Summary
The sterile womb paradigm is an enduring premise in biology that human infants are born sterile. Recent studies suggest that infants incorporate an initial microbiome before birth and receive copious supplementation of maternal microbes through birth and breastfeeding. Moreover, evidence for microbial maternal transmission is increasingly widespread across animals. This collective knowledge compels a paradigm shift—one in which maternal transmission of microbes advances from a taxonomically specialized phenomenon to a universal one in animals. It also engenders fresh views on the assembly of the microbiome, its role in animal evolution, and applications to human health and disease.
Introduction
While the human microbiota comprises only 1–3% of an individual's total body mass, this small percentage represents over 100 trillion microbial cells, outnumbering human cells 10 to 1 and adding over 8 million genes to our set of 22,000 [1],[2]. This complexity establishes a network of interactions between the host genome and microbiome spanning gut development [3], digestion [4],[5], immune cell development [6]–[8], dental health [9],[10], and resistance to pathogens [11],[12]. Recent studies have also provided a greater understanding of how the composition of an individual's microbiota changes throughout development, especially during the first year of life [3],[13]. While the general dogma is that the placental barrier keeps infants sterile throughout pregnancy, increasing evidence suggests that an infant's initial inoculum can be provided by its mother before birth [14]–[18] and is supplemented by maternal microbes through the birthing [19] and breastfeeding [20],[21] processes.
While maternal transmission of microbes in humans has attracted considerable attention in the last few years, nearly a century's worth of research is available for vertical transmission of symbionts in invertebrates [22]. Similar to gut bacteria in humans that assist nutrient intake, many insect-associated bacteria function as nutritional symbionts that supplement the nutrient-poor diet of their host with essential vitamins or amino acids [23],[24]. Since these indispensable symbionts cannot live outside of host cells, they cannot be acquired from the environment and are faithfully transferred from mother to offspring [22],[25]. Maternal transmission in invertebrates has been reviewed elsewhere [22],[26],[27], and Box 1 and Box 2 highlight examples of heritable symbioses across invertebrate phyla.
Box 1. Examples of Maternal Transmission in Marine Invertebrates
Marine Sponges (Phylum Porifera)
Sponges are ancient metazoans that evolved over 600 million years ago as one of the first multicellular animals [83]. In marine sponges, a remarkably large consortium of extracellular microbial symbionts thrives within the sponge's mesohyl, a gelatinous connective tissue located between the external and internal cell layers. Many of these bacterial residents are found in diverse species of sponges with nonoverlapping distributions but not in the surrounding seawater [84]–[86]. These “sponge-specific” microbes are hypothesized to have originated from ancient colonization events before the diversification of marine sponges and are maintained as symbionts through vertical transmission [87]. Independent studies have estimated that up to 33 phylogenetically distinct microbial clusters spanning ten bacterial phyla and one archaeal phylum are vertically transmitted in sponges [41],[84],[86],[88]. Both transmission electron microscopy (TEM) and fluorescent in situ hybridization (FISH) studies have confirmed the presence of microorganisms of different shapes and sizes in the oocytes of oviparous sponges [41] and in the embryos of viviparous sponges [43]–[45].
Vesicomyid Clams (Phylum Mollusca)
Deep-sea hydrothermal vent communities rely upon chemosynthetic bacteria to harness chemical energy stored in reduced sulfur compounds extruding from the vents. Metazoans that live in this extreme environment harbor chemosynthetic endosymbionts in their tissues that provide most, if not all, of the host's nutrition [89]. Somewhat surprisingly, most invertebrates that live near hydrothermal events acquire their endosymbionts anew from the environment each generation [90],[91], even though chemosynthetic bacteria are crucial for survival in such a harsh habitat. A major exception to this trend is found in the Vesicomyidae family of clams [92]. Vesicomyid clams retain a rudimentary gut and rely primarily on sulfur-oxidizing bacteria sequestered intracellularly within specialized host cells called bacteriocytes in the clam's large, fleshy gills [93]. Vertical transmission via transovarial transmission appears to be the dominant mechanism for maintenance of these thioautotrophic bacterial symbionts given that follicle cells surrounding an oocyte and the oocyte itself are heavily infected with the chemosynthetic bacteria [46],[94].
Box 2. Examples of Maternal Transmission in Terrestrial Invertebrates
Insects (Phylum Arthropoda)
Insects that thrive on unbalanced diets such as plant sap, blood, or wood depend upon microbial symbionts for the provision of essential amino acids or vitamins lacking in their food source. In turn, hosts provide a wide range of metabolites to their symbionts as well as protection from environmental stressors. This codependence requires faithful transfer of symbionts to all offspring, usually through transovarial transmission [23],[24]. Reproductive parasites, such as the obligate, intracellular bacteria Wolbachia, are also widespread in insects and hijack maternal transmission routes to ensure their spread within an insect population (reviewed in [95],[96]).
Pea Aphid (Acrythosiphon pisum)
The pea aphid Acrythosiphon pisum (Figure 2A) and its nutritional endosymbiont Buchnera aphidicola are a preeminent example of obligate mutualism in insects. The ancestral Buchnera gammaproteobacteria was acquired by aphids between 160 and 280 million years ago [97] and has since diverged in parallel with its aphid hosts through strict vertical transmission [26],[97]. Buchnera are housed within the cytoplasm of bacteriocytes arranged into dual bacteriome structures located in the aphid body cavity adjacent to the ovaries [98], allowing efficient transfer of Buchnera symbionts to developing oocytes or embryos during the sexual and asexual phases of aphid reproduction, respectively. At the cellular level, symbiont transfer occurs when maternal bacteriocytes release Buchnera symbionts through exocytosis into the extracellular space between the bacteriocyte and oocyte or embryo, which then actively endocytoses the extracellular Buchnera symbionts [51].
