibody functionalized microbeads on a glass surface92 using dot and line patterns of APTES. The APTES pattern was created using a lift-off process on the glass substrate of a hybrid glass-PDMS microfluidic device (Figure 14b). Owing to the use of a lift-off process, the APTES patterns have ridges. Amine groups on APTES are positively charged while streptavidin is negatively charged at the pH of the solution (7.4). The authors observed an exceptionally strong electrostatic interaction between the APTES pattern and the beads, so strong, in fact, that the beads withstood the shear force of hydrodynamic flow and magnetic force, and did not lift off of the surface under the conditions studied. With this assay, mouse IgG was detected down to 15 ng/ml using a sandwich immunoassay. Lee's group reported antibody immobilization on a PMMA surface using TR enzyme (Figure 14c). The PMMA surface of a simple straight channel79 or a CD (compact disc) microfluidic device124 was first treated with oxygen plasma to introduce negative charge, then positive PEI was adsorbed by electrostatic interaction. The PEI provided a plethora of amine functional groups, which were later covalently linked to tyrosine residues of protein A by a TR-catalyzed conversion into reactive O-quinones. Capture antibody to IFN-γ was attached to the protein A to establish a sandwich immunoassay. Compared to GA-mediated immobilization to a PMMA surface, the amount of antibody immobilized using TR was observed to be higher. Use of PEI resulted in low nonspecific protein adsorption. Site-specific attachment of protein A via a tyrosine histidine tag led to an effective orientation for immobilization of capture antibody. Compared to a standard 96-well-plate ELISA, the assay time were observed to be shorter124 as was the detection limit.79
Figure 14.
(a) Sequence-specificity control after immobilization of two different receptor sequences (A and B) onto the COC surface. First, Cy5-labeled oligonucleotide complementary to sequence A and afterward rabbit-IgG conjugated oligonucleotide complementary to sequence B were hybridized. Reprinted with permission from H. Wang et al., Electrochem. Commun. 12, 258 (2010). Copyright 2010 Elsevier. (b) Optical micrographs of double-line bead micropatterns and schematic illustration of the sandwich immunocomplex formation on the streptavidin-coated bead micropattern. The microchannel is filled and incubated, subsequently, with (i) biotin-labeled capture antibody, (ii) target antigen, and (iii) Cy3-labeled detection antibody, forming the sandwich immunocomplex. Reprinted with permission from V. Sivagnanam et al., Anal. Chem. 81, 6509 (2009). Copyright 2009 American Chemical Society. (c) Schematic illustration of the stepwise process involved in the TR-catalyzed protein A-based antibody immobilization. Reprinted with permission from Y. Yuan et al., Biotechnol. Bioeng. 102, 891 (2009). Copyright 2008 Wiley InterScience.
Emneus' group extended an initial study on protein immobilization on porous silicon surfaces using hydrophilic polymer coatings activated with GA.40 In subsequent studies, the group tested DEX, aminodextran (AMD), and PVA, in addition to the original PEI material, as surface coatings for silicon. The study also used an intermediate bioaffinity layer of protein A and protein G, covalently attached to each of the four hydrophilic layers.68 PVA, PEI, and AMD were activated with GA, and DEX was aldehyde-functionalized in situ by oxidizing in sodium periodate. Antibody for environmental-contaminant atrazine was attached to protein A or protein G, and an HRP-based chemiluminescence sandwich immunoassay was demonstrated. Assay performance improved when using protein A and protein G as the active site of IgG was not sterically hindered. An additional benefit of using protein A and protein G was that antibody could be detached by treating with pH 2.2 glycine-HCl buffer. The authors reported that protein G combined with PEI- or DEX-modified surfaces showed the best sensitivity and long-term stability.
SMART IMMOBILIZATION
In order to perform high-sensitivity, multiplexed, high-throughput microfluidic immunoassays or enzyme assays in a reproducible manner, various “smart” methods of immobilizing proteins have been devised. In several studies, the entire microchannel surface supported protein binding (i.e., reactive sites). Some studies have opted to pursue other spatial patterns, as a fully coated channel may not be optimal owing to the difficulty of obtaining a uniform protein coating throughout the microchannel or owing to multiplexing requirements. Protein localization is also important in that each functional element of a microfluidic assay (e.g., separation, mixing, and detection) usually requires a distinct surface chemistry and specific device region. Therefore, the immobilization site should be isolated from other functional sites. Consequently, without localization, it is cumbersome to perform a multistep assay, which is common in protein or enzyme analysis. Unless one has a complicated microfluidic network or a way to isolate the protein binding sites using microfluidic components such as valves, it is not easy to immobilize proteins in a localized fashion. Proteins are generally immobilized in a preparatory step before running an assay. For a reproducible assay, protein activity and protein coverage on the surface should be maintained equally, no matter how much of a time gap exists between the immobilization step and the detection step. Thus, assay reproducibility would be improved if fresh proteins can be immobilized on demand during the assay.192, 195 Taken together, localized, in situ protein immobilization is preferred. However, confining immobilization sites of proteins at an asked time point is not straightforward in a conventional “injection and incubation” microfluidic format. Thus, various localized photochemical (Figure 15a),21, 22, 42, 112, 119, 188, 196 electrochemical (Figure 15b),130, 195, 197 or thermal stimuli (Figure 15c)32, 198 are employed to initiate protein immobilization at a specific location. Reversely, for off-chip assay steps or surface rejuvenation, protein can be detached from local immobilization sites (i.e., elution) upon stimulus (Figure 15d).62, 88
Figure 15.
“Smart immobilization” methods for reproducible, high-sensitivity, multiplexed, high-throughput microfluidic immunoassays, or enzyme assays. (a) Photoactivated protein immobilization, (b) electrochemically activated immobilization, (c) thermally activated immobilization, (d) photoactivated protein elution, (e) spatially addressable multiplexed protein immobilization, (f) reversible protein immobilization for repetitive assay, and (g) multifunctional immobilization surface for simplified immobilization procedure.
As attempts have been made for microfluidics to be used in studying complex biological phenomena, multiplexed and multiparameter capabilities have been pursued.26, 199 Microarray technology is widely used in proteomics200 and genomics167 owing to site-specific immobilization and simultaneous interrogation of numerous of biomolecules on a single substrate. In conjunction with precise and automated fluid-handling capability and enclosed structure (i.e., better protection from environment compared to microarrays), multiplexing capability was sought in microfluidic devices. Thus, we introduce DNA-directed immobilization for multiplexed (i.e., multiple proteins25) and multiparameter (i.e., multiple kinds of biomolecules26) analysis (Figure 15e). Unless designed to be disposable, a microfluidic device has to be cleaned for reuse, and the cleaning was usually done under harsh condition (e.g., strong oxidizing agent at high temperature46). We introduce a smart approach to refresh an immobilization surface in mild condition for repeated use of microfluidic chips (Figure 15f).63 Finally, a complicated multiple-step procedure is usually required to introduce various functions on the immobilization surface (e.g., anchoring to the substrate surface, repelling unwanted biomolecule, and immobilization target proteins). Thus, we introduce “smart” single composite coatings that can perform these various functions (Figure 15g).
Photoactivated immobilization
Spatial control of protein patterning in a microfluidic device is beneficial to multistep, multiplexed immunoassays21, 22 or enzyme reactor design.95, 201 Microcontact printing is a primary method for protein patterning with spatial control, but the method does have constraints as patterning should be done before assembly of the microfluidic chip.201 Introducing heterogeneous surface chemistries and immobilizing multiple proteins for multiplexed and multistep assays in a completely assembled microfluidic device, photopatterning through a high resolution photomask is considered most promising.96, 195, 202 Microfluidic chips are usually transparent (at least one side), so the UV light can penetrate the substrate and photo-initiate protein immobilization. Unlike an electrochemical method, another patterning approach, photoactivated immobilization does not require microfabrication of electrodes.130, 195
Physisorption
Zhan et al. photopatterned PEG hydrogel patches to physically encapsulate enzymes in microfluidic devices consisting of a PDMS substrate and a glass slide.95 First, the glass surface was silanized with TPM [3-(trichlorosilyl)propyl methacrylate], and then PEGDA was photopatterned with the photoinitiator HOMPP (2-hydroxy-2-methylpropiophenone). GOx and HRP were encapsulated in PEG hydrogels having ∼1 nm size pores. After the first PDMS microchannel device was detached from the glass slide, a second PDMS microchannel device with larger width and depth than the first PDMS microchannel was placed to direct fluid flow over and around the patterned hydrogel patches. Glucose penetrated the PEG hydrogel and was detected by GOx-HRP in a two-step enzyme assay. In addition to the enzyme assay, the researchers incorporated a pH-sensitive dye (SNAFL) to measure the pH. Using photopatterning, multiple patches of hydrogel each encapsulated with a different enzyme were created in a single channel to offer a multiplexed enzymatic assay. Koh and Pishko reported a similar PEGDA gel patch for enzyme entrapment to study enzyme kinetics (ALP and urease).57
Bioaffinity interaction
Cremer's group introduced an enzyme photopatterning method in a PDMS microfluidic device using photobleaching of fluorophore-conjugated biotin.201 Bovine fibrinogen was used to passivate the surface of a glass cover slide because the fibrinogen showed a better enzyme binding density and lower nonspecific protein adsorption than BSA, the commonly used blocking material. A commercial fluorophore-conjugated biotin solution was introduced and a 488 nm laser (300 μm beam diameter) irradiated the channel. The light created singlet oxygen, which subsequently attacked the fluorophore to generate free radicals for crosslinking of biotin to fibrinogen.196 A set of streptavidin-conjugated enzymes (GOx, ALP, and HRP) was immobilized in a single channel and the enzyme turnover number was estimated (Figure 16a). Castellana et al. reported UV-light-induced silver nanoparticle patterning and selective protein capture on the pattern.42 A TiO2 film was deposited on a glass wafer, and then the wafer was sealed with a PDMS microfluidic channel (Figure 16b). A unique property of TiO2 is that it creates holes and electrons upon UV exposure. The electrons were used to drive electrodeposition of silver after the introduction of a AgNO3 solution and UV irradiation through a photomask. The researchers used this approach to generate various patterns of silver nanoparticles. A mixture of biotin-PEG-disulfide and PEG-propionate-disulfide was introduced to functionalize the silver pattern with biotin and PEG (i.e., antibiofouling molecules) via thiol chemistry. DNP (N-2,4-Dinitrophenyl)-PEG-disulfide was functionalized on another silver pattern. Streptavidin binding to the biotin-functionalized silver pattern and anti-DNP antibody binding to the DNP-functionalized silver pattern were demonstrated.
Figure 16.
(a) Schematic diagram of the photoimmobilization process of enzyme. (1) Passivation of the surface with a fibrinogen monolayer is followed by (2) biotin-4-fluorescein surface attachment. This is accomplished by photobleaching with a 488-nm laser. (3) Next, the binding of streptavidin-linked enzymes that can be exploited to immobilize catalysts and (4) monitor reaction processes on-chip. Reprinted with permission from M. A. Holden et al., Anal. Chem. 76, 1838 (2004). Copyright 2004 American Chemical Society. (b) Schematic diagram for the protein patterning using a silver nanoparticle film. First, an AgNO3 solution is introduced into the microchannel. Next, UV radiation is passed through a photomask onto the backside of the TiO2 thin film. Ag+ ions adsorbed at the interface are selectively reduced by photoelectrons, which grow into nanoparticle films. Thiol chemistry was used to immobilize streptavidin. Reprinted with permission from E. T. Castellana et al., Anal. Chem. 78, 107 (2006). Copyright 2006 American Chemical Society.
