Abstract
Baculovirus-expressed recombinant Sir3p (rSir3p) has been purified to near homogeneity, and its binding to naked DNA, mononucleosomes, and nucleosomal arrays has been characterized in vitro. At stoichiometric levels rSir3p interacts with intact nucleosomal arrays, mononucleosomes, and naked DNA, as evidenced by formation of supershifted species on native agarose gels. Proteolytic removal of the core histone tail domains inhibits but does not completely abolish rSir3p binding to nucleosomal arrays. The linker DNA in the supershifted complexes remains freely accessible to restriction endonuclease digestion, suggesting that both the tail domains and nucleosomal DNA contribute to rSir3p–chromatin interactions. Together these data indicate that rSir3p cross-links individual nucleosomal arrays into supramolecular assemblies whose physical properties transcend those of typical 10-nm and 30-nm fibers. Based on these data we hypothesize that Sir3p functions, at least in part, by mediating reorganization of the canonical chromatin fiber into functionally specialized higher order chromosomal domains.
Keywords: silencing, nucleosome
Silencing in yeast is a form of transcriptional repression requiring the assembly of a specific heritable chromatin structure analogous to heterochromatin in metazoans (1, 2). Silenced chromatin is transcriptionally repressed throughout the cell cycle (3), distinguishing it from the bulk chromatin encompassing genes that are regulated or constitutively expressed. In the yeast Saccharomyces cerevisiae, the silent information regulator proteins, Sir2p, Sir3p, and Sir4p, are specialized chromatin-associated proteins required for silencing the cryptic mating-type loci and telomeres (2, 4). Genetic and molecular studies have established that Sir3p and Sir4p both are associated with silenced chromatin in vivo, at least in part through interactions with the histone H4 N-termini (5, 6). Furthermore, Sir3p interacts with the N-terminal tails of histone H4 and histone H3 in vitro (7). These data provide evidence supporting a key role for chromatin–Sir protein complexes in establishing and/or maintaining the silenced state. The recent finding that Sir2p is a unique NAD-dependent histone deacetylase (8, 9) further strengthens the potential functional importance of chromatin in transcriptional silencing (see ref. 10).
Although chromatin is a key participant in Sir-mediated transcriptional silencing, the molecular basis of its function is unknown. The unfolded “10-nm” chromatin fiber may simply serve as an anchor for the assembly of a repressive complex of silencing proteins, without any involvement of higher order chromatin structure (e.g., see ref. 7). Another possibility is that proteins such as Sir3p act analogously to the linker histones of higher organisms to stabilize the extensively folded, transcriptionally repressive “30-nm” fiber. Alternatively, by binding to the core histone tail domains (11), proteins such as Sir3p may cause large-scale reorganization of the chromatin fiber into repressive “suprastructures” whose global structural properties transcend those of typical 10-nm and 30-nm fibers. Although each of these models involves a fundamentally different molecular function of Sir3p, they are all compatible with the available genetic and biochemical evidence. Consequently, distinguishing between these possible modes of Sir3p action requires direct characterization of the structural effects caused by Sir3p binding to chromatin.
In this work we have purified baculovirus-expressed recombinant Sir3p (rSir3p) to >90% homogeneity and performed biochemical studies of rSir3p–chromatin interactions in vitro. Our results demonstrate that Sir3p binding to nucleosomal arrays, mononucleosomes, and naked DNA at molar ratios near unity produced a series of very large supershifted complexes on agarose gels. Binding of rSir3p to nucleosomal arrays was significantly inhibited by proteolytic removal of the core histone tail domains but was not abolished completely. The linker DNA in the supershifted complexes remained accessible to restriction endonuclease cleavage after complex formation. Taken together, these results indicate that rSir3p “cross-links” individual nucleosomal arrays into unique supramolecular complexes whose structural properties are fundamentally different from typical 10-nm and 30-nm chromatin fibers. Based on these data we hypothesize that Sir3p functions, at least in part, by mediating global reorganization of the canonical chromatin fiber into specialized higher order chromosomal domains.
Materials and Methods
Sir3p Purification.
Sf9 insect cells were infected with a recombinant baculovirus expressing rSir3p fused to six tandem C-terminal histidines. Forty hours after infection, a nuclear extract was prepared from harvested cells (12). The Sir3p was further purified by ammonium sulfate fractionation, Q-Sepharose chromatography, and Ni affinity chromatography. The purified protein was dialyzed into buffer H as described (12). rSir3p concentration was determined by comparison with known amounts of BSA standards after SDS/PAGE and by the Bradford assay (Bio-Rad). The two methods yielded identical results.
