Abstract
OBJECTIVE
This study assessed the ability to distinguish between type 1 diabetes–affected individuals and their unaffected relatives using HLA and single nucleotide polymorphism (SNP) genotypes.
RESEARCH DESIGN AND METHODS
Eight models, ranging from only the high-risk DR3/DR4 genotype to all significantly associated HLA genotypes and two SNPs mapping to the cytotoxic T-cell–associated antigen-4 gene (CTLA4) and insulin (INS) genes, were fitted to high-resolution class I and class II HLA genotyping data for patients from the Type 1 Diabetes Genetics Consortium collection. Pairs of affected individuals and their unaffected siblings were divided into a “discovery” (n = 1,015 pairs) and a “validation” set (n = 318 pairs). The discriminating performance of various combinations of genetic information was estimated using receiver operating characteristic (ROC) curve analysis.
RESULTS
The use of only the presence or absence of the high-risk DR3/DR4 genotype achieved very modest discriminating ability, yielding an area under the curve (AUC) of 0.62 in the discovery set and 0.59 in the validation set. The full model—which included HLA information from the class II loci DPB1, DRB1, and DQB1; selected alleles from HLA class I loci A and B; and SNPs from the CTLA4 and INS genes—increased the AUC to 0.74 in the discovery set and to 0.71 in the validation set. A cost-effective alternative is proposed, using genotype information equivalent to typing four SNPs (DR3, DR4-DQB1*03:02, CTLA-4, and INS), which achieved an AUC of 0.72 in the discovery set and 0.69 in the validation set.
CONCLUSIONS
Genotyping data sufficient to tag DR3, DR4-DQB1*03:02, CTLA4, and INS were shown to distinguish between subjects with type 1 diabetes and their unaffected siblings adequately to achieve clinically utility to identify children in multiplex families to be considered for early intervention.
Type 1 diabetes is an autoimmune disease that gradually destroys insulin-producing β-cells. Immune interventions for type 1 diabetes are currently being attempted in prevention trials (1,2). Prevention trials assess the risk of type 1 diabetes in relatives based on circulating autoantibodies and evaluation of β-cell function using various methods. Although of clinical value, such dynamic biomarkers are relatively expensive to assay and are sample dependent, so increasing attention is focused on the use of genetic data to predict the risk of disease in asymptomatic individuals (3).
Type 1 diabetes is one of the most widely studied complex genetic disorders, with genes in the HLA region reported to account for ∼40–50% of the familial aggregation (4). The major genetic contribution from within the HLA region is attributable to the class II haplotypes encoded by the DRB1, DQA1, and DQB1 loci (5), with independent risk also demonstrated at HLA-DPB1 and HLA-A, -B, and -C (6,7).
If HLA genotyping were replaced by minimal tag–single nucleotide polymorphism (SNP) genotyping, such a test would represent a cost-effective predictive value that could be applied from birth. In this study, we assess for the first time the ability to distinguish type 1 diabetes status in siblings (sibs) from families with type 1 diabetes based only on genetic information, using full high-resolution HLA genotyping for all classical loci and two SNP markers for the CTLA4 and INS genes. If combined with biomarkers for β-cell function, this test is expected to have utility in the selection of appropriate subjects for prevention trials.
RESEARCH DESIGN AND METHODS
Study subjects
The Type 1 Diabetes Genetics Consortium (T1DGC) is a large, worldwide collaborative study aimed at collecting and genotyping families with type 1 diabetes in a highly standardized fashion from multiple populations to aid in the search for additional type 1 diabetes genes within and outside the HLA region (8). An individual was designated as affected if he or she had documented type 1 diabetes with onset at age ≤37 years, had used insulin within 6 months of diagnosis, and had no concomitant disease or disorder associated with diabetes. Most patients came from families where more than one child was affected, and genotyping and clinical data were also collected for parents and unaffected sibs. High-resolution HLA genotyping was performed at eight classical HLA loci by four genotyping centers using standardized typing protocols, reagents, and quality-control procedures (9). In addition, the T1DGC HLA genotyping protocol, which used sequence-specific oligonucleotide-based linear arrays, also included assays for two type 1 diabetes–associated SNPs: the cytotoxic T-cell–associated antigen-4 gene (CTLA-4) +49 (A/G) (rs3087243) and insulin (INS) −23HphI (A/T) (rs689) (10). These two SNPs were selected for the receiver operating characteristic (ROC) curve analyses because their concomitant genotyping with DRB1 in the T1DGC genotyping protocol assured that genotypes were available for all samples.
