Abstract
Magic-angle-spinning (MAS) solid-state NMR spectroscopy has emerged as a viable method to characterize membrane protein structure and dynamics. Nevertheless, the spectral resolution for uniformly labeled samples is often compromised by redundancy of the primary sequence and the presence of helical secondary structure that results in substantial resonance overlap. The ability to simplify the spectrum in order to obtain unambiguous site-specific assignments is a major bottleneck for structure determination. To address this problem, we used a combination of 15N reverse labeling and afterglow spectroscopic techniques that dramatically improved the ability to resolve peaks in a crowded spectrum. This was demonstrated using the polytopic membrane protein EmrE, an efflux pump involved in multidrug resistance. Residues preceding the 15N reverse labeled amino acid were imaged using a 3D NCOCX afterglow experiment and those following were recorded using a frequency-selective dephasing experiment. Our approach reduced the spectral congestion and provided a sensitive way to obtain chemical shift assignments for a membrane protein where no high-resolution structure is available. This MAS methodology is widely applicable to the study of other polytopic membrane proteins in functional lipid bilayer environments.
Keywords: solid-state NMR, assignment methods, magic-angle-spinning, fast data acquisition, sequential acquisition, detection approaches, EmrE, multidrug resistance, membrane proteins, small multidrug resistance
Introduction
Membrane proteins account for ~30% of all open-reading-frames in eukaryotic genomes (Wallin and Heijne 1998), yet only encompass 1-2% of experimentally determined structures in the protein data bank (Hong et al. 2012). Like soluble proteins, the structure and interactions of membrane proteins with other biomolecules are encoded by the primary sequence. However, unlike soluble proteins, a hydrophobic environment is required to maintain solubility and function (White 2009). Atomic resolution of membrane protein structures has been possible with X-ray crystallography (Rasmussen et al. 2011; Toyoshima et al. 2011; Krishnamurthy and Gouaux 2012; Johnson et al. 2012) and solution NMR (Kim et al. 2009; Verardi et al. 2011; Hiller et al. 2008; Zhou et al. 2008; Liang and Tamm 2007) that rely on detergent or bicelle solubilization. While these methods have been successful for several membrane proteins, crystallization and the presence of non-planar surfaces can result in structural artifacts (Chen et al. 2007; Cross et al. 2011). An alternative methodology is solid-state NMR (SSNMR) spectroscopy that is designed to use or remove the anisotropy of NMR observables and therefore does not have an intrinsic size limitation. This technique has opened the possibility to study membrane proteins under fully functional conditions such as in liposomes (Shi et al. 2009; Gustavsson et al. 2012; Cady et al. 2010; Etzkorn et al. 2007; Bhate and McDermott 2012; Linden et al. 2011; Lehner et al. 2008), aligned bilayers (Park et al. 2006; Traaseth et al. 2009; Muller et al. 2007; Vostrikov et al. 2010; Durr et al. 2007; Sharma et al. 2010; Knox et al. 2010), and in native cell membranes (Renault et al. 2012) (Gustavsson et al. 2012).
Magic-angle-spinning (MAS) has emerged as the most popular technique within SSNMR to study microcrystalline soluble proteins (Gardiennet et al. 2008; Martin and Zilm 2003; Igumenova et al. 2004; Franks et al. 2006; Bertini et al. 2010; Sengupta et al. 2012), amyloid fibrils (Bayro et al. 2012; Jaroniec et al. 2004; Tycko 2011; Li et al. 2011; Paravastu et al. 2008), and membrane proteins reconstituted in liposomes (Shi et al. 2009; Gustavsson et al. 2012; Cady et al. 2010; Etzkorn et al. 2007; Bhate and McDermott 2012; Linden et al. 2011; Lehner et al. 2008; McDermott 2009; Hong and DeGrado 2012; Sackett et al. 2010). One of the major hurdles in this methodology when applied to helical membrane proteins is the spectral crowding in traditional 2D and 3D backbone assignment experiments. This spectral overlap stems from structural heterogeneity of the sample preparation, residual protein dynamics, similarity of the residues within the primary sequence (e.g., Val, Leu, Ile, and Ala), and the degeneracy of chemical shifts for helical secondary structures found in transmembrane domains. Since spectral assignment is the major bottleneck for structure determination, the presence of overlapping resonances significantly slows or halts this process. To overcome the assignment hurdles for membrane proteins (both in MAS SSNMR and oriented SSNMR), a divide and conquer isotopic labeling strategy is often employed to reduce the number of observed spin-systems in recombinant proteins. One of the most useful is the reverse labeling approach, in which the protein is unlabeled at a few, selected residue types (Vuister et al. 1994; Heise et al. 2005; Shi et al. 2009; Krishnarjuna et al. 2011). Other methods include selective labeling, where only a few residues are labeled (Park et al. 2006; Traaseth et al. 2006) and sparse labeling that relies on specifically-labeled precursors to create predictable patterns in the protein (Hong and Jakes 1999; Castellani et al. 2002; Lian and Middleton 2001; Verardi et al. 2012; Jaipuria et al. 2012; Yang et al. 2008; Schuetz et al. 2010; Jehle et al. 2006). These sample-preparation techniques are useful and have aided in the assignment of crystalline proteins and a few membrane proteins examples using MAS (Castellani et al. 2002; Shi et al. 2009; Schmidt-Rohr et al. 2012). In addition, spectroscopic techniques have also been developed to aide in residue identification (Traaseth and Veglia 2011; Schmidt-Rohr et al. 2012). In this Article, we present the application of two spectroscopic techniques, afterglow and frequency-selective dipolar recoupling (FDR), to be used in combination with 15N reverse labeling in order to reduce spectral overlap for polytopic membrane proteins.
Materials and Methods
EmrE Expression and Purification
EmrE was expressed as a fusion protein with maltose-binding protein (MBP) in E. coli BL21(DE3) cells. The [U-13C,15N] sample was grown in minimal media with 13C glucose and 15N ammonium chloride (Sigma-Aldrich). For the RevIL EmrE sample, 15N L-Leu (120 mg/L) and natural abundance L-Ile (120 mg/L) were added to the 13C/15N M9 media. The fusion was bound to amylose resin (New England Biolabs) and eluted using maltose and n-dodecyl-β-D-maltopyranoside (DDM). EmrE was cleaved from MBP using tobacco etch virus protease (TEV) and purified to >95% by size-exclusion chromatography using a Superdex 200 10/300 column (GE Healthcare Life Sciences).
