Abstract
Alcohol use and abuse appear to be related to neuroadaptive changes at functional, neurochemical, and structural levels. Acute and chronic ethanol exposure have been shown to modulate function of the activity-dependent gene transcription factor, cAMP-responsive element binding (CREB) protein in the brain, which may be associated with the development of alcoholism. Study of the downstream effectors of CREB have identified several important CREB-related genes, such as neuropeptide Y, brain-derived neurotrophic factor, activity-regulated cytoskeleton-associated protein, and corticotrophin-releasing factor, that may play a crucial role in the behavioral effects of ethanol and molecular changes in the specific neurocircuitry that underlie both alcohol addiction and a genetic predisposition to alcoholism. Brain chromatin remodeling due to histone covalent modifications may also be involved in mediating the behavioral effects and neuroadaptive changes that occur during ethanol exposure. This review outlines progressive neuroscience research into molecular and epigenetic mechanisms of alcoholism.
Keywords: Alcoholism, Epigenetic, CREB, BDNF, NPY, CRF, Arc, Anxiety, Brain
Introduction
Alcohol addiction is a chronic relapsing disorder and is characterized by repetitive alcohol drinking patterns leading to a loss of control over alcohol consumption [1, 2]. Alcohol and other drugs of abuse have been shown to cause aberrations in synaptic plasticity and related neuronal function [1, 3, 4]. The neuroadaptational changes induced by exposure to alcohol and drugs of abuse may be related to dysregulation of signaling systems, gene transcription, and protein expression at the cellular level [3–5]. Thus, the search for molecular mechanisms that contribute to the initiation and maintenance of alcohol addictive processes has become a major focus of the neuroscience of alcoholism.
Two major psychiatric states have been implicated in the development of alcoholism, namely the positive and negative affective states of alcohol abuse. The positive affective state describes the euphoric effects of alcohol that lead to the promotion of alcohol consumption [1, 5]. The mesolimbic dopaminergic pathway has been shown to be a key mediator in the rewarding effects of alcohol and consists of dopaminergic projections from the ventral tegmental area (VTA) of the midbrain to various areas in the limbic system [1, 4, 6]. Dopaminergic VTA projections to the nucleus accumbens (NAc) have been identified as one of the primary mediators of the rewarding effects of alcohol [4–6]. Molecular and cellular changes in the NAc with acute and repeated alcohol exposure may underlie certain aspects in the development of alcohol addiction [4–6]. The negative affective state of alcohol abuse describes the development of anxiety, depression, and other dysphoric psychiatric sequelae which may be caused by the abrupt cessation of alcohol consumption [5, 7, 8]. The dysphoric state induced during alcohol withdrawal is a robust factor in the maintenance of both alcohol drinking and the eventual development of alcohol addiction [1, 5]. Amygdaloid brain regions, specifically the central nucleus of amygdala (CeA) and medial nucleus of amygdala (MeA), appear to be associated with the dysphoric effects of alcohol withdrawal, particularly the promotion of anxiety-like behaviors [1, 4, 5]. Epidemiological studies have shown that alcohol-use disorders are commonly comorbid with innate mood and anxiety-spectrum disorders, suggesting that a genetic predisposition for anxiety may also predispose an individual to alcoholism [7, 8]. When coupled together, the positive and negative affective states of alcohol abuse provide a convincing psychiatric and neurobiological model for the initiation and maintenance of alcohol consumption leading to addiction. Interestingly, various studies have found that there may be common cellular substrates, particularly the cyclic AMP-responsive element binding protein (CREB) gene transcription factor, which may play a role in both the euphoric and dysphoric pathways regulating the development of alcohol addiction [5, 9, 10].
CREB plays a central role in the process of addiction [3, 5, 9, 10]. Ethanol has a complex pharmacological profile and various signaling systems have been identified as modulators of CREB function that may serve as potential ethanol targets [10–12] (Fig. 1). A great deal of research has focused on the role of CREB and its target genes, such as neuropeptide Y (NPY), brain-derived neurotrophic factor (BDNF), activity-regulated cytoskeleton-associated (Arc) protein, and corticotrophin-releasing factor (CRF) in the development of alcohol addiction [13–16]. In addition, several studies have identified novel epigenetic mechanisms, such as histone modification-induced chromatin remodeling and DNA methylation, in the process of alcohol-related neuroadaptation [17, 18]. In this review, we have focused on evidence that implicates a regulatory role for CREB-related changes and epigenetic mechanisms in the neurocircuitry of the NAc and amygdala that may contribute to alcoholism. To give a comprehensive picture of changes in CREB and related signaling mechanisms, we have also considered findings in other brain regions and cell culture models in relation to alcoholism.
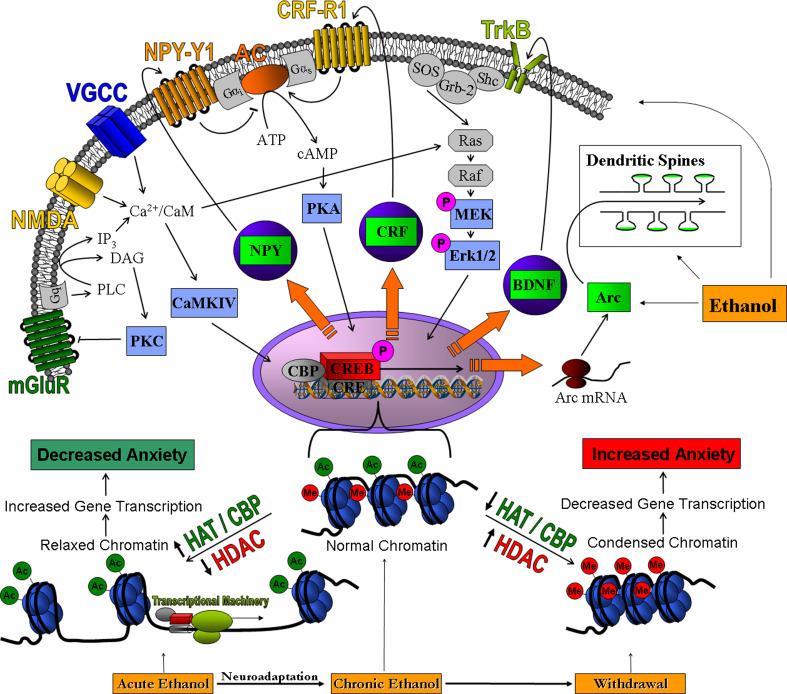
Fig. 1.
Ethanol exposure modulates cyclic AMP responsive element binding (CREB) protein-related cellular processes at the level of receptors and intracellular signaling, gene transcription, and neuronal morphology. Signaling via metabotropic glutamate receptors (mGluR), N-methyl-d-aspartate (NMDA) receptors, voltage-gated calcium channels (VGCC), adenylyl cyclase (AC), neuropeptide Y1 receptors (NPY-Y1), type 1 corticotrophin-releasing factor receptors (CRF-R1) and tyrosine kinase B receptors (TrkB) leads to the modulation of various protein kinases, including protein kinase C (PKC), calcium calmodulin-dependent kinase IV (CaMKIV), protein kinase A (PKA) and extracellular signal regulated kinase 1/2 (Erk1/2). These protein kinases regulate the function of CREB, which recruits and binds CREB binding protein (CBP) and regulates transcription of CREB targets, including neuropeptide Y (NPY), corticotrophin-releasing factor (CRF), brain-derived neurotrophic factor (BDNF) and activity-regulated cytoskeleton-associated protein (Arc), which plays a role in the regulation of dendritic spine morphology. The bottom portion of the figure indicates relaxed and condensed chromatin structures in the amygdaloid circuitry during various states of ethanol exposure (acute, chronic, and withdrawn). CBP contains intrinsic histone acetyltransferase (HAT) activity and works together with histone deacetylases (HDAC) to modify the chromatin architecture. For example, acetylation of histone proteins by HATs results in a relaxed chromatin structure, thereby increasing gene transcription. HDACs enzymatically remove acetyl groups from lysine residues located at the N-terminal tails of histone proteins, allowing for other enzymes to add methyl groups, condensing the chromatin and decreasing gene transcription. HDACs and HATs may serve as potential molecular targets for the action of ethanol in the central and medial nucleus of amygdala [18]
CREB in alcohol addiction
The CREB gene transcription factor has been shown to be involved in many aspects of CNS function including long-term memory formation, synaptic plasticity, and addiction [5, 9, 11, 19, 20]. CREB protein is constitutively expressed and activated by phosphorylation at serine-133 via calcium/calmodulin-dependent protein kinase II and IV (CaMKII and CaMKIV), cAMP-dependent protein kinase A (PKA), and mitogen-activated protein kinase (MAPK) [4, 5, 21]. Phosphorylation of CREB leading to subsequent activation results in the recruitment of CREB-binding protein (CBP) and other transcriptional components, which enables gene transcription to occur at the cAMP-responsive element (CRE) promoter region [9, 21, 22]. The regulation and effects of CREB are widespread and varied, so the scope of this review will be limited to some upstream regulators and downstream effectors that have been shown to be potential targets of ethanol action.
Several studies, as described below, have indicated that deficits in CREB function in the CeA may be involved in alcohol preference and dependence. An early study on CREB function in the amygdala utilized alcohol-preferring (P) and -nonpreferring (NP) rats, selectively bred for high and low alcohol preference, respectively [23–25]. The study found that levels of CREB and phosphorylated CREB (p-CREB), as well as CRE-DNA binding activity, were lower in the amygdala of P rats compared to NP rats [24]. In addition, deficits in the levels of CREB and p-CREB were found in amygdaloid structures of P rats compared to NP rats, specifically in the CeA and MeA, but not in the basolateral amygdala (BLA), which correlated with anxiety-like behaviors and higher ethanol consumption in P rats [26]. Treatment with acute ethanol increased levels of p-CREB and produced anxiolytic effects in P rats and in mice, but not in NP rats [26, 27]. A mechanistic study of CREB function by infusion of a PKA activator (Sp-cAMP) into the CeA increased p-CREB levels and decreased anxiety-like behaviors and ethanol consumption of P rats. In contrast, infusion of a PKA inhibitor (Rp-cAMP) into the CeA decreased p-CREB levels, provoked anxiety-like behaviors, and increased ethanol intake of NP rats [26]. A similar observation of increased anxiety-like behaviors and higher ethanol preference was found in CREB-haplodeficient mice compared to wild-type littermates, further suggesting an important role for CREB in anxiety-like and alcohol drinking behaviors [27]. In order to study the role of CREB in alcohol dependence, Sprague–Dawley (SD) rats were chronically exposed to ethanol and then withdrawn for a 24-h period. Only ethanol-withdrawn SD rats displayed anxiety-like behaviors, which correlated with decreased p-CREB levels in the CeA or MeA, while no change was observed in total CREB protein levels [28]. Increasing levels of p-CREB in the CeA via PKA activator infusion prevented the development of anxiety-like behaviors during ethanol withdrawal in SD rats. In contrast, decreased CREB function via PKA inhibitor infusion into the CeA provoked anxiety-like behaviors and increased ethanol intake of normal SD rats [28]. Taken together, these results suggest that CREB deficits in the CeA may be involved in the process of alcohol preference and dependence (Fig. 2).