Preparation of Naked DNA, Mononucleosomes, and Nucleosomal Arrays.
The 208–12 DNA template containing 12 tandem 208-bp repeats of Lytechinus 5S rDNA was purified from the pPol-I 208–12 plasmid (13), as described (14). The 208–1 DNA fragment was generated by digestion of the 208–12 fragment with 10 units of AvaI/μg of DNA for 60 min at 37°C. The pXP10 plasmid, containing a single Xenopus 5S gene cloned into pUC18 (15), was linearized by digestion with 10 units of EcoRI/μg of DNA for 60 min at 37°C.
Chicken erythrocyte histone octamers were purified as described (14). Tailless histone octamers were generated by limited digestion with immobilized trypsin and purified by hydroxylapatite chromatography as described (16). The extent of digestion was assessed by electrophoresis on a 18% SDS/PAGE gel. Only those preparations consisting of the P1-P5 peptides identified (17, 18) were used. After purification, histone octamers were stored at 4°C in the presence of 20 μg/ml each of aprotinin and leupeptin.
Nucleosomal arrays and mononucleosomes were reconstituted from either intact or trypsinized core histone octamers and their respective DNAs at a ratio of 1.1 mol octamer/mole 208-bp DNA by salt dialysis as described (19). The final dialysis step was against buffer containing 10 mM Tris⋅HCl, 0.25 mM EDTA, and 2.5 mM NaCl at pH 7.8 (TE). The integrity of the reconstitutes was assayed by sedimentation velocity in TE buffer as described (19).
Sir3p Binding Studies.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21 (DE3, pLys E) cells and purified by mixing bacterial lysates with 100 μl of glutathione beads (Amersham Pharmacia) in 25 mM Hepes KOH (pH 7.6), 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, and 0.05% Nonidet P-40 + 0.2 M KCl for 60 min at 4°C. GST fusion products were eluted from the beads with the use of 50 mM Tris⋅HCl (pH 8.0) and 10 mM glutathione (20). The purity of the GST-tail fusions was monitored by SDS/PAGE and Coomassie staining. For fusion protein binding studies, 10 μg of GST or GST fusion protein bound to 100 μl of GST beads in TGD (20 mM Tris, pH 8.0/150 mM NaCl/1 mM DTT/0.1% Triton X-100) were prepared as described (7). Sir3p was added at a ratio of 3 mol Sir3p/mol GST or GST fusion and incubated with rotation at 4°C for 30 min. Beads were washed five times with 1 ml of TGD, and proteins were eluted in SDS/PAGE sample buffer. One hundred percent of the eluate and 50% of the input material were resolved by SDS/PAGE, and the gel was stained with Coomassie blue.
For studies of Sir3p binding to nucleosomal substrates and DNA, 200 ng of 208–12 nucleosomal arrays, mononucleosomes, or pXP10 chromatin was incubated with increasing molar ratios of Sir3p to 208 bp (rSir3p) at room temperature for 30 min. Incubations were performed in buffer containing 10 mM Tris, 0.25 mM EDTA, 7.5 mM NaCl, and 1.75 mM MgCl2. Glycerol was subsequently added to a final percentage of 10%, and samples were electrophoresed at 8 V/cm for 2 h on 1% agarose gel buffered with 1× TBE (30 mM Tris borate/2 mM EDTA, pH 8.0). Gels were stained with SYBR green (Molecular Probes) according to the manufacturer's specifications. For accessibility studies, aliquots of the same samples were digested with 10 units of EcoRI/μg of DNA at 37°C for 90 min. Digestion was stopped by adding EDTA to a final concentration of 15 mM. The native nucleosomal arrays and nucleosomal arrays plus Sir3p complexes were loaded onto 1% agarose gels (1× TBE) and electrophoresed at 8 V/cm for 2 h. To determine the DNA composition of the samples, aliquots were treated with 10 μg of proteinase K at 50°C for 60 min, phenol/chloroform extracted, and ethanol precipitated. The DNA was resuspended in 10 μl of dye solution and electrophoresed on a 1% agarose gel (1× TBE) at 8 V/cm for 2 h.