MAS Sample Preparation
Detergent-solubilized EmrE in DDM at a concentration of 0.85 mg/ml was added to 25 mg DMPC to give a final lipid/protein of 75:1 (mol/mol). Detergent was removed with bio-beads (75 mg/mg detergent) and incubated for 13.5 hr. The proteoliposomes were pelleted by ultracentrifugation for 1.5 hr at 142,414 × g (max force) and packed into a 3.2 mm MAS rotor using sample spacers to prevent dehydration.
Solid-State NMR Experiments
All NMR experiments were carried out using a DDR2 Agilent NMR spectrometer operating at a 1H frequency of 600 MHz. The variable temperature unit was set to -10 °C and the MAS rate was 12.5 kHz. Based on temperature standards, the actual sample was approximately +5 °C. The transfers from 15N to 13CA and 15N to 13CO utilized SPECIFIC-CP (Baldus et al. 1998) where the 15N offset was set to 121 ppm, the 13CA offset to 58 ppm, and the 13CO offset to 176 ppm. The 15N to 13CA transfer used a tangent adiabatic ramp (Baldus et al. 1996) on the 15N channel, while the 13CO SPECIFIC-CP utilized the ramp on the carbon channel. The Δ/2π and β/2π parameters of the adiabatic cross-polarization were set to 1.2 kHz and 0.3 kHz for the 15N to 13CA transfer and 3.7 kHz and 0.9 kHz for the 15N to 13CO transfer (Franks et al. 2007). The total length of the cross-polarization time was empirically optimized and set to 4.5 msec for the 15N to 13CA and 4 msec for the 15N to 13CO transfer. All datasets used a 13C spectral width of 100 kHz and an acquisition time of 20 msec. The [U-13C,15N] sample's indirect 15N dimension was acquired with a spectral width of 3125 Hz and 28 increments and its indirect 13C dimension was acquired with a spectral width of 3125 Hz and 24 increments. A total of 112 scans were used for the [U-13C,15N] sample. The RevIL sample had an indirect 15N spectral width of 3125 Hz and 24 increments, and an indirect 13C dimension spectral width of 3125 Hz and 28 increments. A total of 112 scans were used for the RevIL EmrE sample. For spin diffusion experiments between 13C spins, the DARR condition was set to the n=1 rotational resonance condition on 1H (12.5 kHz) and the mixing time was set to 18 msec (Takegoshi et al. 2001). The afterglow NCOCX followed the acquisition of a standard NCACX with a 50 msec DARR mixing period.
The [U-13C,15N] and RevIL standard NCA experiments were acquired with the SPECIFIC-CP parameters listed above and 36 and 32 points in the indirect 15N dimension, respectively, with a spectral width of 3125 Hz. Both standard NCA experiments were acquired with 128 scans. The FDR-NCA experiment used π/2 (5.5 μsec) pulses centered on 13CO and π (11 μsec) pulses on 15N for a total dephasing time of 1.6 msec (Traaseth and Veglia 2011). The FDR-NCA was acquired with 30 points in the indirect 15N dimension with a spectral width of 3125 Hz and 128 scans. The FDR-NCACX with DARR was acquired with 512 scans and a 25 msec DARR mixing period. All data sets were acquired with the spectral width of the direct 13C dimension set to 100 kHz and an acquisition time of 20 msec. The 13C dimension was referenced to the CH2 resonance of adamantane (40.48 ppm) (Morcombe and Zilm 2003) and indirectly to 15N using the relative frequency ratio between 15N and 13C of 0.402979946 (Wishart et al. 1995).
Results and Discussion
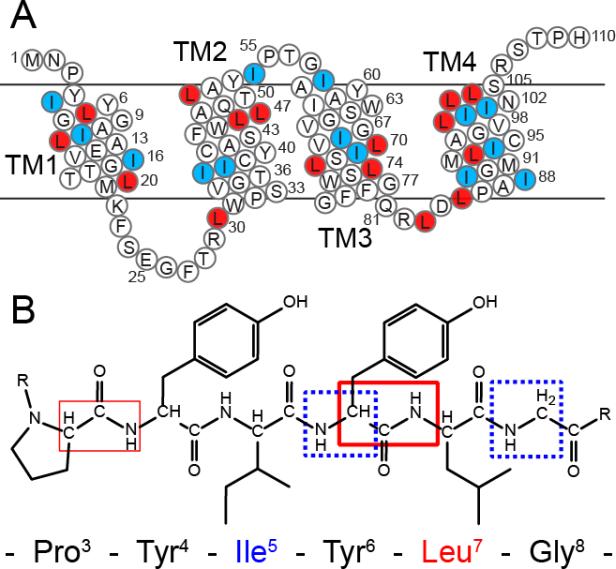
To evaluate the challenge of assigning a polytopic membrane protein by MAS spectroscopy, we carried out experiments on the 110-residue protein EmrE. The prototype for studying the small multidrug resistance (SMR) protein family (Figure 1A), EmrE is comprised of four transmembrane domains and functions as a homo-dimer to efflux a variety of cationic and aromatic molecules by coupling transport with the proton motive force across the inner membrane of bacteria (Schuldiner 2009; Korkhov and Tate 2009). EmrE represents a typical membrane protein in that it contains an abundance of hydrophobic Leu (16) and Ile (15) residues, approximately one-third of the primary sequence and nearly all within the transmembrane domains. In total, 47% of the residues within EmrE are comprised of Leu, Ile, Gly, or Ala residues. Unlike some of the rhodopsins, which naturally form well-ordered arrays in vivo (Neugebauer et al. 1978) and are structurally homogeneous at low lipid to protein ratios (Shi et al. 2009), EmrE and other membrane proteins require dilute lipid conditions to minimize nonspecific interactions between the functional oligomeric states. For this reason EmrE was reconstituted into DMPC lipid vesicles in a 75:1 lipid to protein molar ratio.