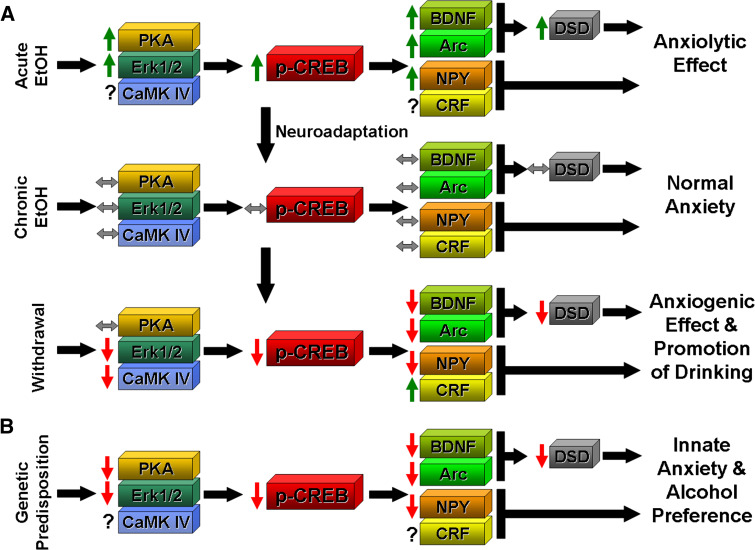
Fig. 2.
a A hypothetical molecular model for the action of ethanol on protein kinases, CREB, and CREB targets within amygdaloid circuitry, specifically the central nucleus of amygdala (CeA), involved in the regulation of anxiety and alcohol drinking behaviors. Acute ethanol elevates phosphorylated CREB (p-CREB) levels by increasing protein kinase function, which leads to the increased expression of CREB-target genes. Increased brain-derived neurotrophic factor (BDNF) and activity-regulated cytoskeleton-associated protein (Arc) expression result in increased dendritic spine density (DSD) which may mediate the anxiolytic effects of ethanol. The anxiolytic effects of acute ethanol may also involve increased neuropeptide Y (NPY) expression in the CeA. Neuroadaptation due to chronic ethanol exposure normalizes protein kinase levels, and p-CREB and CREB target gene expression levels. Withdrawal from chronic ethanol exposure reverses the observed effects of acute ethanol leading to increased anxiety and promotion of alcohol drinking. Corticotrophin-releasing factor (CRF) is also regulated by CREB and hypofunction of NPY and hyperfunction of CRF in the CeA may be involved in the regulation of anxiety and alcohol drinking behaviors during ethanol withdrawal. b Innately low CREB function in the CeA can lead to lower levels of BDNF, Arc, NPY, and DSD, which may contribute to a genetic predisposition to anxiety and alcohol preference. ( ) Decrease; (
) Decrease; ( ) increase; (
) increase; ( ) normal; (
) normal; ( ) unknown or unclear
) unknown or unclear
In addition to amygdaloid brain structures, abnormalities in CREB function in other brain regions such as the NAc, cortex, and cerebellum have been shown to be involved in alcohol addiction. A series of early studies demonstrated that acute ethanol resulted in a rapid increase in p-CREB levels, which was attenuated by chronic ethanol treatment in rat cerebellum and striatum [29–31]. Furthermore, CRE-DNA binding and CREB phosphorylation were not affected by chronic ethanol exposure; however, the levels of both CRE-DNA binding and p-CREB were significantly decreased in cortical brain regions during ethanol withdrawal [32, 33]. Voluntary ethanol intake caused a decrease in CREB phosphorylation in the NAc shell, but not core, of rats [34, 35]. The levels of p-CREB decreased further during 24- and 72-h ethanol withdrawal periods, but were shown to increase toward control levels after 7 days, alluding to the NAc as a brain region involved in alcohol drinking behaviors [34]. CREB expression was also investigated in the brain structures of C57BL/6 and DBA/2 mice [36]. It was shown that C57BL/6 mice displayed a higher ethanol preference compared to DBA/2 mice [36, 37]. Interestingly, levels of CREB and p-CREB have also been shown to be innately lower in the shell, but not in the core, of the NAc or any amygdaloid regions of C57BL/6 mice compared to DBA/2 mice [36]. It is also important to mention that a role for CREB has been identified in various aspects of alcoholism such as the development of tolerance [38, 39], changes in neuronal development, cell death [40, 41], and ethanol sensitivity [42].
In summary, these results suggest that CREB deficits in both the NAc and CeA, either innately or due to withdrawal after ethanol exposure, may promote ethanol intake, and the correction of CREB deficits may decrease ethanol consumption and prevent alcohol addiction. Thus, decreased CREB function may mediate the development of alcohol addiction via the positive and negative affective states of alcoholism. There are several studies that suggest possible mechanisms by which direct ethanol targets may regulate CREB function and the neuroadaptive changes that occur in alcoholism (Fig. 1).
Upstream targets of ethanol-mediated modulation of CREB
Many proteins have been identified as direct molecular targets for ethanol, including neurotransmitter receptors, ion channels, and enzymes [10, 12, 43, 44]. Interestingly, a number of the identified protein targets of ethanol may also play a role in the regulation of CREB and adaptational changes in the brain during ethanol exposure (Fig. 1). Important electrophysiological studies performed over 20 years ago identified the modulating effects of ethanol on γ-aminobutyric acid (GABA) and N-methyl-D-aspartate (NMDA) receptor currents [45, 46]. Early studies in hippocampal cell cultures found that acute ethanol inhibited NMDA receptor-mediated Ca2+ currents [45]. More recently, site-directed mutagenesis identified an ethanol binding pocket in the NMDA receptor transmembrane (TM3) domain. Also, amino acid sequence variation in this domain correlated with variations in ethanol sensitivity [47–49]. NR2B-containing receptors have been shown to exhibit high Ca2+ conductance and may be particularly important in synaptic plasticity [50]. Notably, NR2B-containing receptors displayed the highest sensitivity to ethanol and may also serve as a possible pharmacological target for the treatment of alcohol addiction [51, 52]. A study of mouse cortical cell cultures showed that ethanol-induced increases in NR2B receptor subunit expression were dependent on CREB during ethanol exposure [53]. Metabotropic glutamate receptors (mGluR), particularly Group I mGluRs, may regulate CREB function through increased intracellular Ca2+ and activation of MAPK signaling pathways [54]. Acute ethanol has been shown to inhibit Ca2+ currents induced by protein kinase C (PKC)-dependent phosphorylation of mGluR5, but not mGluR1 [55, 56]. Reduction of mGluR5 activity using the mGluR5 antagonist, 2-methyl-6-(phenylethyl)-pyridine (MPEP), resulted in a dose-dependent decrease in ethanol consumption in various animal models [57–60]. Voltage-gated Ca2+ channels (VGCCs), especially the slow-inactivating L-type VGCC, have been shown to sustain Ca2+ influx resulting in CREB activation and changes in synaptic function [61]. Early studies in PC12 cell cultures showed that ethanol had a significant inhibitory effect on the influx of Ca2+ through L-type VGCCs [62]. Chronic ethanol administration evoked a PKC-dependent upregulation of L-type VGCC expression and elicited changes in channel subunit composition, resulting in dysregulation of VGCC function [63, 64]. Furthermore, antagonism of L-type VGCCs has been shown to reduce ethanol consumption in both ethanol-dependent and genetically altered animal models [65–67]. Taken together, these studies have identified receptors and ion channels that regulate intracellular Ca2+ as direct targets in the effects of ethanol. These ethanol targets may serve as upstream regulators of CREB and may be involved in the regulation of CREB target gene expression (Fig. 1).
Adenylyl cyclase (AC) plays an important role in the cAMP-dependent activation of PKA and in regulating CREB function via phosphorylation. Studies in cell culture and rat cortex demonstrated that acute ethanol exposure could stimulate AC activity and increase cAMP production [68], while chronic ethanol exposure resulted in AC desensitization in mouse cortex [69, 70]. AC sensitivity to ethanol was shown to be isoform-specific in transfected human embryonic kidney (HEK293) cells [71]. Studies using AC isoform chimeras found two ethanol-sensitive domains within each of the cytoplasmic domains [72]. These results demonstrate that there may be a direct interaction between ethanol and AC and that this interaction may be involved in ethanol-mediated regulation of CREB phosphorylation.
Protein kinases in alcohol addiction
Protein kinases that regulate CREB phosphorylation include CaMKII, CaMKIV, PKA, and MAPK (Fig. 1). The functions of these protein kinases may be modulated by ethanol directly or via upstream modulation of ethanol targets [73–75]. CaMKs are regulated by rapid changes in intracellular Ca2+ and have an integral role in activity-dependent regulation of CREB function [76]. Various studies have implicated a role for ethanol-mediated modulation of CaMKIV in the regulation of CREB function. An early study on rats withdrawn from chronic ethanol exposure found a decrease in CaMKIV and p-CREB levels in cortical and amygdaloid structures [28, 33]. Further studies demonstrated that voluntary ethanol intake decreased CaMKIV and p-CREB levels in the NAc, implicating that decreased CaMKIV function may be associated with ethanol dependence [35, 77]. Ethanol has also been shown to activate CaMKII and result in increased phosphorylation of CaMKII targets in rat cortex [78]. In addition, CaMKII has been shown to mediate some of the effects of ethanol by directly phosphorylating channel proteins, namely voltage- and calcium-activated potassium (BK) channels [79]. However, CaMKII has not been implicated in the regulation of acute ethanol sensitivity of NMDA receptors [80].
PKA is a cAMP-activated tetrameric protein kinase consisting of two catalytic subunits and two regulatory subunits [81]. Ethanol-mediated activation of AC and subsequent increases in intracellular cAMP cause the dissociation of PKA regulatory and catalytic subunits. Cell culture studies revealed that acute ethanol treatment resulted in translocation of the PKA catalytic subunit to the nucleus, which correlated with increased CREB phosphorylation [73, 82]. In vivo, chronic ethanol treatment and withdrawal have been shown to have no effect on levels of the α-subunit of the catalytic domain of PKA (PKA-Cα) in either cortical or amygdaloid structures of SD rats [28, 33]. On the other hand, voluntary ethanol intake increased levels of PKA-Cα in the CeA and MeA of P rats [26]. Numerous studies have manipulated PKA activity to show that PKA-mediated changes in CREB function play a role in regulating ethanol consumption through both euphoric and dysphoric pathways [28, 83–85]. Inhibition of PKA function was also shown to reduce the sedative effects of acute ethanol, suggesting that reduced PKA function may increase ethanol tolerance and play a role in the development of ethanol dependence [86]. Transgenic mice lacking the PKA RIIβ regulatory subunit also display decreased ethanol-induced sedation and increased ethanol consumption [87]. A recent study found that chronic intermittent ethanol exposure decreased PKA activity in the NAc and amygdala due to the upregulation of the PKA inhibitor α, suggesting that increased PKA-dependent transcription during acute ethanol consumption may mediate a decrease in PKA activity with repetitive withdrawal [88].