Cockroaches (Order Blattodea)
Just as insects are morphologically diverse, the mechanisms by which insects transport symbionts to oocytes are highly varied. In cockroaches, Blattabacterium-filled bacteriocyte cells migrate from the abdominal fat body to the distantly located ovarioles where they adhere to the oocyte membrane [99],[100]. Interestingly, the bacteriocytes remain associated with the oocyte for eight to nine days before finally expelling their symbionts through exocytosis. The Blattabacterium cells then squeeze between the follicle cells surrounding the oocyte and are engulfed into the oocyte cytoplasm via endocytosis just prior to ovulation [100].
Whiteflies (Family Aleyrodidae)
The whitefly circumvents exocytosis of its intracellular nutritional symbiont, Portiera aleyrodidarum, by depositing entire bacteriocytes into its eggs. These maternal bacteriocytes remain intact yet separate from the developing embryo until the embryonic bacteriomes form, at which point the maternal bacteriocytes deteriorate [22].
Tsetse Flies, Bat Flies, and Louse Flies (Superfamily Hippoboscoidea)
Members of the Hippoboscoidea superfamily (Order Diptera) are obligate blood feeders that have developed a unique reproductive strategy termed adenotrophic viviparity that offers a different solution to internal maternal transfer of symbionts. Females of this superfamily develop a single fertilized embryo at a time within their uterus (modified vaginal canal) until it is deposited as a mature third instar larva immediately preceding pupation. During their internal development, the larvae are nourished with milk produced by modified accessory glands that empty into the uterus [101]. The milk primarily consists of protein and lipids [102], but it also serves as a reservoir for maternally transmitted microbial symbionts [103]. For example, the obligate mutualistic symbiont of tsetse flies, Wigglesworthia glossinidia, is absent from the female germ line and surrounding reproductive tissues but is found extracellularly in the female milk glands and is first detected in tsetse offspring once milk consumption begins during the first larval stage [103].
Stinkbugs (Superfamily Pentatomoidea)
One of the most common mechanisms of external maternal transmission in insects is that of “egg smearing,” which occurs when a female contaminates the surface of her eggs with symbiont-laden feces during oviposition. Upon hatching, offspring probe or consume the discarded egg shells to acquire the maternal bacteria. This mode of transmission is commonly found in plant-sucking stinkbugs, including the Pentatomidae and Acanthosomatidae families [104]. In the Cynidae family of stinkbugs, along with the Coreidae family of leaf-footed bugs, gut symbionts are transferred maternally via coprophagy, in which offspring consume maternal feces, sometimes directly from the mother's anus [22],[104]. Stinkbugs of the Plataspidae family, on the other hand, have developed a unique mode of transmission via a maternally provided “symbiont capsule” deposited on the underside of the egg mass [56]. These capsules are comprised of bacterial cells dispersed throughout a resin-like matrix surrounded by a brown, cuticle-like envelope that protects the symbionts from environmental stressors such as UV irradiation or dissection [57]. After hatching, plataspid nymphs immediately probe the capsules to ingest the symbionts [56],[59].
European Beewolf (Philanthus triangulum)
While nutritional symbionts appear to be the most common type of bacteria transmitted via external maternal transmission in insects, the European beewolf (Philanthus triangulum) instead cultivates a symbiotic bacteria that protects offspring against microbial infection during development. Beewolves are solitary digger wasps that deposit their offspring in moist, underground nests, making them susceptible to fungal and bacterial infections [105]. To combat these pathogens, female beewolves cultivate Streptomyces philanthi bacteria in specialized glands in their antennae, which they copiously spread on the ceiling of the brood cell before oviposition [106]–[108]. After hatching, the larvae take up the bacterial cells and incorporate them into their cocoon that they build before pupation. When adult beewolves emerge from their cocoon in the summer, female beewolves acquire the maternally provided Streptomyces symbiont and house them in the female-specific gland reservoirs along each antenna [108],[109].
By integrating previous studies in invertebrates with recent evidence for maternal microbial transmission in humans and other vertebrates, we contend that maternal provisioning of microbes is a universal phenomenon in the animal kingdom. As a result, a considerable new phase of study in heritable symbiont transmission is underway. Thus, this essay presents current evidence for maternal microbial transmission and provides new insights into its impact on microbiome assembly and evolution, with applications to human health and disease.
Internal Maternal Transmission
At the turn of the twentieth century, French pediatrician Henry Tissier asserted that human infants develop within a sterile environment and acquire their initial bacterial inoculum while traveling through the maternal birth canal [28]. More than a century later, the sterile womb hypothesis remains dogma, as any bacterial presence in the uterus is assumed to be dangerous for the infant. Indeed, studies of preterm deliveries have found a strong correlation between intrauterine infections and preterm labor, especially when birth occurs less than 30 weeks into the pregnancy [29],[30]. Since preterm birth is the leading cause of infant mortality worldwide [31], much attention has focused on identifying the bacterial culprits responsible for spontaneous preterm labor. Surprisingly, most of the bacteria detected in intrauterine infections are commonly found in the female vaginal tract [29], and risk of preterm birth is markedly increased in women diagnosed with bacterial vaginosis during pregnancy [32]. Interestingly, the vaginal microbial community varies significantly among American women of different ethnicities (Caucasian, African-American, Asian, or Hispanic), with African-American and Hispanic women more likely to have a microbiota traditionally associated with bacterial vaginosis (predominance of anaerobic bacteria over Lactobacillus species) [33] and a higher rate of spontaneous preterm deliveries (reviewed in [34]).
While intrauterine infection and inflammation is important in understanding the etiology of preterm birth, relatively few studies have examined the uterine microbiome of healthy, term pregnancies owing to the sterile womb paradigm. Investigations into the potential for bacterial transmission through the placental barrier have detected bacteria in umbilical cord blood [17], amniotic fluid [14],[35], and fetal membranes [35],[36] from babies without any indication of inflammation (Figure 1). Furthermore, an infant's first postpartum bowel movement of ingested amniotic fluid (meconium) is not sterile as previously assumed, but instead harbors a complex community of microbes, albeit less diverse than that of adults [18],[37]. Interestingly, many of the bacterial genera found in the meconium, including Enterococcus and Escherichia, are common inhabitants of the gastrointestinal tract [18],[37]. To test whether maternal gut bacteria can be provisioned to fetuses in utero, Jiménez et al. [18] fed pregnant mice milk inoculated with genetically-labeled Enterococcus faecium and then examined the meconium microbes of term offspring after sterile C-section. Remarkably, E. faecium with the genetic label was cultured from the meconium of pups from inoculated mothers, but not from pups of control mice fed noninoculated milk. Meconium from the treatment group also had a higher abundance of bacteria than that of the control group. Importantly, the study controlled for potential bacterial contamination from contact between skin and the meconium by sampling an internal portion of the meconium [18]. Thus, this study provides foundational evidence for maternal microbial transmission in mammals.