We selected data from families in the T1DGC final dataset with the following inclusion criteria: European ancestry, at least two affected sibs, and at least one unaffected sib. One affected sib and one unaffected sib were selected from each family (n = 1,015) to create the discovery set of affected/unaffected pairs; therefore, only families with DNA for unaffected sibs were used. In addition, 318 of the 1,015 pedigrees contained at least one additional unaffected sib. For these families, the second unaffected sib was paired with the second affected sib (not used for the discovery set), allowing the generation of a second set of 318 pairs of affected and unaffected sibs. These nonoverlapping pairs were used for validation of the risk models. For families with two affected/unaffected pairs, the pairs were randomly assigned to the discovery or validation sets. The discovery set consisted of 50.9% females, the mean age at onset of the affected sibs was 9.9 years (SD 7.8), and the mean age at recruitment was 22.2 years (13.2) for the unaffected sib and 22.7 years (12.7) for the affected sib. The validation set included 50.5% females, the mean age of onset was 9.3 years (7.05), and the mean age at recruitment was 22.6 years (14.2) for unaffected sibs and 23.3 years (14.1) for affected sibs.
Statistical methods
Allele selection and genotype coding.
The HLA alleles and genotypes that were selected for encoding type 1 diabetes risk were selected based on the literature reports for consistent associations with type 1 diabetes. Individuals were classified as 0 or 1, depending on the carriage or not of a given genotype as follows:
DR3/DR4 only: Each individual was coded for the presence or absence of DR3/4 where DR3 = DRB1*03:01-DQB1*02:01 and DR4 = DRB1*04:01/2/4/5/8/13-DQB1*03:02 or *03:04 or *02:01.
DRB1-DQB1 risk genotypes: For each individual, an indicator variable (taking values 0 or 1) was coded for if they carried one of the following genotypes: DR3/DR4, DR3/DR3, DR4/DR4, DR4/DRx, or DR3/DRx where DR3 = DRB1*03:01-DQB1*02:01, DR4 = DRB1*04:01/2/4/5/8/13-DQB1*03:02 or 03:04 or 02:01, and x is any other haplotype, including DRB1*04:03 or any DRB1*04-carrying haplotypes that include DQB1*03:01.
DRB1-DQB1 protective genotypes: Individuals were coded by variables (0 or 1) depending on carriage of one or two copies of DRB1*15:01-DQB1*06:02 (DR2_DQ6), DRB1*14:01-DQB1*05:03 (DR14), and DRB1*07:01-DQB1*03:03 (DR7_DQ9)
DPB1: Carriage of alleles DPB1*04:02 (DP_0402) (protective) and of DPB1*03:01 (DP3) (susceptible) was included.
Class I HLA-A and HLA-B: Carriage of risk alleles B*39:06 and A*24:02 was included.
Polymorphisms in INS (rs689) and CTLA4 (rs3087243): These SNPs were encoded as 0, 1, or 2 copies of the minor allele.
ROC analysis.
The discriminating power of a given diagnostic is usually summarized by a ROC curve. In this type of analysis, subjects are ranked in descending order of their predicted risk, and the cumulative proportion of subjects who develop disease (case subjects) is plotted against the corresponding cumulative proportion of the population; that is, the sensitivity (true-positive fraction) is plotted in the y-axis versus 1-specificity (the false-negative fraction) in the x-axis (11). A perfect diagnostic would be represented by a line that starts at the origin and travels up the y-axis to 1 and then across the origin to an x-axis value of 1, thus having a total area under the curve (AUC) of 1. A test with AUC of 0.5, which would be represented by a diagonal line from the x, y origin to 1 on both axes, has zero diagnostic value (11). A risk score was calculated for each individual using the logit equation:Logit = ln(p/1 − p) = α + β1X1 + … + βiXi where p is the probability of the outcome (type 1 diabetes), α is the constant, and β is the natural logarithm value of the odds ratio for a specific predictor Xi. In this case, Xi is given by the presence or absence of specific genotypes or by the copies of alleles.