The MAPK family of protein kinases, including extracellular signal-regulated kinases 1/2 (Erk1/2), have been shown to be involved in activity-regulated gene transcription mediating synaptic plasticity [89]. Erk1/2 activation is characterized by the initial activation of Ras, a small soluble G-protein, which results in the activation of downstream proteins, Raf and MAPK/Erk1/2 kinase (MEK), and successive phosphorylation of Erk1/2 [90] (Fig. 1). Several studies have identified a relationship between Erk1/2 activation and ethanol exposure that appears to be highly dependent on age, brain region, and ethanol treatment paradigm [41, 91, 92]. Acute ethanol increased p-Erk1/2 levels while chronic ethanol treatment returned p-Erk1/2 to control levels in the CeA and MeA of SD rats. Furthermore, withdrawal from chronic ethanol exposure resulted in decreased p-Erk1/2 levels [16]. These findings, along with other studies, reveal a direct correlation between ethanol-mediated modulation of Erk1/2 and CREB, and further implicate amygdaloid Erk1/2 as a key regulator of ethanol’s effects on CREB [16, 41]. Recent studies have shown that activation of Erk1/2 in the CeA and VTA by BDNF and glial cell line-derived neurotrophic factor infusion decreased voluntary ethanol consumption, highlighting the importance of Erk1/2 signaling in the regulation of alcohol drinking behaviors [93, 94]. Electrophysiological studies in acute brain slice preparations have shown that Erk1/2 modulation by ethanol attenuated long-term potentiation, identifying a direct role for Erk1/2 in ethanol-induced modulation of synaptic plasticity [95, 96]. Taken together, these studies indicate the involvement of various protein kinases in both the acute effects of alcohol, as well as in the development of alcohol addiction.
Downstream effectors of ethanol-mediated modulation of CREB
Modulation of CREB function by ethanol, via the upstream mediators mentioned above, leads to a change in expression of various CREB target genes, which in turn may mediate both functional and structural changes in the brain that may be involved in alcohol addiction (Fig. 2). Many of the upstream and downstream mediators of CREB-related signaling pathways interact with one another, revealing a complex signaling network that is often usurped by ethanol and other drugs of abuse [1, 5, 74, 75]. The following section discusses some CREB targets that are involved in the molecular mechanisms of alcoholism.
BDNF signaling and synaptic plasticity in alcoholism
BDNF is readily inducible by neuronal activity and has been shown to have important implications in development, neurite outgrowth, synaptic plasticity, and regulation of dendritic morphology [97–101]. BDNF triggers cell signaling cascades by binding to high affinity tropomyosin-related kinase B (TrkB) receptors. TrkB activation causes receptor internalization and the recruitment and binding of adaptor proteins, Grb2 and SOS, which interact with Ras protein resulting in MAPK activation [101, 102]. BDNF signaling, especially via MAPK, results in the activation of CREB [103, 104]. In addition, BDNF and TrkB are CREB target genes [27, 104, 105]. Although the molecular mechanisms which underlie BDNF-mediated modulation of synaptic plasticity, such as changes in dendritic spine morphology, are not yet fully understood, it is important to note that numerous mechanisms have been identified which play a role in the extension and retraction of spines [106]. Arc, also known as activity-regulated gene 3.1 (Arg3.1), is an immediate-early gene that is regulated by BDNF- and CREB-dependent signaling [107, 108]. Arc protein acts as a stabilization factor for filamentous-actin (F-actin), which results in the regulation of dendritic spine morphology [109, 110]. Induction of long-term potentiation in the rat dentate gyrus is associated with upregulation of Arc [100, 111]. Also, exogenous BDNF application in vivo rapidly increased Arc protein levels and induced local expansion of the actin cytoskeleton within dendritic spines [107, 111]. Studies have shown that interactions between post-synaptic density (PSD) proteins, such as PSD-95 protein, and NMDA receptors located at the spine tip may also regulate dendritic spine morphology [112, 113].
Aberrant regulation of BDNF has been implicated in the development of psychiatric disorders, including schizophrenia, depression, anxiety, and alcohol addiction [15, 114, 115]. A possible link may exist between decreased BDNF expression in specific brain regions and both ethanol dependence and increased ethanol preference. BDNF-haplodeficient mice have been shown to display a higher preference for ethanol compared to wild-type littermates [116, 117]. It has also been shown that inhibition of the TrkB receptor increased ethanol intake in wild-type, but not in BDNF haplodeficient, mice [116]. In rats, chronic ethanol treatment resulted in decreased BDNF expression in the hippocampus and cortex [32, 118]. Another study showed that ethanol differentially regulated BDNF expression in the NAc of C57/BL6 mice in comparison to DBA/2 mice [119]. A study on the cytotoxic effects of ethanol in cell cultures found that exogenous BDNF appeared to have a cytoprotective role against ethanol-induced damage [120]. Another study by McGough et al. uncovered the importance of BDNF in the processes of ethanol dependence [121]. Investigators found that BDNF expression was increased in the dorsal striatum of mice after acute and self-administered ethanol. In addition, they found that decreasing BDNF levels led to increased ethanol consumption whereas increasing BDNF attenuated ethanol intake in rodents. These results suggest that the BDNF signaling pathway may serve as a homeostatic pathway involved in the regulation of alcohol addiction [121].
A recent study performed in our laboratory using an antisense oligodeoxynucleotide (ODN) strategy demonstrated a mechanistic role for BDNF in ethanol preference. The reduction of BDNF via infusion of BDNF antisense ODNs into the CeA or MeA of SD rats increased ethanol consumption and anxiety-like behaviors [94]. In contrast, BDNF co-infusion with antisense ODNs rescued these behaviors [94]. This notion was further supported by findings that levels of BDNF protein and mRNA were innately lower in the CeA and MeA, but not BLA, of P rats compared to NP rats [122]. In another study examining BDNF protein levels in the NAc of P and NP rats, investigators found that baseline levels of BDNF were lower in P rats compared to NP rats [123]. The results of this study are in marked contrast to the results of our own study where we found that innate BDNF mRNA and protein levels were similar in the shell and core structures of the NAc of P rats compared to NP rats [122]. Despite the discrepancies between the studies, which may be related to differences in the methodologies used, both of the studies emphasized the importance of BDNF in alcohol drinking behaviors. Various studies conducted in the amygdala, as described above, clearly indicate a strong relationship between amygdaloid BDNF expression, and anxiety and ethanol intake (Fig. 2).
Early studies into the effects of ethanol on dendritic spine morphology found that chronic ethanol treatment led to a decrease in dendritic spine density (DSD) in various areas of the brain [124–126]. These findings suggest that changes in DSD may occur due to ethanol exposure; however, the functional implications of ethanol-induced changes in dendritic morphology are not well understood [127]. More recent studies have begun to explore possible mechanisms mediating the effects of ethanol on spine morphology and dynamics in NAc and amygdaloid brain regions. Using two-photon microscopy to analyze dendritic spines in the NAc of P rats, a recent study found that chronic ethanol exposure resulted in abnormal dendritic morphology and decreased DSD [128]. This finding correlated with a spine-localized upregulation of a truncated NR1 subunit implicated in the regulation of spines [128]. A recent series of studies in our laboratory explored the interaction between ethanol, BDNF, Arc, and DSD [16]. Acute ethanol exposure in SD rats increased Arc expression and DSD in the CeA and MeA, which was linked to increased BDNF signaling and CREB activation. Conversely, withdrawal from chronic ethanol decreased BDNF signaling, CREB activation, Arc expression, and DSD, which was associated with anxiety-like behaviors in SD rats [16]. Employing a mechanistic approach, the study showed that BDNF infusion into the CeA of ethanol withdrawn rats increased BDNF signaling and CREB activation, ultimately leading to increased Arc mRNA and protein levels and decreased anxiety-like behaviors. In addition, infusion of Arc antisense ODNs into the CeA of normal SD rats decreased Arc and DSD while increasing anxiety-like behaviors and ethanol intake [16]. Another study in our laboratory showed that, in comparison to NP rats, P rats had innately lower DSD in the CeA and MeA which was increased by acute ethanol treatment [129]. These data provide a convincing role for Arc-mediated regulation of DSD in the process of ethanol preference and dependence. In hippocampal cell cultures, increased NR2B receptor subunit expression due to chronic ethanol exposure increased NMDA clustering with the post-synaptic density protein, PSD-95, which correlated with an increase in dendritic spine size [130]. This study provided an attractive model linking ethanol-mediated changes in NMDA subunit composition to dendritic morphology. Taken together, these studies identify possible molecular players involved in the complex regulation of structural changes of dendritic spines in various brain regions during ethanol exposure.
NPY signaling in alcoholism
Neuropeptide Y (NPY) is a highly conserved 36 amino acid neuromodulator implicated in the regulation of a wide variety of biological and behavioral functions, including the regulation of food intake [131], neurogenesis [132], and stress and anxiety [133]. NPY binds to Y1, Y2, Y4, and Y5 receptors, which are G-protein coupled receptors (GPCR) associated with Gαi, resulting in decreased cAMP formation [134]. Although NPY receptor signaling is primarily linked to inhibition of cAMP production, studies have also shown that activation of the Y1 receptor may couple with other protein kinases, such as CaMKs, and increase p-CREB [135]. Since NPY is also a CREB-target gene, both NPY and CREB can reciprocally regulate one another [27, 136, 137]. In addition, CREB activation can also increase NPY-Y1 receptor expression [138].
The NPY system has been shown to be related to anxiety-like and alcohol drinking behaviors [14, 26, 139, 140]. Studies have shown increased anxiety-like behaviors and ethanol consumption in NPY knockout mice, while transgenic mice overexpressing NPY consumed lower amounts of ethanol [141, 142]. NPY levels are lower in the CeA of both P and high alcohol drinking (HAD) rats compared to NP and low alcohol drinking (LAD) rat lines, respectively [26, 139]. Intracerebroventricular (ICV) administration of NPY was shown to elicit an anxiolytic effect, which was reversed by infusion of antisense ODNs for the NPY-Y1 receptor into the amygdala [143, 144]. These findings provide evidence suggesting a role for NPY signaling in the regulation of anxiety-like behaviors and ethanol consumption [14]. Additional studies have implicated specific NPY receptors, such as NPY-Y1, NPY-Y2, and NPY-Y5 in regulating ethanol consumption. NPY-Y1 receptor knockout mice consumed higher amounts of ethanol and had a lower sensitivity threshold to the sedative effects of ethanol compared to wild-type littermates [145]. ICV infusion of the Y2 receptor antagonist, BIIE0246, demonstrated that Y2 receptor blockade suppressed ethanol consumption in rats following chronic ethanol treatment [146]. NPY-Y2 receptor knockout mice also displayed a decrease in ethanol consumption in comparison to wild-type littermates [147]. Other studies have implicated the involvement of NPY-Y5 receptors in alcohol drinking behaviors [148]. These studies suggest that Y1, Y2, and Y5 receptors may serve as possible targets for the development of drugs to treat alcohol addiction.
Some studies have specifically looked at the role of NPY in the NAc and amygdaloid brain structures with regard to ethanol consumption. Studies comparing C57/BL6 and DBA/2 mice found that high ethanol preference in C57/BL6 mice correlated with low NPY levels in the shell of the NAc [36]. In a mechanistic study of SD rats, infusion of a PKA inhibitor, Rp-cAMP, into the NAc shell decreased NPY protein expression and increased ethanol preference. Co-infusion of NPY with Rp-cAMP attenuated increased ethanol preference [83]. Importantly, modification of NPY signaling in the NAc shell had no effect on anxiety-like behaviors [83]. Studies performed in P and NP rats, as well as HAD and LAD rats, have found correlations between high ethanol preference and low NPY expression levels in the CeA [139]. In our laboratory, it was shown that deficits in NPY in the CeA and MeA of P rats may be associated with a genetic predisposition to ethanol preference and anxiety [26]. Infusion of NPY or a PKA activator into the CeA reduced ethanol consumption and anxiety-like behaviors of P rats [26]. Other investigators have found that ICV infusion of NPY attenuated ethanol intake in P rats, but not in an unselected stock of rats [149, 150]. In a related study, viral vector-mediated overexpression of NPY in the CeA decreased anxiety-like and alcohol-drinking behaviors of rats predisposed to high anxiety, but had no effect on normal rats [151].