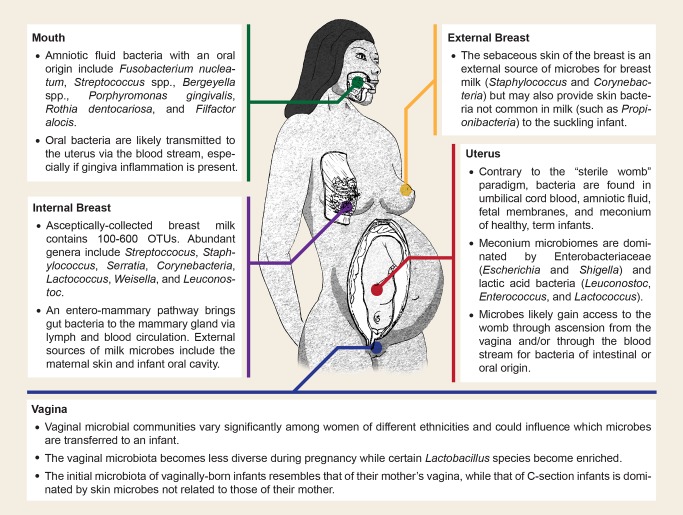
Figure 1. Sources of microbial transmission in humans from mother to child.
Cut-away diagram highlighting the various internal and external sources of maternal microbial transmission as well as the species that are commonly associated with transfer from those regions. Regions discussed include the oral cavity [14],[16], the mammary glands [40],[78],[79], the sebaceous skin surrounding the breast [78],[80], the vaginal tract [19],[33],[73], and the intrauterine environment [14],[15],[17],[18],[29],[35]–[37],[40]. Illustration by Robert M. Brucker.
Other than ascension of vaginal microbes associated with preterm births, the mechanisms by which gut bacteria gain access to the uterine environment are not well understood. One possibility is that bacteria travel to the placenta via the bloodstream after translocation of the gut epithelium. While the intestinal epithelial barrier generally prevents microbial entry into the circulatory system, dendritic cells can actively penetrate the gut epithelium, take up bacteria from the intestinal lumen, and transport the live bacteria throughout the body as they migrate to lymphoid organs [38],[39]. Interestingly, microbial translocation may even increase during pregnancy, as one study showed that pregnant mice were 60% more likely to harbor bacteria in their mesenteric lymph node (presumably brought there by dendritic cells) than nonpregnant mice [40]. Bacterial species normally found in the human oral cavity have also been isolated from amniotic fluid and likely enter the bloodstream during periodontal infections, facilitated by gingiva inflammation [14],[16] (Figure 1).
Overall, the study of internal maternal transmission of microbes in mammals is in its infancy due to the enduring influence of the sterile womb paradigm and to the ethical and technical difficulties of collecting samples from healthy pregnancies before birth. Thus, we still know very little about the number and identity of innocuous microbes that traverse the placenta, whether they persist in the infant, or whether their presence has long-term health consequences for the child. Similarly, we know almost nothing about nonpathogenic viruses or archaea that may be transferred from mother to child alongside their bacterial counterparts. Fortunately, the advent of culture-independent, high-throughput sequencing will serve as a tremendous resource for this field and will hopefully lead to a characterization of the “fetal microbiome” in utero.
Maternal provisioning of microbes to developing offspring is widespread in animals, with evidence of internal microbial transmission in animal phyla as diverse as Porifera [41]–[45] (Box 1), Mollusca [46]–[49] (Box 1), Arthropoda [50]–[52] (Box 2, Figure 2), and Chordata [19],[53],[54] (Box 3, Figure 2). The presence of maternal transmission at the base of the Animalia kingdom and the surprising plasticity by which microbes gain access to germ cells or embryos in these systems signifies that maternal symbiont transmission is an ancient and evolutionarily advantageous mechanism inherent in animals, including humans. Therefore, we can no longer ignore the fact that exposure to microbes in the womb is likely and may even be a universal part of human pregnancy, serving as the first inoculation of beneficial microbes before birth.
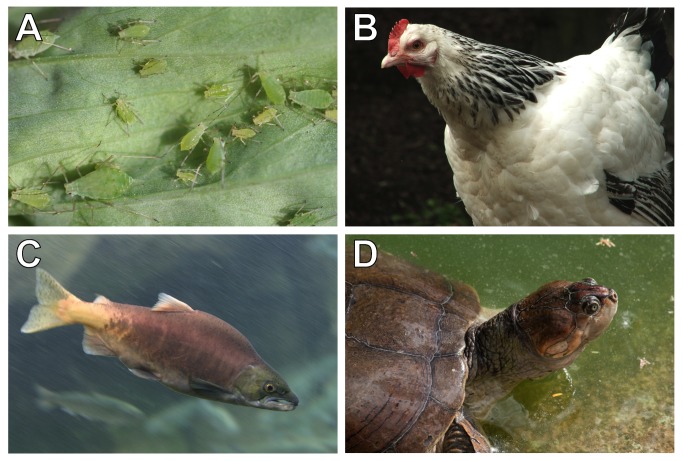
Figure 2. Examples of animals that exhibit microbial maternal transmission.