Covalent immobilization
Wang et al. embedded PAA on a PDMS surface by photopatterning using “surface diffused” benzophenone.119 PDMS was submerged in acetone to cause swelling of the material, so that benzophenone could penetrate into the swollen PDMS. With acrylic acid monomer and UV exposure, PAA was formed as deep as 50 μm underneath the surface (Figure 17a). PAA was tightly anchored to the PDMS surface, and carboxyl functional groups on PAA were further activated with EDC in order to covalently bind antibody directly or to bind antibody through immobilized protein A. As protein A can bind up to four antibodies, fluorescence signal was improved 3 folds over directly attaching antibody to the surface without protein A. Fiddes reported protein immobilization on a PDMS surface via photografting of PA.188 Upon UV radiation, benzophenone abstracts hydrogen from methyl group of the PDMS to produce radicals so that graft polymerization of acrylamide monomer is initiated (Figure 17b). By activating with EDC and sulfo-NHS, fluorescently labeled casein and collagen were immobilized on a grafted PA layer (200 nm thickness). Various protein patterns including spots of casein, linear gradients of casein and collagen, and multiple spots of casein, collagen, and IgG were demonstrated. The activity of immobilized trypsin was also examined. Park et al. used an approach similar to Crooks' work95 in photopatterning a hydrogel patch to immobilize antibodies in a hybrid PDMS-glass slide microdevice.96 Glass was silanized with TPM, and then GMA (glycidyl methacrylate) was photopatterned with HOMPP. Anti-human IgG and anti-mouse IgG were covalently immobilized to a GMA patch via glycidyl (epoxide) functional groups on the surface after a 20-min incubation (Figure 17c). A downside of the GMA patch is that protein is immobilized on the surface and does not penetrate into the material, which may reduce assay sensitivity. Sebra et al. immobilized PEG-conjugated antibodies by graft polymerization on a DTC (dithiocarbamate)-based “photoreactive polymer substrate.”101 First, antibody was acrylate-functionalized by reacting with NHS-PEG-acrylate. Thus, the acrylate is covalently attached to the antibodies with an intermediate PEG spacer. The substrate for the microfluidic device was made by UV polymerization of urethane diacrylate (UDA) and triethylene glycol diacrylate (TEGDA) in the presence of tetraethylthiuram disulfide (TED) and photoinitiator 2,2-dimethoxy-2-phenylacetophenone (DMPA). Because of the TED, this substrate has photoreactive carbamate for subsequent photografting reactions. PEG monoacrylate and acrylated antibody were photografted upon UV exposure (Figure 17d). The hormone glucagon was detected using a sandwich immunoassay with picomolar sensitivity. The authors claimed that antibody copolymerized into the PEG polymer improved the antibody activity owing to a solvated and mobile polymer environment. They further observed reduced nonspecific protein binding.
Figure 17.
(a) Cross-linking of proteins to PDMS micropatterned with PAA. The PDMS is photopatterned with PAA. In a subsequent step, amide bonds are formed between the carboxyl groups of the PAA and amino groups of proteins. Reprinted with permission from Y. Wang et al., Anal. Chem. 77, 7539 (2005). Copyright 2005 American Chemical Society. (b) PAA-grafted PDMS for protein immobilization. (1) Native PDMS, (2) PDMS with a grafted layer of PAA, and (3) PDMS with a grafted layer of PAA that has been conjugated with FITC-casein. Reprinted with permission from L. K. Fiddes et al., Biomaterials 31, 315 (2010). Copyright 2010 Elsevier. (c) GMA photopolymerization and protein immobilization in a specific region on a glass slide using glycidyl functionalized hydrogel. Adapted from K. H. Park et al., Biosens. Bioelectron. 22, 613 (2006) with permission from Elsevier. (d) Detailed schematic of how covalently attached, photografted, antibody-containing tethers can be used to provide rapid detection of a specific antigen using a sandwich immunoassay approach. Adapted with permission from R. P. Sebra et al., Anal. Chem. 78, 3144 (2006). Copyright 2006 American Chemical Society.
Smart materials have also been used for photoactivated protein immobilization after protein analysis, such as a separation step.21, 22 Probed IEF and Western blot are workhorse protein-analysis techniques comprised of multiple biochemical assays. Each assay consists of a separation step, a transfer/immobilization (blotting) step, and a probing step (with immunoreagent). Inspired by photoactivated immobilization on a glass capillary surface,203 the two multistep assays are realized in a simple microfluidic device (i.e., straight microchannel). Owing to simplicity and the small footprint of the microfluidic device, multiple assays were integrated on a single chip, and thus parallel analysis of multiple targets in a short duration assay was reported. In one example, IEF and photoactivated protein immobilization were conducted, followed by antibody probing of the separated proteins (Figure 18a).21 To prepare the channel to conduct all assay stages, the surface of the glass channel was first silanized and acrylated using 3-(trimethoxysilyl) propyl methacrylate. Then, the benzophenone-functionalized monomer N-[3-[(4-benzoylphenyl)formamido]propyl] methacrylamide (BPMAC, synthesized in house) and acrylamide monomer were polymerized via a free-radical process using the initiator system of TEMED (N,N,N',N'-tetramethylethylenediamine) and APS (ammonium persulfate). After completion of IEF, the entire separation channel was irradiated with UV light, which initiated covalent bonding of proteins to the benzophenone copolymerized in the channel-filling PA gel. In this way, the isoelectric-point-based protein separation pattern was immobilized on the PA gel. PSA (prostate specific antigen) in human prostate cancer cell lysate and crude sera from metastatic prostate cancer patients were analyzed via IEF, photo-immobilized, and immunostained using fluorescently-labeled primary and secondary antibodies. Compared to immobilization of separated proteins in a capillary-based probed IEF assay,203 the channel-filling PA gel approach reported a 2× improvement on assay speed and a 180× improvement of protein capture efficiency. Use of microfluidic design with purely electrophoretic control of material introduction eliminated the need for fluidic components such as valves and pumps, thus simplifying the external hardware. In following studies, Western blotting was realized in a single microfluidic channel using a similar benzophenone-based covalent protein photoimmobilization strategy in a 3-D channel-filling gel (Figure 18b).22 In this assay, gel polymerization was initiated using a photo-initiator based system. PA gel was photopolymerized inside microchannels using riboflavin 5′ monophosphate, TEMED, and a blue LED light source (470 nm) to have more spatial control than previous APS-based chemical polymerization.21 Riboflavin was selected, as exposure of the gel to blue light does not appreciably activate benzophenone, thus preserving the photocapture function. After high-resolution protein sizing (SDS-PAGE), proteins were immobilized on the BPMAC-copolymerized PA gel using UV light. The capture efficiency was >75%, rivaling capture of conventional Western blotting membranes. After immobilization, target proteins were detected using primary antibody and ALP-conjugated secondary antibody. Multiplexed detection of ovalbumin, trypsin inhibitor (TI), and β-gal was demonstrated. Two biologically relevant systems were also reported: blotting of NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells) from cell lysate and detection of human antibodies against gp120 and p24 proteins in human sera. In total, 48 parallel Western blots were demonstrated in a single 24-minute assay, with probing for two targets per microfluidic Western blot. The microfluidic format used in conjunction with soft materials was demonstrated to yield 10–60 min total assay times, a 103-fold reduction of antibody and buffer consumption, 5 pM detection limits, and a 3.6 log dynamic range of detection.
Figure 18.
(a) Design and operation of the microfluidic IEF assay. (1) Glass microfluidic device (scale bar: 2 mm), (2) the 80-min five-step assay is completed in a single microchannel, (3) schematic of microchannel cross-section depicting photoactivated protein immobilization: analytes are electrophoresed through the PA gel, exposed to UV, and covalently immobilized (scale bar: 5 μm), (4) schematic of reaction between polypeptide backbone and benzophenone copolymerized in the PA gel. Ph denotes phenyl group. Reprinted from permission from A. J. Hughes and A. E. Herr, Proc. Natl. Acad. Sci. U.S.A. 109, 5972 (2012). Copyright 2012 National Academy of Sciences, USA. (b) Single-channel microfluidic Western blotting. The microfluidic Western blotting step is comprised of: (1) analyte stacking and SDS-PAGE within the PA gel; (2) capture of separated protein bands (“blotting”) onto the benzophenone-copolymerized PA gel under UV exposure; (3) electrophoretic introduction of fluorescently labeled detection antibodies for the target analyte; and (4) standard microscope-slide-sized chips with a scalable electrode array, accommodating 48 blots per chip in triplicate (144 microchannels). Reprinted from permission from A. J. Hughes and A. E. Herr, Proc. Natl. Acad. Sci. U.S.A. 109, 21450 (2012). Copyright 2012 National Academy of Sciences, USA.
Covalent immobilization—bioaffinity interaction
Lee et al. reported means to protect immobilized protein patterns and “expose” the pattern on demand under harsh lithographic condition (Figure 19).112 First, thermal oxide was grown on a silicon surface. The oxide surface was successively treated with APTES and GA. Protein A was immobilized via GA-mediated covalent linkage to the silicon surface. Second, antibody was immobilized to protein A and low-melting-point agarose (LMPA) was spin-coated to form an agarose-gel protective layer. Then, photoresist was spin-coated, followed by photolithography of the protein pattern. Third, the photomask pattern was transferred via UV exposure, and then photoresist was developed using tetramethyl ammonium hydroxide. Exposed LMPA and antibody were etched by oxygen plasma. In the last step, a second antibody was selectively immobilized using the same APTES and GA chemistry on the etched spots. The LMPA gel protecting the first immobilized antibody was removed by enzyme digestion of agarose (GELase). Antibody was active even after the harsh photoresist development process, baking, and the plasma etching process. The activity was confirmed via a fluorescently labeled antibody to the first and second patterned antibodies. The activity of trypsin, protected and exposed by the same process, was also tested with 70% of enzyme activity maintained. Sung et al. reported photopatterned 3-D hydrogel plugs for antibody immobilization on a PDMS surface.97 In order to anchor PA gel to the PDMS (which lacks functional groups), the PDMS was first treated with oxygen plasma. Then, PEI and PAA solutions were injected to yield a multilayer coating on the PDMS surface. Subsequently, an EDC and NHS chemistry was used to covalently bind the PEI and PAA layers together. The photoinitiator benzophenone was injected and diffused through the PDMS surface. Protein G was acrylate-functionalized using NHS-PEG-acrylate. A hydrogel precursor consisting of acrylamide/bis-acrylamide and acrylated-PEG-protein G was introduced. Upon exposure to UV light using a 40× objective lens, a cylindrical hydrogel plug was formed in the microfluidic channel via free-radical polymerization initiated from the surface-buried benzophenone. Mouse IgG was immobilized to the protein G and detected using HRP conjugated anti-mouse IgG. Also, ERγ (estrogen receptor γ) was detected in the same format, showing a sensitivity 13 times higher than with ERγ adsorbed on the bare PDMS surface.
Figure 19.
Patterning two different antibody layers on the same silicon surface: (1) immobilization of mouse IgG with LMPA protection layer, (2) photoresist patterning, (3) oxygen plasma etching followed by photoresist strip, and (4) immobilization of human IgG and removal of protective LMPA pattern using GELase. Adapted with permission from W.-C. Sung et al., Anal. Chem. 81, 7967 (2009). Copyright 2009 American Chemical Society.