Several studies have also characterized a role for amygdaloid NPY signaling in preventing the onset of the negative affective aspects of alcohol dependence. The anxiolytic effects of acute ethanol have been associated with increased NPY levels in the CeA and MeA, but not the BLA, of SD and P rats [18, 26]. Levels of amygdaloid NPY have been shown to be decreased in both withdrawal- and stress-induced anxiety states, which correlated with increased ethanol intake [14]. Furthermore, viral-vector mediated NPY overexpression was able to prevent increased ethanol intake in Wistar rats subjected to repetitive withdrawal [152]. In our laboratory, it was shown that infusion of a specific activator of PKA, Sp-cAMP, into the CeA normalized NPY levels and decreased anxiety-like behaviors of ethanol-withdrawn SD rats, which have been shown to have decreased NPY mRNA and protein levels in the CeA and MeA. Conversely, infusion of an inhibitor of PKA, Rp-cAMP, increased anxiety-like behaviors and ethanol intake, which were rescued by co-infusion of NPY [28, 85, 140]. Ethanol deprivation in P rats exposed to ethanol resulted in an increase in ethanol intake, which was attenuated by infusion with NPY into the CeA [153]. Similarly, NPY infusion into the CeA abolished dependence-induced increases in ethanol consumption in Wistar rats [154]. In summary, many of these studies depict a common theme by which increases in NPY expression can attenuate ethanol consumption and anxiety-like behaviors, primarily in animals with genetic or ethanol-induced deficits in NPY (Fig. 2). Thus, NPY signaling may be important in the development of pharmacological agents for the treatment of anxiety and alcohol addiction.
CRF signaling in alcoholism
Corticotrophin-releasing factor (CRF) is a 41 amino acid polypeptide that is ubiquitously expressed throughout the central nervous system (CNS). CRF acts at two GPCRs (type 1 and type 2 CRF receptors), CRF-R1 and CRF-R2, both of which are coupled to Gαs and can increase intracellular cAMP levels [155]. The amygdala is believed to play a particularly important role in the central function of CRF. Amygdaloid CRF neurons have been shown to project widely to regions of the basal forebrain and brainstem [156]. CRF-R1 appears to be associated with stress and anxiety [157]. CRF expression in response to stressors has been shown to be dependent on CREB binding to the CRE site in the promoter region of the CRF gene, suggesting that CRF may also be a CREB-target gene [158]. CRF plays an important role during the development of alcohol addiction and stress-induced relapses to alcohol drinking [1]. An early study showed that anxiety-like behaviors caused by ethanol withdrawal were rescued by infusion of a CRF-R1 antagonist, alpha-helical CRF, directly into the CeA, but not by ICV infusion [159]. A more recent study utilizing another CRF-R1 antagonist, D-Phe-CRF(12-41), showed that this effect was only seen in ethanol-dependent rats, indicating that the anxiolytic effects of CRF-R1 antagonism may rely on a pathologic increase of CRF [160]. In addition, antagonism of CRF receptors in the CeA decreased ethanol consumption in withdrawn rats [161]. Interestingly, infusion of a CRF-R2 agonist also decreased ethanol consumption, indicating that CRF-R1 and -R2 receptors may have opposing roles in the amygdala [162]. Direct measurement of endogenous CRF levels by in vivo microdialysis showed an increase in CRF in the amygdala of both rats withdrawn from chronic ethanol treatment and rats subjected to acute stress [163]. These findings were further supported by a study of Sardinian alcohol-preferring rats (sP) that displayed increased CRF release, which correlated with a high anxiety state [164]. Interestingly, there were lower CRF mRNA and protein levels in the CeA of P rats compared to NP rats [165]. Yet, exogenous administration of CRF increased electroencephalographic responses, indicating that decreased CRF expression may be coupled with a reciprocal increase in CRF-R1 receptor function or expression [165, 166]. Notably, various studies using either ethanol-treated or genetic animal models of post-dependence have shown that a persistent upregulation of amygdaloid CRF-R1 receptors plays a role in the post-dependent phenotype characterized by increased stress sensitivity and propensity to ethanol relapse [13]. Recent studies examining the interactive role of CRF and GABA receptors found that CRF can regulate the effects of ethanol by PKCε-mediated GABA potentiation [167, 168]. The evidence linking CRF to stress, anxiety, and the amygdala have suggested that CRF could serve as a possible pharmacological target for the treatment of alcohol addiction. It is worth noting that interactive dysregulation of CRF and NPY signaling (Fig. 2) in the amygdala may contribute to the development of alcohol dependence [169]. Future studies on the interactive role of these peptides within the amygdaloid circuitry may provide insight into the complex neurobiological system regulating alcohol drinking in response to stress and anxiety.
Epigenetic mechanisms in alcoholism
Brain chromatin remodeling regulates gene expression via enzymatic restructuring of histone proteins and DNA without altering the primary genetic sequence [170]. The nucleosome is the structural unit of chromatin and is composed of 147 base pairs of DNA wrapped around a histone octamer of the basic histone proteins (H2A, H2B, H3, and H4) [170, 171]. Initiation or inhibition of gene transcription depends on the accessibility of the chromatin to gene transcriptional machinery, such as transcription factors and RNA polymerases, and the binding ability of these effectors to the DNA [170, 172]. Covalent modifications of histone proteins occur at N-terminal tail regions and alter histone-DNA and histone-histone linkages through acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation, and SUMOylation [171, 173, 174]. Enzymes such as histone acetyltransferases (HATs), histone deacetylases (HDACs), methyltransferases, and protein kinases have been implicated in the regulation of gene transcription via chromatin remodeling [172–176]. HATs add acetyl groups to specific lysine residues which causes relaxation of the chromatin structure, allowing for increased binding of transcription factors to the DNA and increased gene expression [171, 174]. Interestingly, recruitment of CBP by CREB plays a role in chromatin remodeling (Fig. 1), as CBP has been shown to have intrinsic HAT activity [171, 175, 176]. In contrast to HATs, HDACs can remove acetyl groups, resulting in condensation of chromatin coils and decreased gene transcription [177, 178]. Pharmacological inhibition of HDACs can reinstate transcriptional activity by preventing histone deacetylation and promoting a relaxed chromatin structure [177, 178]. Several studies have implicated a role for epigenetic mechanisms, especially chromatin remodeling, in neurodegenerative and psychiatric disorders and in the development of drug addiction [172, 176, 179, 180]. The role of epigenetic mechanisms in neuropsychiatric disease and alcoholism is underlined by the study of the pharmacotherapeutic effect of HDAC inhibitors, which has recently become an important area of research [18, 179–181].
A recent study in our laboratory investigated brain chromatin remodeling in alcoholism. We found that acute ethanol exposure in SD rats elicited anxiolytic effects that were associated with decreased activity of HDACs and increased histone acetylation in the CeA and MeA. Furthermore, withdrawal from chronic ethanol treatment had the opposite effect by increasing HDAC activity and decreasing histone acetylation, which correlated with increased anxiety-like behaviors [18]. The observed changes in histone acetylation also correlated with changes in CBP levels and NPY expression in amygdaloid regions [18]. Treatment with trichostatin A (TSA), an HDAC inhibitor, was able to reverse the effects that ethanol withdrawal had on chromatin remodeling, NPY expression, and the precipitation of anxiety-like behaviors [18]. Another study demonstrated that TSA also reversed ethanol-related brain damage by restoring neural stem cell differentiation [182]. These results suggest that chromatin remodeling may play a role in the development of alcoholism and also introduce HDAC inhibitors as possible pharmacological agents for treating alcoholism. An earlier study using Western blot analysis of rat brains (cerebral hemispheres) showed that acetylated histone levels were unaffected by ethanol exposure [183]. However, it is possible that ethanol-induced chromatin remodeling occurs in only certain brain regions, as recently observed in amygdaloid structures during ethanol exposure [18]. Interestingly, adolescent, but not adult, rats intermittently treated with ethanol demonstrated significantly increased levels of histone H3 and H4 acetylation in brain regions associated with reward, such as the frontal cortex and NAc [184]. These results indicate that alcohol exposure during adolescence may contribute to an increased vulnerability to alcoholism, which may be caused by changes in brain chromatin remodeling.
Methylation of the genome can also impart either transcriptional activation or repression, depending on the histone protein and the specific amino acid residue that becomes methylated. Histone methyltransferases and histone demethylases work in cooperation with one another to correspondingly add or remove methylated groups to specific lysine or arginine residues located at the N-terminal tail regions of histone proteins [185–188]. The degree of methylation of these specific amino acid residues, which can be either mono-, di-, or tri-methylated, corresponds to the overall chromatin structure. Condensed chromatin, or heterochromatin, is often highly methylated and transcriptionally inactive, whereas relaxed chromatin, or euchromatin, has lower levels of methylation (Fig. 1) and higher transcriptional activity [185, 187, 189]. DNA methylation is also important in regulating gene expression and is facilitated by DNA methyltransferases and DNA demethylases that respectively add and remove methyl groups on DNA [190, 191]. Methylation of CpG islands in DNA is another epigenetic mechanism, and hypermethylation of DNA results in the reduction of gene expression [192]. Recently, the role of DNA methylation in ethanol-related changes in NMDA receptor (NR2B subunit) expression was explored. Chronic ethanol treatment was shown to upregulate NR2B gene expression; however, after 48-h of ethanol withdrawal NR2B levels were similar to control levels [53, 193, 194]. Furthermore, it was found that upregulation of NR2B subunit expression correlated to a reduction in CpG island methylation within the NR2B gene after chronic ethanol exposure [194]. Acute ethanol treatment neither affected DNA methylation nor changed the NR2B gene expression [194]. A study in humans showed that acute alcohol intake increased DNA methylation in the promoter region of the α-synuclein gene [195]. This hypermethylated state may be responsible for the satiated response to alcohol exposure by alcoholics [195].
In addition to classically defined epigenetic mechanisms as described above, microRNAs (miRNAs) can also convey epigenetic-like characteristics through post-transcriptional regulation of gene expression [173]. miRNAs can rapidly regulate gene expression by targeting certain mRNAs for degradation or through specific inhibition of mRNA translation. Alcohol has been shown to modulate BK channel expression, which may lead to tolerance [38, 79]. Recent studies on alcohol tolerance have implicated the importance of miRNAs in alcohol-induced changes in BK channel function [196, 197]. A study by Pietrzykowski et al. showed a prompt downregulation of BK channel expression upon acute alcohol exposure, which was accompanied by upregulated expression of miRNA-9 [196]. Interestingly, miRNA-9 post-transcriptionally regulated BK mRNA splice variants encoding BK channel isoforms that exhibited different ethanol sensitivities [196]. These findings suggest that the rapid onset of alcohol tolerance may occur due to post-translational epigenetic modifications involving miRNAs. Recently, ethanol exposure was also found to suppress four miRNAs, miRNA-21, -335, -9, and -153, which may be involved in ethanol-related teratogenicity [198]. Future studies are needed to shed light on how other miRNAs may be involved in the development of alcoholism.