(A) Pea aphid (Acyrthosiphon pisum), photo credit: Whitney Cranshaw, Colorado State University/©Bugwood.org/CC-BY-3.0-US; (B) Domesticated chicken hen (Gallus gallus domesticus), photo credit: Ben Scicluna; (C) Sockeye salmon (Oncorhynchus nerka), photo credit: Cacophony; (D) South American river turtle (Podocnemis expansa), photo credit: Wilfredor. All photos were obtained from Wikimedia Commons (www.commons.wikimedia.org).
Box 3. Examples of Maternal Transmission in Vertebrates
Aside from studies in human and mouse models, very little is known about maternal transmission of microbial communities in vertebrates, especially outside Class Mammalia. Furthermore, research on vertical transmission in nonmammalians has largely focused on maternally transmitted pathogens, especially in animals of agricultural importance like chickens and fish.
Domesticated Chickens (Gallus gallus domesticus)
Zoonotic Salmonella infections acquired from contaminated chicken eggs is estimated to cause more than 100,000 illnesses each year in the United States [110]. In addition to horizontal transmission of Salmonella on eggs through surface contamination, direct transovarial transmission also occurs when Salmonella colonizes the reproductive tissues of hens (Figure 2B). Depending on the infection location within the female reproductive tract, the bacteria are deposited into the yolk, albumen, eggshell membrane, and/or eggshell of the developing egg before oviposition (reviewed in [111]). Other poultry pathogens, such as Mycoplasma synoviae in chickens [112] and M. gallisepticum, M. cloacale, and M. anatis in ducks [113], have also been cultured from the yolk of embryonated eggs, though whether commensal flora are incorporated into the egg is not known.
Ray-Finned Fish (Class Actinopterygii)
Several bacterial pathogens of economically important fish are transmitted transovarially in the egg yolk including Renibacterium salmoninarum, the agent of bacterial kidney disease in salmonids (Figure 2C), and Flavobacterium psychrophilum, which causes bacterial cold water disease in salmonids and rainbow trout fry disease in trout (reviewed in [114]). F. psychrophilum has also been found in ovarian fluid and on the surface of eggs of steelhead trout [115]. Additionally, an obligate, intracellular eukaryotic parasite, Pseudoloma neurophilia, is a common pathogen found in zebrafish (Danio rerio) facilities and has been observed in spores of the ovarian stroma and within developing follicle cells of spawning females, suggesting that it can be vertically transmitted, though it is primarily spread from fish to fish in contaminated water (reviewed in [116]).
Turtles (Order Chelonii)
The formation of egg components in the uterine tube and uterus of turtles takes approximately two weeks, providing ample opportunity for maternal transmission of intestinal or reproductive microbes to the egg [117]. One study of unhatched (dead) eggs from loggerhead sea turtle (Caretta caretta) nests found several potential pathogens, including Pseudomonas aeruginosa and Serratia marcesans, in fluid from the interior of the eggs, though environmental contamination of the eggs cannot be ruled out [118]. A similar study of eggs from two species of South American river turtles, Podocnemis expansa (Figure 2D) and P. unifilis, identified several Enterobacteriaceae species, including Escherichia coli, Shigella flexneri, and Salmonella cholerasuis, in the eggs but not in the environmental samples taken from the turtle nests [119], suggesting that they may have a maternal origin. In support of this hypothesis, a separate study in green turtles (Chelonia mydas) that collected eggs directly from the maternal cloacal opening during egg laying isolated Pseudomonas, Salmonella, Enterobacter, and Citrobacter from the eggshell, albumen, and yolk. In fact, the yolk was the egg component most heavily infected with bacteria [120]. Altogether, many potentially pathogenic species have been isolated from turtle eggs, but whether these bacteria actually cause disease in turtles or are part of their natural flora remains to be determined.
External Maternal Transmission
External maternal transmission encompasses any transfer of maternal symbionts to offspring during or after birth. In invertebrates, it is often accomplished by “egg smearing,” in which females coat eggs with microbes as they are deposited [55], or through the provision of a microbe-rich maternal fecal pellet that is consumed by larval offspring upon hatching [56]–[59] (see Box 2). Similarly, human infants are “smeared” with maternal vaginal and fecal microbes as they exit the birth canal [60]–[62] (Figure 1). Several studies have shown that the human neonatal microbiota across all body habitats (skin, oral, nasopharyngeal, and gut) is influenced by their mode of delivery [19],[63]–[65], with infants born vaginally acquiring microbes common in the female vagina while C-section infants display a microbiota more similar to that of human skin [19]. Furthermore, while the microbiota of a vaginally delivered infant clusters with the vaginal bacteria of its mother, the microbiota of C-section babies is no more related to the skin flora of its mother than that of a stranger, indicating that most microbes are transmitted to the neonate from those handling the infant [19]. Importantly, epidemiological data suggest that a Cesarean delivery can have long-term consequences on the health of a child, especially concerning immune-mediated diseases. For example, children born via C-section are significantly more likely to develop allergic rhinitis [66], asthma [66], celiac disease [67], type 1 diabetes [68], and inflammatory bowel disease [69]. These statistics are alarming given that 32.8% of all births in the United States in 2010 were delivered via C-section with similar rates on the rise in most developed countries [70].
The higher rate of immune-mediated diseases in C-section children may indicate that maternally transferred vaginal or fecal microbes are unique in their ability to elicit immune maturation in the neonate. Development of the intestinal mucosa and secondary lymphoid tissues in the gut is contingent upon recognition of microbial components by pattern-recognition receptors on intestinal epithelial cells (reviewed in [71],[72]). It is possible that these receptors cannot properly interact with the community of microbes acquired during Cesarean deliveries, leading to disrupted immune development and an increased risk for immune-mediated disorders in C-section children. Conversely, transmission for thousands of years of vaginal and fecal microbes at birth has likely produced specific human-microbe interactions important for neonatal gut development. In fact, a recent study found that the vaginal microbial community changes during pregnancy, becoming less diverse as the pregnancy progresses [73]; yet, in spite of the general decrease in richness, certain Lactobacillus bacterial species are enriched in the vaginal community during pregnancy and are hypothesized to be important for establishing the neonatal upper GI microbiota after vaginal delivery [73].