Figure 1.
Primary sequence of EmrE with the reverse labels that will act as filters in the spectroscopic approaches highlighted. (A) Sequence of EmrE with white circles highlighting [U-13C,15N] labeled residues, red circles indicating [15N-Leu] residues, and blue circles showing Ile residues labeled at natural abundance. (B) The afterglow NCOCX is expected to give strong peaks for residues preceding Leu (thick red box) with weaker signals for the other spin systems within the protein (thinner red box). The FDR experiment gives a filtered NCA correlation spectrum, where only residues appearing after the unlabeled residue give rise to peaks in an NCA spectrum (dotted-blue boxes).
Standard MAS Backbone Experiments Applied to a Polytopic Membrane Protein
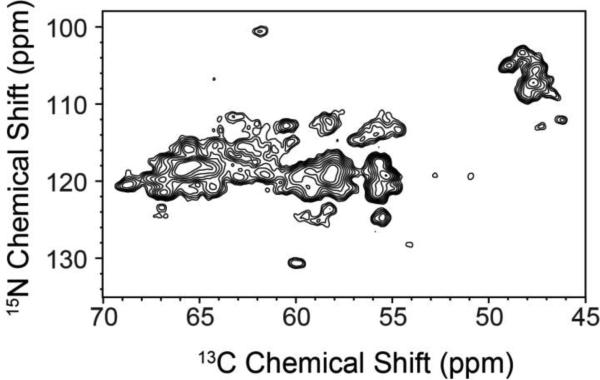
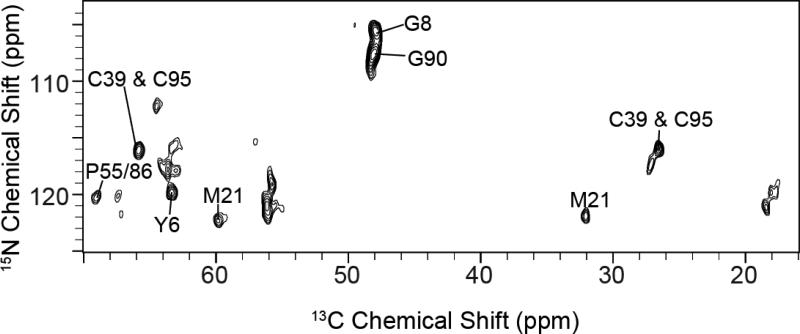
Figure 2 shows a 2D NCA correlation MAS spectrum obtained using SPECIFIC-CP from 15N to 13CA. The 13C linewidths for resolved peaks were measured to be ~0.8-0.9 ppm, reflecting relatively narrow resonances for a non-crystalline membrane protein in lipid bilayers. As observed within the spectrum, the 15N resolution results in spectral overlap spanning ~10 ppm for the majority of non-Gly peaks. 3D experiments such as NCACX and NCOCX with DARR mixing improved the resolution (Figure 3A; spectra in black), but alone were insufficient to resolve all spin systems given the 15N spectral congestion. These results are consistent with those reported by Reif and co-workers for [U-13C,15N] EmrE (Agarwal et al. 2007) and demonstrate the need for alternative approaches to supplement standard backbone experiments.
Figure 2.
Standard 2D NCA of [U-13C,15N] EmrE in DMPC liposomes. Due to the sequence homogeneity of EmrE (47% of the residues are either Leu, Ile, Gly, or Ala) and secondary structure similarity (Fleishman et al. 2006), the NCA spectrum is congested with few peaks fully resolved. The NCA is plotted at 7.1σ.
Figure 3.
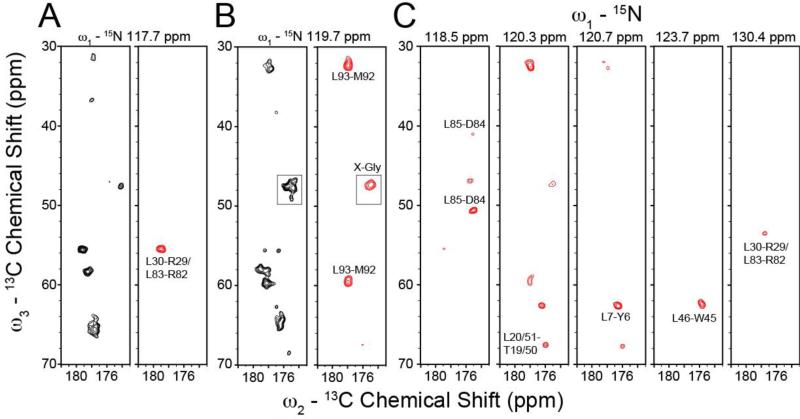
Comparison of standard and afterglow NCOCX experiments applied to [U-13C,15N] and RevIL, respectively. The afterglow approach is used as an isotopic filter to aide in the assignment of EmrE. (A, B) Strip plots from the standard 3D NCOCX acquired using [U-13C,15N] EmrE (black peaks) and the afterglow 3D NCOCX spectrum acquired on RevIL EmrE (red peaks). Note that the afterglow 3D NCOCX was acquired with 18 msec DARR and after an NCACX experiment with 50 msec DARR. Only Leu-X spin-systems were observed in the afterglow dataset with the exception of one Gly-X peak shown in panel B (boxed region; due to overlapping resonances). At a lower contour level, the Gly-X region in the boxed region appears the same as the standard NCOCX on [U-13C,15N] EmrE. (C) Additional strip plots from the 3D NCOCX afterglow dataset on RevIL EmrE. The standard NCOCX strips are plotted at 6.3σ and the afterglow NCOCX is shown at 6.6σ.
In order to reduce the spectral overlap to identify individual peaks, we prepared a reverse labeled EmrE sample by supplementing the 13C-glucose/15N-ammonium chloride media with 15N-Leu and natural abundance Ile (referred to as RevIL). This labeling scheme (Figure 1A) resulted in complete 13C and 15N incorporation at all sites except Leu (15N labeled) and Ile residues, and created inter-residue pairwise connectivity that we exploited in a novel manner to identify residues preceding Leu and those following Leu and Ile in the primary sequence (Figure 1B).
