Abstract
The matrix (M) protein of vesicular stomatitis virus inhibits both nuclear import and export. Here, we demonstrate that this inhibitory property is conserved between the M proteins from two other vesiculoviruses, chandipura virus and spring viremia carp virus. All three M proteins completely block nuclear transport of spliced mRNA, small nuclear RNAs, and small nuclear ribonucleoproteins and slow the nuclear transport of many other cargoes. In all cases where transport was merely slowed by the M proteins, the chandipura virus M protein had the strongest inhibitory activity. When expressed in transfected HeLa cells, active M proteins displayed prominent association with the nuclear rim. Moreover, mutation of a conserved methionine abolished both the inhibitory activity and efficient targeting of the M proteins to the nuclear rim. We propose that all of the vesiculoviral M proteins associate with the same nuclear target, which is likely to be a component of the nuclear pore complex.
Keywords: nucleocytoplasmic transport, vesiculoviruses, RNA transport
Trafficking of macromolecules between the nucleus and the cytoplasm occurs through nuclear pore complexes (NPCs), large proteinaceous structures (>50 different proteins) that perforate the nuclear envelope. Many small molecules (<40 kDa) can diffuse through NPCs, but large molecules must be transported across NPCs via carrier-mediated and signal-dependent processes. Much of the import and export of molecules across NPCs involves the interaction of transport receptors with their cargoes, the RanGTPase, and components of the NPC (1, 2).
Transport receptors, termed importins and exportins (or karyopherins), bind their appropriate cargoes directly or via specialized adaptor proteins (3). Once these complexes have formed, movement through the NPCs proceeds by a process involving sequential interactions of the receptor–cargo complexes with docking sites on the nuclear pore proteins (nucleoporins). A number of nucleoporins, particularly those containing phenylalanine–glycine (FG) repeat motifs, have been shown to interact directly with transport receptors (4). RanGTPase, which binds to transport receptors, plays a critical role in transport by promoting the association of cargo with export receptors as well as the dissociation of cargo from import receptors. Hydrolysis of RanGTP in the cytoplasm and regeneration of RanGTP in the nucleus sustains a gradient of RanGTP across the nuclear envelope, resulting in delivery of the transport cargoes to the appropriate cell compartments (5, 6).
Carrier-mediated movement across NPCs can be blocked in a variety of ways. Inactivation of RanGTPase leads to a block of most nucleocytoplasmic transport (7). Also, interference with the interactions between receptor–cargo complexes and nucleoporins inhibits nuclear transport. The lectin wheat germ agglutinin, which binds to O-glycosylated nucleoporins, blocks both import and export across NPCs (8), and antibodies to Nup98 or Nup153, two FG repeat-containing components of the NPC, block the export of small nuclear RNAs (snRNAs) and mRNA (9, 10). Likewise, the isolated nucleoporin binding domains of the transport factors importin β and TAP inhibit the export of mRNA and snRNAs (11, 12). This domain of importin β is also an efficient inhibitor of protein import.
Infection of eukaryotic cells by viruses can affect the nucleocytoplasmic transport of host-cell proteins and RNAs (13–16). Previously, we and others have demonstrated that the matrix (M) protein of vesicular stomatitis virus (VSV) is a potent inhibitor of nuclear transport (16–18). M protein, a structural component of VSV virions, blocks the nuclear export of snRNAs and spliced mRNAs as well as the nuclear import of small nuclear ribonucleoproteins (snRNPs) (16, 17). This inhibition of nuclear transport requires that M protein be in the nucleus (17). When expressed in transfected HeLa cells, a green fluorescent protein (GFP)-tagged M protein colocalized with antinucleoporin antibodies at the nuclear rim, whereas an inactive mutant of M protein, M51R, did not, suggesting that association with a component of the NPC is necessary for M activity (17).
Here, we report that the M proteins of several vesiculoviruses, VSV, chandipura virus (CV), and spring viremia of carp virus (SVCV), share significant sequence similarity. All of these M proteins reduced the rate of nuclear transport of many cargoes and blocked transport of spliced mRNA, snRNAs, and snRNPs in Xenopus oocytes. We show that a hierarchy of inhibitory activities exists among the M proteins, with CV M protein being the strongest inhibitor of transport. In all cases, inhibition requires a conserved methionine, and the active M proteins associate efficiently with the nuclear rim, suggesting that the vesiculoviral M proteins interact with the same nuclear target, which is likely to be a component of the NPC.
Materials and Methods
Sequence and Secondary Structure Analysis.
Sequence similarity searches were performed with the BLAST program against the nonredundant database with the BLOSUM62 scoring matrix (19). The multiple sequence alignment was constructed by using ClustalW (20). Secondary structure predictions for the individual M proteins were carried out by using the Ph.D. program and a consensus generated for the multiple sequence alignment (21). The PREDATOR program was used to generate a secondary structure prediction based on the multiple alignment (22, 23).