Results
Purification and Characterization of rSir3p.
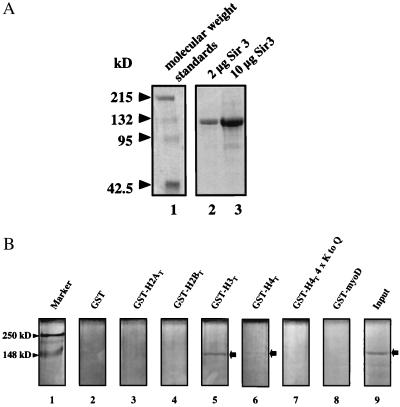
Recombinant full-length Sir3p (rSir3p) was purified to near-homogeneity from Baculovirus-infected insect cells with the use of a combination of ion exchange and affinity chromatography. SDS/PAGE of the purified protein is shown in Fig. 1A. Greater than 90% of the sample migrated with an apparent molecular mass of ≈120 kDa, close to the predicted molecular mass of 113 kDa based on its primary sequence. Western analysis with antibodies against rSir3p yielded a banding pattern identical to that of the Coomassie-stained gel (data not shown), strongly suggesting that the minor bands are rSir3p degradation products. Initial characterization of purified rSir3p by gel filtration on Superdex 2000 yielded major elution peaks at ∼170 kDa and ∼350 kDa (data not shown), suggesting that a significant fraction of the rSir3p exists as an oligomer under the conditions used in our experiments. This finding is consistent with yeast two-hybrid experiments indicating that Sir3p is capable of self-association (21, 22). Rigorous analysis of the solution behavior of rSir3p by analytical ultracentrifugation has yet to be completed.
Figure 1.
(A) Purified rSir3p was analyzed by SDS/PAGE as described in Materials and Methods. (B) GST-fusion pull-down experiments were performed with rSir3p and GST alone (lane 2), GST-MyoD (lane 8), or GST fused to the N termini of histones H2A (lane 3), H2B (lane 4), H3 (lane 5), H4 (lane 6), and H4 with glutamine substitutions at positions K5, K8, K12, and K16 (lane 7) as described in Materials and Methods. The arrow indicates the location of Sir3p after SDS/PAGE of the eluate.
To determine whether recombinant rSir3p possesses the ability to bind the histone H3 and H4 N-terminal tails, rSir3p was incubated with several GST fusion proteins, including GST-H3T and GST-H4T (Fig. 1B). Under these conditions, Sir3p bound to GST-H3T and, to a lesser extent, to GST-H4T, but not GST, GST-myoD, GST-H2AT, GST-H2BT, or a mutant GST-H4T (Fig. 1B). Thus, purified rSir3p bound the histone H3 and H4 tail domains in vitro, as expected from previous genetic and molecular observations (5–7).
Sir3p Binding to Mononucleosomes, Nucleosomal Arrays, and Naked DNA.
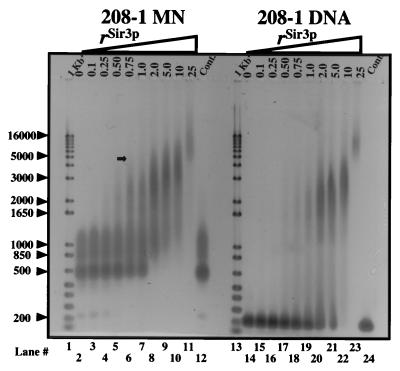
To address whether rSir3p interacts with nucleosomes in vitro, mononucleosomes reconstituted onto a 208-bp Lytechinus 5S rDNA fragment (208–1 MN) were mixed with rSir3p at ratios (rSir3p) of 0.1–25 mol rSir3p/mol 208-bp DNA. rSir3p bound to 208–1 MN at rSir3p = 1.0–10, as indicated by extensive mobility shifts in a native 1.0% agarose gel (Fig. 2). Shifted species were first observed at rSir3p = 0.5, whereas virtually all 208–1 MN existed in supershifted complexes at rSir3p = 2.0–5.0. It is noteworthy that the supershifted species assembled at rSir3p = 0.5–5.0 yielded “smears” on native agarose gels. Smearing indicates that multiple intermediate-sized rSir3p-208–1 MN complexes are present under these conditions. Importantly, at rSir3p = 2.0–10, Sir3p-208–1 MN complexes with an upper size limit equal to that of 4- to 5-kb linear DNA fragments were observed (Fig. 2, arrow). These data indicate that incubation of rSir3p with 208–1 MN at low rSir3p produces a series of intermediate-sized nucleoprotein structures, whereas a larger, size-limited complex is formed at rSir3p ≥ 1. A more detailed analysis will be required to determine the precise number and stoichiometry of the nucleoprotein complexes present at low and high rSir3p.