The discrimination ability of the models was examined in a test (n = 1,015 affected/unaffected sib pairs) and validation set (n = 318 pairs) using the PredictABEL package for R software (http://cran.r-project.org/web/packages/PredictABEL/index.html). All eight models were applied to the “discovery” and “validation” sets.
RESULTS
A logistic regression on type 1 disease status was fitted in the discovery set of 1,015 pairs of affected/unaffected sibs. Details for the eight models obtained are included in the Supplementary Data.
The AUC and the corresponding 95% CIs for each of these models were obtained in the discovery and validation sets (Table 1). To put these results in context, AUC values from 0.5 to 0.7 for a diagnostic/prognostic test represent low accuracy, values from 0.7 to 0.9 represent tests that are useful for some purposes, and values >0.9 represent diagnostic/prognostic tests with high accuracy (12).
Table 1.
AUC estimate (95% CI) for the performance of genetic markers in predicting type 1 diabetes using data from affected/unaffected sib pairs from the T1DGC
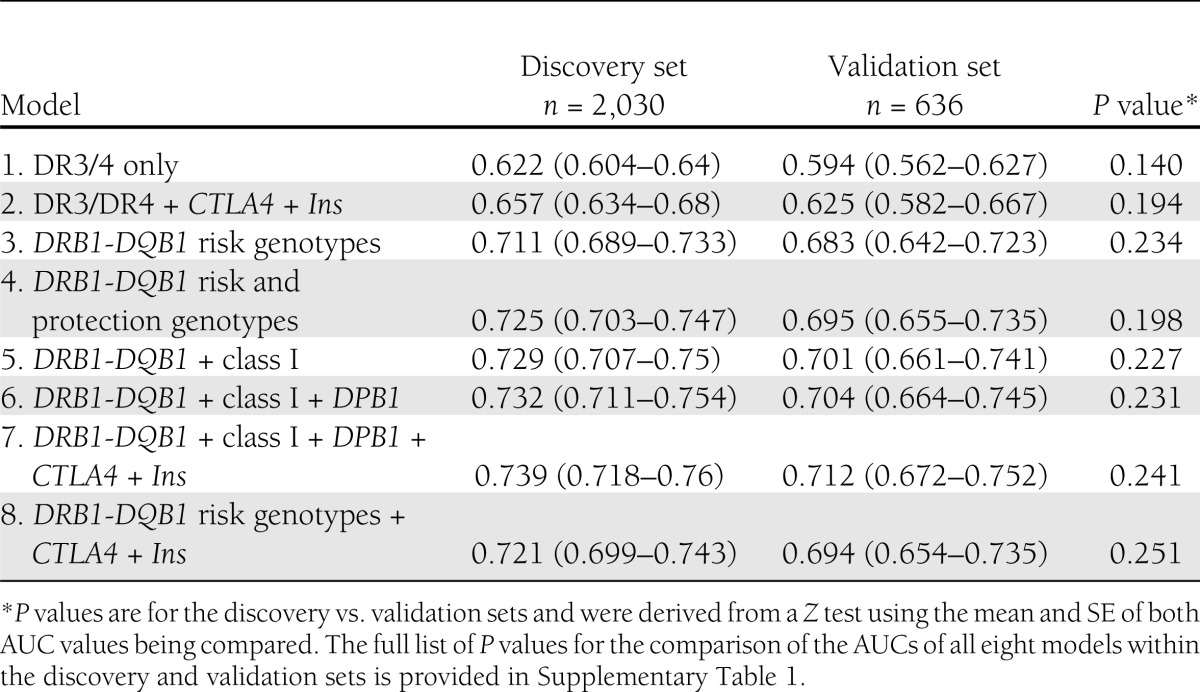
The use of only the presence/absence of the high-risk DR3/DR4 genotype (model 1) achieves very modest discriminating results, yielding an AUC of 0.62 (95% CI 0.60–0.64) in the discovery set and 0.59 (0.56–0.63) in the validation set. Increasing levels of HLA risk (models 3, 4, and 5) and susceptibility genotypes progressively add to the AUC obtained to reach an AUC of 0.73 (0.71–0.75) in the discovery set and 0.70 (0.66–0.74) in the validation set for DRB1-DQB1 risk and protection genotypes, HLA class I, and HLA*DPB1 (model 6; Table 1). Full HLA genotyping plus CTLA-4 and INS provided the highest discriminating model; the AUC was 0.74 (0.72–0.76) in the discovery set and 0.71 (0.67–0.75) in the validation set (model 7).