DNA Plasmids, Mutagenesis, Recombinant Protein Expression, and Purification.
The pSP64poly(A)-VSV-M, pGEX-VSV-M, and pEGFP-VSV-M (Orsay strain) DNAs have been described (17). The pBSK plasmid encoding the CV M gene was kindly provided by A. C. Marriott (University of Warwick, Warwick, U.K.). To generate pSP64poly(A)CV-M, an RsaI fragment of pBSK-CV-M containing the CV M coding region was ligated to SmaI cut pSP64poly(A). To generate pGEX-CV-M, the CV M coding region was PCR amplified by using primers that contained SmaI restriction sites. The resulting PCR product was cut and ligated into the similarly cut pGEX vector. pEGFP-CV-M DNA was generated by subcloning the coding region of the CV M protein-containing fragment from pGEX-CV-M into the pEGPF-C1 vector (CLONTECH). DNA encoding the SVCV M protein was obtained by PCR amplification of an SVCV cDNA library kindly provided by J. C. Leong (Oregon State University, Corvallis, OR). To generate pSP64poly(A)-SVCV-M, the resulting PCR product was cleaved with XbaI and SmaI and ligated to SmaI cut pSP64poly(A) DNA. To generate pGEX-SVCV-M, the SVCV M coding region was PCR amplified by using primers that contained EcoRI and BamHI restriction sites. The resulting PCR product was cut and ligated into the similarly cut pGEX vector. pEGFP-SVCV-M DNA was generated by subcloning the coding region of the SVCV M protein-containing fragment from pGEX-SVCV-M into the pEGPF-C1 vector.
All point mutations were generated by two-step PCR as described (17). The mutations were introduced into the M genes in the pGEX-M vectors and then subsequently subcloned into the pEGPF-C1 vector. Mutations were confirmed by DNA sequencing.
Recombinant glutathione S-transferase (GST)-M proteins were prepared as described (17).
In Vitro Transcription.
DNA templates for in vitro transcription of U1, U1Sm−, U5, and U6 snRNAs, U3 small nucleolar RNA (snoRNA), adenovirus major late mRNA, U6 RRE, ET202 RNA, and tRNAMet were generated as described (17, 24, 25). The template for transcription of constitutive transport element (CTE) RNA (CTE250, MPMV nucleotides 8007–8240) is described (24, 26). In vitro synthesis of [α-32P]GTP-labeled RNAs was performed in 20-μl reactions as detailed elsewhere (27). For in vitro synthesis of poly(A)+ mRNAs encoding the various M proteins, plasmid DNAs were linearized with EcoRI and used in large-scale transcription reactions with SP6 polymerase according to the protocol of Promega.
Expression and Detection of M Proteins in Xenopus laevis Oocytes.
For in vivo expression and labeling of M proteins, mRNAs encoding M proteins were injected into the cytoplasms of stage VI oocytes and incubated for 16–24 h in MBS-H containing 0.25 μCi/μl (in ≈100 μl for ≈10 oocytes; 1 Ci = 37 GBq) of [35S]methionine (Amersham Pharmacia) (28). The nuclear and cytoplasmic fractions from such oocytes were analyzed as described (17).
Analysis of RNA and Protein Transport in X. laevis Oocytes.
Preparation and injection of X. laevis oocytes were as described (28). Approximately 20 fmol of mRNAs encoding the various M proteins were injected into the cytoplasm ≈18 h before the injection of import or export substrates. In other experiments, purified GST-M proteins (10 nl at 100 μg/ml) were injected directly into the nucleus, as indicated.
RNA mixtures (15 nl) containing ≈5 fmol of 32P-labeled import or export substrates were injected into either the cytoplasm or nucleus of oocytes, respectively. GST-Rev protein (10 nl at 100 μg/ml) was injected into the nuclei of oocytes. GST-SV40 nuclear localization signal (NLS)-GFP and GST-nucleoplasmin (NP) NLS-GFP were kindly provided by S. Adam (Northwestern University) and were injected (10 nl at 100 μg/ml) into the cytoplasm of oocytes. Blue dextran and U3 snoRNA were included in all injection mixtures as controls for injection and dissection accuracy. At the indicated time points, the oocytes were dissected into cytoplasmic and nuclear fractions and analyzed by PAGE followed by autoradiography or Western blotting as described (17).
Antibodies and Western Blotting.
