Abstract
Signaling through the Toll receptor is required for dorsal/ventral polarity in Drosophila embryos, and also plays an evolutionarily conserved role in the immune response. Upon ligand binding, Toll appears to multimerize and activate the associated kinase, Pelle. However, the immediate downstream targets of Pelle have not been identified. Here we show that Drosophila tumor necrosis factor receptor-associated factor 2 (dTRAF2), a homologue of human TRAF6, physically and functionally interacts with Pelle, and is phosphorylated by Pelle in vitro. Importantly, dTRAF2 and Pelle cooperate to activate Dorsal synergistically in cotransfected Schneider cells. Deletion of the C-terminal TRAF domain of dTRAF2 enhances Dorsal activation, perhaps reflecting the much stronger interaction of the mutant protein with phosphorylated, active Pelle. Taken together, our results indicate that Pelle and dTRAF2 physically and functionally interact, and that the TRAF domain acts as a regulator of this interaction. dTRAF2 thus appears to be a downstream target of Pelle. We discuss these results in the context of Toll signaling in flies and mammals.
The Toll receptor pathway is evolutionarily conserved from insects to mammals. It is responsible for establishing dorsal/ventral polarity of Drosophila embryos (1), functions in the insect anti-microbial immune response (2, 3), and plays a key role in mammalian innate immunity (4, 5). It is now clear that there are multiple Toll-like receptors (TLRs) that contain a related intracellular domain (Tir domain) (6). Upon ligand binding, a signal is transduced through the Tir domain to an associated complex containing a responsive kinase, Pelle in Drosophila and an IL-1 receptor-associated kinase (IRAK) in mammals. Although the proximal targets of Pelle/IRAK are unknown, signaling eventually leads to activation of a Dorsal/nuclear factor (NF)-κB transcription factor, by inducing degradation of the inhibitory protein Cactus/I-κB.
In Drosophila, Toll interacts with a complex containing at least Pelle and an associated protein, Tube. Tube/Pelle recruitment to the membrane is required for activation of signaling (7, 8). Concomitant with signaling, Pelle becomes transiently autophosphorylated and activated by a mechanism that involves concentration-dependent self-association (B.S. and J.L.M., unpublished data). Phosphorylated Pelle then appears to be released from the complex, presumably to phosphorylate downstream targets (see below).
It is not yet clear how Pelle activates downstream signaling, and what its targets might be. One candidate is Pellino, which was identified as a Pelle-binding protein by yeast two-hybrid assays (9). Pellino interacts only with phosphorylated wild-type Pelle, not with an unphosphorylated kinase-inactive mutant, suggesting a phosphorylation-related interaction. Other evidence seems to link Pelle with the Cactus/Dorsal complex. First, Pelle can phosphorylate Cactus in vitro (7), just as mouse Pelle (IRAK) can phosphorylate I-κBα (10). Second, Pelle was shown to interact with Dorsal (e.g., ref. 11), again in a phosphorylation-dependent manner. However, it is not yet known whether Cactus or Dorsal is a physiological substrate of Pelle. It is possible that Pelle indirectly activates Cactus through other unknown intermediates. In mammals, considerable evidence suggests that I-κB is phosphorylated on physiologically significant sites by I-κB kinase (IKK; see ref. 12).
In mammalian IL-1R signaling, IRAK is recruited to the IL-1R complex and phosphorylated upon ligand binding (13). TRAF6, a member of the tumor necrosis factor (TNF) receptor-associated factor (TRAF) family, associates with IRAK upon IL-1 induction. Overexpression of TRAF6 can lead to nuclear factor (NF)-κB activation, and a dominant-negative mutant of TRAF6 inhibits IL-1-induced NF-κB activation (14). TRAF6 is also involved in human Toll-induced NF-κB activation (15). These findings suggest that TRAF6 is a downstream mediator of IRAK. Importantly, TRAF6 is capable of interacting with several downstream factors that appear to participate in activation of NF-κB. One of these is NIK, a MAP3K-like kinase required for TNF-, CD95-, and IL-1-induced NF-κB activation (16), and which activates IKKα by phosphorylation (17, 18). TRAF6 also interacts with the TAB2/TAK1 complex, such that activated TAK1 phosphorylates NIK and stimulates IKK-α activity, thereby linking TRAF6 to the NIK-IKK cascade in the IL-1 signaling pathway (19). TRAF6 is also capable of interacting with evolutionarily conserved signaling intermediate in Toll pathways (ECSIT) (20). Overexpression of ECSIT accelerates activation of MEKK-1, and dominant negative ECSIT blocks MEKK-1 processing and activation of NF-κB. MEKK-1, a component of the c-Jun-N-terminal kinase (JNK) pathway, was shown to activate NF-κB by phosphorylating IKKβ (18). Taken together, the evidence suggests that TRAF6 is a mediator linking IRAK with NF-κB activation, although the underlying mechanisms are unknown.