Conclusion
The numerous molecular mechanisms involved in the development of alcohol addiction are complex and intertwined. CREB appears to be an important molecular mediator of neuroadaptational changes during alcohol exposure. There are several upstream and downstream regulators of CREB function during alcohol exposure (Fig. 1). Available evidence on CREB target genes clearly indicates that hypofunction of BDNF and NPY and hyperfunction of CRF in the CeA may be involved in the molecular mechanisms associated with the process of alcohol addiction (Fig. 2). In addition, epigenetic mechanisms, such as chromatin remodeling via histone acetylation and DNA methylation, as well as miRNAs, are not only important in regulating gene expression but also in establishing neuronal homeostasis during alcohol exposure and in the development of alcoholism. Finally, it is important to note that, while this review has described various molecular and epigenetic mechanisms of alcoholism, it is by no means exhaustive. Further studies are necessary to understand the mechanisms of alcohol addiction in order to improve current pharmacotherapeutics for alcoholism and related disorders.
Acknowledgments
The studies conducted in the laboratory of Dr. S.C. Pandey were supported by the grants from the National Institute on Alcohol Abuse and Alcoholism (AA-010005; AA-013341; AA-016690; AA-015626) and the Department of Veterans Affairs (Merit Review Grant; Research Career Scientist award).
References
- 1.Koob GF. Alcoholism: allostasis and beyond. Alcohol Clin Exp Res. 2003;27:232–243. doi: 10.1097/01.ALC.0000057122.36127.C2. [DOI] [PubMed] [Google Scholar]
- 2.Andreasen NC, Black DW. Introductory textbook of psychiatry. Washington: American Psychiatric Publication; 2006. [Google Scholar]
- 3.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 4.Koob GF, Sanna PP, Bloom FE. Neuroscience of addiction. Neuron. 1998;21:467–476. doi: 10.1016/s0896-6273(00)80557-7. [DOI] [PubMed] [Google Scholar]
- 5.Pandey SC. The gene transcription factor cyclic AMP-responsive element binding protein: role in positive and negative affective states of alcohol addiction. Pharmacol Ther. 2004;104:47–58. doi: 10.1016/j.pharmthera.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Gonzales RA, Job MO, Doyon WM. The role of mesolimbic dopamine in the development and maintenance of ethanol reinforcement. Pharmacol Ther. 2004;103:121–146. doi: 10.1016/j.pharmthera.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Conway KP, Compton W, Stinson FS, Grant BF. Lifetime comorbidity of DSM-IV mood and anxiety disorders and specific drug use disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. J Clin Psychiatry. 2006;67:247–257. doi: 10.4088/jcp.v67n0211. [DOI] [PubMed] [Google Scholar]
- 8.Schuckit MA, Hesselbrock V. Alcohol dependence and anxiety disorders: what is the relationship? Am J Psychiatry. 1994;151:1723–1734. doi: 10.1176/ajp.151.12.1723. [DOI] [PubMed] [Google Scholar]
- 9.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- 11.Morrow AL, Ferrani-Kile K, Davis MI, Shumilla JA, Kumar S, Maldve R, Pandey SC. Ethanol effects on cell signaling mechanisms. Alcohol Clin Exp Res. 2004;28:217–227. doi: 10.1097/01.alc.0000113439.97498.ac. [DOI] [PubMed] [Google Scholar]
- 12.Harris RA, Trudell JR, Mihic SJ (2008) Ethanol’s molecular targets. Sci Signal 1, re7 [DOI] [PMC free article] [PubMed]
- 13.Heilig M, Koob GF. A key role for corticotropin-releasing factor in alcohol dependence. Trends Neurosci. 2007;30:399–406. doi: 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorsell A. Central neuropeptide Y in anxiety- and stress-related behavior and in ethanol intake. Ann N Y Acad Sci. 2008;1148:136–140. doi: 10.1196/annals.1410.083. [DOI] [PubMed] [Google Scholar]
- 15.Davis MI. Ethanol-BDNF interactions: still more questions than answers. Pharmacol Ther. 2008;118:36–57. doi: 10.1016/j.pharmthera.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene Arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008;28:2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32:1525–1534. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 18.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abel T, Kandel E. Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res Brain Res Rev. 1998;26:360–378. doi: 10.1016/s0165-0173(97)00050-7. [DOI] [PubMed] [Google Scholar]
- 20.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 22.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 23.Li TK, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23:163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- 24.Pandey SC, Mittal N, Lumeng L, Li TK. Involvement of the cyclic AMP-responsive element binding protein gene transcription factor in genetic preference for alcohol drinking behavior. Alcohol Clin Exp Res. 1999;23:1425–1434. [PubMed] [Google Scholar]
- 25.Bell RL, Rodd ZA, Lumeng L, Murphy JM, McBride WJ. The alcohol-preferring P rat and animal models of excessive alcohol drinking. Addict Biol. 2006;11:270–288. doi: 10.1111/j.1369-1600.2005.00029.x. [DOI] [PubMed] [Google Scholar]
- 26.Pandey SC, Zhang H, Roy A, Xu T. Deficits in amygdaloid cAMP-responsive element-binding protein signaling play a role in genetic predisposition to anxiety and alcoholism. J Clin Invest. 2005;115:2762–2773. doi: 10.1172/JCI24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey SC, Roy A, Zhang H, Xu T. Partial deletion of the cAMP response element-binding protein gene promotes alcohol-drinking behaviors. J Neurosci. 2004;24:5022–5030. doi: 10.1523/JNEUROSCI.5557-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey SC, Roy A, Zhang H. The decreased phosphorylation of cyclic adenosine monophosphate (cAMP) response element binding (CREB) protein in the central amygdala acts as a molecular substrate for anxiety related to ethanol withdrawal in rats. Alcohol Clin Exp Res. 2003;27:396–409. doi: 10.1097/01.ALC.0000056616.81971.49. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Horn K, Wand GS. Chronic ethanol exposure impairs phosphorylation of CREB and CRE-binding activity in rat striatum. Alcohol Clin Exp Res. 1998;22:382–390. [PubMed] [Google Scholar]
- 30.Yang X, Diehl AM, Wand GS. Ethanol exposure alters the phosphorylation of cyclic AMP responsive element binding protein and cyclic AMP responsive element binding activity in rat cerebellum. J Pharmacol Exp Ther. 1996;278:338–346. [PubMed] [Google Scholar]
- 31.Yang X, Horn K, Baraban JM, Wand GS. Chronic ethanol administration decreases phosphorylation of cyclic AMP response element-binding protein in granule cells of rat cerebellum. J Neurochem. 1998;70:224–232. doi: 10.1046/j.1471-4159.1998.70010224.x. [DOI] [PubMed] [Google Scholar]
- 32.Pandey SC, Zhang D, Mittal N, Nayyar D. Potential role of the gene transcription factor cyclic AMP-responsive element binding protein in ethanol withdrawal-related anxiety. J Pharmacol Exp Ther. 1999;288:866–878. [PubMed] [Google Scholar]
- 33.Pandey SC, Roy A, Mittal N. Effects of chronic ethanol intake and its withdrawal on the expression and phosphorylation of the CREB gene transcription factor in rat cortex. J Pharmacol Exp Ther. 2001;296:857–868. [PubMed] [Google Scholar]
- 34.Li J, Li YH, Yuan XR. Changes of phosphorylation of cAMP response element binding protein in rat nucleus accumbens after chronic ethanol intake: naloxone reversal. Acta Pharmacol Sin. 2003;24:930–936. [PubMed] [Google Scholar]
- 35.Misra K, Roy A, Pandey SC. Effects of voluntary ethanol intake on the expression of Ca2+/calmodulin-dependent protein kinase IV and on CREB expression and phosphorylation in the rat nucleus accumbens. Neuroreport. 2001;12:4133–4137. doi: 10.1097/00001756-200112210-00054. [DOI] [PubMed] [Google Scholar]
- 36.Misra K, Pandey SC. Differences in basal levels of CREB and NPY in nucleus accumbens regions between C57BL/6 and DBA/2 mice differing in inborn alcohol drinking behavior. J Neurosci Res. 2003;74:967–975. doi: 10.1002/jnr.10831. [DOI] [PubMed] [Google Scholar]
- 37.Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Ghezzi A, Yin JC, Atkinson NS. CREB regulation of BK channel gene expression underlies rapid drug tolerance. Genes Brain Behav. 2009;8:369–376. doi: 10.1111/j.1601-183X.2009.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Oswald L, Wand G. The cyclic AMP/protein kinase A signal transduction pathway modulates tolerance to sedative and hypothermic effects of ethanol. Alcohol Clin Exp Res. 2003;27:1220–1225. doi: 10.1097/01.ALC.0000081626.02910.19. [DOI] [PubMed] [Google Scholar]
- 40.Zou J, Crews F. CREB and NF-κB transcription factors regulate sensitivity to excitotoxic and oxidative stress induced neuronal cell death. Cell Mol Neurobiol. 2006;26:385–405. doi: 10.1007/s10571-006-9045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chandler LJ, Sutton G. Acute ethanol inhibits extracellular signal-regulated kinase, protein kinase B, and adenosine 3′:5′-cyclic monophosphate response element binding protein activity in an age- and brain region-specific manner. Alcohol Clin Exp Res. 2005;29:672–682. doi: 10.1097/01.alc.0000158935.53360.5f. [DOI] [PubMed] [Google Scholar]
- 42.Acquaah-Mensah GK, Misra V, Biswal S. Ethanol sensitivity: a central role for CREB transcription regulation in the cerebellum. BMC Genomics. 2006;7:308. doi: 10.1186/1471-2164-7-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narahashi T, Kuriyama K, Illes P, Wirkner K, Fischer W, Muhlberg K, Scheibler P, Allgaier C, Minami K, Lovinger D, Lallemand F, Ward RJ, DeWitte P, Itatsu T, Takei Y, Oide H, Hirose M, Wang XE, Watanabe S, Tateyama M, Ochi R, Sato N. Neuroreceptors and ion channels as targets of alcohol. Alcohol Clin Exp Res. 2001;25:182S–188S. doi: 10.1097/00000374-200105051-00030. [DOI] [PubMed] [Google Scholar]
- 44.Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br J Pharmacol. 2008;154:299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lovinger DM, White G, Weight FF. Ethanol inhibits NMDA-activated ion current in hippocampal neurons. Science. 1989;243:1721–1724. doi: 10.1126/science.2467382. [DOI] [PubMed] [Google Scholar]
- 46.Nestoros JN. Ethanol specifically potentiates GABA-mediated neurotransmission in feline cerebral cortex. Science. 1980;209:708–710. doi: 10.1126/science.7394531. [DOI] [PubMed] [Google Scholar]