Afterglow Experiments Identify Residues Preceding the Reverse Labeled 15N Amino Acid
To image the residues prior to Leu in EmrE, we used the recently developed afterglow method (Banigan and Traaseth 2012). This pulse sequence uses the residual 15N magnetization from the cross-polarization element to acquire an additional 2D NCO (or 3D NCOCX) dataset in a sequential manner following the standard 2D NCA (or 3D NCACX) acquisition period (Banigan and Traaseth 2012). Since both acquisitions are carried out on 13C, only one receiver is necessary and therefore can be carried out on all standard SSNMR spectrometers. The beauty of this approach is that the NCACX is identical to the standard acquisition, which results in no loss in sensitivity, and the ability to acquire a second dataset with negligible increase in data acquisition time (< 2%).
Experiments were acquired on RevIL EmrE using a 3D NCACX acquisition followed by the afterglow 3D NCOCX. The NCACX experiment utilized a 50 msec DARR mixing period while the NCOCX had an 18 msec DARR. In our original implementation, the afterglow NCO dataset gave ~32% of the sensitivity with respect to the standard NCO experiment for [U-13C,15N] ubiquitin (Banigan and Traaseth 2012). For the RevIL EmrE sample, Leu residues do not have 13C incorporation at the CA position, resulting in minimal loss of 15N magnetization during the initial NCACX experiment. For these reasons, transfers from the 15N amide of Leu to the preceding 13C carbonyl of residue i-1 will result in more intense peaks in the afterglow NCOCX dataset than non-Leu labeled sites. Based on the labeling pattern and primary sequence, we expected to observe 12 Leu-related spin systems (there are 3 Leu-Leu pairs and an Ile-Leu pair in EmrE that would not be observed in the RevIL sample). Overall we identified eight strongspin-system peaks (shown in Figure 3), with seven related to Leu residues. The strong peaks were defined as those having peak intensities greater than 7σ (σ is the standard deviation of the noise in the spectrum). The missing five spin systems are likely due to overlap in the 3D strips resulting from sequence redundancies (e.g., T18-L19 and T50-L51; R29-L30 and R82-L83; V69-L70 and V98-L99), but can also arise from small variations in peak intensity across the primary sequence (e.g., T1ρ spin relaxation rates). While the storage of 15N magnetization along the z-axis for the afterglow method will have a negligible impact on signal intensities due to long T1 times (Gopinath and Veglia 2012; Giraud et al. 2004), differences in T1ρ can impact observed signal intensities during the cross-polarization periods. The one non-Leu spin system observed in the afterglow spectrum resulted from overlapping X-Gly residues (i.e., peak clustering gave increased intensity). This region is shown in the boxed areas in Figure 3B. Upon reduction of the noise floor in the afterglow spectrum, we observed extra peaks in this region that were also seen in the standard NCOCX spectrum acquired on [U-13C,15N] labeled EmrE. This is related to the discussion above that the afterglow approach when applied to [U-13C,15N] samples gives the same spectral features (albeit at 32% of the signal intensity) compared to the standard NCOCX experiment (Banigan and Traaseth 2012). Nevertheless, the Gly peaks in the afterglow NCOCX do not pose a problem for identifying spin systems involving Leu residues due to the absence of Gly-Leu pairs in the primary sequence and the fact that Gly residues have distinct 13CA chemical shifts. In the case of proteins having Gly-Leu pairs, these peaks in the afterglow NCOCX will be stronger than the background Gly signals due to the lack of 15N Leu magnetization transfer to 13CA.
Identification of Residues Following the Reverse Labeled Amino Acid
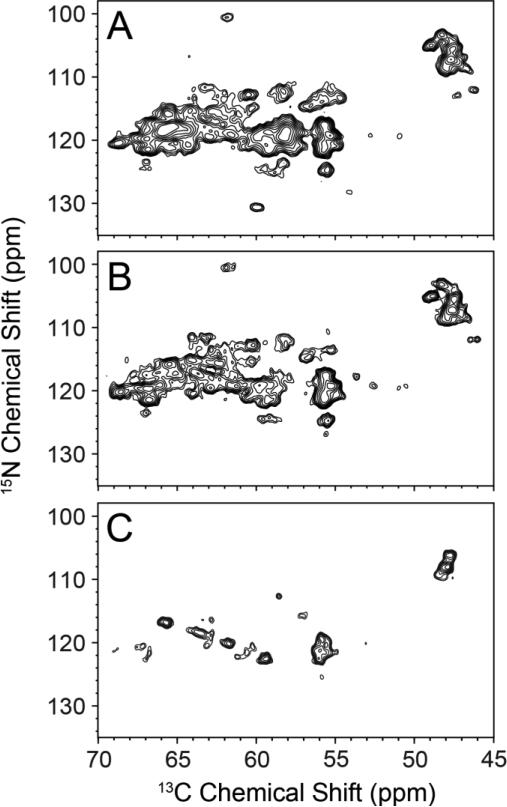
As seen in Figure 4A and B, the standard NCA spectra for [U-13C,15N] and RevIL EmrE both are substantially overlapped, which complicates data analysis. We note that a previous MAS-SSNMR study on [U-13C,15N] labeled EmrE reported similar spectral crowding to that we observed (Agarwal et al. 2007). With the goal of reducing this congestion, we acquired a 2D FDR-NCA experiment with the RevIL EmrE sample that was designed to identify residues following the reverse labeled sites in EmrE (i.e., Leu and Ile) (Figure 1B, blue boxes). This experiment was previously implemented with model samples that did not suffer from chemical shift overlap (Traaseth and Veglia 2011). The FDR-NCA experiment selectively recouples 15N with 13CO in a frequency selective manner followed by a SPECIFIC-CP transfer from 15N to 13CA. The result is an NCA-like spectrum, which preferentially dephases one-bond 15N-13CO dipolar couplings without perturbing multiple bond couplings. In contrast to the NCA spectrum for RevIL EmrE, we observed 15 clearly resolved peaks using the FDR-NCA experiment (Figure 4C). When taking into account the appearance of additional shoulders, the observed number of peaks closely matches the 21 expected resonances based on the primary sequence and isotopic labeling pattern for RevIL. Identification of the residue-types were carried out using FDR-NCA in conjunction with a short DARR mixing period (FDR-NCACX; Figure 5). The identified residue types are consistent with the known residues that follow Leu and Ile in the primary sequence (Figure 1A). Using the combination of 15N reverse labeling with the afterglow and FDR spectroscopic techniques, we were able to reduce the spectral congestion leading to the identification of discrete peaks and the assignments reported in Figures 3 and 5 for EmrE.