Figure 2.
rSir3p assembles mononucleosomes and naked DNA into defined, high-molecular-weight complexes. 208–1 MN (lanes 3–11) and 208–1 DNA (lanes 15–23) were incubated at the indicated rSir3p, and samples were electrophoresed on a 1.0% agarose gel (1× TBE) as described in Materials and Methods. Lanes 2, 12, 14, and 24 contain mock-incubated (no rSir3p) 208–1 MN and 208–1 DNA controls. Lanes 1 and 13 contain 1 Kb+ DNA size markers (GIBCO/BRL). Arrowheads indicate the size of selected markers in base pairs.
The small amount of naked DNA present after 208–1 MN reconstitution (Fig. 2, lane 2) was not observed at rSir3p > 0.5, suggesting that Sir3p also binds to naked DNA. To examine this possibility directly, the rSir3p binding experiments were repeated with 208–1 DNA. As with 208–1 MN, large shifted 208–1 species were apparent at rSir3p > 2.0. However, the pattern of rSir3p binding to 208-MN was different from that of rSir3p binding to 208–1 DNA. For example, at rSir3p = 2.0 all 208–1 MN formed large complexes, compared with only ∼50% of the 208–1 DNA (Fig. 2, lanes 8 and 20). In addition, no intermediate-sized rSir3p-naked DNA complexes were observed at molar ratios less than unity. Thus, rSir3p was able bind to naked DNA, albeit with somewhat different characteristics than rSir3p binding to mononucleosomes.
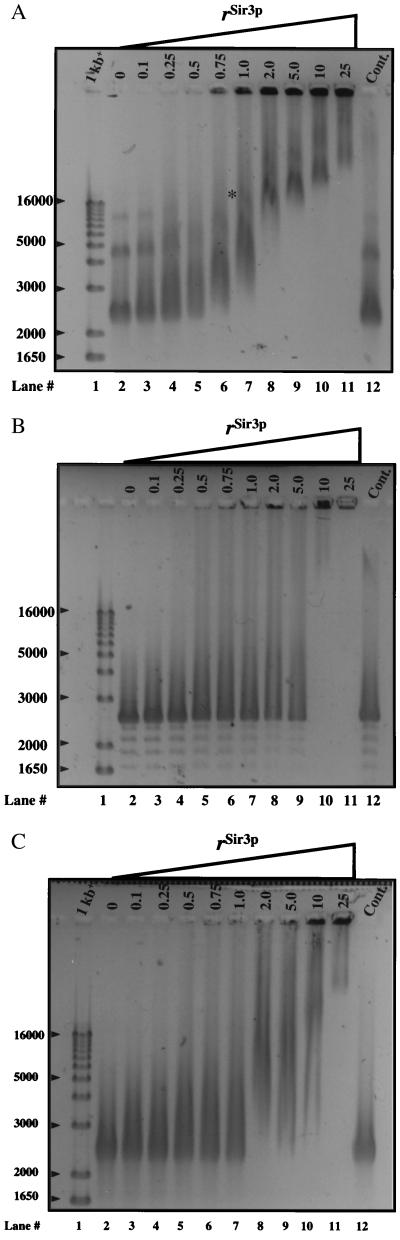
We next examined Sir3p binding to a more physiologically relevant substrate, nucleosomal arrays. The 12-mer nucleosomal arrays (208–12 NA) used in these experiments were obtained by reconstituting chicken erythrocyte histone octamers onto a DNA template (208–12 DNA) consisting of 12 tandem repeats of 208–1 DNA (23). The intrinsic structural dynamics of 208–12 NA have been characterized extensively both in the absence (24, 25) and presence (26, 27) of linker histones, providing a firm foundation for interpreting the effects of Sir3p binding on higher order chromatin structure. Incubation of Sir3p with 208–12 NA at rSir3p = 0.1–25 is shown in Fig. 3A. At rSir3p = 1.0 all 208–12 NA exhibited a shifted mobility, with a significant fraction of the sample forming complexes so large that they were barely able to migrate into a 1.0% agarose gel. In addition, at rSir3p = 1.0 an intermediate-sized shifted band was observed (Fig. 3A, asterisk) that became prominent at rSir3p = 2.0–5.0. Because the largest rSir3p nucleosomal array complexes present at rSir3p = 0.75–10 could barely enter the gel, it was not possible to determine whether a limiting high-molecular-weight