Breastfeeding provides a secondary route of maternal microbial transmission as shown in humans (reviewed in [74], Figure 1) and nonhuman primates such as rhesus monkeys [75]. In humans, maternal milk microbes are implicated in infant immune system development [76], resistance against infection [77], and protection against the development of allergies and asthma later in childhood [74]. High-throughput sequencing of breast milk from 16 healthy women identified 100–600 species of bacteria in each sample with nine genera present in every sample: Staphylococcus, Streptococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Propionibacterium, Sphingomonas, and Bradyrhizobiaceae [78]. This “core” milk microbiome represented approximately 50% of all bacteria in each sample, with the other half representing individual variation in microbial composition [78]. A similar study found that the bacterial composition in breast milk changes over time: milk produced immediately after labor harbored more lactic acid bacteria along with Staphylococcus, Streptococcus, and Lactococcus, while breast milk after six months of lactation had a significant increase in typical inhabitants of the oral cavity, such as Veillonella, Leptotrichia, and Prevotella [79], perhaps to prime the infant for the switch to solid food. However, as with any DNA-based, culture-independent study that does not discriminate between live and dead bacteria, the number and identity of bacteria detected in these studies should be interpreted with some caution.
Given that milk is only produced temporarily in a woman's life, the origin of milk microbes is still somewhat of a mystery. Breast milk was traditionally thought to be sterile; however, colostrum (the first milk produced after delivery) collected aseptically already harbors hundreds of bacterial species [79]. Breast milk does share many taxa with the microbiota found on sebaceous skin tissue around the nipple [78],[80], and high levels of Streptococcus in breast milk may be a result of retrograde flow from an infant's oral cavity back to the milk ducts during suckling [81] since Streptococcus is the dominant phylotype in infant saliva [82]. However, the presence of anaerobic gut bacteria in human milk suggests that an entero-mammary route of transfer also exists that may utilize phagocytic dendritic cells to traffic gut microbes to the mammary glands, similar to microbial transfer to amniotic fluid as discussed earlier. To support this hypothesis, Perez et al. [40] found identical strains of bacteria in milk cells, blood cells, and fecal samples from lactating women, but more work is needed to directly connect bacterial translocation in the gut to incorporation in breast milk.
Overall, maternal transmission of beneficial microbes in humans has widespread relevance for human health. Evolution with these microbes has resulted in our dependence on them for the proper maturation and development of the immune system and gastrointestinal tract. Somewhat paradoxically, modern medicine designed to prevent infant mortality (such as emergency Cesarean sections and formula feeding) has likely contributed to the rise in immune-mediated diseases in developed countries due to the inherent lack of exposure to maternal microbes associated with these practices. Fortunately, biomedicine is also making strides in finding effective probiotic supplements to promote immune development and ameliorate some of the risks that C-section or formula-fed infants face as children and adults. Hopefully, as we gain understanding of the diversity and function of maternally transmitted microbes in humans, more complete and effective probiotic blends will recapitulate the microbial communities found in vaginally delivered, breast-fed infants and restore the microbe-host interactions that humans depend upon for proper development.
Conclusions
Since the early twentieth century, the study of maternal microbial transmission has focused heavily on animal systems in which maternal transmission maintains sophisticated partnerships with one or two microbial species. However, with the development of high-throughput sequencing technologies, it is now possible to identify entire microbiomes that are transferred from mother to offspring in systems not traditionally considered to exhibit maternal transmission, such as humans. By expanding the definition of maternal transmission to include all internal and external microbial transfers from mother to offspring, we contend that maternal transmission is universal in the animal kingdom and is used to provision offspring with important microbes at birth, rather than leave their acquisition to chance.
Finally, with microbes contributing 99% of all unique genetic information present in the human body, maternal microbial transmission should be viewed as an additional and important mechanism of genetic and functional change in human evolution. Similar to deleterious mutations in our genetic code, disruption of maternal microbial acquisition during infancy could “mutate” the composition of the microbial community, leading to improper and detrimental host-microbe interactions during development. Maternal transmission is also a key factor in shaping the structure of the microbiome in animal species over evolutionary time, since microbes that promote host fitness, especially in females, will simultaneously increase their odds of being transferred to the next generation. Thus, whether internal or external, the universality and implications of maternal microbial transmission are nothing short of a paradigm shift for the basic and biomedical life sciences.
Abbreviations
- FISH
fluorescent in situ hybridization
- GI
gastrointestinal tract
- TEM
transmission electron microscopy
References
- 1. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. (2006) Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murgas Torrazza R, Neu J (2011) The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol 31 Suppl 1: S29–34. [DOI] [PubMed] [Google Scholar]
- 4. Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- 5. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 6. Round JL, Lee SM, Li J, Tran G, Jabri B, et al. (2011) The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332: 974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, et al. (2008) Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. (2009) Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ling Z, Kong J, Jia P, Wei C, Wang Y, et al. (2010) Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol 60: 677–690. [DOI] [PubMed] [Google Scholar]
- 10. Colombo AV, Silva CM, Haffajee A, Colombo AP (2006) Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol 55: 609–615. [DOI] [PubMed] [Google Scholar]
- 11. Candela M, Perna F, Carnevali P, Vitali B, Ciati R, et al. (2008) Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol 125: 286–292. [DOI] [PubMed] [Google Scholar]
- 12. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, et al. (2011) Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469: 543–547. [DOI] [PubMed] [Google Scholar]
- 13. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO (2007) Development of the human infant intestinal microbiota. PLoS Biol 5: e177 doi:10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bearfield C, Davenport ES, Sivapathasundaram V, Allaker RP (2002) Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG 109: 527–533. [DOI] [PubMed] [Google Scholar]
- 15. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, et al. (2008) Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE 3: e3056 doi:10.1371/journal.pone.0003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DiGiulio DB (2012) Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med 17: 2–11. [DOI] [PubMed] [Google Scholar]
- 17. Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, et al. (2005) Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol 51: 270–274. [DOI] [PubMed] [Google Scholar]
- 18. Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, et al. (2008) Is meconium from healthy newborns actually sterile? Res Microbiol 159: 187–193. [DOI] [PubMed] [Google Scholar]
- 19. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, et al. (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107: 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gronlund MM, Gueimonde M, Laitinen K, Kociubinski G, Gronroos T, et al. (2007) Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy 37: 1764–1772. [DOI] [PubMed] [Google Scholar]
- 21. Martin V, Maldonado-Barragan A, Moles L, Rodriguez-Banos M, Campo RD, et al. (2012) Sharing of bacterial strains between breast milk and infant feces. J Hum Lact 28: 36–44. [DOI] [PubMed] [Google Scholar]
- 22.Buchner P (1965) Endosymbiosis of animals with plant microorganisms. New York: Interscience Publishers.