Figure 4.
Resolution improvement for EmrE using the reverse labeling approach in conjunction with 2D FDR-NCA experiments. (A) [U-13C,15N] EmrE standard NCA 2D dataset using SPECIFIC-CP (same as in Figure 2). (B) RevIL EmrE standard NCA 2D dataset using SPECIFIC-CP and acquired in a similar manner as that in panel A. (C) FDR-NCA spectrum of RevIL EmrE. The spectrum in panel C shows only peaks for residues immediately following Ile and Leu in the primary sequence. Panels A, B, and C are plotted at 7.1σ, 5.8σ, and 7.0σ, respectively.
Figure 5.
FDR-NCACX spectrum with 25 msec DARR mixing to assign residue types in the 2D spectrum. This spectrum is plotted at 7.0σ.
Implications of the Method for EmrE
The improved resolution offered by the afterglow and FDR-NCA spectra allowed us to significantly reduce the spectral congestion relative to [U-13C,15N] EmrE samples (Agarwal et al. 2007). Based on the spectra in Figures 3 and 5, we found that some peaks showed one apparent population while others had broader spectral features indicative of heterogeneity (i.e., multiple populations) for the ligand free form of EmrE. This is relevant in light of the recent discovery that the EmrE dimer is comprised of two exchanging asymmetric monomer populations when bound to tetraphenylphosphonium (TPP+) in isotropic bicelles using [1H-15N] TROSY-HSQC solution NMR (Morrison et al. 2012). Additional studies are needed to reduce the carbon and nitrogen line widths in order to resolve the monomers in the dimer that have very similar secondary structures as depicted in the cryo-electron microscopy and X-ray structural models (Chen et al. 2007; Fleishman et al. 2006). The lack of extensive inter-monomer packing other than transmembrane-4 in the structural models suggests that the ligand-free form of the protein undergoes larger conformational fluctuations leading to a less compact structure than the TPP+ bound form. This finding is supported by EPR experiments carried out in liposomes (Amadi et al. 2010). Furthermore, the presence of TPP+ binding in the solution NMR studies (Morrison et al. 2012) may induce chemical shift dispersion of the two monomers when imaged by NMR methods that detect the isotropic component of the shift (Morrison et al. 2012). Future efforts are needed to characterize the tilt angles of the dimer helices with respect to the lipid membrane for both the apo and ligand-bound forms using oriented SSNMR (Cross et al. 2011; Traaseth et al. 2009; De Angelis et al. 2006). This combination of MAS and oriented techniques promises to reveal high-resolution and complementary structural information that cannot be easily measured from isotropic chemical shifts (Cross et al. 2011; Traaseth et al. 2009; De Angelis et al. 2006).
Conclusion
The ability to unambiguously assign membrane protein spectra using MAS will significantly advance the understanding of structure and dynamics in lipid membranes. Through a combination of spectroscopic techniques with 15N reverse labeling, our approach will be a powerful tool for obtaining unambiguous assignments for polytopic membrane proteins by MAS and reducing the time needed for data acquisition. Our method is able to selectively identify resonances that preceded and followed 15N amino acids incorporated into [U-13C,15N] membrane protein samples. Future improvements would be the use of inexpensive precursors such as α-ketoisovalerate and phenylpyruvate at natural abundance that will reduce costs and achieve the same isotopic labeling pattern (Rasia et al. 2012), extension of the afterglow method to additional nuclei as recently reported (Bellstedt et al. 2012), and incorporation of triple cross-polarization elements (Gopinath and Veglia 2012; Akbey et al. 2011; Herbst et al. 2008) with the afterglow experiment. Finally, using the outlined methodology, it will also be possible to select two or three isotopically labeled species at the same time as in solution NMR (Masterson et al. 2008) for detecting binary or ternary complexes using SSNMR spectroscopy.
Acknowledgement
This work was supported by NIH grant 5K22AI083745 and start-up funds from New York University.