complex had been formed. Nevertheless, other than the absolute size of the complexes, the patterns of binding of rSir3p to 208–1 MN (Fig. 2A) and 208–12 NA (Fig. 3A) generally appeared to be quite similar. Sir3p also bound to 208–12 DNA (Fig. 3B). As with 208–1 DNA, Sir3p bound differently to 208–12 DNA than it did to 208–12 NA, based on the different pattern of shifted species (Fig. 3 A and B). Incubation of Sir3p with nucleosomal arrays reconstituted onto pXP10 DNA, or with naked pXP10 DNA alone, yielded the same pattern of supershifted species on agarose gels as seen with 208–12 NA and DNA (data not shown). This result indicates that Sir3p binding to nucleosomal substrates and naked DNA was not DNA sequence-dependent. Importantly, rSir3p-dependent assembly of supershifted species also occurred in 50–150 mM NaCl and 2 mM MgCl2 (data not shown), demonstrating that Sir3p can bind to nucleosomal arrays under ionic conditions where the arrays are extensively folded (24–27).
Figure 3.
rSir3p assembles 12-mer nucleosomal arrays into supramolecular nucleoprotein complexes. (A) Intact 208–12 NA, (B) 208–12 DNA, and (C) trypsinized 208–12 NA were incubated at the indicated rSir3p, and samples were electrophoresed on a 1.0% agarose gel (1× TBE) as described under Materials and Methods. Arrowheads indicate the size of selected markers in base pairs.
Given that Sir3p interacts with the histone H3 and H4 N-terminal tails of GST-fusion proteins in vitro (7) (Fig. 1B), and genetic evidence indicates that these interactions are important for rSir3p's ability to bind and silence chromatin in vivo (5, 6), we tested whether the core histone tails played a role in Sir3p binding to 208–12 NA in vitro (Fig. 3C). “Tailless” 208–12 NA assembled from trypsinized histone octamers was mixed with rSir3p at rSir3p = 0.1–25 under the same conditions used for intact 208–12 NA and 208–12 DNA and products electrophoresed on a 1% agarose gel. In the absence of the tail domains the pattern of shifted bands closely resembled that observed for 208–12 DNA, with one exception. At rSir3p = 2.0–10, a moderately shifted species was present after incubation with tailless 208–12 NA but not with 208–12 DNA. Thus removal of the core histone tail domains abolished most, but not all, of the interactions of rSir3p with 208–12 NA. This result suggests that rSir3p interacts with both the tail domains and other chromatin components while mediating the assembly of supramolecular complexes.
Accessibility of Supershifted Complexes to Restriction Digestion.