Notably, no significant difference in the AUC values achieved is observed among any of the models 3 to 7 (see Supplementary Table 1 for a full list of P values), but the AUC achieved by models 1 and 2 is significantly lower than that achieved by models 3 to 7, which include full modeling of the DR-DQ T1D risk, and not just the DR3/DR4 genotype, in both the discovery and validation sets, reaching P values as low as P < 2.2 × 10−16 for the comparison of the full model (model 7) with the only DR3/DR4 model (model 1) in the discovery set (Supplementary Table 1)
We found that minimal genotyping of DR3 and DR4-DQB1*03:02 genotypes plus CTLA-4 and INS provided discriminating values of AUC of 0.72 in the discovery set and 0.69 in the validation set (model 8). This model is only marginally less informative, but not significantly so, than the full model of DRB1-DQB1 risk and protection genotypes + HLA class I + DPB1 + CTLA-4 + INS (Table 1) but requires considerably less genotype data.
CONCLUSIONS
In this study, we used the T1DGC data to investigate the discriminating performance of high-resolution HLA susceptible and protective genotypes, in combination with two additional validated genetic variants, to distinguish between type 1 diabetes patients and their affected sibs. We find that the use of genotypic information summarizing HLA-DRB1-DQB1, DPB1, HLA-A, and HLA-B alleles achieves discriminating levels of classification between affected individuals and their sibs that can be of clinical utility (AUC >0.70). Because full HLA genotyping is currently too costly for large prevention trials, we have investigated the ROCs of a model that uses only information to derive the presence of combinations of the high-risk DR3 and DR4-DQB1*03:02 haplotypes in combination with the type 1 diabetes–associated SNPs for CTLA-4 and INS that were typed concurrently with the HLA. Simplified SNP-based genotyping represents a plausible, cost-effective alternative to full HLA genotyping for this model. For example, a rapid test to identify DR3/DR4 subjects analyzed two SNPs, specifically, rs2040410 and rs7454108, which are associated with DR3-DQB1*02:01 and DR4-DQB1*03:02. These SNPs were analyzed in samples from 143 HLA-typed children who participated in the Diabetes Autoimmunity Study of the Young (DAISY) and in 5,019 subjects from the T1DGC. In the T1DGC samples, the two SNPs had a sensitivity of 98.5% (1,173 of 1,191) and a specificity of 99.7% for DR3⁄4-DQB1*03:02. In the DAISY population, the test was 100% sensitive and 100% specific (1).
Model 8 represents a minimal genotyping model of the presence or absence of the DR3- and DR4-DQB1*03:02–carrying genotypes, with appropriate weights for each genotype, together with the CTLA-4 and INS SNPs. Model 8 achieved a substantial level of prediction (AUC = 0.72 in the discovery set and 0.69 in the validation set), comparable to model 7, which included full HLA genotyping data for multiple loci. A test such as model 8 would require genotyping of only four to six SNPs and might be further improved by the addition of SNP genotypes from other non-HLA, type 1 diabetes–associated loci (e.g., PTPN22, IL2RA, or IFIH1) (13) to provide a simple diagnostic test from a single sample that may be collected at birth.
The diagnostic may then be further improved by the use of dynamic biomarkers, such as autoantibodies and evaluation of β-cell function, to achieve optimal prediction. Indeed, a recent independent study has shown that among children who carry a high-risk DR-DQ genotype, the greatest diabetes discrimination was obtained by the sum of risk alleles for eight genes (IFIH1, CTLA4, PTPN22, IL18RAP, SH2B3, KIAA0350, COBL, and ERBB3) for progression from islet autoimmunity to type 1 diabetes (14).