- 47.Peoples RW, Weight FF. Cutoff in potency implicates alcohol inhibition of n-methyl-d-aspartate receptors in alcohol intoxication. Proc Natl Acad Sci USA. 1995;92:2825–2829. doi: 10.1073/pnas.92.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren H, Salous AK, Paul JM, Lipsky RH, Peoples RW. Mutations at F637 in the NMDA receptor NR2A subunit M3 domain influence agonist potency, ion channel gating and alcohol action. Br J Pharmacol. 2007;151:749–757. doi: 10.1038/sj.bjp.0707254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronald KM, Mirshahi T, Woodward JJ. Ethanol inhibition of n-methyl-d-aspartate receptors is reduced by site-directed mutagenesis of a transmembrane domain phenylalanine residue. J Biol Chem. 2001;276:44729–44735. doi: 10.1074/jbc.M102800200. [DOI] [PubMed] [Google Scholar]
- 50.Paoletti P, Neyton J. NMDA receptor subunits: function and pharmacology. Curr Opin Pharmacol. 2007;7:39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 51.Smothers CT, Clayton R, Blevins T, Woodward JJ. Ethanol sensitivity of recombinant human n-methyl-d-aspartate receptors. Neurochem Int. 2001;38:333–340. doi: 10.1016/s0197-0186(00)00094-2. [DOI] [PubMed] [Google Scholar]
- 52.Nagy J. The NR2B subtype of NMDA receptor: a potential target for the treatment of alcohol dependence. Curr Drug Targets CNS Neurol Disord. 2004;3:169–179. doi: 10.2174/1568007043337409. [DOI] [PubMed] [Google Scholar]
- 53.Rani CS, Qiang M, Ticku MK. Potential role of cAMP response element-binding protein in ethanol-induced n-methyl-d-aspartate receptor 2B subunit gene transcription in fetal mouse cortical cells. Mol Pharmacol. 2005;67:2126–2136. doi: 10.1124/mol.104.007872. [DOI] [PubMed] [Google Scholar]
- 54.Wang JQ, Fibuch EE, Mao L. Regulation of mitogen-activated protein kinases by glutamate receptors. J Neurochem. 2007;100:1–11. doi: 10.1111/j.1471-4159.2006.04208.x. [DOI] [PubMed] [Google Scholar]
- 55.Netzeband JG, Gruol DL. Modulatory effects of acute ethanol on metabotropic glutamate responses in cultured Purkinje neurons. Brain Res. 1995;688:105–113. doi: 10.1016/0006-8993(95)00517-t. [DOI] [PubMed] [Google Scholar]
- 56.Minami K, Gereau RW, 4th, Minami M, Heinemann SF, Harris RA. Effects of ethanol and anesthetics on type 1 and 5 metabotropic glutamate receptors expressed in Xenopus laevis oocytes. Mol Pharmacol. 1998;53:148–156. doi: 10.1124/mol.53.1.148. [DOI] [PubMed] [Google Scholar]
- 57.Gass JT, Olive MF. Role of protein kinase C epsilon (PKCε) in the reduction of ethanol reinforcement due to mGluR5 antagonism in the nucleus accumbens shell. Psychopharmacology (Berl) 2009;204:587–597. doi: 10.1007/s00213-009-1490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroeder JP, Overstreet DH, Hodge CW. The mGluR5 antagonist MPEP decreases operant ethanol self-administration during maintenance and after repeated alcohol deprivations in alcohol-preferring (P) rats. Psychopharmacology (Berl) 2005;179:262–270. doi: 10.1007/s00213-005-2175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMillen BA, Crawford MS, Kulers CM, Williams HL. Effects of a metabotropic, mGlu5, glutamate receptor antagonist on ethanol consumption by genetic drinking rats. Alcohol Alcohol. 2005;40:494–497. doi: 10.1093/alcalc/agh200. [DOI] [PubMed] [Google Scholar]
- 60.Hodge CW, Miles MF, Sharko AC, Stevenson RA, Hillmann JR, Lepoutre V, Besheer J, Schroeder JP. The mGluR5 antagonist MPEP selectively inhibits the onset and maintenance of ethanol self-administration in C57BL/6J mice. Psychopharmacology (Berl) 2006;183:429–438. doi: 10.1007/s00213-005-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mermelstein PG, Bito H, Deisseroth K, Tsien RW. Critical dependence of cAMP response element-binding protein phosphorylation on L-type calcium channels supports a selective response to EPSPs in preference to action potentials. J Neurosci. 2000;20:266–273. doi: 10.1523/JNEUROSCI.20-01-00266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullikin-Kilpatrick D, Mehta ND, Hildebrandt JD, Treistman SN. Gi is involved in ethanol inhibition of l-type calcium channels in undifferentiated but not differentiated PC-12 cells. Mol Pharmacol. 1995;47:997–1005. [PubMed] [Google Scholar]
- 63.Katsura M, Shibasaki M, Hayashida S, Torigoe F, Tsujimura A, Ohkuma S. Increase in expression of α1 and α2/δ1 subunits of l-type high voltage-gated calcium channels after sustained ethanol exposure in cerebral cortical neurons. J Pharmacol Sci. 2006;102:221–230. doi: 10.1254/jphs.fp0060781. [DOI] [PubMed] [Google Scholar]
- 64.Walter HJ, McMahon T, Dadgar J, Wang D, Messing RO. Ethanol regulates calcium channel subunits by protein kinase C δ-dependent and -independent mechanisms. J Biol Chem. 2000;275:25717–25722. doi: 10.1074/jbc.M910282199. [DOI] [PubMed] [Google Scholar]
- 65.Gardell LR, Reid LD, Boedeker KL, Liakos TM, Hubbell CL. Isradipine and naltrexone in combination with isradipine interact with a period of abstinence to reduce rats’ intakes of an alcoholic beverage. Alcohol Clin Exp Res. 1997;21:1592–1598. [PubMed] [Google Scholar]
- 66.Rezvani AH, Janowsky DS. Decreased alcohol consumption by verapamil in alcohol preferring rats. Prog Neuropsychopharmacol Biol Psychiatry. 1990;14:623–631. doi: 10.1016/0278-5846(90)90013-7. [DOI] [PubMed] [Google Scholar]
- 67.De Beun R, Schneider R, Klein A, Lohmann A, De Vry J. Effects of nimodipine and other calcium channel antagonists in alcohol-preferring AA rats. Alcohol. 1996;13:263–271. doi: 10.1016/0741-8329(95)02054-3. [DOI] [PubMed] [Google Scholar]
- 68.Hatta S, Saito T, Ohshika H. Effects of ethanol on the function of G proteins in rat cerebral cortex membranes. Alcohol Alcohol Suppl. 1994;29:45–51. [PubMed] [Google Scholar]
- 69.Mochly-Rosen D, Chang FH, Cheever L, Kim M, Diamond I, Gordon AS. Chronic ethanol causes heterologous desensitization of receptors by reducing αs messenger RNA. Nature. 1988;333:848–850. doi: 10.1038/333848a0. [DOI] [PubMed] [Google Scholar]
- 70.Tabakoff B, Whelan JP, Ovchinnikova L, Nhamburo P, Yoshimura M, Hoffman PL. Quantitative changes in G proteins do not mediate ethanol-induced downregulation of adenylyl cyclase in mouse cerebral cortex. Alcohol Clin Exp Res. 1995;19:187–194. doi: 10.1111/j.1530-0277.1995.tb01491.x. [DOI] [PubMed] [Google Scholar]
- 71.Yoshimura M, Tabakoff B. Selective effects of ethanol on the generation of cAMP by particular members of the adenylyl cyclase family. Alcohol Clin Exp Res. 1995;19:1435–1440. doi: 10.1111/j.1530-0277.1995.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 72.Yoshimura M, Pearson S, Kadota Y, Gonzalez CE. Identification of ethanol responsive domains of adenylyl cyclase. Alcohol Clin Exp Res. 2006;30:1824–1832. doi: 10.1111/j.1530-0277.2006.00219.x. [DOI] [PubMed] [Google Scholar]
- 73.Constantinescu A, Diamond I, Gordon AS. Ethanol-induced translocation of cAMP-dependent protein kinase to the nucleus. Mechanism and functional consequences. J Biol Chem. 1999;274:26985–26991. doi: 10.1074/jbc.274.38.26985. [DOI] [PubMed] [Google Scholar]
- 74.Ron D, Jurd R (2005) The “ups and downs” of signaling cascades in addiction. Sci STKE 2005, re14 [DOI] [PubMed]
- 75.Lee AM, Messing RO. Protein kinases and addiction. Ann N Y Acad Sci. 2008;1141:22–57. doi: 10.1196/annals.1441.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wayman GA, Lee YS, Tokumitsu H, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, Bian WL, Xie GQ, Cui SZ, Wu ML, Li YH, Que LL, Yuan XR. Chronic ethanol intake-induced changes in open-field behavior and calcium/calmodulin-dependent protein kinase IV expression in nucleus accumbens of rats: naloxone reversal. Acta Pharmacol Sin. 2008;29:646–652. doi: 10.1111/j.1745-7254.2008.00805.x. [DOI] [PubMed] [Google Scholar]
- 78.Mahadev K, Chetty CS, Vemuri MC. Effect of prenatal and postnatal ethanol exposure on Ca2+/calmodulin-dependent protein kinase II in rat cerebral cortex. Alcohol. 2001;23:183–188. doi: 10.1016/s0741-8329(01)00133-1. [DOI] [PubMed] [Google Scholar]
- 79.Liu J, Asuncion-Chin M, Liu P, Dopico AM. CaM kinase II phosphorylation of slo Thr107 regulates activity and ethanol responses of BK channels. Nat Neurosci. 2006;9:41–49. doi: 10.1038/nn1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu M, Chandler LJ, Woodward JJ. Ethanol inhibition of recombinant NMDA receptors is not altered by coexpression of CaMKII-α or CaMKII-β. Alcohol. 2008;42:425–432. doi: 10.1016/j.alcohol.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brandon EP, Idzerda RL, McKnight GS. PKA isoforms, neural pathways, and behaviour: making the connection. Curr Opin Neurobiol. 1997;7:397–403. doi: 10.1016/s0959-4388(97)80069-4. [DOI] [PubMed] [Google Scholar]
- 82.Dohrman DP, Diamond I, Gordon AS. Ethanol causes translocation of cAMP-dependent protein kinase catalytic subunit to the nucleus. Proc Natl Acad Sci USA. 1996;93:10217–10221. doi: 10.1073/pnas.93.19.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Misra K, Pandey SC. The decreased cyclic-AMP dependent-protein kinase A function in the nucleus accumbens: a role in alcohol drinking but not in anxiety-like behaviors in rats. Neuropsychopharmacology. 2006;31:1406–1419. doi: 10.1038/sj.npp.1300900. [DOI] [PubMed] [Google Scholar]
- 84.Yao L, Arolfo MP, Dohrman DP, Jiang Z, Fan P, Fuchs S, Janak PH, Gordon AS, Diamond I. βγ Dimers mediate synergy of dopamine D2 and adenosine A2 receptor-stimulated PKA signaling and regulate ethanol consumption. Cell. 2002;109:733–743. doi: 10.1016/s0092-8674(02)00763-8. [DOI] [PubMed] [Google Scholar]
- 85.Zhang H, Pandey SC. Effects of PKA modulation on the expression of neuropeptide Y in rat amygdaloid structures during ethanol withdrawal. Peptides. 2003;24:1397–1402. doi: 10.1016/j.peptides.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 86.Lai CC, Kuo TI, Lin HH. The role of protein kinase A in acute ethanol-induced neurobehavioral actions in rats. Anesth Analg. 2007;105:89–96. doi: 10.1213/01.ane.0000263030.13249.36. [DOI] [PubMed] [Google Scholar]
- 87.Thiele TE, Willis B, Stadler J, Reynolds JG, Bernstein IL, McKnight GS. High ethanol consumption and low sensitivity to ethanol-induced sedation in protein kinase A-mutant mice. J Neurosci. 2000;20:RC75. doi: 10.1523/JNEUROSCI.20-10-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Repunte-Canonigo V, Lutjens R, van der Stap LD, Sanna PP. Increased expression of protein kinase A inhibitor α (PKI-α) and decreased PKA-regulated genes in chronic intermittent alcohol exposure. Brain Res. 2007;1138:48–56. doi: 10.1016/j.brainres.2006.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 90.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 91.Sanna PP, Simpson C, Lutjens R, Koob G. ERK regulation in chronic ethanol exposure and withdrawal. Brain Res. 2002;948:186–191. doi: 10.1016/s0006-8993(02)03191-8. [DOI] [PubMed] [Google Scholar]