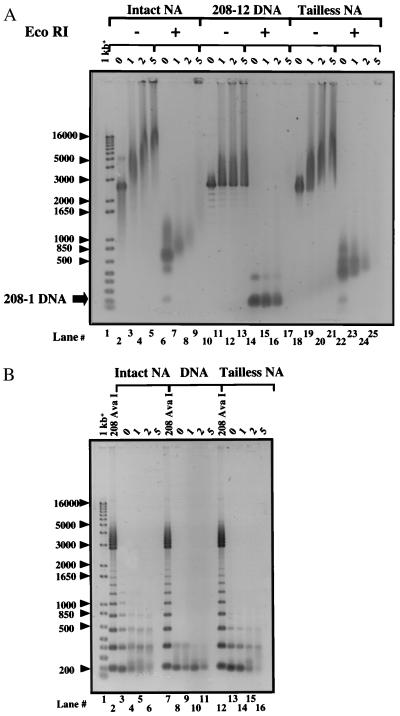
Each 208-bp repeat in the 208–12 DNA template is linked by DNA containing two closely spaced EcoRI restriction sites (23). After reconstitution into nucleosomal arrays, most EcoRI sites are present in the linker DNA region connecting adjacent nucleosomes and are accessible to digestion. A fraction of the sites are blocked by alternatively positioned histone octamers, producing a ladder of bands after EcoRI digestion (28, 29) (see Fig. 4C). To determine whether formation of the supershifted complexes was associated with a rSir3p-dependent reduction in EcoRI accessibility, intact or tailless 208–12 NA were mixed at rSir3p = 0–5.0 as in Fig. 3 and incubated with enzyme. Nondigested controls (Fig. 4A, lanes 2–5, 10–13, 18–21) were incubated under identical conditions, except that no enzyme was present. Samples were subsequently electrophoresed on a 1.0% agarose gel. All supershifted species formed at rSir3p = 1.0–2.0 were digested into small nucleosomal particles after incubation with EcoRI, including those complexes that were unable to migrate through the agarose gel before digestion (Fig. 4A). Similarly, EcoRI digestion of rSir3p-naked DNA complexes formed under the same conditions exclusively yielded 208–1 fragments. The intermediate shifted complexes formed by the intact and tailless 208–12 NA samples at rSir3p = 5.0 also remained completely accessible to EcoRI digestion, although the largest complexes present at rSir3p = 5.0 failed to migrate through the gel after restriction digestion (Fig. 4A, lane 9). Controls in which 208–12 NA was reconstituted at supersaturating molar histone-to-DNA ratios, which produces nonspecific high-molecular-weight aggregates containing sterically inaccessible EcoRI binding sites (19), were refractory to digestion by EcoRI, as expected (data not shown).
Figure 4.
208–12 NA linker DNA is accessible after assembly into supramolecular nucleoprotein complexes. (A) Aliquots of the intact 208–12 NA, 208–12 DNA, and trypsinized 208–12 NA complexes assembled at the indicated rSir3p were incubated with EcoRI at 37°C for 90 min. Undigested (−) and digested (+) samples were subsequently electrophoresed on a 1.0% agarose gel (1× TBE) as described in Materials and Methods. (B) Aliquots of the identical EcoRI-digested samples of intact 208–12 NA, 208–12 DNA, and trypsinized 208–12 NA from A were phenol-chloroform extracted to remove bound proteins and ethanol precipitated. Resuspended DNA was electrophoresed as in A. Lanes 2, 7, and 12 contain partial AvaI digests of pPol I 208–12, which generates a 208-bp DNA ladder. Arrowheads indicate the size of selected markers in base pairs.
The 208–1 DNA fragments produced by EcoRI digestion showed no evidence of reduced mobility. In contrast, for both intact and tailless 208–12 NA, the species liberated by digestion of the supershifted complexes formed at rSir3p = 1.0–2.0 showed a progressive reduction in mobility compared with controls (Fig. 4A, compare lanes 7–8 and 23–24 with 6 and 22, respectively). Within the resolution of a 1.0% agarose gel, this result could be due to rSir3p blockage of a small number of consecutive EcoRI sites, thereby producing increased amounts of di- and trinucleosomes. Alternatively, the reduced mobilities could result from binding of ≥1 rSir3p protein per nucleosome. To distinguish between these possibilities, aliquots of the EcoRI-digested complexes produced at rSir3p = 0–5.0 were deproteinized, and the resulting DNA was electrophoresed on a 1.0% agarose gel (Fig. 4B). Note the 208-bp ladder of bands produced by incomplete digestion of the control 208–12 NA (lanes 3 and 13). Densitometric quantitation indicated that the pattern and abundance of DNA bands produced by digestion of the supershifted complexes formed at rSir3p = 1.0–2.0 did not differ significantly from those of digests of 208–12 NA, although a small increase in the ratio of dimer to monomer bands was detected at rSir3p = 5.0. Thus, no substantive protection of additional EcoRI sites was associated with rSir3p-mediated complex formation under stoichiometric binding conditions, suggesting, in turn, that the particles liberated by EcoRI digestion of rSir3p-208–12 NA complexes (Fig. 4A, lanes 6–8) are 208–1 MN containing increasing amounts of bound rSir3p.