Strengths of the current study include the large sample size, the use of internal validation, and the first-time use of high-resolution HLA genotyping. Another important strength is the use of sibs of affected individuals matched for environmental, social, and lifestyle factors that may influence type 1 diabetes risk. Moreover, sibs of patients with type 1 diabetes represent a high-risk population that is commonly targeted by prevention trials.
The study has some limitations. The data used are cross-sectional rather than longitudinal, so the current study does not assess a risk for onset of diabetes over a given period of time. These data apply only to populations of European origin, and the discriminating value of HLA genotypes may not necessarily apply to other ethnic groups. Importantly, the data are derived from multiplex families where the genetic contribution to type 1 diabetes is particularly high and which are, therefore, primary targets for enrollment for prevention trials.
With these caveats in mind, the current study represents the first effort to systematically quantify the discriminating value of full HLA genotyping. In combination with autoantibodies, glucose, and C-peptide, data generated by this method may provide a high-accuracy prognostic tool for risk of type 1 diabetes in a high-risk population.
Acknowledgments
This work was supported by R01 DK-61722 (J.A.N., A.M.V.). This research uses resources provided by the T1DGC, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development, and JDRF and is supported by U01 DK-062418.
No potential conflicts of interest relevant to this article were reported.
A.M.V. carried out statistical analyses, researched data, and wrote the manuscript. M.D.V. and J.A.N. researched data, contributed to discussion, and reviewed and edited the manuscript. H.A.E. contributed to discussion and reviewed and edited the manuscript. A.M.V. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc12-2284/-/DC1.
References
- 1.Zhang L, Eisenbarth GS. Prediction and prevention of type 1 diabetes mellitus. J Diabetes 2011;3:48–57 [DOI] [PubMed] [Google Scholar]
- 2.Gupta S. Immunotherapies in diabetes mellitus type 1. Med Clin North Am 2012;96:621–634 [DOI] [PubMed] [Google Scholar]
- 3.Janssens AC, Ioannidis JP, van Duijn CM, Little J, Khoury MJ, GRIPS Group Strengthening the reporting of Genetic Risk Prediction Studies: the GRIPS statement. Genet Med 2011;13:453–456 [DOI] [PubMed] [Google Scholar]
- 4.Erlich H, Valdes AM, Noble J, et al. Type 1 Diabetes Genetics Consortium HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble JA, Valdes AM. Genetics of the HLA region in the prediction of type 1 diabetes. Curr Diab Rep 2011;11:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varney MD, Valdes AM, Carlson JA, et al. Type 1 Diabetes Genetics Consortium HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with type 1 diabetes: analysis of the type 1 diabetes genetics consortium families. Diabetes 2010;59:2055–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noble JA, Valdes AM, Varney MD, et al. Type 1 Diabetes Genetics Consortium HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes 2010;59:2972–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett JC, Clayton DG, Concannon P, et al. Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mychaleckyj JC, Noble JA, Moonsamy PV, et al. T1DGC HLA genotyping in the international Type 1 Diabetes Genetics Consortium. Clin Trials 2010;7(Suppl):S75–S87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pociot F, Akolkar B, Concannon P, et al. Genetics of type 1 diabetes: what’s next? Diabetes 2010;59:1561–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altman DG, Bland JM. Diagnostic tests 3: receiver operating characteristic plots. BMJ 1994;309:188 [DOI] [PMC free article] [PubMed]
- 12.Wians FH. Clinical laboratory tests: which, why, and what do the results mean? Lab Med 2009;40:105–113 [Google Scholar]
- 13.Julier C, Akolkar B, Concannon P, Morahan G, Nierras C, Pugliese A, Type I Diabetes Genetics Consortium The Type I Diabetes Genetics Consortium ‘Rapid Response’ family-based candidate gene study: strategy, genes selection, and main outcome. Genes Immun 2009;10(Suppl. 1):S121–S127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler C, Krumsiek J, Lempainen J, et al. A strategy for combining minor genetic susceptibility genes to improve prediction of disease in type 1 diabetes. Genes Immun 2012;13:549–555 [DOI] [PubMed] [Google Scholar]