- 23. Douglas AE (1998) Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera . Annu Rev Entomol 43: 17–37. [DOI] [PubMed] [Google Scholar]
- 24. Feldhaar H, Gross R (2009) Insects as hosts for mutualistic bacteria. Int J Med Microbiol 299: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Douglas AE (1989) Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc 64: 409–434. [DOI] [PubMed] [Google Scholar]
- 26. Baumann P (2005) Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59: 155–189. [DOI] [PubMed] [Google Scholar]
- 27. Bright M, Bulgheresi S (2010) A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8: 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tissier H (1900) Recherches sur la flore intestinale des nourrissons (état normal et pathologique). Paris: G. Carre and C. Naud.
- 29. Goldenberg RL, Hauth JC, Andrews WW (2000) Intrauterine infection and preterm delivery. N Engl J Med 342: 1500–1507. [DOI] [PubMed] [Google Scholar]
- 30. Goncalves LF, Chaiworapongsa T, Romero R (2002) Intrauterine infection and prematurity. Ment Retard Dev Disabil Res Rev 8: 3–13. [DOI] [PubMed] [Google Scholar]
- 31. Lawn JE, Cousens S, Zupan J (2005) 4 million neonatal deaths: when? Where? Why? Lancet 365: 891–900. [DOI] [PubMed] [Google Scholar]
- 32. Fiscella K (1996) Racial disparities in preterm births. The role of urogenital infections. Public Health Rep 111: 104–113. [PMC free article] [PubMed] [Google Scholar]
- 33. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, et al. (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108 Suppl 1: 4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menon R, Dunlop AL, Kramer MR, Fortunato SJ, Hogue CJ (2011) An overview of racial disparities in preterm birth rates: caused by infection or inflammatory response? Acta Obstet Gynecol Scand 90: 1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rautava S, Collado MC, Salminen S, Isolauri E (2012) Probiotics modulate host-microbe interaction in the placenta and fetal gut: a randomized, double-blind, placebo-controlled trial. Neonatology 102: 178–184. [DOI] [PubMed] [Google Scholar]
- 36. Steel JH, Malatos S, Kennea N, Edwards AD, Miles L, et al. (2005) Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr Res 57: 404–411. [DOI] [PubMed] [Google Scholar]
- 37. Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, et al. (2013) Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy 43: 198–211. [DOI] [PubMed] [Google Scholar]
- 38. Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, et al. (1999) Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401: 804–808. [DOI] [PubMed] [Google Scholar]
- 39. Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, et al. (2001) Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2: 361–367. [DOI] [PubMed] [Google Scholar]
- 40. Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, et al. (2007) Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119: e724–732. [DOI] [PubMed] [Google Scholar]
- 41. Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008) Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74: 7694–7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Enticknap JJ, Kelly M, Peraud O, Hill RT (2006) Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol 72: 3724–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmitt S, Weisz JB, Lindquist N, Hentschel U (2007) Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl Environ Microbiol 73: 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ereskovsky AV, Gonobobleva E, Vishnyakov A (2005) Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarcida). Marine Biol 146: 869–875. [Google Scholar]
- 45. Sharp KH, Eam B, Faulkner DJ, Haygood MG (2007) Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl Environ Microbiol 73: 622–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cary SC, Giovannoni SJ (1993) Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc Natl Acad of Sci U S A 90: 5695–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peek AS, Feldman RA, Lutz RA, Vrijenhoek RC (1998) Cospeciation of chemoautotrophic bacteria and deep sea clams. Proc Natl Acad Sci U S A 95: 9962–9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stewart FJ, Cavanaugh CM (2006) Bacterial endosymbioses in Solemya (Mollusca: Bivalvia)–model systems for studies of symbiont-host adaptation. Antonie van Leeuwenhoek 90: 343–360. [DOI] [PubMed] [Google Scholar]
- 49. Stewart FJ, Young CR, Cavanaugh CM (2008) Lateral symbiont acquisition in a maternally transmitted chemosynthetic clam endosymbiosis. Mol Biol Evol 25: 673–687. [DOI] [PubMed] [Google Scholar]
- 50. Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42: 165–190. [DOI] [PubMed] [Google Scholar]
- 51. Koga R, Meng XY, Tsuchida T, Fukatsu T (2012) Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci U S A 109: E1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Balmand S, Lohs C, Aksoy S, Heddi A (2013) Tissue distribution and transmission routes for the tsetse fly endosymbionts. J Invertebr Pathol 112: S116–S122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Inoue R, Ushida K (2003) Vertical and horizontal transmission of intestinal commensal bacteria in the rat model. FEMS Microbiol Ecol 46: 213–219. [DOI] [PubMed] [Google Scholar]
- 54. Carlier Y, Truyens C, Deloron P, Peyron F (2012) Congenital parasitic infections: a review. Acta Trop 121: 55–70. [DOI] [PubMed] [Google Scholar]
- 55. Kaltenpoth M, Winter SA, Kleinhammer A (2009) Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol Ecol 69: 373–383. [DOI] [PubMed] [Google Scholar]
- 56. Fukatsu T, Hosokawa T (2002) Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl Environ Microbiol 68: 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hosokawa T, Kikuchi Y, Meng XY, Fukatsu T (2005) The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol Ecol 54: 471–477. [DOI] [PubMed] [Google Scholar]