Discussion
rSir3p has been purified to near-homogeneity, and its interactions with mononucleosomes, nucleosomal arrays, and naked DNA have been characterized in vitro. rSir3p binds to the core histone tail domains and possibly nucleosomal DNA in a way that cross-links mononucleosomes and short nucleosomal arrays into large defined “supramolecular” complexes (see Fig. 5). These results demonstrate that rSir3p has a profound effect on higher order chromatin structure in vitro. However, rSir3p affects chromatin structural dynamics in a way quite different from that of linker histones, which stabilize the canonical chromatin fiber in the locally folded “30-nm” conformation (26, 27). For example, linker histone binding to 208–12 NA at molar ratios between 0 and 2.0 leads to formation of a slightly retarded complex in a 1.0% agarose gel (26), as opposed to the series of supershifted complexes caused by rSir3p binding under the same conditions. Furthermore, unlike that of Sir3p, linker histone binding to 208–12 NA is independent of the core histone tail domains (27). Instead, these data indicate that rSir3p assembles nucleosomal templates into novel high-molecular-weight complexes whose structural properties transcend those of canonical 10-nm and 30-nm chromatin fibers. The ability of Sir3p to cross-link nucleosomal templates into large assemblies presumably originates from the fact that each rSir3p monomer can interact with both the H3 and H4 tail domains, and thus each nucleosome contains up to four tail domain binding sites for rSir3p (Fig. 5). Given that the patterns of binding of rSir3p to trypsinized nucleosomal arrays and naked DNA are not identical (Figs. 2B and 3B) and that there is no rSir3p-dependent protection of EcoRI sites in the linker DNA of the supramolecular complexes formed at rSir3p ≥ 1 (Fig. 4), we hypothesize that interaction of rSir3p with nucleosome-bound DNA also may contribute to complex formation. Finally, rSir3p may interact with additional components of the nucleosome. It should be noted that oligomerization of rSir3p will serve to accentuate the extent of rSir3p–nucleosome interactions. Internucleosome cross-linking appears to be a common feature of other silencing and/or heterochromatin-associated proteins such as yeast Tup1/Ssn6 (R.T. Simpson, personal communication) and metazoan MENT (30, 31), although MENT appears to act through a mechanism (30) that is different from that of the yeast proteins.
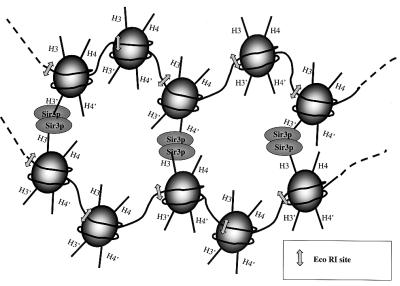
Figure 5.
Schematic model of rSir3p-dependent cross-linking of chromatin fibers into defined supramolecular structures. Shown is a portion of two nucleosomal arrays bridged together through interactions of rSir3p with the core histone H3 and/or H4 tail domains. Sir3p is drawn as an oligomer for the purposes of illustration only. H3 and H4 tail domains of each nucleosome are indicated by straight lines. Arrows indicate the positions of the EcoRI sites. Such a mechanism is proposed to facilitate both the formation of specific, transcriptionally silent higher order chromosomal domains associated with heterochromatin and telomeric clustering in vivo (see text).
Yeast telomeres and mating type loci are widely used as potential molecular models of silenced chromatin (see ref. 2). In yeast, silenced chromatin consists of nucleosomal arrays (32, 33) and many specialized nonhistone proteins, including Sir proteins (2). The results presented here represent initial studies focused on characterizing the effects of a purified Sir protein on chromatin structure in vitro. The relevance to chromatin fiber structure–function relationships is discussed above. In terms of silencing, we have started with yeast Sir3p in part because several in vivo studies indicate that Sir3p alone can modulate both silencing (34) and chromatin-mediated effects on replication (35), in a manner that is at least somewhat independent of the other Sir proteins (6). Although the following hypotheses remain to be tested directly, our results are consistent with the idea that Sir3-mediated assembly of specialized higher order chromatin structures at specific loci in yeast is a key component of achieving a transcriptionally silenced state. The specific structure of the putative repressive chromosomal domain(s) likely will be dependent on the unique nucleosome positioning configuration at silent loci (32, 33), as well as the presence of silencing proteins other than Sir3p. Clustering of telomeres and other heterochromatic regions in vivo (36), spreading of silencing from the telomeres inward (6, 34), and Sir3p-mediated chromosomal fiber “looping” (37) all can be explained in principle by the ability of Sir3p and analogous proteins to promote fiber–fiber interactions through internucleosomal cross-linking. Protein-mediated internucleosomal bridging (Fig. 5) (30, 31) also provides one type of general molecular mechanism that can explain both the functional and structural aspects of heterochromatin. Tests of these hypotheses with purified native and mutant silencing proteins will reveal additional biochemical insights relevant to the assembly, maintenance, and inheritance of specific heterochromatic states in vivo.