Mouse monoclonal anti-GST and anti-GFP antibodies were from Amersham Pharmacia and Santa Cruz Biotechnology, respectively. For Western blot analysis, extracts of oocytes or HeLa cells were fractionated by SDS/PAGE, and the proteins were transferred to Immobilon-P poly(vinylidene difluoride) membranes (Millipore). Membranes were probed with antibodies in TBS-T (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/1 mM EDTA/0.25% Tween 20) containing 5% powdered milk.
DNA Transfections.
For transient transfections of GFP-M DNAs into tissue culture cells, 4 × 105 HeLa cells in MEM containing 15% FCS were seeded onto coverslips 24 h before use. Transfections were carried out with 0.5–1 μg of pEGFP-M DNAs and 10 μl of Lipofectamine according to the protocol of Life Technologies (Grand Island, NY); 24 h later, cells were processed for immunofluorescence.
Immunofluoresence.
To process cells for immunofluorescence, cells were either fixed with 2% paraformaldehyde for 15 min before permeabilization with 0.5% Triton X-100 or extracted first with 0.5% Triton X-100 for 3 min followed by paraformaldehyde fixation (17). The activities of the GFP-M protein chimeras were determined after injection of the mRNAs encoding these proteins into oocyte cytoplasms.
Results
Sequence Comparison of the Vesiculoviral M Proteins.
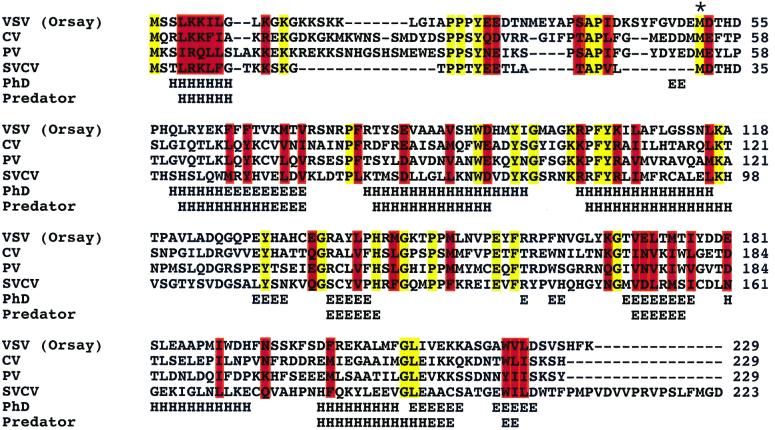
We showed previously that VSV M protein in the nucleus of X. laevis oocytes is a potent inhibitor of snRNA and spliced mRNA export and of snRNP import (17). To discover other proteins that might have similar activity, we searched published databases for proteins with overall sequence homology to the VSV M protein. Significant similarities were detected (Fig. 1) between the M proteins of VSV and other sequenced vesiculoviruses including CV, SVCV, and piry virus (PV). Similar homologies have been noted by others (29, 30). We detected no significant sequence similarities between the VSV M protein and cellular proteins.
Figure 1.
Sequence alignment and predicted secondary structure of M proteins from VSV (Orsay strain), CV, PV, and SVCV. The CLUSTALW program was used for the alignment. Identical amino acids are indicated in yellow, and conserved amino acids are shown in red. The M proteins from CV:PV, CV:VSV, CV:SVCV, PV:VSV, PV:SVCV, and VSV:SVCV are 70.3%, 51.5%, 42.3%, 48.9%, 46.3%, and 49.8% similar, respectively. Secondary structure predictions from the PREDATOR and PH.D. programs are indicated below the alignment as alpha helices (H) and beta strands (E). * indicates a conserved methionine that is essential for the inhibitory function of the M proteins.
Sequence comparison using the CLUSTALW alignment program showed that the vesiculoviral M proteins share 10.9% identity and 29% similarity (Fig. 1). With respect to the M protein of VSV, the M proteins of CV, SVCV, and PV are 51.5%, 49%, and 49.8% similar. Sequence relatedness is highest between the M proteins from CV and PV (70.3%) and lowest between the M proteins from CV and SVCV (48%). The resemblance of the M proteins to one another encouraged us to test whether M proteins from other vesiculoviruses could inhibit nucleocytoplasmic transport.
The Vesiculoviral M Proteins Block Nucleocytoplasmic Transport of snRNAs, Spliced mRNA, and snRNPs.