References
- Agarwal V, Fink U, Schuldiner S, Reif B. MAS solid-state NMR studies on the multidrug transporter EmrE. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2007;1768(12):3036–3043. doi: 10.1016/j.bbamem.2007.09.012. doi: http://dx.doi.org/10.1016/j.bbamem.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Akbey Ü , Camponeschi F, van Rossum B-J, Oschkinat H. Triple Resonance Cross-Polarization for More Sensitive 13C MAS NMR Spectroscopy of Deuterated Proteins. ChemPhysChem. 2011;12(11):2092–2096. doi: 10.1002/cphc.201100084. doi:10.1002/cphc.201100084. [DOI] [PubMed] [Google Scholar]
- Amadi ST, Koteiche HA, Mishra S, Mchaourab HS. Structure, Dynamics, and Substrate-induced Conformational Changes of the Multidrug Transporter EmrE in Liposomes. Journal of Biological Chemistry. 2010;285(34):26710–26718. doi: 10.1074/jbc.M110.132621. doi:10.1074/jbc.M110.132621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus M, Geurts DG, Hediger S, Meier BH. Efficient N-15-C-13 polarization transfer by adiabatic-passage Hartmann-Hahn cross polarization. J Magn Reson Ser A. 1996;118(1):140–144. [Google Scholar]
- Baldus M, Petkova AT, Herzfeld JH, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95:1197–1207. (Journal Article) [Google Scholar]
- Banigan JR, Traaseth NJ. Utilizing Afterglow Magnetization from Cross-Polarization Magic-Angle-Spinning Solid-State NMR Spectroscopy to Obtain Simultaneous Heteronuclear Multidimensional Spectra. The Journal of Physical Chemistry B. 2012;116(24):7138–7144. doi: 10.1021/jp303269m. doi:10.1021/jp303269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayro MJ, Daviso E, Belenky M, Griffin RG, Herzfeld J. An Amyloid Organelle, Solid-state NMR Evidence for Cross-β Assembly of Gas Vesicles. Journal of Biological Chemistry. 2012;287(5):3479–3484. doi: 10.1074/jbc.M111.313049. doi:10.1074/jbc.M111.313049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellstedt P, Herbst C, Häfner S, Leppert J, Görlach M, Ramachandran R. Solid state NMR of proteins at high MAS frequencies: symmetry-based mixing and simultaneous acquisition of chemical shift correlation spectra. Journal of biomolecular NMR. 2012;54(4):325–335. doi: 10.1007/s10858-012-9680-z. doi:10.1007/s10858-012-9680-z. [DOI] [PubMed] [Google Scholar]
- Bertini I, Emsley L, Lelli M, Luchinat C, Mao JF, Pintacuda G. Ultrafast MAS Solid-State NMR Permits Extensive C-13 and H-1 Detection in Paramagnetic Metalloproteins. Journal of the American Chemical Society. 2010;132(16):5558–+. doi: 10.1021/ja100398q. doi:Doi 10.1021/Ja100398q. [DOI] [PubMed] [Google Scholar]
- Bhate MP, McDermott AE. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(38):15265–15270. doi: 10.1073/pnas.1211900109. doi:DOI 10.1073/pnas.1211900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463(7281):689–U127. doi: 10.1038/nature08722. doi:Doi 10.1038/Nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420(6911):98–102. doi: 10.1038/nature01070. doi:10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- Chen Y-J, Pornillos O, Lieu S, Ma C, Chen AP, Chang G. X-ray structure of EmrE supports dual topology model. Proceedings of the National Academy of Sciences. 2007;104(48):18999–19004. doi: 10.1073/pnas.0709387104. doi:10.1073/pnas.0709387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Sharma M, Yi M, Zhou H-X. Influence of solubilizing environments on membrane protein structures. Trends in Biochemical Sciences. 2011;36(2):117–125. doi: 10.1016/j.tibs.2010.07.005. doi: http://dx.doi.org/10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis AA, Howell SC, Nevzorov AA, Opella SJ. Structure Determination of a Membrane Protein with Two Trans-membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy. Journal of the American Chemical Society. 2006;128(37):12256–12267. doi: 10.1021/ja063640w. doi:10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr UHN, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b(5). Journal of the American Chemical Society. 2007;129(21):6670–+. doi: 10.1021/ja069028m. doi:Doi 10.1021/Ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzkorn M, Martell S, Andronesi OC, Seidel K, Engelhard M, Baldus M. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew Chem Int Edit. 2007;46(3):459–462. doi: 10.1002/anie.200602139. doi:DOI 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. Quasi-symmetry in the Cryo-EM Structure of EmrE Provides the Key to Modeling its Transmembrane Domain. Journal of Molecular Biology. 2006;364(1):54–67. doi: 10.1016/j.jmb.2006.08.072. doi:10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. Journal of Biomolecular NMR. 2007;39(2):107–131. doi: 10.1007/s10858-007-9179-1. doi:10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- Franks WT, Wylie BJ, Stellfox SA, Rienstra CM. Backbone conformational constraints in a microcrystalline U-N-15-Labeled protein by 3D dipolar-shift solid-state NMR spectroscopy. Journal of the American Chemical Society. 2006;128(10):3154–3155. doi: 10.1021/ja058292x. doi:Doi 10.1021/Ja058292x. [DOI] [PubMed] [Google Scholar]
- Gardiennet C, Loquet A, Etzkorn M, Heise H, Baldus M, Böckmann A. Structural constraints for the Crh protein from solid-state NMR experiments. Journal of biomolecular NMR. 2008;40(4):239–250. doi: 10.1007/s10858-008-9229-3. doi:10.1007/s10858-008-9229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud N, Böckmann A, Lesage A, Penin F, Blackledge M, Emsley L. Site-Specific Backbone Dynamics from a Crystalline Protein by Solid-State NMR Spectroscopy. Journal of the American Chemical Society. 2004;126(37):11422–11423. doi: 10.1021/ja046578g. doi:10.1021/ja046578g. [DOI] [PubMed] [Google Scholar]
- Gopinath T, Veglia G. Dual Acquisition Magic-Angle Spinning Solid-State NMR-Spectroscopy: Simultaneous Acquisition of Multidimensional Spectra of Biomacromolecules. Angewandte Chemie International Edition. 2012;51(11):2731–2735. doi: 10.1002/anie.201108132. doi:10.1002/anie.201108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson M, Traaseth NJ, Veglia G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Bba-Biomembranes. 2012;1818(2):146–153. doi: 10.1016/j.bbamem.2011.07.040. doi:DOI 10.1016/j.bbamem.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15871–15876. doi: 10.1073/pnas.0506109102. doi:10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst C, Riedel K, Ihle Y, Leppert J, Ohlenschläger O, Görlach M, Ramachandran R. MAS solid state NMR of RNAs with multiple receivers. Journal of biomolecular NMR. 2008;41(3):121–125. doi: 10.1007/s10858-008-9247-1. doi:10.1007/s10858-008-9247-1. [DOI] [PubMed] [Google Scholar]
- Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–1210. doi: 10.1126/science.1161302. doi:DOI 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, DeGrado WF. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Science. 2012;21(11):1620–1633. doi: 10.1002/pro.2158. doi:10.1002/pro.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Jakes K. Selective and extensive 13C labeling of a membrane protein for solid-state NMR investigations. Journal of biomolecular NMR. 1999;14(1):71–74. doi: 10.1023/a:1008334930603. doi:10.1023/a:1008334930603. [DOI] [PubMed] [Google Scholar]
- Hong M, Zhang Y, Hu F. Membrane Protein Structure and Dynamics from NMR Spectroscopy. Annual Review of Physical Chemistry. 2012;63(1):1–24. doi: 10.1146/annurev-physchem-032511-143731. doi:doi:10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ. Assignments of carbon NMR resonances for microcrystalline ubiquitin. Journal of the American Chemical Society. 2004;126(21):6720–6727. doi: 10.1021/ja030547o. doi:Doi 10.1021/Ja030547o. [DOI] [PubMed] [Google Scholar]
- Jaipuria G, Lobo NP, Shet D, Atreya HS. High resolution methyl selective (1)(3)C-NMR of proteins in solution and solid state. Journal of biomolecular NMR. 2012;54(1):33–42. doi: 10.1007/s10858-012-9647-0. doi:10.1007/s10858-012-9647-0. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):711–716. doi: 10.1073/pnas.0304849101. doi:DOI 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle S, Rehbein K, Diehl A, van Rossum BJ. Amino-acid selective experiments on uniformly 13C and 15N labeled proteins by MAS NMR: Filtering of lysines and arginines. J Magn Reson. 2006;183(2):324–328. doi: 10.1016/j.jmr.2006.08.015. doi:10.1016/j.jmr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Johnson E, Nguyen PT, Yeates TO, Rees DC. Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition. Protein Science. 2012;21(1):84–96. doi: 10.1002/pro.765. doi:10.1002/pro.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Recent advances in the application of solution NMR spectroscopy to multi-span integral membrane proteins. Progress in Nuclear Magnetic Resonance Spectroscopy. 2009;55(4):335–360. doi: 10.1016/j.pnmrs.2009.07.002. doi:DOI 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox RW, Lu GJ, Opella SJ, Nevzorov AA. A Resonance Assignment Method for Oriented-Sample Solid-State NMR of Proteins. Journal of the American Chemical Society. 2010;132(24):8255–8257. doi: 10.1021/ja102932n. doi:10.1021/ja102932n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhov VM, Tate CG. An emerging consensus for the structure of EmrE. Acta Crystallogr D. 2009;65:186–192. doi: 10.1107/S0907444908036640. doi:Doi 10.1107/S0907444908036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. doi: http://www.nature.com/nature/journal/v481/n7382/abs/nature10737.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnarjuna B, Jaipuria G, Thakur A, D'Silva P, Atreya H. Amino acid selective unlabeling for sequence specific resonance assignments in proteins. Journal of biomolecular NMR. 2011;49(1):39–51. doi: 10.1007/s10858-010-9459-z. doi:10.1007/s10858-010-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner I, Basting D, Meyer B, Haase W, Manolikas T, Kaiser C, Karas M, Glaubitz C. The Key Residue for Substrate Transport (Glu14) in the EmrE Dimer Is Asymmetric. Journal of Biological Chemistry. 2008;283(6):3281–3288. doi: 10.1074/jbc.M707899200. doi:10.1074/jbc.M707899200. [DOI] [PubMed] [Google Scholar]
- Li J, Hoop CL, Kodali R, Sivanandam VN, van der Wel PCA. Amyloid-like Fibrils from a Domain-swapping Protein Feature a Parallel, in-Register Conformation without Native-like Interactions. Journal of Biological Chemistry. 2011;286(33):28988–28995. doi: 10.1074/jbc.M111.261750. doi:DOI 10.1074/jbc.M111.261750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian L-Y, Middleton DA. Labelling approaches for protein structural studies by solution-state and solid-state NMR. Progress in Nuclear Magnetic Resonance Spectroscopy. 2001;39(3):171–190. doi:10.1016/s0079-6565(01)00034-6. [Google Scholar]
- Liang BY, Tamm LK. Structure of outer membrane protein G by solution NMR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(41):16140–16145. doi: 10.1073/pnas.0705466104. doi:DOI 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden AH, Lange S, Franks WT, Akbey U, Specker E, van Rossum BJ, Oschkinat H. Neurotoxin II Bound to Acetylcholine Receptors in Native Membranes Studied by Dynamic Nuclear Polarization NMR. Journal of the American Chemical Society. 2011;133(48):19266–19269. doi: 10.1021/ja206999c. doi:Doi 10.1021/Ja206999c. [DOI] [PubMed] [Google Scholar]
- Martin RW, Zilm KW. Preparation of protein nanocrystals and their characterization by solid state NMR. Journal of Magnetic Resonance. 2003;165(1):162–174. doi: 10.1016/s1090-7807(03)00253-2. doi:Doi 10.1016/S1090-7807(03)00253-2. [DOI] [PubMed] [Google Scholar]
- Masterson LR, Tonelli M, Markley JL, Veglia G. Simultaneous Detection and Deconvolution of Congested NMR Spectra Containing Three Isotopically Labeled Species. Journal of the American Chemical Society. 2008;130(25):7818–7819. doi: 10.1021/ja802701w. doi:10.1021/ja802701w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott A. Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annual Review of Biophysics. 2009;38(1):385–403. doi: 10.1146/annurev.biophys.050708.133719. doi:doi:10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. Journal of Magnetic Resonance. 2003;162(2):479–486. doi: 10.1016/s1090-7807(03)00082-x. doi: http://dx.doi.org/10.1016/S1090-7807(03)00082-X. [DOI] [PubMed] [Google Scholar]
- Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T, Henzler-Wildman KA. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2012;481(7379):45–50. doi: 10.1038/nature10703. doi: http://www.nature.com/nature/journal/v481/n7379/abs/nature10703.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller SD, De Angelis AA, Walther TH, Grage SL, Lange C, Opella SJ, Ulrich AS. Structural characterization of the pore forming protein TatA(d) of the twin-arginine translocase in membranes by solid-state N-15-NMR. Bba-Biomembranes. 2007;1768(12):3071–3079. doi: 10.1016/j.bbamem.2007.09.008. doi:DOI 10.1016/j.bbamem.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Neugebauer D-C, Zingsheim HP, Oesterhelt D. Recrystallization of the purple membrane in vivo and in vitro. Journal of Molecular Biology. 1978;123(2):247–257. doi: 10.1016/0022-2836(78)90324-8. doi: http://dx.doi.org/10.1016/0022-2836(78)90324-8. [DOI] [PubMed] [Google Scholar]