- 58. Hosokawa T, Kikuchi Y, Fukatsu T (2007) How many symbionts are provided by mothers, acquired by offspring, and needed for successful vertical transmission in an obligate insect-bacterium mutualism? Mol Ecol 16: 5316–5325. [DOI] [PubMed] [Google Scholar]
- 59. Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T (2008) Symbiont acquisition alters behaviour of stinkbug nymphs. Biol Lett 4: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bager P, Wohlfahrt J, Westergaard T (2008) Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy 38: 634–642. [DOI] [PubMed] [Google Scholar]
- 61. Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR (2008) A meta-analysis of the association between Caesarean section and childhood asthma. Clin Exp Allergy 38: 629–633. [DOI] [PubMed] [Google Scholar]
- 62. Huh SY, Rifas-Shiman SL, Zera CA, Edwards JW, Oken E, et al. (2012) Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child 97: 610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li Y, Caufield PW, Dasanayake AP, Wiener HW, Vermund SH (2005) Mode of delivery and other maternal factors influence the acquisition of Streptococcus mutans in infants. J Dent Res 84: 806–811. [DOI] [PubMed] [Google Scholar]
- 64. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, et al. (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118: 511–521. [DOI] [PubMed] [Google Scholar]
- 65. Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G (2008) Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr 138: 1796S–1800S. [DOI] [PubMed] [Google Scholar]
- 66. Renz-Polster H, David MR, Buist AS, Vollmer WM, O'Connor EA, et al. (2005) Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy 35: 1466–1472. [DOI] [PubMed] [Google Scholar]
- 67. Decker E, Engelmann G, Findeisen A, Gerner P, Laass M, et al. (2010) Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics 125: e1433–1440. [DOI] [PubMed] [Google Scholar]
- 68. Cardwell CR, Stene LC, Joner G, Cinek O, Svensson J, et al. (2008) Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia 51: 726–735. [DOI] [PubMed] [Google Scholar]
- 69. Bager P, Simonsen J, Nielsen NM, Frisch M (2012) Cesarean section and offspring's risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis 18: 857–862. [DOI] [PubMed] [Google Scholar]
- 70.Gibbons L, Belizan JM, Lauer JA, Betran AP, Merialdi M, et al.. (2010) The global numbers and costs of additionally needed and unnecessary Cesarean sections performed by year: overuse as a barrier to universal coverage. World Health Report (2010) Background Paper 30.
- 71. McElroy SJ, Weitkamp JH (2011) Innate immunity in the small intestine of the preterm infant. Neoreviews 12: e517–e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maynard CL, Elson CO, Hatton RD, Weaver CT (2012) Reciprocal interactions of the intestinal microbiota and immune system. Nature 489: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aagaard K, Riehle K, Ma J, Segata N, Mistretta T-A, et al. (2012) A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE 7: e36466 doi:10.1371/journal.pone.0036466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, et al. (2013) The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 69: 1–10. [DOI] [PubMed] [Google Scholar]
- 75. Jin L, Hinde K, Tao L (2011) Species diversity and relative abundance of lactic acid bacteria in the milk of rhesus monkeys (Macaca mulatta). J Med Primatol 40: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Diaz-Ropero MP, Martin R, Sierra S, Lara-Villoslada F, Rodriguez JM, et al. (2007) Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J Appl Microbiol 102: 337–343. [DOI] [PubMed] [Google Scholar]
- 77. Maldonado J, Canabate F, Sempere L, Vela F, Sanchez AR, et al. (2012) Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J Pediatr Gastroenterol Nutr 54: 55–61. [DOI] [PubMed] [Google Scholar]
- 78. Hunt KM, Foster JA, Forney LJ, Schütte UM, Beck DL, et al. (2011) Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 6: e21313 doi:10.1371/journal.pone.0021313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, et al. (2012) The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96: 544–551. [DOI] [PubMed] [Google Scholar]
- 80. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, et al. (2009) Topographical and temporal diversity of the human skin microbiome. Science 324: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ramsay DT, Kent JC, Owens RA, Hartmann PE (2004) Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 113: 361–367. [DOI] [PubMed] [Google Scholar]
- 82. Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, et al. (2011) Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS ONE 6: e23503 doi:10.1371/journal.pone.0023503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li CW, Chen JY, Hua TE (1998) Precambrian sponges with cellular structures. Science 279: 879–882. [DOI] [PubMed] [Google Scholar]
- 84. Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, et al. (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68: 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fieseler L, Horn M, Wagner M, Hentschel U (2004) Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl Environ Microbiol 70: 3724–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71: 295–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wilkinson CR (1984) Immunological evidence for the Precambrian origin of bacterial symbioses in marine sponges. Proc R Soc Lond 220: 509–517. [Google Scholar]
- 88. Webster NS, Taylor MW, Behnam F, Lucker S, Rattei T, et al. (2010) Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol 12: 2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cavanaugh CM, McKiness ZP, Newton ILG, Stewart FJ (2006) Marine chemosynthetic symbioses. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes. 3rd edition. New York: Springer.