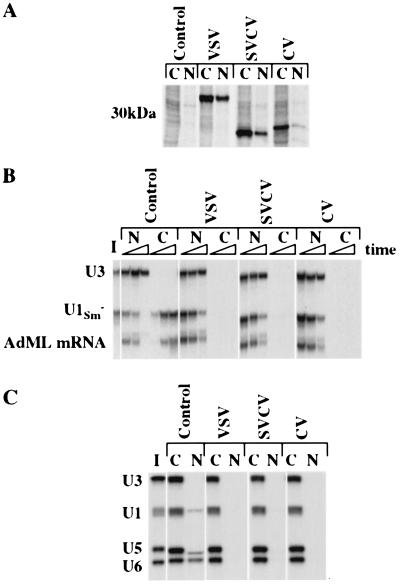
To express the vesiculoviral M proteins in Xenopus oocytes, DNAs encoding the M proteins of VSV, SVCV, and CV (PV cDNA was not available) were used as templates for in vitro synthesis of mRNAs. Upon injection into the cytoplasms of Xenopus oocytes, these mRNAs directed synthesis of 35S-methionine-labeled proteins with molecular weights in the range expected (Fig. 2A). Like the VSV M protein, the SVCV and CV M proteins were present in both the cytoplasms and the nuclei of the oocytes. The overall incorporation of labeled methionine into CV M protein was lower than that of either VSV or SVCV M protein, although the three proteins have comparable numbers of methionines (Fig. 1); thus, CV M protein accumulates to a lower level, under these conditions.
Figure 2.
The vesiculoviral M proteins block nuclear transport of spliced mRNA, snRNA, and snRNPs. (A) Expression and distribution of M proteins. Messenger RNAs encoding M proteins of VSV, SVCV, and CV were injected into the cytoplasms of oocytes, and the newly synthesized proteins were labeled by the addition of [35S]methionine to the incubation medium. Eighteen hours later, nuclear (N) and cytoplasmic (C) extracts were prepared, and 0.5 oocyte equivalents of protein were analyzed by SDS/PAGE followed by autoradiography. Note that the mobility of the VSV M protein is reduced because of the presence of a 6× histidine tag. (B and C) RNA export and import. Oocytes were preinjected with M protein mRNA of VSV, SVCV, and CV into the cytoplasm, and 16 h later the indicated 32P-labeled RNAs were injected into either the nuclei (B) or cytoplasms (C). Upon further incubation, oocytes were dissected; RNAs were isolated from nuclei (N) and cytoplasms (C) fractions, and 0.5 oocyte equivalents of each were separated by electrophoresis on 8% denaturing polyacrylamide gels and detected by autoradiography. The time points for oocyte dissection were 1, 3, and 22 h (B) and 24 h (C).
The abilities of the vesiculoviral M proteins to inhibit nuclear export and import were assayed by using several types of transport cargoes. All three M proteins made in oocytes blocked nuclear export of both U1 snRNA and spliced adenovirus major late mRNA for at least 20 h (Fig. 2B). Similarly, they all blocked export of the RNAs upon injection into oocyte nuclei as recombinant GST fusion proteins (see below, Fig. 4A; data not shown). The M proteins also blocked the import of U1, U5, and U6 snRNPs, which use different import pathways (Fig. 2C). Thus, the ability to inhibit nucleocytoplasmic transport is conserved between the M proteins of the vesiculoviruses.
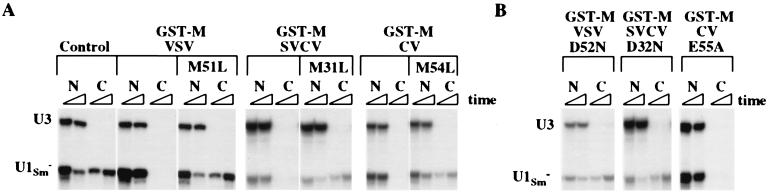
Figure 4.
A conserved methionine residue is essential for the activity of the vesiculoviral M proteins. Wild-type or mutant GST-M proteins were injected into the nucleus 1 h before injection of export substrates. Export of 32P-labeled U1Sm− snRNA was monitored 1 and 3 h after RNA injection. (A) RNA export was analyzed in the absence and presence of GST-tagged M proteins of VSV, VSV(M51L), SVCV, SVCV(M31L), CV, and CV(M54L). (B) RNA export was analyzed as in A in the presence of GST-tagged M proteins of VSV(D52N), SVCV(D32N), and CV(E55A).
The Vesiculoviral M Proteins Do Not Inactivate Transport Receptors.
The effect of the M proteins on additional transport cargoes was monitored to determine whether the M proteins block movement of snRNAs, spliced mRNAs, and snRNPs by disabling required transport receptors or factors. CRM1 is the receptor that is responsible for the export of both snRNAs and proteins containing leucine-rich export signals such as the HIV-1 Rev protein (31). The factor TAP has been implicated in the export of both spliced mRNA and RNAs containing the CTE of Mason–Pfizer monkey virus (24, 26, 32, 33). The import receptor importin β, in conjunction with cargo-specific adaptors, mediates import of both snRNPs and NLS-containing proteins (34, 35).