Photoactivated detachment
Light-activated spatial control of proteins is not limited to site-specific patterning, but amenable to site-specific “de-patterning” (i.e., photocleaving), useful for on-demand protein elution. Kim's group reported on aptamer-based microfluidic affinity chromatography using a photocleavable linker for concentration and elution of proteins.62, 88 First, magnetic beads were prepared by coating a nanomagnetite core with PS and divinylbenzene polymer. Amine-functionalized PEG, copolymerized in the bead shell, helped ensure bead solubility, reduced nonspecific protein binding, and allowed aptamer crosslinking. An in-house synthesized photocleavable linker (4-(4-(1-hydroxyethyl)-2-methoxy-5-nitrophenoxy butanoic acid) was immobilized on the amino functionalized beads using NHS and carbodiimide (1,3-diisopropylcarbodiimide, DIC) chemistry. Aminated aptamer for HCV (hepatitis C virus) RNA polymerase was immobilized via an NHS functional group of the photocleavable linker. After magnetic beads were captured in a central chamber of a microfluidic device using a permanent magnet, HCV RNA polymerase in patient serum was captured, concentrated, and eluted upon UV exposure. Eluted enzyme was digested off-chip by trypsin and analyzed by MALDI-TOF/MS with an estimated detection limit of 9.6 fmol. The authors observed improved assay performance, as compared to antibody-based capture, because site-specific immobilization of aptamers through amination at the 3′- and 5′-positions maximizes antigen binding activity. Further, aptamers were not appreciably degraded by protease in serum. The linker showed 70% cleaving activity. In follow-up work, a similar photoelution method was used to analyze FITC-labeled proteins.88 In a microfluidic chip made by anodic boding of glass and silicon, magnetic beads functionalized with a photocleavable linker [4-(4-(1-(9-fluorenylmethoxycarbonylamino)ethyl)-2-methoxy-5-nitrophenoxy)-butanoic acid] were pseudo-immobilized via microposts. Two types of photocleavable linkers were prepared; one with biotin and one with NHS ester (Figure 20a). Biotin was conjugated to the bead with the BOP [benzotriazol-1-yloxytris(dimethylamino)phosphonium hexafluorophosphate] chemistry. Aminated aptamer for HCV RNA replicase was attached to the magnetic beads using NHS and DIC chemistry. Biotin-conjugated beads were used for purification of streptavidin and aptamer conjugated beads were used for purification of the HCV RNA replicase. Target proteins were mixed with model proteins (HSA, ovalbumin), injected into the packed bed, concentrated, and then photoeluted upon UV exposure (Figure 20b). The fluorescence signal was monitored for detection.
Figure 20.
(a) Structures of the photo-cleavable sites on the bead. (1) Biotinylated bead with a short spacer and (2) active ester containing bead with a long spacer for aptamer coupling. (b) Schematic view of microaffinity purification process. (1) Injection of the protein mixture into the microchip packed with microbeads, (2) purification of the target protein, (3) UV irradiation, and (4) analysis of the photolytically eluted protein. Reprinted with permission from W. J. Chung et al., Electrophoresis 26, 694 (2005). Copyright 2005 Wiley InterScience.
Thermally activated immobilization
PNIPAM [poly(N-isopropylacrylamide)] is a hydrophilic polymer at room temperature that becomes insoluble and hydrophobic in aqueous phase at ∼35 °C.204 Locally heating and cooling of a PNIPAM-coated surface actuates protein adsorption and desorption. Huber et al. published a thermally activated protein immobilization strategy using temperature-responsive PNIPAM.204 A silicon surface was silanized with MPTMS (3-mercaptopropyl trimethoxysilane), then NIPAM monomer was injected and polymerized using the azo-radical initiator AIBN [azobis(isobutyronitrile)]. Using a microheater integrated into a silicon-glass microfluidic chip, surface-grafted PNIPAM was heated for protein adsorption and cooled for protein desorption. The surface property was switched in <1 s. Adsorption/desorption of BSA, myoglobin, hemoglobin, and cytochrome C were tested. Stayton's group published work on PNIPAM-based microfluidic affinity chromatography198 and immunoassays.32 PNIPAM- and biotin-conjugated PEG were covalently attached to aminated PS latex beads using NHS chemistry. The microfluidic device consisted of stacked PET sheets integrated with a thin-film microheater. Upon heating application, PNIPAM-functionalized beads aggregated on the surface owing to hydrophobic interactions. Subsequently, streptavidin was introduced to physisorb to the bead surface. After cooling, streptavidin-captured beads were eluted.198 In follow-up studies, streptavidin was first immobilized on the PNIPAM-coated PS beads to capture biotinylated antibodies (Figure 21a).32 In a similar microfluidic device integrated with a heater, a competitive immunoassay of cardiac glycoside digoxin was demonstrated (Figure 21b). The authors observed that the reversible immobilization approach allowed the device to be used over several weeks with freshly prepared antibody-conjugated beads.
Figure 21.
Schematic representations of the temperature-responsive bead immunoassay system. (a) A 100 nm diameter latex nanobead is surface-conjugated with biotin–PEG and PNIPAM. Streptavidin is bound to the exposed biotin, providing binding sites for the biotinylated anti-digoxin IgG, and (b) a schematic of the experimental protocol. (1) Suspended beads are loaded into the microfluidic channel, (2) the temperature in the channel is then increased from room temperature to 37 °C, resulting in aggregation and adhesion of the beads to the channel wall, (3) flow is initiated, washing unadsorbed beads out of the channel, (4) a mixture of fluorescently labeled digoxigenin and digoxin is flowed into the channel, (5) components of this mixture that fail to bind the immobilized antibodies are washed through, and (6) finally, the temperature in the channel is reduced, and the aggregation-absorption process is reversed as antigen-bound beads leave the channel with the flow stream. Reprinted with permission from N. Malmstadt et al., Lab Chip 4, 412 (2004). Copyright 2004 The Royal Society of Chemistry.
Electrochemically activated protein immobilization
Protein is also immobilized on surfaces of microfluidic devices by electrochemical means through a process termed “electrochemical biolithography.”197 Electrochemically generated radicals or pH changes initiate protein immobilization. Using electrochemical stimuli, protein is immobilized at a specific location, typically around the electrode. One disadvantage is that electrodes must therefore be integrated on the surface of the microfluidic device. Nishizawa's group used electrochemically generated hydrobromous acid (HBrO) to immobilize proteins on a glass microfluidic device (Figure 22a).195 Pt microelectrodes were patterned on the glass substrate and silanized with 2-methacryloyloxyethyl phosphorylcholine (MPC). MPC has phosphorylcholine that mimics a cellular surface and minimizes protein adsorption. A microchannel was formed by attaching the electrode-patterned glass substrate to a blank substrate using stencil-patterned silicone rubber rim. PEI (positively charged) and heparin (negatively charged) were introduced to the channel surface to form electrostatically assembled layers with protein antifouling function. KBr in phosphate-buffered saline (PBS) buffer was injected and HBrO was generated electrochemically. HBrO diffused from the electrode surface and locally detached the PEI/heparin by oxidative reaction. Then, injected protein A was adsorbed to the “exposed” glass surface. Subsequently, analyte (mouse IgG) was introduced and an immunoassay was performed. Two capture antibodies (anti-C3 and anti-C4) were immobilized in a single channel and a multiplexed immunoassay was demonstrated. Protein attachment is believed to be weak (physisorption). In follow-up work, spatial control of protein attachment was improved using a PEGDA antifouling layer.197 PEGDA was photopatterned using microcontact printing. First, a glass substrate was silanized with 3-(trichlorosilyl)propyl methacrylate for protein adsorption. A solution of PEGDA monomer and photoinitiator (2,2-dimethoxy-2-phenylacetophenone) was inked to a PDMS stamp. After the stamp was placed on the glass surface, a UV flood exposure selectively polymerized PEGDA gel on the glass surface. Then, an unpolymerized region was coated by PEI-heparin multilayers. Electrochemically-generated HBrO detached a patch of the PEI-heparin multilayers from the glass surface. Protein A and subsequently antibody were immobilized to the “exposed” patch. Using the same electrode, DEP (dielectrophoresis) was performed to attract leucocytes to the antibody patch for cell capture. Owing to the antifouling PEGDA layer, the cell pattern was well confined within the patch. Multiplexed capture of lucocytes containing neutrophils and eosinophils was demonstrated. Rubloff's group used chitosan (pH sensitive aminopolysaccharide) to immobilize proteins upon electrochemical stimuli (Figure 22b).130 Chitosan is soluble at low pH, but becomes insoluble at neutral to high pH.130, 185 Thus, when the pH was increased by hydrolysis (OH− generation) on the electrode surface, chitosan was assembled into a hydrogel network. The microfluidic device was made by clamping a glass wafer housing SU-8 microchannels to a PDMS-coated Plexiglas wafer. Gold electrodes were patterned on the glass wafer. After the chitosan solution was injected, the gold electrode was negatively biased for 240 s. Negatively charged gold electrodes electrostatically attracted positively charged chitosan, and a local alkaline pH turned chitosan insoluble, thus depositing the species on the electrode surface. Amine groups, rich on chitosan, were activated with GA, and then protein was immobilized in a site-specific manner. GFP was immobilized and the fluorescence image indicated that GFP was active in this chitosan hydrogel environment. In follow-up work, an enzyme was immobilized on an in situ assembled chitosan hydrogel in a reversible manner.186 First, pro (tyrosine) tag was genetically fused to the enzyme Pfs (S-adenosylhomocysteine nucleosidase). In the presence of tyrosinase and pro-tagged Pfs, Pfs was covalently bound to chitosan (i.e., through generation of reactive O-quinone). Covalent bonds were further stabilized by NaBH3CN. Upon biasing, Pfs-chitosan was deposited onto the gold electrode to form a hydrogel. The enzymatic activity of Pfs was characterized, and the activity in the chitosan hydrogel was higher than that of enzyme in solution. The chitosan assembly disintegrated with a mild acid treatment allowing renewal of the electrode surface.
Figure 22.
(a) Electrochemically activated protein immobilization within a sealed microchannel. (1) Introduce a blocking agent (polyethyleneimine (PEI) and heparin) through the microchannel for antibiofouling, (2) generate HBrO local to the microelectrode, which removes a portion of blocking agent, thus making this part of the channel bottom available to protein physisorption, and (3) introduce proteins into the microchannel for immobilization. Reprinted with permission from H. Kaji et al., Anal. Chem. 78, 5469 (2006). Copyright 2006 American Chemical Society. (b) Chitosan-based electrochemically activated protein immobilization on gold electrode. (1) pH dependent protonation/deprotonation of the chitosan molecule, (2) schematic view of chitosan deposition, and (3) schematic view of chitosan deposition in a microfluidic channel. Reprinted with permission from J. J. Park et al., Lab Chip 6, 1315 (2006). Copyright 2006 The Royal Society of Chemistry.
Selective immobilization for multiplexed assay
Multiplexed assays are important, especially when measuring protein expression in biological systems. Multiple analytes in a single sample are detected in a multiplexed immunoassay.8, 205 Further, multistep enzymatic assays (e.g., GOx and HRP)95 or simultaneous characterization of enzyme kinetics can be done in a single microfluidic device.201 Here, recent studies on multiplexed immobilization via DNA hybridization are introduced. DNA microarrays, a multiplexed DNA platform, are a well-established technique.167 Protein microarray techniques stem from this earlier work of DNA microarrays.187, 206, 207 Thus, adaptation of DNA-directed protein immobilization for multiplexed microfluidic protein assays is attractive. A prerequisite for such immobilization is patterning of capture ssDNA of a specific sequence on a predetermined location of surface. Patterning is often done using a spot printer.25, 26 Possible drawbacks for spot printing are the high cost and the need to print before a chip is assembled, which may denature proteins and limit chip substrate material depending on the chip finishing processes needed.