- 90. Di Meo CA, Wilbur AE, Holben WE, Feldman RA, Vrijenhoek RC, et al. (2000) Genetic variation among endosymbionts of widely distributed vestimentiferan tubeworms. Appl Environ Microbiol 66: 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Laue BE, Nelson DC (1997) Sulfur-oxidizing symbionts have not co-evolved with their hydrothermal vent tube worm hosts: an RFLP analysis. Mol Mar Biol Biotechnol 6: 180–188. [PubMed] [Google Scholar]
- 92. Goffredi SK, Barry JP (2002) Species-specific variation in sulfide physiology between closely-related Vesicomyid clams. Mar Ecol Prog Ser 225: 227–238. [Google Scholar]
- 93. Cavanaugh CM (1983) Symbiotic chemoautotrophic bacteria in marine invertebrates from sulfide-rich habitats. Nature 302: 58–61. [Google Scholar]
- 94. Endow K, Ohta S (1990) Occurrence of bacteria in the primary oocytes of vesicomyid clam Calyptogena soyoae . Mar Ecol Prog Ser 64: 309–311. [Google Scholar]
- 95. LePage D, Bordenstein SR (2013) Wolbachia, can we save lives with a great pandemic? Trends Parasitol In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Saridaki A, Bourtzis K (2010) Wolbachia: more than just a bug in insects genitals. Curr Opin Microbiol 13: 67–72. [DOI] [PubMed] [Google Scholar]
- 97. Moran NA, Munson MA, Baumann P, Ishikawa H (1993) A molecular clock in endosymbiotic bacteria is calibrated using insect hosts. Proc R Soc Lond 253: 167–171. [Google Scholar]
- 98. Baumann P, Baumann L, Lai CY, Rouhbakhsh D, Moran NA, et al. (1995) Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol 49: 55–94. [DOI] [PubMed] [Google Scholar]
- 99. Sacchi L, Grigolo A, Laudani U, Ricevuti G, Dealessi F (1985) Behavior of symbionts during oogenesis and early stages of development in the German cockroach, Blatella germanica (Blattodea). J Invertebr Pathol 46: 139–152. [DOI] [PubMed] [Google Scholar]
- 100. Sacchi L, Grigolo A, Mazzini M, Bigliardi E, Baccetti B, et al. (1988) Symbionts in the oocytes of Blattella germanica (L.) (Dictyoptera: Blattellidae): their mode of transmission. Int J Insect Morphol Embryol 17: 437–446. [Google Scholar]
- 101. Tobe SS (1978) Reproductive physiology of Glossina. Annu Rev Entomol 23: 283–307. [DOI] [PubMed] [Google Scholar]
- 102. Cmelik SHW, Bursell E, Slack E (1969) Composition of gut contents of third-instar tsetse larvae (Glossina morsitans Westwood). Comp Biochem and Physiol 29: 447. [Google Scholar]
- 103. Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, et al. (2008) Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol 54: 1236–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Prado SS, Zucchi TD (2012) Host-symbiont interactions for potentially managing Heteropteran pests. Psyche 2012: 1–9. [Google Scholar]
- 105. Strohm E, Linsenmair KE (1995) Leaving the cradle: how beewolves (Philanthus triangulum F.) obtain the necessary spatial information for emergence. Zoology (Jena) 98: 137–146. [Google Scholar]
- 106. Kaltenpoth M, Gottler W, Herzner G, Strohm E (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15: 475–479. [DOI] [PubMed] [Google Scholar]
- 107. Kaltenpoth M, Goettler W, Dale C, Stubblefield JW, Herzner G, et al. (2006) ‘Candidatus Streptomyces philanthi’, an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol 56: 1403–1411. [DOI] [PubMed] [Google Scholar]
- 108. Goettler W, Kaltenpoth M, Herzner G, Strohm E (2007) Morphology and ultrastructure of a bacteria cultivation organ: the antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae). Arthropod Struct Dev 36: 1–9. [DOI] [PubMed] [Google Scholar]
- 109. Kaltenpoth M, Goettler W, Koehler S, Strohm E (2010) Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol Ecol 24: 463–477. [Google Scholar]
- 110. Schroeder CM, Naugle AL, Schlosser WD, Hogue AT, Angulo FJ, et al. (2005) Estimate of illnesses from Salmonella enteritdis in eggs, United States, 2000. Emerg Infect Dis 11: 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Gast R, et al. (2009) Mechanisms of egg contamination by Salmonella Enteritidis. FEMS Microbiol Rev 33: 718–738. [DOI] [PubMed] [Google Scholar]
- 112. MacOwan KJ, Atkinson MJ, Bell MA, Brand TF, Randall CJ (1984) Egg transmission of a respiratory isolate of Mycoplasma synoviae and infection of the chicken embryo. Avian Pathol 13: 51–58. [DOI] [PubMed] [Google Scholar]
- 113. Bencina D, Tadina T, Dorrer D (1988) Natural infection of ducks with Mycoplasma synoviae and Mycoplasma gallisepticum and Mycoplasma egg transmission. Avian Pathol 17: 441–449. [DOI] [PubMed] [Google Scholar]
- 114. Brock JA, Bullis R (2001) Disease prevention and control for gametes and embryos of fish and marine shrimp. Aquaculture 197: 137–159. [Google Scholar]
- 115. Brown LL, Cox WT, Levine RP (1997) Evidence that the causal agent of bacterial cold-water disease Flavobacterium psychrophilum is transmitted within salmonid eggs. Dis Aquat Org 29: 213–218. [Google Scholar]
- 116. Sanders JL, Watral V, Kent ML (2012) Microsporidiosis in zebrafish research facilities. ILAR J 53: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Alkindi AY, Mahmoud IY, Woller MJ, Plude JL (2006) Oviductal morphology in relation to hormonal levels in the snapping turtle, Chelydra serpentina. Tissue Cell 38: 19–33. [DOI] [PubMed] [Google Scholar]
- 118. Craven KS, Awong-Taylor J, Griffiths L, Bass C, Muscarella M (2007) Identification of bacterial isolates from unhatched loggerhead (Caretta caretta) sea turtle eggs in Georgia, USA. Marine Turtle Newsletter 115: 9–11. [Google Scholar]
- 119. Benevides de Morais P, Wessel de Oliveira K, Malvasio A, Gomes de Ataide A, Pimenta RS (2010) Enterobacteriaceae associated with eggs of Podocnemis expansa and Podocnemis unifilis (Testudines: Chelonia) in nonpolluted sites of National Park of Araguaia Plains, Brazil. J Zoo Wildl Med 41: 656–661. [DOI] [PubMed] [Google Scholar]
- 120. Al-Bahry S, Mahmoud I, Elshafie A, Al-Harthy A, Al-Ghafri S, et al. (2009) Bacterial flora and antibiotic resistance from eggs of green turtles Chelonia mydas: an indication of polluted effluents. Mar Pollut Bull 58: 720–725. [DOI] [PubMed] [Google Scholar]