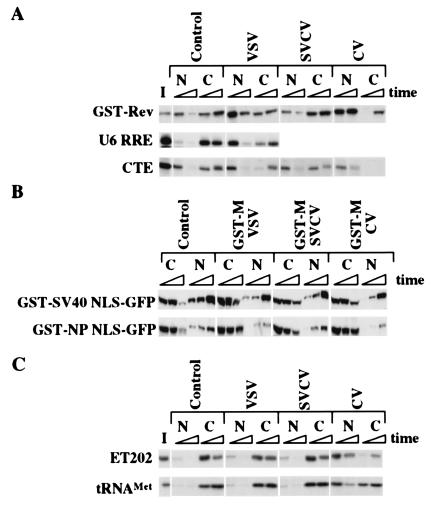
The M proteins slowed, but did not block, the transport of other cargoes that use these transport receptors and factors. For example, CRM1-dependent export of Rev protein continued at a reduced rate in the presence of the M proteins (Fig. 3A Top). Likewise, Rev-dependent export of U6 RNA containing the Rev-responsive element (U6 RRE) was only slowed by VSV M protein (Fig. 3A Middle). TAP-mediated export of CTE RNA was slowed but not blocked by VSV and SVCV M proteins (Fig. 3A Bottom); at early times, export of CTE RNA was also prevented by CV M protein (Fig. 3A Bottom), but, because of the nuclear instability of CTE RNA, we could not determine whether export would have occurred at time points later than 8 h. Similarly, importin β-dependent import of proteins containing either canonical or bipartite NLSs was slowed but not blocked by each of the M proteins when injected into the nucleus as purified recombinant GST-fusion proteins (Fig. 3B). The continued function of transport pathways dependent on CRM1, TAP, and importin β shows that the block to transport snRNAs, spliced mRNAs, and snRNPs by the M proteins is unlikely to result from the inactivation of these nuclear transport receptors and factors.
Figure 3.
The vesiculoviral M proteins slow, but do not block, multiple transport pathways. (A) Nuclear export. mRNAs encoding M proteins of VSV, SVCV, and CV were injected into the cytoplasm of oocytes, and 16 h later the indicated recombinant proteins or 32P-labeled RNAs were injected into the nuclei. At the indicated times, nuclear (N) and cytoplasmic (C) fractions were isolated, and the RNAs (U6 RRE or CTE) or proteins (GST-Rev) were separated by electrophoresis. The U6 RRE (Middle) and CTE (Bottom) RNAs were detected by autoradiography. GST-Rev (Top) protein was visualized by Western blotting with αGST antibody. Export of GST-Rev protein was monitored 1 and 3.5 h (Top) after being injected. U6 RRE RNA export was monitored at 1 and 4 h after RNA injection (Middle). Export of CTE RNA was monitored 4 and 8 h after RNA injection (Bottom). Note that by 8 h, most of the CTE RNA in the nucleus was degraded. (B) Nuclear import. Recombinant GST-tagged M proteins of VSV, SVCV, and CV were injected into the nucleus of oocytes, and 1 h later either GST-SV40 NLS-GFP (Upper) or GST-NP NLS-GFP (Lower) protein was injected into the cytoplasm. At 1, 4, and 24 h later, nuclear and cytoplasmic extracts were prepared and analyzed by SDS/PAGE followed by Western blotting with αGST antibody. (C) Nuclear Export. Export of 32P-labeled ET-202 RNA and tRNA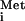 was monitored in the presence of the M proteins as in A at 1 and 4.5 h and at 1 and 3 h, respectively.
was monitored in the presence of the M proteins as in A at 1 and 4.5 h and at 1 and 3 h, respectively.
A Hierarchy of Inhibitory Activities Exists Between the Vesiculoviral M Proteins.
Quantitative differences between the abilities of the vesiculoviral M proteins to inhibit transport became apparent when we assayed the movement of cargoes whose transport was slowed but not blocked (see Fig. 3 A and B). These differences also were observed with the export of several other RNAs that use other transport receptors (25, 36, 37). ET-202 is an artificial RNA molecule selected for its ability to be exported in the presence of the VSV M protein; it has been shown to be transported by a pathway distinct from tRNA export (25). The rate of export of ET-202 RNA was affected more by the CV M protein than the VSV and SVCV M proteins (Fig. 3C Upper). Similarly, VSV and SVCV M proteins made in the oocytes had only a very small effect on the export of tRNA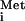 and tRNATyr compared with that of CV M protein (Fig. 3C Lower; data not shown). The potency of CV M protein was not simply due to increased amounts of proteins because the CV M protein accumulated to the lowest protein levels when expressed in oocytes (Fig. 2A). Thus, a gradient of inhibitory activities exists with the M protein of CV>VSV>SVCV.
and tRNATyr compared with that of CV M protein (Fig. 3C Lower; data not shown). The potency of CV M protein was not simply due to increased amounts of proteins because the CV M protein accumulated to the lowest protein levels when expressed in oocytes (Fig. 2A). Thus, a gradient of inhibitory activities exists with the M protein of CV>VSV>SVCV.