Schroeder et al. reported on microfluidic reaction vessels for DNA-directed multiplexed immunoassays (Figure 23).25 The PMMA chip consists of 12 wells, in which 4 × 4 microarrays for capture cDNAs are housed. Each well has microfluidic access for reagent injection and washing. First, a carboxylic acid group of the photolinker 4-benzoylbenzoic acid was activated with carbodiimide (1,3-dicyclohexylcarbodiimide) and NHS. Then, cDNAs were crosslinked to the PMMA surface using UV exposure via hydrogen abstraction of benzophenone from C-H bonds on the PMMA surface.75 The four cDNAs were spot-patterned in the chip wells. Using the activated photolinker, four different amino-modified cDNA oligonucleotides were functionalized with benzophenone. Second, streptavidin was covalently attached to DNAs complementary to the surface-immobilized cDNAs. Streptavidin was conjugated with sulfo-SMPB, which then covalently linked to the thiolated DNA. Four different biotinylated antibodies to protein targets PSA, TNF-α, interleukin 6 (IL6), and interleukin 23 (IL23) were attached to each streptavidin-functionalized DNA molecule via bioaffinity interaction. The antibody mixture was conjugated with DNA and injected into the chip wells to selectively immobilize each antibody on a specific cDNA spot. Then, a multiplexed sandwich immunoassay was demonstrated. Gold-nanoparticle conjugated antibodies were used for light-transmission detection via catalytic silver deposition.
Figure 23.
Schematic drawing of multiplexed immunoassay performed in the wells of a disposable microarray. The different sandwich assays were assembled by site-specific DNA-directed immobilization to the dedicated capture probes cD-cG, illustrated in the scheme. Reprinted with permission from H. Schroeder et al., Anal. Chem. 81, 1275 (2009). Copyright 2009 American Chemical Society.
Extension of DNA-directed protein immobilization was conducted to realize the DEAL (DNA-encoded antibody libraries) platform by Heath's group. DEAL simultaneously detects ssDNA and proteins and can sort cells for multiparameter analysis.26 The rationale for DNA immobilization on the DEAL platform stems from the realization that surface chemistry for immobilization of different classes of biomolecules may not be compatible. Thus, by using DNA-mediated immobilization, such incompatibility issues could be alleviated. Antibodies are conjugated with SANH (succinimidyl 4-hydrazinonicotinate acetone hydrazine) in order to introduce hydrazide groups. Aminated DNA was aldehyde-functionalized using AFB (succinimidyl 4-formylbenzoate). Aldehyde-functionalized DNA and hydrazide-functionalized antibody were incubated overnight to form covalent hydrazone bond.125 A glass slide was coated with PLL after hydrophilization in oxygen plasma. Capture cDNA was printed on the glass slide and a PDMS microfluidic chip was bonded to the slide. Then, DNA-encoded capture antibodies were immobilized on the slide via DNA hybridization. The cytokines IFN-γ, TNF-α, and IL-2 were detected using a sandwich immunoassay. Using antibodies against cell markers, CD4+ and CD8 + T cells were sorted. Multiparameter assays of DNA, TNF-α protein, and B cells were also demonstrated.
Reversible immobilization
Covalent bonds are usually irreversible, meaning that microfluidic chips relying on covalent immobilization strategies are typically used once and then discarded. Alternatively, the immobilization surface can be refreshed using a strong oxidizer.46 Bioaffinity interactions like DNA hybridization or protein A/G binding to antibody are reversible using strong acids/bases, concentrated salts, or heat treatments. Physisorption, especially electrostatic interactions, can be reversed by increasing salt concentrations.20 The cost of the assay can be reduced and multiple assays can be performed in a single device if the surface can be regenerated reproducibly.
Li et al. used a chelating agent to refresh an enzyme-immobilized surface on magnetic beads.63 The concept employed is that trypsin can be immobilized via a Lewis acid-base interaction between metal ions and an electron acceptor group on the protein surface (e.g., imidazole of histidine).208, 209 For elution, a competitive reaction with EDTA can break the bond to release the enzyme. First, metal-ion chelated magnetic beads were prepared. Magnetic cores were synthesized using a solvothermal reaction of FeCl3·6H2O in ethylene glycol. Then, the magnetic core was coated with silica via sol-gel formation of tetraethyl orthosilicate (TEOS). Second, iminodiacetic acid (IDA), a metal chelating agent, was covalently attached to the silanizing agent 3-glycidoxypropyltrimethoxysilane agent GLYMO (i.e., GOPTS) via reaction of the glycidyl group with secondary amines. GLYMO-IDA was then attached to the silica shell of the magnetic beads by a silane-coupling reaction. Copper ions (Cu2+) and TPCK (tosyl phenylalanyl chloromethyl ketone)-treated trypsin were incubated with the magnetic beads. The enzyme was bound to the bead via the Lewis acid-base interaction with the divalent cation chelator, IDA (Figure 24). The magnetic beads were captured in a microfluidic channel by a permanent magnet. Cytochrome c and BSA were digested by trypsin, and analyzed with MALDI-TOF/MS. The magnetic beads were readily packed, flushed, and repacked by magnetic-field and fluidic control. Compared to covalently bound enzymes, IDA-metal chelated adsorption was regenerated easily by adding competing EDTA solution to remove metal ions. Regenerated enzyme showed reproducible results up to 4 runs. Bead-based solid-phase digestion was faster than in-solution trypsin digestion.
Figure 24.
Process of Cu-IDA-GLYMO-MS microspheres preparation and trypsin immobilization. Reprinted with permission from Y. Li et al., J. Proteome Res. 6, 2367 (2007). Copyright 2007 American Chemical Society.
Multi-functional immobilization surface
Stable immobilization of highly active proteins and minimization of nonspecific biomolecular adsorption are keys to high performance microfluidic protein assays. Here, rationally designed multifunctional polymers that include different functional groups for protein immobilization, antibiofouling, and surface anchoring are detailed.210, 211, 212 Using these polymers, multiple surface-coating steps35, 40, 68, 191 that lead to increased assay preparation times and reduced assay reproducibility are eliminated.
Sakai-Kato reported water-soluble multifunctional phospholipid polymers for anchoring to PMMA surfaces, covalently linking trypsin and repelling co-existing proteins.212 First, the researchers synthesize MEONP (p-nitrophenyloxycarbonyl polyethyleneglycol methacrylate), a reactive p-nitrophenyl-ester-functionalized acrylate monomer with a PEG spacer.184 Then, the group prepared amphiphilic phospholipid polymer PMBN [MPC (phospholipid 2-methacryloyloxyethyl phosphorylcholine)—BMA (butyl methacrylate)—MEONP] using radical polymerization with initiator AIBN. BMA is hydrophobic and used to anchor to the PMMA surface. MPC is a phospholipid, which prevents nonspecific adsorption of proteins and cells to the surface. MEONP covalently links to amine groups of trypsin via hydrolysis. Trypsin activity was tested via digestion of ArgOEt (arginine ethyl ester) to Arg, and electrophoretic separation of the substrate and product. Note that the process required almost two days to completely immobilize trypsin to the PMBN polymer. Jon's group prepared amphiphilic polymers with hydrophobic groups anchoring to COC surfaces, PEG protein-repelling group, and NHS-ester groups for covalent protein immobilization (Figure 25).210 The multifunctional polymers poly(DMA-r-PEGMA-r-NAS) and poly(BMA-r-PEGMA-r-NAS) were prepared; hydrophobic dodecyl methacrylate (DMA) or hydrophobic benzyl methacrylate (BMA), hydrophilic polyethylene glycol methacrylate (PEGMA), and N-acryloylsuccinimide (NAS, a reactive NHS ester of acrylic monomer) were radical-copolymerized using AIBN. Incubation of an injection-molded COC device in the polymer solution for 1 h completed surface coating. The surface anchoring properties, protein repelling functionality, and covalent immobilization of antibodies were tested. Poly(BMA-r-PEGMA-r-NAS) showed better antibiofouling and higher antibody immobilization capacity. Therefore, sandwich immunoassays of troponin I were demonstrated on poly(BMA-r-PEGMA-r-NAS) surfaces, resulting in a 10 ng/ml detection limit. In another study, the authors also used a similar multifunctional surface to immobilize proteins on a PS surface.211 A major difference from Ref. 210 was that an additional step for activating methacrylate with EDC and NHS was used before protein immobilization. Amine-functionalized biotin was covalently linked to the activated surface to attach biotinylated antibodies via streptavidin-biotin linkage. In addition, protein A was covalently linked to the surface to attach multiple antibodies for immunoassays.
Figure 25.
(a) Chemical structures of the multifunctional amphiphilic polymers, and (b) schematic representation of the procedure for immobilizing biomolecules onto a polymer-modified COC surface with antibiofouling properties. Reprinted with permission from D. Sung et al., Biosens. Bioelectron. 26, 3967 (2011). Copyright 2011 Elsevier.
CONCLUSION AND OUTLOOK
Immobilization of antigens, antibodies, or enzymes in a microfluidic device is critical for functions ranging from immunoassays to enzyme activity assays. Intriguing ideas on effective protein immobilization are being generated at an amazing pace. The basis for most strategies originates in three general immobilization mechanisms (physisorption, bioaffinity interaction, and covalent bonding), and a combination of the three mechanisms. A key aim of immobilization strategies is improving assay sensitivity, specificity, reproducibility, and sometimes throughput. To meet this goal, researchers are focused on (1) forming stable and strong bonds between protein and immobilization surface, (2) minimizing nonspecific adsorption of biomacromolecules or cells, (3) keeping the protein in an active state, and (4) orienting protein for unhindered access of binding partners. In some instances, the use of physisorption, bioaffinity interaction, or covalent bonding alone does not render meeting this goal possible. Consequently, various combinations of these immobilization mechanisms are frequently used with important outcomes.
Regardless of a weak and irreproducible nature, physisorption is often employed in protein immobilization, especially when quickly proving a new assay concept. Nevertheless, physisorption, especially electrostatic interaction, can be strong enough that immobilized proteins remain attached even in the presence of strong external forces, such as the hydrodynamic shear force, magnetic forces, or electrostatic forces. In some cases, physisorption is achieved almost instantly, compared to covalent interactions usually requiring long incubation periods. Thus, multistep assays such as Western blotting are possible using the physisorption (e.g., electrostatic protein blotting right after electrophoretic separation without degradation of protein separation performance).
Bioaffinity interactions are extensively used in literature owing to its relatively strong, high specific, oriented protein immobilization, and in some instances, the possibility of reversing linkage. However, we noted that bioaffinity interaction was generally used in conjunction with covalent linkage chemistries and/or physisorption. The streptavidin-biotin interaction is widely used in bioanalytical chemistry, owing to exceptional binding strength, specificity, and multiple binding sites for increased binding capacity. Protein A and protein G are also popular binding agents, especially in immunoassays, because both bind to the Fc region of IgG and do not block the Fab region, consequently improving the antigen-binding capability of immobilized IgG. Another benefit is that protein A and G binding can be reversed using suitable chemical treatments.
DNA hybridization is a promising bioaffinity strategy for immobilization, especially for multiplexed or multiparameter assays. DNA-encoded immobilization relies on a specific hybridization between a single-stranded DNA (ssDNA) sequence and its complementary sequence. DNA-directed immobilization originates from DNA microarray technology, which offers an enormous multiplexing capability. After an ssDNA-encoded protein is attached to its complementary DNA, a “protein microarray” is formed in a microfluidic device for a multiplexed assay. Although analysis of only a handful of species has been demonstrated, a strong potential exists for this bioaffinity-based multiplexed immobilization scheme. Numerous efforts have been undertaken to replace antibody with aptamers for capture of antigen. The benefits of aptamers are potentially numerous compared to those of antibodies and include: smaller size for less steric hindrance, no need for animals in production, lower costs to produce, longer shelf life, and less sensitivity to environmental conditions. Nevertheless, antibodies are still the dominant capture reagent.