Acknowledgments
GST-myoD was a gift from Dr. Bruce Patterson (National Institutes of Health, National Cancer Institute). The plasmids coding for the GST-yeast histone tail fusions were provided by Dr. Michael Grunstein (University of California, Los Angeles). This work was supported by National Institutes of Health Grants GM-56890 (to C.A.F.) and GM45916 (to J.C.H.). This paper is dedicated to the memory of Alan P. Wolffe.
Abbreviations
- rSir3p
recombinant Sir3p
- GST
glutathione S-transferase
References
- 1.Hendrich B D, Willard H F. Hum Mol Genet. 1995;4:1765–1777. doi: 10.1093/hmg/4.suppl_1.1765. [DOI] [PubMed] [Google Scholar]
- 2.Pillus L, Grunstein M. In: Chromatin Structure and Gene Expression. Elgin S C R, editor. New York: Oxford Univ. Press; 1995. pp. 123–146. [Google Scholar]
- 3.Miller A M, Nasmyth K A. Nature (London) 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- 4.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 5.Johnson L M, Kayne P S, Kahn E S, Grunstein M. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hecht A, Strahl-Bolsinger S, Grunstein M. Nature (London) 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 7.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 8.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. . (First Published May 16, 2000; 10.1073/pnas.110148297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner K G, Landry J, Sternglanz R, Denu J M. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. . (First Published December 5, 2000; 10.1073/pnas.250422697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottschling D E. Curr Biol. 2000;10:R708–R711. doi: 10.1016/s0960-9822(00)00714-4. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher T M, Hansen J C. Crit Rev Eukaryotic Gene Expression. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 12.Bell S P, Mitchell J, Leber J, Kobayashi R, Stilmann B. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 13.Georgel P, Demeler B, Terpening C, Paule M R, van Holde K E. J Biol Chem. 1993;268:1947–1954. [PubMed] [Google Scholar]
- 14.Hansen J C, Ausio J, Stanik V H, van Holde K E. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 15.Wolffe A P, Jordan E, Brown D D. Cell. 1986;44:381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher T M, Hansen J C. J Biol Chem. 1995;270:25359–25362. doi: 10.1074/jbc.270.43.25359. [DOI] [PubMed] [Google Scholar]
- 17.Bohm L, Crane-Robinson C. Biosci Rep. 1984;4:365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- 18.Tse C, Sera T, Wolffe A P, Hansen J C. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen J C, Lohr D. J Biol Chem. 1993;268:5840–5848. [PubMed] [Google Scholar]
- 20.Georgel P T, Tsukiyama T, Wu C. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moretti P, Freeman K, Coodly L, Shore D. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto S, Johnston S D, Berman J. Genetics. 2000;155:523–538. doi: 10.1093/genetics/155.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson R T, Thoma F, Brubaker J M. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz P M, Hansen J C. J Biol Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 25.Tse C, Hansen J C. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 26.Carruthers L M, Bednar J, Woodcock C L, Hansen J C. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 27.Carruthers L M, Hansen J C. J Biol Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 28.Dong F, Hansen J C, van Holde K E. Proc Natl Acad Sci USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pennings S C, Meersseman G, Bradbury E M. J Mol Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 30.Grigoryev S A, Woodcock C L. J Biol Chem. 1998;273:3082–3089. doi: 10.1074/jbc.273.5.3082. [DOI] [PubMed] [Google Scholar]
- 31.Grigoryev S A, Bednar J, Woodcock C L. J Biol Chem. 1999;274:5626–5636. doi: 10.1074/jbc.274.9.5626. [DOI] [PubMed] [Google Scholar]
- 32.Weiss K, Simpson R T. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravindra A, Weiss K, Simpson R T. Mol Cell Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renauld H, Apparicio O M, Zierath P D, Billington B L, Chablani S K, Gottschling D E. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 35.Stevenson J B, Gottschling D E. Genes Dev. 1999;15:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 37.de Bruin D, Zaman Z, Liberatore R A, Ptashne M. Nature (London) 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]