Conservation of an Essential Methionine in the Vesiculoviral M Proteins.
As we demonstrated recently, the ability of VSV M protein to inhibit transport requires a methionine at position 51 (Met-51), and even a conservative change to leucine abolishes this inhibitory activity (17). Because a methionine is present in comparable locations of all of the vesiculoviral M proteins (Fig. 1), we tested whether these residues were functionally important by changing them to leucines in the context of the GST-M fusion proteins. Upon injection into oocyte nuclei, none of these mutant proteins blocked snRNA export (Fig. 4A), even though they were stable and distributed to both the nucleus and cytoplasm (data not shown). Thus, the same essential function is probably served by Met-51 of VSV, Met-31 of SVCV, and Met-54 of CV.
Adjacent to the essential methionine in each of the M proteins is an acidic amino acid (Fig. 1), and VSV M protein maintained its inhibitory activity when the aspartic acid was changed to glutamic acid (data not shown). We tested the importance of these residues in the context of GST-M fusion proteins by changing the charged amino acids to their respective amide amino acids or to alanine. Both VSV and SVCV M proteins were inactivated when this aspartic acid was neutralized (Fig. 4B). In contrast, the M protein of CV appeared to be active when the acidic amino acid Glu-55 was neutralized by mutation to glutamine (data not shown) or alanine (Fig. 4B). Because the CV M protein is such a potent inhibitor of transport, it is possible that a moderate decrease in activity might have escaped detection under our assay conditions.
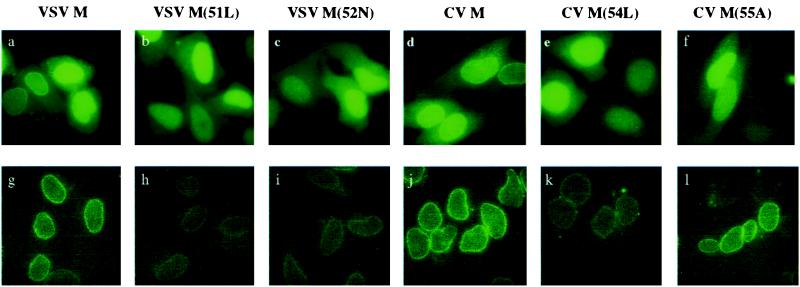
Active M Proteins Can Associate with the Nuclear Rim.
Previously, we showed that in transfected HeLa cells the wild-type VSV M protein associates with the nuclear rim but an inactive mutant protein does not, suggesting that a component of the NPC is a target for the M protein (17). Here, we monitored the intracellular localization of the different M proteins by transfecting HeLa cells with equivalent amounts of DNAs encoding the various wild-type and mutant GFP-M proteins (Fig. 5a). The cells were either fixed directly with paraformaldehyde (Fig. 5a Upper) or treated with Triton X-100 before fixation (Fig. 5a Lower); the latter treatment allowed for visualization of proteins associated with the nuclear rim. As we observed for GFP-tagged VSV M protein (Fig. 5a; ref. 17), GFP-tagged CV M protein was detected in the nucleus, in the cytoplasm, and at the nuclear rim (Fig. 5d). In contrast, GFP-tagged SVCV M protein was not found at the nuclear rim (data not shown); however, this fusion protein was inactive as an inhibitor of RNA export in oocyte assays (data not shown), indicating that the GFP tag interferes with the function of the SVCV M protein.
Figure 5.
Active M proteins associate prominently with the nuclear rim. Plasmid DNAs encoding wild-type and mutant GFP-tagged M proteins of VSV (a–c, g–i) or CV (d–f, j–l) were transfected into HeLa cells. Then, 24 h later the cells were either fixed with formaldehyde (a–f) or extracted with 0.5% Triton X-100 before fixation (g–l) and visualization by fluorescence microscopy. To capture the images in g–l, identical camera settings were used. In a–f, the camera setting was set to auto exposure to obtain a comparable signal for the active and inactive M proteins because the inactive M proteins are expressed at levels that are ≈10-fold higher.