Covalent bonds are the strongest and most frequently employed protein immobilization mechanism. Amine-based covalent linking is widely employed for protein immobilization owing to rich lysine residues on protein surfaces. Glutaraldehyde activation of amine is popular technique in microfluidics, followed by NHS ester and carbodiimide. A downside of amine-based chemistry is that multiple bonds can be formed, since numerous lysine residues exist on a protein surface, which may block active sites of the protein or cause conformational change. Thus, more specific, simple, and protein-friendly covalent linking methods would fill an existing gap. Covalent linkage is often used with a spacer molecule like PEG or hydrophilic polymer coatings to reduce steric hindrance and the possibility of protein denaturation.
Most microfluidic protein assays find immobilization of analyte on the inner surface of a microfluidic channel, which is rational given that the core of the microchannels are used for transport of reagent, analytes, substrate, and sample. Immobilization methods depend on the immobilization surface. More and more efforts are devoted to identifying strategies for immobilizing proteins on plastic surfaces, as plastics are becoming widely used in microfluidic devices owing to the low cost and mass-production potential. PDMS remains a popular substrate for rapid prototyping of new devices. Thus, new immobilization methods for PDMS surfaces have been an active area of research. Owing to the inert surfaces (i.e., lacking functional groups), immobilization chemistries for plastic and PDMS are not straightforward. In contrast to plastic and PDMS, the formation of silanol groups is a routine starting point of the immobilization process on glass or silicon surfaces. Oxygen-plasma treatment or less frequently solution-phase oxidation are used to prepare plastics and PDMS, but as evidenced by many studies, the linkages formed on the oxidized polymer surface are weak and unstable. Consequently, a surface-grafted polymer or physisorbed functional layer on the channel surface is a major immobilization strategy being pursued for plastics or PDMS substrates. Exploiting the hydrophobicity of PDMS or plastics is a straightforward way to immobilize proteins having hydrophobic patches on the biomolecular surface. Although hydrophobic interaction is still used often in benchtop biochemical assays such as Western blotting and dot blots, hydrophobic interactions are not generally recommended if tight reproducibility and quantitative assays are needed.
Various surface coatings including inorganic (e.g., sol-gel, gold/silver nanoparticle), polymeric (e.g., PEI, PAA, PDADMAC, PVA, PEG gel, and polyacrylamide gel), and biopolymeric (dextran, chitosan, and protein aggregates) coatings, help to minimize nonspecific adsorption and improve the activity of immobilized proteins. A recent trend has been to incorporate multiple functions (e.g., activated functional group for protein immobilization, antibiofouling layer for reduced nonspecific protein adsorption, and surface anchoring group) in a single copolymer, to obviate the time-consuming and error-prone layering of multiple functional coatings. Thus, devising effective and smart coating layers are emerging as a major research topic.
Three-dimensional solid supports like hydrogels, packed beads, sol-gels, and polymer monoliths yield high surface-area-to-volume ratios that underpin efficient material capture, improved binding kinetics owing to short diffusion lengths in the materials, and in some cases provide water-retaining protein-friendly environments (e.g., hydrogels). Devising immobilization chemistries that are compatible with these solid supports will continue to be a worthy research topic.
Researchers are adding more and more functionalities to microfluidic devices to meet the ambitious goal of microfluidics: a “truly integrated, samples-in-answers-out stand-alone platform.” Various “smart” immobilization techniques have been introduced. One prominent smart immobilization method is light-directed immobilization. Multiple proteins can be patterned with excellent spatial control (depending on the exposure system used) after a microfluidic chip is completely sealed. Microcontact printing, spot printing, and other soft lithography techniques are limited to analyte patterning before assembly of the chip. Early literature employed reversible sealing of PDMS on glass in order to perform multiplexing-oriented patterning methods (e.g., the μFN platform by Delamarche's group). However, leakage or cross-talk between microchannels could be a challenge of this approach. An advantage of light-activated immobilization over electrochemistry-mediated protein patterning is that electrodes do not need to be patterned inside the microfluidic chip, which simplifies design and fabrication of the microfluidic device. Light-activated immobilization is facile, compared to covalent chemistries in which a long incubation period is required. Thus, light-activated immobilization was used in microfluidic Western blotting in which protein should be immobilized as soon as electrophoretic separation is completed in order to maintain high separation resolution. A downside could be that the light-activated immobilization may require expensive optics and motorized stages to precisely pattern protein patches in such small microfluidic channels. Light has also been successfully employed to cleave a bond to release biomolecules from the surface. A thermal transition between hydrophobic to hydrophilic surfaces may provide a quick and easy way to capture and release proteins. Heat may not need to be transferred to the temperature responsive polymer using microfabricated heaters. Instead, infrared radiation or injection of hot fluid could also be used to activate temperature-responsive immobilization surfaces, thus simplifying device fabrication and assay operation. Reversible immobilization has been realized by simply introducing chelating agents like EDTA, allows the immobilized protein to be eluted from an immobilization surface. Even though the reproducibility of protein capture efficiency on a rejuvenated surface is limited, continued studies may be worthwhile because covalently attached proteins can only be removed by completely renewing the immobilization surface (e.g., using a strong oxidizing agent).
Many smart immobilization strategies could be combined for more sophisticated multi-analyte assays required for complex sample analyses. For example, light, electrochemical, or thermal stimuli are “orthogonal,” meaning that they can be used simultaneously to address a specific site at a certain time point without affecting other responsive functions. Ultimately, for a highly integrated multiparameter assay, biomolecules such as proteins and oligonucleotides as well as cells could be injected into the microfluidic device, immobilized on different spots, and released on demand using these mutually independent stimuli. Clearly, protein immobilization strategies play a critical role in the measurement of biological systems and processes. We look forward to continued substantial innovation in this area.
References
- Gauci V., Wright E., and Coorssen J., J. Chem. Biol. 4, 3 (2011). 10.1007/s12154-010-0043-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricoin E. F., Zoon K. C., Kohn E. C., Barrett J. C., and Liotta L. A., Nat. Rev. Drug Discovery 1, 683 (2002). 10.1038/nrd891 [DOI] [PubMed] [Google Scholar]
- Borrebaeck C. A., Immunol. Today 21, 379 (2000). 10.1016/S0167-5699(00)01683-2 [DOI] [PubMed] [Google Scholar]
- Ludwig J. A. and Weinstein J. N., Nat. Rev. Cancer 5, 845 (2005). 10.1038/nrc1739 [DOI] [PubMed] [Google Scholar]
- Koeller K. M. and Wong C.-H., Nature 409, 232 (2001). 10.1038/35051706 [DOI] [PubMed] [Google Scholar]
- van Beilen J. B. and Li Z., Curr. Opin. Biotechnol. 13, 338 (2002). 10.1016/S0958-1669(02)00334-8 [DOI] [PubMed] [Google Scholar]
- Rasor J. P. and Voss E., Appl. Catal. A 221, 145 (2001). 10.1016/S0926-860X(01)00804-3 [DOI] [Google Scholar]
- Ng A. H., Uddayasankar U., and Wheeler A. R., Anal. Bioanal. Chem. 397, 991 (2010). 10.1007/s00216-010-3678-8 [DOI] [PubMed] [Google Scholar]
- Henares T. G., Mizutani F., and Hisamoto H., Anal. Chim. Acta 611, 17 (2008). 10.1016/j.aca.2008.01.064 [DOI] [PubMed] [Google Scholar]
- Bange A., Halsall H. B., and Heineman W. R., Biosens. Bioelectron. 20, 2488 (2005). 10.1016/j.bios.2004.10.016 [DOI] [PubMed] [Google Scholar]
- Křenková J. and Foret F., Electrophoresis 25, 3550 (2004). 10.1002/elps.200406096 [DOI] [PubMed] [Google Scholar]
- Asanomi Y., Yamaguchi H., Miyazaki M., and Maeda H., Molecules 16, 6041 (2011). 10.3390/molecules16076041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M. and Maeda H., Trends Biotechnol. 24, 463 (2006). 10.1016/j.tibtech.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Whitesides G. M., Nature 442, 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- Burns M. A., Johnson B. N., Brahmasandra S. N., Handique K., Webster J. R., Krishnan M., Sammarco T. S., Man P. M., Jones D., Heldsinger D., Mastrangelo C. H., and Burke D. T., Science 282, 484 (1998). 10.1126/science.282.5388.484 [DOI] [PubMed] [Google Scholar]