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18349–18354. doi: 10.1073/pnas.0806270105. doi:DOI 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Prytulla S, De Angelis AA, Brown JM, Kiefer H, Opella SJ. High-Resolution NMR Spectroscopy of a GPCR in Aligned Bicelles. Journal of the American Chemical Society. 2006;128(23):7402–7403. doi: 10.1021/ja0606632. doi:10.1021/ja0606632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasia RM, Brutscher B, Plevin MJ. Selective Isotopic Unlabeling of Proteins Using Metabolic Precursors: Application to NMR Assignment of Intrinsically Disordered Proteins. ChemBioChem. 2012;13(5):732–739. doi: 10.1002/cbic.201100678. doi:10.1002/cbic.201100678. [DOI] [PubMed] [Google Scholar]
- Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the [bgr]2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. doi: http://www.nature.com/nature/journal/v477/n7366/abs/nature10361.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault M, Tommassen-van Boxtel R, Bos MP, Post JA, Tommassen J, Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proceedings of the National Academy of Sciences. 2012 doi: 10.1073/pnas.1116478109. doi:10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett K, Nethercott MJ, Epand RF, Epand RM, Kindra DR, Shai Y, Weliky DP. Comparative analysis of membrane-associated fusion peptide secondary structure and lipid mixing function of HIV gp41 constructs that model the early pre-hairpin intermediate and final hairpin conformations. J Mol Biol. 2010;397(1):301–315. doi: 10.1016/j.jmb.2010.01.018. doi:10.1016/j.jmb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Rohr K, Fritzsching KJ, Liao SY, Hong M. Spectral editing of two-dimensional magic-angle-spinning solid-state NMR spectra for protein resonance assignment and structure determination. Journal of biomolecular NMR. 2012;54(4):343–353. doi: 10.1007/s10858-012-9676-8. doi:10.1007/s10858-012-9676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Wasmer C, Habenstein B, Verel R, Greenwald J, Riek R, Bockmann A, Meier BH. Protocols for the sequential solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s(1-227). Chembiochem. 2010;11(11):1543–1551. doi: 10.1002/cbic.201000124. doi:10.1002/cbic.201000124. [DOI] [PubMed] [Google Scholar]
- Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2009;1794(5):748–762. doi: 10.1016/j.bbapap.2008.12.018. doi:10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Sengupta I, Nadaud PS, Helmus JJ, Schwieters CD, Jaroniec CP. Protein fold determined by paramagnetic magic-angle spinning solid-state NMR spectroscopy. Nat Chem. 2012;4(5):410–417. doi: 10.1038/nchem.1299. doi:Doi 10.1038/Nchem.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Yi MG, Dong H, Qin HJ, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the Mechanism of the Influenza A Proton Channel from a Structure in a Lipid Bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. doi:DOI 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ahmed MAM, Zhang W, Whited G, Brown LS, Ladizhansky V. Three-Dimensional Solid-State NMR Study of a Seven-Helical Integral Membrane Proton Pump—Structural Insights. Journal of Molecular Biology. 2009;386(4):1078–1093. doi: 10.1016/j.jmb.2009.01.011. doi:10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. (Journal Article) [Google Scholar]
- Toyoshima C, Kanai R, Cornelius F. First Crystal Structures of Na+,K+-ATPase: New Light on the Oldest Ion Pump. Structure. 2011;19(12):1732–1738. doi: 10.1016/j.str.2011.10.016. doi: http://dx.doi.org/10.1016/j.str.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Structural dynamics and topology of phospholamban in oriented lipid bilayers using multidimensional solid-state NMR. Biochemistry. 2006;45(46):13827–13834. doi: 10.1021/bi0607610. doi:Doi 10.1021/Bi0607610. [DOI] [PubMed] [Google Scholar]
- Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(25):10165–10170. doi: 10.1073/pnas.0904290106. doi:DOI 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traaseth NJ, Veglia G. Frequency-selective heteronuclear dephasing and selective carbonyl labeling to deconvolute crowded spectra of membrane proteins by magic angle spinning NMR. Journal of Magnetic Resonance. 2011;211(1):18–24. doi: 10.1016/j.jmr.2011.03.013. doi:10.1016/j.jmr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62(1):279–299. doi: 10.1146/annurev-physchem-032210-103539. doi:10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proceedings of the National Academy of Sciences. 2011;108(22):9101–9106. doi: 10.1073/pnas.1016535108. doi:10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardi R, Traaseth NJ, Masterson LR, Vostrikov VV, Veglia G. Isotope labeling for solution and solid-state NMR spectroscopy of membrane proteins. Advances in experimental medicine and biology. 2012;992:35–62. doi: 10.1007/978-94-007-4954-2_3. doi:10.1007/978-94-007-4954-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov VV, Daily AE, Greathouse DV, Koeppe RE. Charged or Aromatic Anchor Residue Dependence of Transmembrane Peptide Tilt. Journal of Biological Chemistry. 2010;285(41):31723–31730. doi: 10.1074/jbc.M110.152470. doi:DOI 10.1074/jbc.M110.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister GW, Kim S-J, Wu C, Bax A. 2D and 3D NMR Study of Phenylalanine Residues in Proteins by Reverse Isotopic Labeling. Journal of the American Chemical Society. 1994;116(20):9206–9210. doi:10.1021/ja00099a041. [Google Scholar]
- Wallin E, Heijne GV. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Science. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. doi:10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH. Biophysical dissection of membrane proteins. Nature. 2009;459(7245):344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- Wishart D, Bigam C, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley J, Sykes B. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. Journal of biomolecular NMR. 1995;6(2):135–140. doi: 10.1007/BF00211777. doi:10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Yang J, Tasayco ML, Polenova T. Magic angle spinning NMR experiments for structural studies of differentially enriched protein interfaces and protein assemblies. J Am Chem Soc. 2008;130(17):5798–5807. doi: 10.1021/ja711304e. doi:10.1021/ja711304e. [DOI] [PubMed] [Google Scholar]
- Zhou YP, Cierpicki T, Jimenez RHF, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31(6):896–908. doi: 10.1016/j.molcel.2008.08.028. doi:DOI 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]