Consistent with our previous report, the distributions of all GFP-tagged inactive mutant proteins (Fig. 5 b, c, and e) derived from the VSV or CV M proteins differed from those of the active proteins (Fig. 5 a, d, and f) in that they did not exhibit a readily detectable association with the nuclear rim. Upon extraction with Triton X-100, differences between active and inactive M proteins were more pronounced, with active M proteins displaying prominent nuclear rim association (Fig. 5 g, j, and l). In contrast, only small amounts of the inactive M proteins could be detected at the nuclear rim (Fig. 5 h, i, and k), even though the inactive M proteins were present in ≈10-fold higher amounts, as assayed by Western blotting (data not shown). These results suggest that the inactive M proteins display a greatly reduced affinity for a component of the nuclear rim. Thus, prominent association with the nuclear rim correlates with the inhibitory activities of all three M proteins.
Discussion
We have shown that the M proteins of three different vesiculoviruses: CV, VSV, and SVCV, slow the transport of several cargoes and completely block transport of snRNAs, spliced mRNAs, and snRNPs. It is likely that all three M proteins recognize the same intranuclear target because each M protein loses its ability to block nuclear transport when a conserved methionine residue is mutated. Moreover, prominent association of the M proteins with the nuclear rim correlates with their inhibitory activities, suggesting that the nuclear target of the vesiculoviral M proteins is likely to be a component of the NPC.
We previously proposed that an essential methionine in VSV M protein, Met-51, is important for interaction with its target (17). The M proteins of CV and SVCV have methionines in comparable locations, and all three proteins are inactivated by substitution of the methionine with leucine (Fig. 4A). These methionine residues are in regions predicted to form loop or turn structures (Fig. 1), and in VSV M protein this amino acid contributes to the epitope that is recognized by a mAb capable of reversing transport inhibition (17). In the case of CV M protein, an adjacent methionine residue (Met-53) is unable to compensate for mutation of the essential methionine (Met-54), demonstrating that precise positioning of this residue is required for inhibitory activity.
The functional target of the M proteins remains to be identified. Active GFP-tagged M proteins associate prominently with the nuclear rim, whereas the inactive mutant proteins do not (Fig. 5; ref. 17). Recently, it was reported that the target of the VSV M protein is the nucleoporin Nup98, but a direct, functional association was not demonstrated, and some M protein was observed at the nuclear rim even in cells lacking Nup98 (18). Our identification of two additional M proteins, from CV and SVCV, that are likely to interact with the same nuclear target should aid in the detection of the NPC-associated target.
The different M proteins display a hierarchy of inhibitory activities, with the order from the most to the least effective being the M protein of CV, VSV, and SVCV (Fig. 3). Our sequence comparison of the M proteins supports this observed hierarchy; the M protein from CV is more closely related to VSV M protein than to SVCV M protein. The resemblance in the sequences of the M proteins of CV and PV, including the essential methionine residue, leads us to predict that the PV M protein also is a strong inhibitor of nuclear transport. Because it is likely that all three M proteins interact with the same nuclear target, we propose that differences in the affinities of the M proteins for this target contribute to their hierarchy of inhibitory activities.
The observation that export of tRNA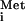 is sensitive to inhibition by the M protein of CV (Fig. 3C) was unexpected, as we previously had not observed inhibition of tRNA
is sensitive to inhibition by the M protein of CV (Fig. 3C) was unexpected, as we previously had not observed inhibition of tRNA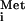 export by the M protein of VSV (16, 17). In the experiments presented here, we observed a slowing of tRNA
export by the M protein of VSV (16, 17). In the experiments presented here, we observed a slowing of tRNA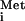 export in the presence of the M proteins of VSV and SVCV at very early times after tRNA
export in the presence of the M proteins of VSV and SVCV at very early times after tRNA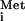 injection (Fig. 3C, compare VSV and SVCV columns with the control column). Thus, the rate of transport of all cargoes whose movement we have monitored can be slowed to some extent by the M proteins. The effect on the rate of tRNA transport by CV M protein became even more apparent when we monitored export of tRNAPro (J.M.P., unpublished data), a tRNA that normally is exported more slowly than tRNA
injection (Fig. 3C, compare VSV and SVCV columns with the control column). Thus, the rate of transport of all cargoes whose movement we have monitored can be slowed to some extent by the M proteins. The effect on the rate of tRNA transport by CV M protein became even more apparent when we monitored export of tRNAPro (J.M.P., unpublished data), a tRNA that normally is exported more slowly than tRNA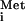 (C. R. Trotta, personal communication). Thus, sensitivity to inhibition by the M proteins may be affected by the efficiency with which a cargo is transported.
(C. R. Trotta, personal communication). Thus, sensitivity to inhibition by the M proteins may be affected by the efficiency with which a cargo is transported.