- Easley C. J., Karlinsey J. M., Bienvenue J. M., Legendre L. A., Roper M. G., Feldman S. H., Hughes M. A., Hewlett E. L., Merkel T. J., and Ferrance J. P., Proc. Natl. Acad. Sci. U.S.A. 103, 19272 (2006). 10.1073/pnas.0604663103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li X., Greenspoon S. A., Scherer J. R., and Mathies R. A., Lab Chip 11, 1041 (2011). 10.1039/c0lc00533a [DOI] [PubMed] [Google Scholar]
- He M. and Herr A. E., J. Am. Chem. Soc. 132, 2512 (2010). 10.1021/ja910164d [DOI] [PubMed] [Google Scholar]
- Tia S. Q., He M., Kim D., and Herr A. E., Anal. Chem. 83, 3581 (2011). 10.1021/ac200322z [DOI] [PubMed] [Google Scholar]
- Kim D., Karns K., Tia S. Q., He M., and Herr A. E., Anal. Chem. 84, 2533 (2012). 10.1021/ac3000013 [DOI] [PubMed] [Google Scholar]
- Hughes A. J., Lin R. K., Peehl D. M., and Herr A. E., Proc. Natl. Acad. Sci. U.S.A. 109, 5972 (2012). 10.1073/pnas.1108617109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. J. and Herr A. E., Proc. Natl. Acad. Sci. U.S.A. 109, 21450 (2012). 10.1073/pnas.1207754110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Novak J., Julian B. A., and Herr A. E., J. Am. Chem. Soc. 133, 19610 (2011). 10.1021/ja207963f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M., Kim D., and Herr A. E., “ Microchamber Western blotting using poly-L-lysine conjugated polyacrylamide gel for blotting of SDS coated proteins,” Anal. Chem. (to be published). 10.1021/ac401012j [DOI] [PubMed]
- Schroeder H., Adler M., Gerigk K., Muüller-Chorus B., Goütz F., and Niemeyer C. M., Anal. Chem. 81, 1275 (2009). 10.1021/ac802228k [DOI] [PubMed] [Google Scholar]
- Bailey R. C., Kwong G. A., Radu C. G., Witte O. N., and Heath J. R., J. Am. Chem. Soc. 129, 1959 (2007). 10.1021/ja065930i [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M., Juncker D., Michel B., Hunziker P., and Delamarche E., Biosens. Bioelectron. 19, 1193 (2004). 10.1016/j.bios.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A. and Klapperich C., Biomed. Microdevices 9, 245 (2007). 10.1007/s10544-006-9026-2 [DOI] [PubMed] [Google Scholar]
- Jönsson C., Aronsson M., Rundström G., Pettersson C., Mendel-Hartvig I., Bakker J., Martinsson E., Liedberg B., MacCraith B., Öhman O., and Melin J., Lab Chip 8, 1191 (2008). 10.1039/b800297e [DOI] [PubMed] [Google Scholar]
- Yang Y.-N., Lin H.-I., Wang J.-H., Shiesh S.-C., and Lee G.-B., Biosens. Bioelectron. 24, 3091 (2009). 10.1016/j.bios.2009.03.034 [DOI] [PubMed] [Google Scholar]
- Shin K.-S., Lee S. W., Han K.-C., Kim S. K., Yang E. K., Park J. H., Ju B.-K., Kang J. Y., and Kim T. S., Biosens. Bioelectron. 22, 2261 (2007). 10.1016/j.bios.2006.11.011 [DOI] [PubMed] [Google Scholar]
- Malmstadt N., Hoffman A. S., and Stayton P. S., Lab Chip 4, 412 (2004). 10.1039/b315394k [DOI] [PubMed] [Google Scholar]
- Yang D., Niu X., Liu Y., Wang Y., Gu X., Song L., Zhao R., Ma L., Shao Y., and Jiang X., Adv. Mater. 20, 4770 (2008). 10.1002/adma.200801302 [DOI] [Google Scholar]
- Xiang Q., Hu G., Gao Y., and Li D., Biosens. Bioelectron. 21, 2006 (2006). 10.1016/j.bios.2005.09.019 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu Y., Liu C., Huang J., Yang P., and Liu B., Electrochem. Commun. 12, 258 (2010). 10.1016/j.elecom.2009.12.008 [DOI] [Google Scholar]
- Tan S. N., Ge L., Tan H. Y., Loke W. K., Gao J., and Wang W., Anal. Chem. 84, 10071 (2012). 10.1021/ac302537r [DOI] [PubMed] [Google Scholar]
- Schult K., Katerkamp A., Trau D., Grawe F., Cammann K., and Meusel M., Anal. Chem. 71, 5430 (1999). 10.1021/ac9907686 [DOI] [PubMed] [Google Scholar]
- Dong Y., Phillips K., and Cheng Q., Lab Chip 6, 675 (2006). 10.1039/b514902a [DOI] [PubMed] [Google Scholar]
- Yang T., Jung S.-Y., Mao H., and Cremer P. S., Anal. Chem. 73, 165 (2001). 10.1021/ac000997o [DOI] [PubMed] [Google Scholar]
- Yakovleva J., Davidsson R., Lobanova A., Bengtsson M., Eremin S., Laurell T., and Emnéus J., Anal. Chem. 74, 2994 (2002). 10.1021/ac015645b [DOI] [PubMed] [Google Scholar]
- Cannon B., Weaver N., Pu Q., Thiagarajan V., Liu S., Huang J., Vaughn M. W., and Cheng K. H., Langmuir 21, 9666 (2005). 10.1021/la0502645 [DOI] [PubMed] [Google Scholar]
- Castellana E. T., Kataoka S., Albertorio F., and Cremer P. S., Anal. Chem. 78, 107 (2006). 10.1021/ac051288j [DOI] [PubMed] [Google Scholar]
- Hou C. and Herr A. E., Anal. Chem. 82, 3343 (2010). 10.1021/ac100182j [DOI] [PubMed] [Google Scholar]
- Herr A. E., Hatch A. V., Throckmorton D. J., Tran H. M., Brennan J. S., Giannobile W. V., and Singh A. K., Proc. Natl. Acad. Sci. U.S.A. 104, 5268 (2007). 10.1073/pnas.0607254104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karns K. and Herr A. E., Anal. Chem. 83, 8115 (2011). 10.1021/ac202061v [DOI] [PubMed] [Google Scholar]
- He M. and Herr A. E., Nat. Protoc. 5, 1844 (2010). 10.1038/nprot.2010.142 [DOI] [PubMed] [Google Scholar]
- Markov D. A., Swinney K., and Bornhop D. J., J. Am. Chem. Soc. 126, 16659 (2004). 10.1021/ja047820m [DOI] [PubMed] [Google Scholar]
- Gerber D., Maerkl S. J., and Quake S. R., Nat. Methods 6, 71 (2009). 10.1038/nmeth.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet E., Lausted C., Lin T., Yang C. W. T., Hood L., and Lagally E. T., Lab Chip 10, 581 (2010). 10.1039/b920589f [DOI] [PubMed] [Google Scholar]
- Chen X., Kapil M. A., Hughes A. J., and Herr A. E., Anal. Chem. 83, 6573 (2011). 10.1021/ac200982j [DOI] [PubMed] [Google Scholar]
- Tia S. Q., Brown K., Chen D., and Herr A. E., Anal. Chem. 85, 2882 (2013). 10.1021/ac3035053 [DOI] [PubMed] [Google Scholar]
- Thomsen M. S., Pölt P., and Nidetzky B., Chem. Commun. 2007, 2527. 10.1039/B702115A [DOI] [PubMed] [Google Scholar]
- Hughes A. J. and Herr A. E., Anal. Chem. 82, 3803 (2010). 10.1021/ac100201z [DOI] [PubMed] [Google Scholar]
- Baeza M., López C., Alonso J., López-Santín J., and Álvaro G., Anal. Chem. 82, 1006 (2010). 10.1021/ac902267f [DOI] [PubMed] [Google Scholar]
- Mao H., Yang T., and Cremer P. S., Anal. Chem. 74, 379 (2002). 10.1021/ac010822u [DOI] [PubMed] [Google Scholar]
- He P., Greenway G., and Haswell S. J., Microfluid. Nanofluid. 8, 565 (2010). 10.1007/s10404-009-0476-8 [DOI] [Google Scholar]
- Koh W.-G. and Pishko M., Sens. Actuators B 106, 335 (2005). 10.1016/j.snb.2004.08.025 [DOI] [Google Scholar]
- Brueggemeier S. B., Kron S. J., and Palecek S. P., Anal. Biochem. 329, 180 (2004). 10.1016/j.ab.2004.02.012 [DOI] [PubMed] [Google Scholar]
- Köstler S., Ribitsch V., Stana-Kleinschek K., Jakopic G., and Strnad S., Colloids Surf., A 270–271, 107 (2005). 10.1016/j.colsurfa.2005.05.049 [DOI] [Google Scholar]
- Ekström S., Önnerfjord P., Nilsson J., Bengtsson M., Laurell T., and Marko-Varga G., Anal. Chem. 72, 286 (2000). 10.1021/ac990731l [DOI] [PubMed] [Google Scholar]
- Peterson D. S., Rohr T., Svec F., and Fréchet J. M., Anal. Chem. 74, 4081 (2002). 10.1021/ac020180q [DOI] [PubMed] [Google Scholar]
- Cho S., Lee S. H., Chung W. J., Kim Y. K., Lee Y. S., and Kim B. G., Electrophoresis 25, 3730 (2004). 10.1002/elps.200406103 [DOI] [PubMed] [Google Scholar]
- Li Y., Xu X., Yan B., Deng C., Yu W., Yang P., and Zhang X., J. Proteome Res. 6, 2367 (2007). 10.1021/pr060558r [DOI] [PubMed] [Google Scholar]
- Wu H., Tian Y., Liu B., Lu H., Wang X., Zhai J., Jin H., Yang P., Xu Y., and Wang H., J. Proteome Res. 3, 1201 (2004). 10.1021/pr049889z [DOI] [PubMed] [Google Scholar]
- Jo K., Heien M. L., Thompson L. B., Zhong M., Nuzzo R. G., and Sweedler J. V., Lab Chip 7, 1454 (2007). 10.1039/b706940e [DOI] [PubMed] [Google Scholar]
- Park S. S., Cho S. I., Kim M. S., Kim Y. K., and Kim B. G., Electrophoresis 24, 200 (2003). 10.1002/elps.200390015 [DOI] [PubMed] [Google Scholar]
- Gao J., Xu J., Locascio L. E., and Lee C. S., Anal. Chem. 73, 2648 (2001). 10.1021/ac001126h [DOI] [PubMed] [Google Scholar]
- Yakovleva J., Davidsson R., Bengtsson M., Laurell T., and Emnéus J., Biosens. Bioelectron. 19, 21 (2003). 10.1016/S0956-5663(03)00126-X [DOI] [PubMed] [Google Scholar]
- Huh Y. S., Chung A. J., and Erickson D., Microfluid. Nanofluid. 6, 285 (2009). 10.1007/s10404-008-0392-3 [DOI] [Google Scholar]
- Choi J.-W., Oh K. W., Thomas J. H., Heineman W. R., Halsall H. B., Nevin J. H., Helmicki A. J., Henderson H. T., and Ahn C. H., Lab Chip 2, 27 (2002). 10.1039/b107540n [DOI] [PubMed] [Google Scholar]
- Anderson J. R., Chiu D. T., Wu H., Schueller O. J., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- Ng J. M. K., Gitlin I., Stroock A. D., and Whitesides G. M., Electrophoresis 23, 3461 (2002). [DOI] [PubMed] [Google Scholar]
- Sia S. K. and Whitesides G. M., Electrophoresis 24, 3563 (2003). 10.1002/elps.200305584 [DOI] [PubMed] [Google Scholar]
- Becker H. and Locascio L. E., Talanta 56, 267 (2002). 10.1016/S0039-9140(01)00594-X [DOI] [PubMed] [Google Scholar]
- Dankbar D. M. and Gauglitz G., Anal. Bioanal. Chem. 386, 1967 (2006). 10.1007/s00216-006-0871-x [DOI] [PubMed] [Google Scholar]
- Becker H. and Gärtner C., Electrophoresis 21, 12 (2000). [DOI] [PubMed] [Google Scholar]
- Laib S. and MacCraith B. D., Anal. Chem. 79, 6264 (2007). 10.1021/ac062420y [DOI] [PubMed] [Google Scholar]
- Bai Y., Koh C. G., Boreman M., Juang Y.-J., Tang I.-C., Lee L. J., and Yang S.-T., Langmuir 22, 9458 (2006). 10.1021/la061123l [DOI] [PubMed] [Google Scholar]