- 92.Kalluri HS, Ticku MK. Regulation of ERK phosphorylation by ethanol in fetal cortical neurons. Neurochem Res. 2003;28:765–769. doi: 10.1023/a:1022822119560. [DOI] [PubMed] [Google Scholar]
- 93.Carnicella S, Kharazia V, Jeanblanc J, Janak PH, Ron D. GDNF is a fast-acting potent inhibitor of alcohol consumption and relapse. Proc Natl Acad Sci USA. 2008;105:8114–8119. doi: 10.1073/pnas.0711755105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pandey SC, Zhang H, Roy A, Misra K. Central and medial amygdaloid brain-derived neurotrophic factor signaling plays a critical role in alcohol-drinking and anxiety-like behaviors. J Neurosci. 2006;26:8320–8331. doi: 10.1523/JNEUROSCI.4988-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie GQ, Wang SJ, Li J, Cui SZ, Zhou R, Chen L, Yuan XR. Ethanol attenuates the HFS-induced, ERK-mediated LTP in a dose-dependent manner in rat striatum. Alcohol Clin Exp Res. 2009;33:121–128. doi: 10.1111/j.1530-0277.2008.00818.x. [DOI] [PubMed] [Google Scholar]
- 96.Roberto M, Nelson TE, Ur CL, Brunelli M, Sanna PP, Gruol DL. The transient depression of hippocampal CA1 LTP induced by chronic intermittent ethanol exposure is associated with an inhibition of the MAP kinase pathway. Eur J Neurosci. 2003;17:1646–1654. doi: 10.1046/j.1460-9568.2003.02614.x. [DOI] [PubMed] [Google Scholar]
- 97.Kuczewski N, Porcher C, Lessmann V, Medina I, Gaiarsa JL. Activity-dependent dendritic release of BDNF and biological consequences. Mol Neurobiol. 2009;39:37–49. doi: 10.1007/s12035-009-8050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Horch HW. Local effects of BDNF on dendritic growth. Rev Neurosci. 2004;15:117–129. doi: 10.1515/revneuro.2004.15.2.117. [DOI] [PubMed] [Google Scholar]
- 99.Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- 100.Messaoudi E, Ying SW, Kanhema T, Croll SD, Bramham CR. Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J Neurosci. 2002;22:7453–7461. doi: 10.1523/JNEUROSCI.22-17-07453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 102.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pizzorusso T, Ratto GM, Putignano E, Maffei L. Brain-derived neurotrophic factor causes cAMP response element-binding protein phosphorylation in absence of calcium increases in slices and cultured neurons from rat visual cortex. J Neurosci. 2000;20:2809–2816. doi: 10.1523/JNEUROSCI.20-08-02809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 105.Deogracias R, Espliguero G, Iglesias T, Rodriguez-Pena A. Expression of the neurotrophin receptor TrkB is regulated by the cAMP/CREB pathway in neurons. Mol Cell Neurosci. 2004;26:470–480. doi: 10.1016/j.mcn.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 106.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 107.Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neurosci. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene Arc/Arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo . J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang F, Chotiner JK, Steward O. Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J Neurosci. 2007;27:9054–9067. doi: 10.1523/JNEUROSCI.2410-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Soule J, Messaoudi E, Bramham CR. Brain-derived neurotrophic factor and control of synaptic consolidation in the adult brain. Biochem Soc Trans. 2006;34:600–604. doi: 10.1042/BST0340600. [DOI] [PubMed] [Google Scholar]
- 112.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 113.Vickers CA, Stephens B, Bowen J, Arbuthnott GW, Grant SG, Ingham CA. Neurone specific regulation of dendritic spines in vivo by post synaptic density 95 protein (PSD-95) Brain Res. 2006;1090:89–98. doi: 10.1016/j.brainres.2006.03.075. [DOI] [PubMed] [Google Scholar]
- 114.Starkman BG, Pandey SC. Brain-derived neurotrophic factor and mental illness: an epigenetic approach. Proc Natl Acad Sci India. 2007;77(B):105–113. [Google Scholar]
- 115.Angelucci F, Brene S, Mathe AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 116.Jeanblanc J, He DY, McGough NN, Logrip ML, Phamluong K, Janak PH, Ron D. The dopamine D3 receptor is part of a homeostatic pathway regulating ethanol consumption. J Neurosci. 2006;26:1457–1464. doi: 10.1523/JNEUROSCI.3786-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hensler JG, Ladenheim EE, Lyons WE. Ethanol consumption and serotonin-1A (5-HT1A) receptor function in heterozygous BDNF (+/−) mice. J Neurochem. 2003;85:1139–1147. doi: 10.1046/j.1471-4159.2003.01748.x. [DOI] [PubMed] [Google Scholar]
- 118.MacLennan AJ, Lee N, Walker DW. Chronic ethanol administration decreases brain-derived neurotrophic factor gene expression in the rat hippocampus. Neurosci Lett. 1995;197:105–108. doi: 10.1016/0304-3940(95)11922-j. [DOI] [PubMed] [Google Scholar]
- 119.Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sakai R, Ukai W, Sohma H, Hashimoto E, Yamamoto M, Ikeda H, Saito T. Attenuation of brain derived neurotrophic factor (BDNF) by ethanol and cytoprotective effect of exogenous BDNF against ethanol damage in neuronal cells. J Neural Transm. 2005;112:1005–1013. doi: 10.1007/s00702-004-0246-4. [DOI] [PubMed] [Google Scholar]
- 121.McGough NN, He DY, Logrip ML, Jeanblanc J, Phamluong K, Luong K, Kharazia V, Janak PH, Ron D. RACK1 and brain-derived neurotrophic factor: a homeostatic pathway that regulates alcohol addiction. J Neurosci. 2004;24:10542–10552. doi: 10.1523/JNEUROSCI.3714-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Prakash A, Zhang H, Pandey SC. Innate differences in the expression of brain-derived neurotrophic factor in the regions within the extended amygdala between alcohol preferring and nonpreferring rats. Alcohol Clin Exp Res. 2008;32:909–920. doi: 10.1111/j.1530-0277.2008.00650.x. [DOI] [PubMed] [Google Scholar]
- 123.Yan QS, Feng MJ, Yan SE. Different expression of brain-derived neurotrophic factor in the nucleus accumbens of alcohol-preferring (P) and -nonpreferring (NP) rats. Brain Res. 2005;1035:215–218. doi: 10.1016/j.brainres.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 124.Lescaudron L, Jaffard R, Verna A. Modifications in number and morphology of dendritic spines resulting from chronic ethanol consumption and withdrawal: a Golgi study in the mouse anterior and posterior hippocampus. Exp Neurol. 1989;106:156–163. doi: 10.1016/0014-4886(89)90089-7. [DOI] [PubMed] [Google Scholar]
- 125.Lee K, Dunwiddie T, Deitrich R, Lynch G, Hoffer B. Chronic ethanol consumption and hippocampal neuron dendritic spines: a morphometric and physiological analysis. Exp Neurol. 1981;71:541–549. doi: 10.1016/0014-4886(81)90031-5. [DOI] [PubMed] [Google Scholar]
- 126.Riley JN, Walker DW. Morphological alterations in hippocampus after long-term alcohol consumption in mice. Science. 1978;201:646–648. doi: 10.1126/science.566953. [DOI] [PubMed] [Google Scholar]
- 127.Chandler LJ. Ethanol and brain plasticity: receptors and molecular networks of the postsynaptic density as targets of ethanol. Pharmacol Ther. 2003;99:311–326. doi: 10.1016/s0163-7258(03)00096-2. [DOI] [PubMed] [Google Scholar]
- 128.Zhou FC, Anthony B, Dunn KW, Lindquist WB, Xu ZC, Deng P. Chronic alcohol drinking alters neuronal dendritic spines in the brain reward center nucleus accumbens. Brain Res. 2007;1134:148–161. doi: 10.1016/j.brainres.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moonat S, Sakharkar A, Zhang H, Pandey SC. Effects of acute ethanol exposure on amygdaloid dendritic morphology and anxiety-like behaviors in P and NP rats. J Neurochem. 2009;108(Suppl 1):96–97. [Google Scholar]
- 130.Carpenter-Hyland EP, Chandler LJ. Homeostatic plasticity during alcohol exposure promotes enlargement of dendritic spines. Eur J Neurosci. 2006;24:3496–3506. doi: 10.1111/j.1460-9568.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- 131.Halford JC, Cooper GD, Dovey TM. The pharmacology of human appetite expression. Curr Drug Targets. 2004;5:221–240. doi: 10.2174/1389450043490541. [DOI] [PubMed] [Google Scholar]
- 132.Hansel DE, Eipper BA, Ronnett GV. Regulation of olfactory neurogenesis by amidated neuropeptides. J Neurosci Res. 2001;66:1–7. doi: 10.1002/jnr.1191. [DOI] [PubMed] [Google Scholar]
- 133.Sajdyk TJ, Shekhar A, Gehlert DR. Interactions between NPY and CRF in the amygdala to regulate emotionality. Neuropeptides. 2004;38:225–234. doi: 10.1016/j.npep.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 134.Fetissov SO, Kopp J, Hokfelt T. Distribution of NPY receptors in the hypothalamus. Neuropeptides. 2004;38:175–188. doi: 10.1016/j.npep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 135.Sheriff S, Qureshy AF, Chance WT, Kasckow JW, Balasubramaniam A. Predominant role by CaM kinase in NPY Y1 receptor signaling: involvement of CREB. Peptides. 2002;23:87–96. doi: 10.1016/s0196-9781(01)00583-6. [DOI] [PubMed] [Google Scholar]
- 136.Chance WT, Sheriff S, Peng F, Balasubramaniam A. Antagonism of NPY-induced feeding by pretreatment with cyclic AMP response element binding protein antisense oligonucleotide. Neuropeptides. 2000;34:167–172. doi: 10.1054/npep.2000.0807. [DOI] [PubMed] [Google Scholar]
- 137.Higuchi H, Yang HY, Sabol SL. Rat neuropeptide Y precursor gene expression. mRNA structure, tissue distribution, and regulation by glucocorticoids, cyclic AMP, and phorbol ester. J Biol Chem. 1988;263:6288–6295. [PubMed] [Google Scholar]
- 138.Sheriff S, Dayal R, Kasckow J, Regmi A, Chance W, Fischer J, Balasubramaniam A. NPY upregulates genes containing cyclic AMP response element in human neuroblastoma cell lines bearing Y1 and Y2 receptors: involvement of CREB. Regul Pept. 1998;75–76:309–318. doi: 10.1016/s0167-0115(98)00083-4. [DOI] [PubMed] [Google Scholar]
- 139.Hwang BH, Zhang JK, Ehlers CL, Lumeng L, Li TK. Innate differences of neuropeptide Y (NPY) in hypothalamic nuclei and central nucleus of the amygdala between selectively bred rats with high and low alcohol preference. Alcohol Clin Exp Res. 1999;23:1023–1030. [PubMed] [Google Scholar]
- 140.Roy A, Pandey SC. The decreased cellular expression of neuropeptide Y protein in rat brain structures during ethanol withdrawal after chronic ethanol exposure. Alcohol Clin Exp Res. 2002;26:796–803. [PubMed] [Google Scholar]
- 141.Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:366–369. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- 142.Palmiter RD, Erickson JC, Hollopeter G, Baraban SC, Schwartz MW. Life without neuropeptide Y. Recent Prog Horm Res. 1998;53:163–199. [PubMed] [Google Scholar]
- 143.Heilig M, Soderpalm B, Engel JA, Widerlov E. Centrally administered neuropeptide Y (NPY) produces anxiolytic-like effects in animal anxiety models. Psychopharmacology (Berl) 1989;98:524–529. doi: 10.1007/BF00441953. [DOI] [PubMed] [Google Scholar]