The question arises as to why the M proteins block transport of snRNAs, spliced mRNAs, and snRNPs, but only reduce the rate of transport of other cargoes, even when the same transport receptors or factors (CRM1, TAP, and importin β) are used. We note that all of the cargoes whose transport is blocked are large multiprotein/RNA complexes. Thus, the M proteins might block transport by influencing the size and/or flexibility of the NPCs, thereby setting an upper limit on the size of complexes that can be accommodated. Alternatively, because the M proteins slow both import and export, they would exert a compound effect when transport of the cargo requires regeneration of a pool of shuttling adaptors. For example, the M proteins could affect snRNA and spliced mRNA export both directly by inhibiting export per se and indirectly by slowing the reimport of the shuttling adaptor proteins.
It is curious that several different vesiculoviruses, all of which replicate in the cytoplasm, retain the ability to inhibit nucleocytoplasmic transport. One consequence of inhibiting both import and export might be the reduction of the host cell's ability to produce interferons in response to viral infection, thereby allowing more productive viral replication. In addition, inhibition of cellular mRNA export could lead to increased amounts of ribosomes available for the translation of viral mRNAs. Whatever the reason for the conservation of this function within the vesiculoviral M proteins, these proteins promise to be valuable tools in our study of nucleocytoplasmic transport.
Acknowledgments
We thank Elsebet Lund, Christopher Trotta, Doreen Glodowski, and Marie-Louise Hammarskjöld for comments, suggestions, and critical reading of the manuscript, and Suzanne Imboden and Corey Slominski for technical assistance. We thank Steve Adam, Anthony Marriott, and J. C. Leong for kindly supplying reagents. This work was supported by National Institutes of Health Grant GM-30220 and Defense Advanced Research Planning Agency Grant MDA972-97-1-0005 (to J.E.D.) and a Burroughs Wellcome Fund of the Life Sciences Research Foundation fellowship (to J.M.P.).
Abbreviations
- NPC
nuclear pore complex
- snRNA
small nuclear RNA
- VSV
vesicular stomatitis virus
- snRNP
small nuclear ribonucleoprotein
- GFP
green fluorescent protein
- CV
chandipura virus
- SVCV
spring viremia carp virus
- GST
glutathione S-transferase
- PV
piry virus
- CTE
constitutive transport element
- NLS
nuclear localization signal
- M protein
matrix protein
- NP
nucleoplasmin
References
- 1.Görlich D, Kutay U. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 2.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 3.Weis K. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 4.Wente S R. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- 5.Moore M S. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 6.Dahlberg J E, Lund E. Curr Opin Cell Biol. 1998;10:400–408. doi: 10.1016/s0955-0674(98)80017-3. [DOI] [PubMed] [Google Scholar]
- 7.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlberg J E, Lund E. Semin Cell Dev Biol. 1997;8:65–70. doi: 10.1006/scdb.1996.0123. [DOI] [PubMed] [Google Scholar]
- 9.Powers M A, Forbes D J, Dahlberg J E, Lund E. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullman K S, Shah S, Powers M A, Forbes D J. Mol Biol Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Görlich D. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachi A, Braun I C, Rodrigues J P, Pante N, Ribbeck N, von Kobbe C, Kutay U, Wilm M, Görlich D, Carmo-Fonseca M, Izaurralde E. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Li Y, Krug R M. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobbelstein M, Roth J W, Kimberly T, Levine A J, Shenk T. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustin K E, Sarnow P. EMBO J. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Her L-S, Lund E, Dahlberg J E. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 17.Petersen J M, Her L-S, Varvel V, Lund E, Dahlberg J E. Mol Cell Biol. 2000;20:8590–8601. doi: 10.1128/mcb.20.22.8590-8601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Kobbe C, van Deursen J M, Rodrigues J P, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo-Fonseca M, Izaurralde E. Mol Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rost B, Sander C, Schneider R. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 22.Frishman D, Argos P. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 23.Frishman D, Argos P. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M L, Dahlberg J E. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm C, Lund E, Dahlberg J E. Proc Natl Acad Sci USA. 1997;94:10122–10127. doi: 10.1073/pnas.94.19.10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ernst R K, Bray M, Rekosh D, Hammarskjöld M L. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 27.Pasquinelli A E, Dahlberg J E, Lund E. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- 28.Gurdon J B, Wickens W P. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- 29.Barge A, Gagnon J, Chaffotte A, Timmins P, Langowski J, Ruigrok R W, Gaudin Y. Virology. 1996;219:465–470. doi: 10.1006/viro.1996.0272. [DOI] [PubMed] [Google Scholar]
- 30.Taylor A, Easton A J, Marriott A C. Virus Genes. 1999;19:223–228. doi: 10.1023/a:1008188730975. [DOI] [PubMed] [Google Scholar]
- 31.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 32.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 33.Kang Y, Cullen B R. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palacios I, Hetzer M, Adam S A, Mattaj I W. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi N C, Adam E J, Adam S A. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arts G J, Fornerod M, Mattaj I W. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 37.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Gorlich D. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]