- Yuan Y., He H., and Lee L. J., Biotechnol. Bioeng. 102, 891 (2009). 10.1002/bit.22136 [DOI] [PubMed] [Google Scholar]
- Wen X., He H., and Lee L. J., J. Immunol. Methods 350, 97 (2009). 10.1016/j.jim.2009.07.011 [DOI] [PubMed] [Google Scholar]
- Rubina A. Y., Kolchinsky A., Makarov A. A., and Zasedatelev A. S., Proteomics 8, 817 (2008). 10.1002/pmic.200700629 [DOI] [PubMed] [Google Scholar]
- Lee A. G., Arena C. P., Beebe D. J., and Palecek S. P., Biomacromolecules 11, 3316 (2010). 10.1021/bm100792y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. G., Beebe D. J., and Palecek S. P., Biomed. Microdevices 14, 247 (2012). 10.1007/s10544-011-9602-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong J., Lammertink R., and Wessling M., Lab Chip 6, 1125 (2006). 10.1039/b603275c [DOI] [PubMed] [Google Scholar]
- Peterson D. S., Lab Chip 5, 132 (2005). 10.1039/b405311g [DOI] [PubMed] [Google Scholar]
- Seong G. H. and Crooks R. M., J. Am. Chem. Soc. 124, 13360 (2002). 10.1021/ja020932y [DOI] [PubMed] [Google Scholar]
- Wang Y.-C. and Han J., Lab Chip 8, 392 (2008). 10.1039/b717220f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. J., Kim M. S., Cho S., Park S. S., Kim J. H., Kim Y. K., Kim B. G., and Lee Y. S., Electrophoresis 26, 694 (2005). 10.1002/elps.200410005 [DOI] [PubMed] [Google Scholar]
- Sato K., Yamanaka M., Takahashi H., Tokeshi M., Kimura H., and Kitamori T., Electrophoresis 23, 734 (2002). [DOI] [PubMed] [Google Scholar]
- Tennico Y. H., Hutanu D., Koesdjojo M. T., Bartel C. M., and Remcho V. T., Anal. Chem. 82, 5591 (2010). 10.1021/ac101269u [DOI] [PubMed] [Google Scholar]
- Richter T., Shultz-Lockyear L. L., Oleschuk R. D., Bilitewski U., and Harrison D. J., Sens. Actuators B 81, 369 (2002). 10.1016/S0925-4005(01)00963-7 [DOI] [Google Scholar]
- Sivagnanam V., Song B., Vandevyver C., and Gijs M. A., Anal. Chem. 81, 6509 (2009). 10.1021/ac9009319 [DOI] [PubMed] [Google Scholar]
- Sato K., Tokeshi M., Odake T., Kimura H., Ooi T., Nakao M., and Kitamori T., Anal. Chem. 72, 1144 (2000). 10.1021/ac991151r [DOI] [PubMed] [Google Scholar]
- He M. and Herr A. E., Anal. Chem. 81, 8177 (2009). 10.1021/ac901392u [DOI] [PubMed] [Google Scholar]
- Zhan W., Seong G. H., and Crooks R. M., Anal. Chem. 74, 4647 (2002). 10.1021/ac020340y [DOI] [PubMed] [Google Scholar]
- Park K. H., Park H. G., Kim J.-H., and Seong K. H., Biosens. Bioelectron. 22, 613 (2006). 10.1016/j.bios.2006.01.029 [DOI] [PubMed] [Google Scholar]
- Sung W.-C., Chen H.-H., Makamba H., and Chen S.-H., Anal. Chem. 81, 7967 (2009). 10.1021/ac901138w [DOI] [PubMed] [Google Scholar]
- Sakai-Kato K., Kato M., and Toyo'oka T., Anal. Chem. 75, 388 (2003). 10.1021/ac026240+ [DOI] [PubMed] [Google Scholar]
- Ji J., Zhang Y., Zhou X., Kong J., Tang Y., and Liu B., Anal. Chem. 80, 2457 (2008). 10.1021/ac702218v [DOI] [PubMed] [Google Scholar]
- Hisamoto H., Shimizu Y., Uchiyama K., Tokeshi M., Kikutani Y., Hibara A., and Kitamori T., Anal. Chem. 75, 350 (2003). 10.1021/ac025794+ [DOI] [PubMed] [Google Scholar]
- Sebra R. P., Masters K. S., Cheung C. Y., Bowman C. N., and Anseth K. S., Anal. Chem. 78, 3144 (2006). 10.1021/ac052246y [DOI] [PubMed] [Google Scholar]
- Zhang S., Nature Mater. 3, 7 (2004). 10.1038/nmat1047 [DOI] [PubMed] [Google Scholar]
- Martinez A. W., Phillips S. T., Whitesides G. M., and Carrilho E., Anal. Chem. 82, 3 (2010). 10.1021/ac9013989 [DOI] [PubMed] [Google Scholar]
- Cheng C. M., Martinez A. W., Gong J., Mace C. R., Phillips S. T., Carrilho E., Mirica K. A., and Whitesides G. M., Angew. Chem., Int. Ed. 49, 4771 (2010). 10.1002/anie.201001005 [DOI] [PubMed] [Google Scholar]
- Osborn J. L., Lutz B., Fu E., Kauffman P., Stevens D. Y., and Yager P., Lab Chip 10, 2659 (2010). 10.1039/c004821f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusmini F., Zhong Z., and Feijen J., Biomacromolecules 8, 1775 (2007). 10.1021/bm061197b [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Sakiyama T., Kumada Y., Imamura K., and Imanaka H., Curr. Proteomics 5, 161 (2008). 10.2174/157016408785909622 [DOI] [Google Scholar]
- Petersen K. E., Proc. IEEE 70, 420 (1982). 10.1109/PROC.1982.12331 [DOI] [Google Scholar]
- Terry S. C., Jerman J. H., and Angell J. B., IEEE Trans. Electron Devices 26, 1880 (1979). 10.1109/T-ED.1979.19791 [DOI] [Google Scholar]
- Li M., Sci. China, Ser. G 55, 2316 (2012). 10.1007/s11433-012-4930-3 [DOI] [Google Scholar]
- Wang Z. H., Meng Y. H., Ying P. Q., Qi C., and Jin G., Electrophoresis 27, 4078 (2006). 10.1002/elps.200500956 [DOI] [PubMed] [Google Scholar]
- Lee L. M., Heimark R. L., Guzman R., Baygents J. C., and Zohar Y., Lab Chip 6, 1080 (2006). 10.1039/b603095e [DOI] [PubMed] [Google Scholar]
- Lee J. A., Hwang S., Kwak J., Park S. I., Lee S. S., and Lee K.-C., Sens. Actuators B 129, 372 (2008). 10.1016/j.snb.2007.08.034 [DOI] [Google Scholar]
- Iles A., Oki A., and Pamme N., Microfluid. Nanofluid. 3, 119 (2007). 10.1007/s10404-006-0101-z [DOI] [Google Scholar]
- Harrison D. J., Manz A., Fan Z., Lüdi H., and Widmer H. M., Anal. Chem. 64, 1926 (1992). 10.1021/ac00041a030 [DOI] [Google Scholar]
- Bousse L., Mouradian S., Minalla A., Yee H., Williams K., and Dubrow R., Anal. Chem. 73, 1207 (2001). 10.1021/ac0012492 [DOI] [PubMed] [Google Scholar]
- Glass N. R., Tjeung R., Chan P., Yeo L. Y., and Friend J. R., Biomicrofluidics 5, 036501 (2011). 10.1063/1.3625605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T., Maerkl S. J., and Quake S. R., Science 298, 580 (2002). 10.1126/science.1076996 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lai H.-H., Bachman M., Sims C. E., Li G., and Allbritton N. L., Anal. Chem. 77, 7539 (2005). 10.1021/ac0509915 [DOI] [PubMed] [Google Scholar]
- Sui G., Wang J., Lee C.-C., Lu W., Lee S. P., Leyton J. V., Wu A. M., and Tseng H.-R., Anal. Chem. 78, 5543 (2006). 10.1021/ac060605z [DOI] [PubMed] [Google Scholar]
- Chin C. D., Linder V., and Sia S. K., Lab Chip 7, 41 (2007). 10.1039/b611455e [DOI] [PubMed] [Google Scholar]
- Chin C. D., Linder V., and Sia S. K., Lab Chip 12, 2118 (2012). 10.1039/c2lc21204h [DOI] [PubMed] [Google Scholar]
- Kagebayashi C., Yamaguchi I., Akinaga A., Kitano H., Yokoyama K., Satomura M., Kurosawa T., Watanabe M., Kawabata T., and Chang W., Anal. Biochem. 388, 306 (2009). 10.1016/j.ab.2009.02.030 [DOI] [PubMed] [Google Scholar]
- He H., Yuan Y., Wang W., Chiou N.-R., Epstein A. J., and Lee L. J., Biomicrofluidics 3, 022401 (2009). 10.1063/1.3116665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson G. T., Bioconjugate Techniques, 2nd ed. (Academic Press, San Diego, 2008). [Google Scholar]
- Wang C., Oleschuk R., Ouchen F., Li J., Thibault P., and Harrison D. J., Rapid Commun. Mass Spectrom. 14, 1377 (2000). [DOI] [PubMed] [Google Scholar]
- Page J., Derango R., and Huang A., Colloids Surf., A 132, 193 (1998). 10.1016/S0927-7757(97)00176-3 [DOI] [Google Scholar]
- Guisán J., Enzyme Microb. Technol. 10, 375 (1988). 10.1016/0141-0229(88)90018-X [DOI] [Google Scholar]
- Drury J. L. and Mooney D. J., Biomaterials 24, 4337 (2003). 10.1016/S0142-9612(03)00340-5 [DOI] [PubMed] [Google Scholar]
- Park J. J., Luo X., Yi H., Valentine T. M., Payne G. F., Bentley W. E., Ghodssi R., and Rubloff G. W., Lab Chip 6, 1315 (2006). 10.1039/b603101c [DOI] [PubMed] [Google Scholar]
- Herr A. E. and Singh A. K., Anal. Chem. 76, 4727 (2004). 10.1021/ac049686u [DOI] [PubMed] [Google Scholar]
- Hou C. and Herr A. E., Analyst 138, 158 (2013). 10.1039/c2an36033k [DOI] [PubMed] [Google Scholar]
- Beebe D. J., Moore J. S., Yu Q., Liu R. H., Kraft M. L., Jo B.-H., and Devadoss C., Proc. Natl. Acad. Sci. U.S.A. 97, 13488 (2000). 10.1073/pnas.250273097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Nam S.-W., Lee N. Y., Kim Y. S., and Park S., Ultramicroscopy 108, 1384 (2008). 10.1016/j.ultramic.2008.04.044 [DOI] [PubMed] [Google Scholar]
- Sommer G. J. and Hatch A. V., Electrophoresis 30, 742 (2009). 10.1002/elps.200800598 [DOI] [PubMed] [Google Scholar]
- Liu Y., Yang D., Yu T., and Jiang X., Electrophoresis 30, 3269 (2009). 10.1002/elps.200900128 [DOI] [PubMed] [Google Scholar]
- Harvey M., Optimization of Nitrocellulose Membrane-Based Immunoassays (Schleicher & Schuell, Keene, 1991). [Google Scholar]
- Li X., Ballerini D. R., and Shen W., Biomicrofluidics 6, 011301 (2012). 10.1063/1.3687398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane A., Dronov R., Hodges A., and Voelcker N. H., Trends Biotechnol. 27, 230 (2009). 10.1016/j.tibtech.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Israelachvili J. N., Intermolecular and Surface Forces, 3rd ed. (Academic Press, Waltham, 2010). [Google Scholar]
- Schreiber G. and Fersht A. R., Nat. Struct. Mol. Biol. 3, 427 (1996). 10.1038/nsb0596-427 [DOI] [PubMed] [Google Scholar]
- Gong P. and Levicky R., Proc. Natl. Acad. Sci. U.S.A. 105, 5301 (2008). 10.1073/pnas.0709416105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson B. S. and Branton D., Science 195, 302 (1977). 10.1126/science.831278 [DOI] [PubMed] [Google Scholar]
- Gershoni J. M. and Palade G. E., Anal. Biochem. 124, 396 (1982). 10.1016/0003-2697(82)90056-2 [DOI] [PubMed] [Google Scholar]
- Zhou H. X., Biopolymers 59, 427 (2001). [DOI] [PubMed] [Google Scholar]
- Franchina J. G., Lackowski W. M., Dermody D. L., Crooks R. M., Bergbreiter D. E., Sirkar K., Russell R. J., and Pishko M. V., Anal. Chem. 71, 3133 (1999). 10.1021/ac981300q [DOI] [PubMed] [Google Scholar]