- 144.Heilig M. Antisense inhibition of neuropeptide Y (NPY)-Y1 receptor expression blocks the anxiolytic-like action of NPY in amygdala and paradoxically increases feeding. Regul Pept. 1995;59:201–205. doi: 10.1016/0167-0115(95)00103-i. [DOI] [PubMed] [Google Scholar]
- 145.Thiele TE, Koh MT, Pedrazzini T. Voluntary alcohol consumption is controlled via the neuropeptide Y Y1 receptor. J Neurosci. 2002;22(RC208):1–6. doi: 10.1523/JNEUROSCI.22-03-j0006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rimondini R, Thorsell A, Heilig M. Suppression of ethanol self-administration by the neuropeptide Y (NPY) Y2 receptor antagonist BIIE0246: evidence for sensitization in rats with a history of dependence. Neurosci Lett. 2005;375:129–133. doi: 10.1016/j.neulet.2004.10.084. [DOI] [PubMed] [Google Scholar]
- 147.Thiele TE, Naveilhan P, Ernfors P. Assessment of ethanol consumption and water drinking by NPY Y2 receptor knockout mice. Peptides. 2004;25:975–983. doi: 10.1016/j.peptides.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 148.Schroeder JP, Iller KA, Hodge CW. Neuropeptide-Y Y5 receptors modulate the onset and maintenance of operant ethanol self-administration. Alcohol Clin Exp Res. 2003;27:1912–1920. doi: 10.1097/01.ALC.0000098873.80433.BA. [DOI] [PubMed] [Google Scholar]
- 149.Katner SN, Slawecki CJ, Ehlers CL. Neuropeptide Y administration into the amygdala does not affect ethanol consumption. Alcohol. 2002;28:29–38. doi: 10.1016/s0741-8329(02)00235-5. [DOI] [PubMed] [Google Scholar]
- 150.Badia-Elder NE, Stewart RB, Powrozek TA, Roy KF, Murphy JM, Li TK. Effect of neuropeptide Y (NPY) on oral ethanol intake in Wistar, alcohol-preferring (P), and -nonpreferring (NP) rats. Alcohol Clin Exp Res. 2001;25:386–390. [PubMed] [Google Scholar]
- 151.Primeaux SD, Wilson SP, Bray GA, York DA, Wilson MA. Overexpression of neuropeptide Y in the central nucleus of the amygdala decreases ethanol self-administration in “anxious” rats. Alcohol Clin Exp Res. 2006;30:791–801. doi: 10.1111/j.1530-0277.2006.00092.x. [DOI] [PubMed] [Google Scholar]
- 152.Thorsell A, Repunte-Canonigo V, O’Dell LE, Chen SA, King AR, Lekic D, Koob GF, Sanna PP. Viral vector-induced amygdala NPY overexpression reverses increased alcohol intake caused by repeated deprivations in Wistar rats. Brain. 2007;130:1330–1337. doi: 10.1093/brain/awm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gilpin NW, Stewart RB, Badia-Elder NE. Neuropeptide Y administration into the amygdala suppresses ethanol drinking in alcohol-preferring (P) rats following multiple deprivations. Pharmacol Biochem Behav. 2008;90:470–474. doi: 10.1016/j.pbb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gilpin NW, Misra K, Koob GF. Neuropeptide Y in the central nucleus of the amygdala suppresses dependence-induced increases in alcohol drinking. Pharmacol Biochem Behav. 2008;90:475–480. doi: 10.1016/j.pbb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Perrin MH, Vale WW. Corticotropin releasing factor receptors and their ligand family. Ann N Y Acad Sci. 1999;885:312–328. doi: 10.1111/j.1749-6632.1999.tb08687.x. [DOI] [PubMed] [Google Scholar]
- 156.Gray TS. Amygdaloid CRF pathways. Role in autonomic, neuroendocrine, and behavioral responses to stress. Ann N Y Acad Sci. 1993;697:53–60. doi: 10.1111/j.1749-6632.1993.tb49922.x. [DOI] [PubMed] [Google Scholar]
- 157.Koob GF, Heinrichs SC. A role for corticotropin releasing factor and urocortin in behavioral responses to stressors. Brain Res. 1999;848:141–152. doi: 10.1016/s0006-8993(99)01991-5. [DOI] [PubMed] [Google Scholar]
- 158.Yao M, Stenzel-Poore M, Denver RJ. Structural and functional conservation of vertebrate corticotropin-releasing factor genes: evidence for a critical role for a conserved cyclic AMP response element. Endocrinology. 2007;148:2518–2531. doi: 10.1210/en.2006-1413. [DOI] [PubMed] [Google Scholar]
- 159.Rassnick S, Heinrichs SC, Britton KT, Koob GF. Microinjection of a corticotropin-releasing factor antagonist into the central nucleus of the amygdala reverses anxiogenic-like effects of ethanol withdrawal. Brain Res. 1993;605:25–32. doi: 10.1016/0006-8993(93)91352-s. [DOI] [PubMed] [Google Scholar]
- 160.Funk CK, O’Dell LE, Crawford EF, Koob GF. Corticotropin-releasing factor within the central nucleus of the amygdala mediates enhanced ethanol self-administration in withdrawn, ethanol-dependent rats. J Neurosci. 2006;26:11324–11332. doi: 10.1523/JNEUROSCI.3096-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Finn DA, Snelling C, Fretwell AM, Tanchuck MA, Underwood L, Cole M, Crabbe JC, Roberts AJ. Increased drinking during withdrawal from intermittent ethanol exposure is blocked by the CRF receptor antagonist D-Phe-CRF(12-41) Alcohol Clin Exp Res. 2007;31:939–949. doi: 10.1111/j.1530-0277.2007.00379.x. [DOI] [PubMed] [Google Scholar]
- 162.Funk CK, Koob GF. A CRF2 agonist administered into the central nucleus of the amygdala decreases ethanol self-administration in ethanol-dependent rats. Brain Res. 2007;1155:172–178. doi: 10.1016/j.brainres.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Merlo Pich E, Lorang M, Yeganeh M, Rodriguez de Fonseca F, Raber J, Koob GF, Weiss F. Increase of extracellular corticotropin-releasing factor-like immunoreactivity levels in the amygdala of awake rats during restraint stress and ethanol withdrawal as measured by microdialysis. J Neurosci. 1995;15:5439–5447. doi: 10.1523/JNEUROSCI.15-08-05439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Richter RM, Zorrilla EP, Basso AM, Koob GF, Weiss F. Altered amygdalar CRF release and increased anxiety-like behavior in Sardinian alcohol-preferring rats: a microdialysis and behavioral study. Alcohol Clin Exp Res. 2000;24:1765–1772. [PubMed] [Google Scholar]
- 165.Hwang BH, Stewart R, Zhang JK, Lumeng L, Li TK. Corticotropin-releasing factor gene expression is down-regulated in the central nucleus of the amygdala of alcohol-preferring rats which exhibit high anxiety: a comparison between rat lines selectively bred for high and low alcohol preference. Brain Res. 2004;1026:143–150. doi: 10.1016/j.brainres.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 166.Ehlers CL, Chaplin RI, Wall TL, Lumeng L, Li TK, Owens MJ, Nemeroff CB. Corticotropin releasing factor (CRF): studies in alcohol preferring and non-preferring rats. Psychopharmacology (Berl) 1992;106:359–364. doi: 10.1007/BF02245418. [DOI] [PubMed] [Google Scholar]
- 167.Nie Z, Schweitzer P, Roberts AJ, Madamba SG, Moore SD, Siggins GR. Ethanol augments GABAergic transmission in the central amygdala via CRF1 receptors. Science. 2004;303:1512–1514. doi: 10.1126/science.1092550. [DOI] [PubMed] [Google Scholar]
- 168.Bajo M, Cruz MT, Siggins GR, Messing R, Roberto M. Protein kinase C epsilon mediation of CRF- and ethanol-induced GABA release in central amygdala. Proc Natl Acad Sci USA. 2008;105:8410–8415. doi: 10.1073/pnas.0802302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Valdez GR, Koob GF. Allostasis and dysregulation of corticotropin-releasing factor and neuropeptide Y systems: implications for the development of alcoholism. Pharmacol Biochem Behav. 2004;79:671–689. doi: 10.1016/j.pbb.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 170.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 171.Smith MM. Histone structure and function. Curr Opin Cell Biol. 1991;3:429–437. doi: 10.1016/0955-0674(91)90070-f. [DOI] [PubMed] [Google Scholar]
- 172.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 173.Saetrom P, Snove O, Jr, Rossi JJ. Epigenetics and microRNAs. Pediatr Res. 2007;61:17R–23R. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- 174.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 175.Chandramohan Y, Droste SK, Arthur JS, Reul JM. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-d-aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur J Neurosci. 2008;27:2701–2713. doi: 10.1111/j.1460-9568.2008.06230.x. [DOI] [PubMed] [Google Scholar]
- 176.Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14:341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 178.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 179.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nestler EJ. Epigenetic mechanisms in psychiatry. Biol Psychiatry. 2009;65:189–190. doi: 10.1016/j.biopsych.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 181.Kalsi G, Prescott CA, Kendler KS, Riley BP. Unraveling the molecular mechanisms of alcohol dependence. Trends Genet. 2009;25:49–55. doi: 10.1016/j.tig.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 182.Ishii T, Hashimoto E, Ukai W, Tateno M, Yoshinaga T, Ono T, Watanabe K, Saito S, Saito T. Epigenetic regulation in alcohol-related brain damage. Nihon Arukoru Yakubutsu Igakkai Zasshi. 2008;43:705–713. [PubMed] [Google Scholar]
- 183.Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- 184.Pascual M, Boix J, Felipo V, Guerri C. Repeated alcohol administration during adolescence causes changes in the mesolimbic dopaminergic and glutamatergic systems and promotes alcohol intake in the adult rat. J Neurochem. 2009;108:920–931. doi: 10.1111/j.1471-4159.2008.05835.x. [DOI] [PubMed] [Google Scholar]
- 185.Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 186.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 187.Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19:563–573. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- 188.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 189.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 190.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 191.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 192.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kalluri HS, Mehta AK, Ticku MK. Up-regulation of NMDA receptor subunits in rat brain following chronic ethanol treatment. Brain Res Mol Brain Res. 1998;58:221–224. doi: 10.1016/s0169-328x(98)00112-0. [DOI] [PubMed] [Google Scholar]
- 194.Marutha Ravindran CR, Ticku MK. Role of CpG islands in the up-regulation of NMDA receptor NR2B gene expression following chronic ethanol treatment of cultured cortical neurons of mice. Neurochem Int. 2005;46:313–327. doi: 10.1016/j.neuint.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 195.Bonsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- 196.Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59:274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29:1777–1786. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- 198.Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27:8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]