- Roth C. M. and Lenhoff A. M., Langmuir 9, 962 (1993). 10.1021/la00028a015 [DOI] [Google Scholar]
- Lan Q., Bassi A. S., Zhu J. X. J., and Margaritis A., Chem. Eng. J. 81, 179 (2001). 10.1016/S1385-8947(00)00197-2 [DOI] [Google Scholar]
- Tsougeni K., Petrou P. S., Papageorgiou D. P., Kakabakos S. E., Tserepi A., and Gogolides E., Sens. Actuators B 161, 216 (2012). 10.1016/j.snb.2011.10.022 [DOI] [Google Scholar]
- Sia S. K., Linder V., Parviz B. A., Siegel A., and Whitesides G. M., Angew. Chem., Int. Ed. 43, 498 (2004). 10.1002/anie.200353016 [DOI] [PubMed] [Google Scholar]
- Wang R., Yang Y.-L., Qin M., Wang L.-K., Yu L., Shao B., Qiao M.-Q., Wang C., and Feng X.-Z., Chem. Mater. 19, 3227 (2007). 10.1021/cm070445n [DOI] [Google Scholar]
- Qin M., Wang L.-K., Feng X.-Z., Yang Y.-L., Wang R., Wang C., Yu L., Shao B., and Qiao M.-Q., Langmuir 23, 4465 (2007). 10.1021/la062744h [DOI] [PubMed] [Google Scholar]
- Chiu D. T., Jeon N. L., Huang S., Kane R. S., Wargo C. J., Choi I. S., Ingber D. E., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 97, 2408 (2000). 10.1073/pnas.040562297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Delamarche E., Schmid H., Michel B., Bosshard H. R., and Biebuyck H., Langmuir 14, 2225 (1998). 10.1021/la980037l [DOI] [Google Scholar]
- Delamarche E., Bernard A., Schmid H., Bietsch A., Michel B., and Biebuyck H., J. Am. Chem. Soc. 120, 500 (1998). 10.1021/ja973071f [DOI] [Google Scholar]
- Delamarche E., Bernard A., Schmid H., Michel B., and Biebuyck H., Science 276, 779 (1997). 10.1126/science.276.5313.779 [DOI] [PubMed] [Google Scholar]
- Bernard A., Michel B., and Delamarche E., Anal. Chem. 73, 8 (2001). 10.1021/ac0008845 [DOI] [PubMed] [Google Scholar]
- Foley J., Schmid H., Stutz R., and Delamarche E., Langmuir 21, 11296 (2005). 10.1021/la0518142 [DOI] [PubMed] [Google Scholar]
- Starita-Geribaldi M. and Sudaka P., Bioseparation 1, 111 (1990). [PubMed] [Google Scholar]
- Lu Y., Shi W., Qin J., and Lin B., Anal. Chem. 82, 329 (2010). 10.1021/ac9020193 [DOI] [PubMed] [Google Scholar]
- Handman E. and Jarvis H., J. Immunol. Methods 83, 113 (1985). 10.1016/0022-1759(85)90064-X [DOI] [PubMed] [Google Scholar]
- Peters E. C., Svec F., and Fréchet J., Adv. Mater. 11, 1169 (1999). [DOI] [Google Scholar]
- Bhatia R. B., Brinker C. J., Gupta A. K., and Singh A. K., Chem. Mater. 12, 2434 (2000). 10.1021/cm000260f [DOI] [Google Scholar]
- Smith C. L., Milea J. S., and Nguyen G. H., in Immobilisation of DNA on Chips II (Springer, 2005), p. 63. [Google Scholar]
- Knecht S., Ricklin D., Eberle A. N., and Ernst B., J. Mol. Recogn. 22, 270 (2009). 10.1002/jmr.941 [DOI] [PubMed] [Google Scholar]
- Wacker R. and Niemeyer C. M., ChemBioChem 5, 453 (2004). 10.1002/cbic.200300788 [DOI] [PubMed] [Google Scholar]
- Ramsay G., Nat. Biotechnol. 16, 40 (1998). 10.1038/nbt0198-40 [DOI] [PubMed] [Google Scholar]
- Larsson A., Karlsson-Parra A., and Sjöquist J., Clin. Chem. 37, 411 (1991). [PubMed] [Google Scholar]
- Bunka D. H. and Stockley P. G., Nat. Rev. Microbiol. 4, 588 (2006). 10.1038/nrmicro1458 [DOI] [PubMed] [Google Scholar]
- Xu Y., Yang X., and Wang E., Anal. Chim. Acta 683, 12 (2010). 10.1016/j.aca.2010.10.007 [DOI] [PubMed] [Google Scholar]
- Mosing R. K. and Bowser M. T., J. Sep. Sci. 30, 1420 (2007). 10.1002/jssc.200600483 [DOI] [PubMed] [Google Scholar]
- Thierry B., Jasieniak M., de Smet L. C., Vasilev K., and Griesser H. J., Langmuir 24, 10187 (2008). 10.1021/la801140u [DOI] [PubMed] [Google Scholar]
- MacBeath G. and Schreiber S. L., Science 289, 1760 (2000). 10.1126/science.289.5485.1760 [DOI] [PubMed] [Google Scholar]
- Xiong L. and Regnier F. E., J. Chromatogr. A 924, 165 (2001). 10.1016/S0021-9673(01)00904-9 [DOI] [PubMed] [Google Scholar]
- Yu L., Li C. M., Zhou Q., and Luong J. H., Bioconjugate Chem. 18, 281 (2007). 10.1021/bc060108p [DOI] [PubMed] [Google Scholar]
- Belder D., Deege A., Husmann H., Kohler F., and Ludwig M., Electrophoresis 22, 3813 (2001). [DOI] [PubMed] [Google Scholar]
- Didar T. F., Foudeh A. M., and Tabrizian M., Anal. Chem. 84, 1012 (2012). 10.1021/ac2025877 [DOI] [PubMed] [Google Scholar]
- Hu M., Yan J., He Y., Lu H., Weng L., Song S., Fan C., and Wang L., ACS Nano 4, 488 (2010). 10.1021/nn901404h [DOI] [PubMed] [Google Scholar]
- He Y., Lu H.-T., Sai L.-M., Lai W.-Y., Fan Q.-L., Wang L.-H., and Huang W., J. Phys. Chem. B 110, 13370 (2006). 10.1021/jp057498h [DOI] [PubMed] [Google Scholar]
- Mao X., Luo Y., Dai Z., Wang K., Du Y., and Lin B., Anal. Chem. 76, 6941 (2004). 10.1021/ac049270g [DOI] [PubMed] [Google Scholar]
- Thierry B., Kurkuri M., Shi J. Y., Lwin L. E. M. P., and Palms D., Biomicrofluidics 4, 032205 (2010). 10.1063/1.3480573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Svec F., and Fréchet J. M., Biotechnol. Bioeng. 62, 30 (1999). [DOI] [PubMed] [Google Scholar]
- Ericson C., Liao J.-L., i. Nakazato K., and Hjertén S., J. Chromatogr. A 767, 33 (1997). 10.1016/S0021-9673(97)00008-3 [DOI] [Google Scholar]
- Konno T., Watanabe J., and Ishihara K., Biomacromolecules 5, 342 (2004). 10.1021/bm034356p [DOI] [PubMed] [Google Scholar]
- Chen T., Embree H. D., Wu L. Q., and Payne G. F., Biopolymers 64, 292 (2002). 10.1002/bip.10196 [DOI] [PubMed] [Google Scholar]
- Luo X., Lewandowski A. T., Yi H., Payne G. F., Ghodssi R., Bentley W. E., and Rubloff G. W., Lab Chip 8, 420 (2008). 10.1039/b713756g [DOI] [PubMed] [Google Scholar]
- Niemeyer C. M., Sano T., Smith C. L., and Cantor C. R., Nucleic Acids Res. 22, 5530 (1994). 10.1093/nar/22.25.5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes L. K., Chan H. K. C., Lau B., Kumacheva E., and Wheeler A. R., Biomaterials 31, 315 (2010). 10.1016/j.biomaterials.2009.09.040 [DOI] [PubMed] [Google Scholar]
- Eteshola E. and Leckband D., Sens. Actuators B 72, 129 (2001). 10.1016/S0925-4005(00)00640-7 [DOI] [Google Scholar]
- Stults N., Lin P., Hardy M., Lee Y., Uchida Y., Tsukada Y., and Sugimori T., Anal. Biochem. 135, 392 (1983). 10.1016/0003-2697(83)90701-7 [DOI] [PubMed] [Google Scholar]
- Linder V., Verpoorte E., Thormann W., de Rooij N. F., and Sigrist H., Anal. Chem. 73, 4181 (2001). 10.1021/ac010421e [DOI] [PubMed] [Google Scholar]
- Mäntymaa A., Halme J., Välimaa L., and Kallio P., Biomicrofluidics 5, 046504 (2011). 10.1063/1.3668261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikotila J., Välimaa L., Takalo H., and Pettersson K., Colloids Surf., B 70, 271 (2009). 10.1016/j.colsurfb.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Dodge A., Fluri K., Verpoorte E., and de Rooij N. F., Anal. Chem. 73, 3400 (2001). 10.1021/ac0015366 [DOI] [PubMed] [Google Scholar]
- Kaji H., Hashimoto M., and Nishizawa M., Anal. Chem. 78, 5469 (2006). 10.1021/ac060304p [DOI] [PubMed] [Google Scholar]
- Holden M. A. and Cremer P. S., J. Am. Chem. Soc. 125, 8074 (2003). 10.1021/ja035390e [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Kaji H., and Nishizawa M., Biosens. Bioelectron. 24, 2892 (2009). 10.1016/j.bios.2009.02.025 [DOI] [PubMed] [Google Scholar]
- Malmstadt N., Yager P., Hoffman A. S., and Stayton P. S., Anal. Chem. 75, 2943 (2003). 10.1021/ac034274r [DOI] [PubMed] [Google Scholar]
- Qavi A. J., Washburn A. L., Byeon J.-Y., and Bailey R. C., Anal. Bioanal. Chem. 394, 121 (2009). 10.1007/s00216-009-2637-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G., Nat. Genet. 32, 526 (2002). 10.1038/ng1037 [DOI] [PubMed] [Google Scholar]
- Holden M. A., Jung S.-Y., and Cremer P. S., Anal. Chem. 76, 1838 (2004). 10.1021/ac035234q [DOI] [PubMed] [Google Scholar]
- Priest C., Biomicrofluidics 4, 032206 (2010). 10.1063/1.3493643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill R. A., Bhamidipati A., Bi X., Deb-Basu D., Cahill L., Ferrante J., Gentalen E., Glazer M., Gossett J., Hacker K., Kirby C., Knittle J., Loder R., Mastroieni C., MacLaren M., Mills T., Nguyen U., Parker N., Rice A., Roach D., Suich D., Voehringer D., Voss K., Yang J., Yang T., and Vander Horn P. B., Proc. Natl. Acad. Sci. U.S.A. 103, 16153 (2006). 10.1073/pnas.0607973103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D. L., Manginell R. P., Samara M. A., Kim B.-I., and Bunker B. C., Science 301, 352 (2003). 10.1126/science.1080759 [DOI] [PubMed] [Google Scholar]
- Kingsmore S. F., Nat. Rev. Drug Discovery 5, 310 (2006). 10.1038/nrd2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeva S. I., Lee J.-S., Smith J. E., Rosen S. T., and Mirkin C. A., J. Am. Chem. Soc. 128, 8378 (2006). 10.1021/ja0613106 [DOI] [PubMed] [Google Scholar]
- Boozer C., Ladd J., Chen S., and Jiang S., Anal. Chem. 78, 1515 (2006). 10.1021/ac051923l [DOI] [PubMed] [Google Scholar]
- Birger Anspach F., J. Chromatogr. A 672, 35 (1994). 10.1016/0021-9673(94)80592-X [DOI] [Google Scholar]
- Akgöl S. and Denizli A., J. Mol. Catal. B: Enzyme 28, 7 (2004). 10.1016/j.molcatb.2003.12.010 [DOI] [Google Scholar]
- Sung D., Shin D. H., and Jon S., Biosens. Bioelectron. 26, 3967 (2011). 10.1016/j.bios.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Sung D., Park S., and Jon S., Langmuir 25, 11289 (2009). 10.1021/la902784g [DOI] [PubMed] [Google Scholar]
- Sakai-Kato K., Kato M., Ishihara K., and Toyo'oka T., Lab Chip 4, 4 (2004). 10.1039/b310932a [DOI] [PubMed] [Google Scholar]