The involvement of TRAFs in Toll signaling in Drosophila is less well documented. Recently, two homologues of mammalian TRAFs, dTRAF1 and dTRAF2 (Drosophila TRAFs), were isolated by Liu et al. (21). Coexpression of dTRAF1 and the Ste20 kinase (Misshapen, Msn) was shown to result in synergistic activation of JNK in mammalian cultured cells. Sequence comparison indicated that both dTRAFs contain a zinc finger domain and a C-terminal TRAF domain. dTRAF2 has, in addition, an N-terminal RING finger domain, and the TRAF domain of dTRAF2 is more closely related to that of mammalian TRAF6. Like TRAF6, dTRAF2 also interacts with dECSIT, and overexpression of dECSIT can induce an anti-microbial response in Drosophila (20). It is unknown whether dTRAF2 plays a role in Toll-directed Dorsal activation.
We have studied the possible function of dTRAF2 in the Toll pathway. We first show that dTRAF2 activates Dorsal when both are coexpressed in Drosophila Schneider cells. Importantly, we found a weak but direct interaction between Pelle and dTRAF2 in vitro, and showed that dTRAF2 can be phosphorylated by Pelle. Consistent with this finding, dTRAF2 and Pelle synergistically activate Dorsal in cotransfected cells, in a manner that requires Pelle kinase activity. Domain mapping showed that deletion of the C-terminal TRAF domain results in enhanced ability to activate Dorsal, and a much higher affinity for Pelle in vitro. These results suggest that Pelle and dTRAF2 interact through the dTRAF2 N-terminal region, and that the TRAF domain regulates this interaction. Our results suggest that dTRAF2 is a direct downstream target of Pelle.
Materials and Methods
Plasmids and Antibodies.
AcDTRAF2 was made by inserting a DNA fragment treated with AvaII and the Klenow fragment of DNA polymerase (Klenow) and BamHI from Bs-dTRAF2 (21), into the AcPA vector (22) cut with EcoRV and BamHI. Two primers were used in PCR to amplify the dTRAF2 coding sequence from AcDTRAF2. The 1.5-kb PCR fragment was digested with MluI and BglII, then was inserted into AcHAPelle (24) between MluI and BglII sites, to make AcHAdTRAF2. AcHAdTRAF2C(265–463) was constructed by ligating the large Mlu I/Klenow–BglII fragment from AcHAdTRAF2 with an ≈700-bp NdeI/Klenow–BglII fragment from AcHAdTRAF2. dTRAF2N(1–156), dTRAF2N(1–264), and dTRAF2N(1–309) fragments were amplified by PCR with the same 5′-primer. PCR fragments were digested with MluI and BglII, and inserted into AcHAPelle to produce the hemagglutinin (HA)-tagged dTRAF2 deletion constructs.
GST-dTRAF2 was made by ligating the MluI/Klenow–BglII dTRAF2 fragment, the large BamHI/Klenow–PstI fragment, and the small BamHI/PstI fragment from pGEX-2TK together (GST, glutathione S-transferase). GST-dTRAF2C(265–463) was constructed by ligating the NdeI/Klenow–BglII dTRAF2C fragment, the large EcoRI/Klenow–PstI fragment, and the small BamHI-PstI fragment from pGEX-2TK. GST-dTRAF2N(1–264) was obtained by self-ligating the NdeI/SmaI/Klenow large fragment from GST-dTRAF2. All other plasmids were as described (23, 24).
Mouse anti-HA (influenza virus epitope) monoclonal antibody was from Babco (Richmond, CA).
Transient Transfection Assays.
SL2 cells were transfected with a total of 10 μg of DNA mixture containing 2 μg of an internal control plasmid, copia-βGal, 1 μg of zen-CAT reporter DNA, 2 μg of pGEM3, and 5 μg of Act5C expression plasmids (22, 24–26). The indicated amounts of Ac-dTRAF2 DNA, AcPelle, and 0.4 μg of AcDorsal were mixed with different amounts of AcPA (empty vector) to bring total Act5C promoter-containing plasmids to 5 μg. Chloramphenicol acetyltransferase (CAT) activities were normalized by β-galactosidase(β-gal) levels.
In Vitro Protein Binding Assays.
Binding assays were performed essentially as described (23). Briefly, 2–4 μg of purified GST or GST-fusion proteins were immobilized on glutathione beads at 4°C for 1 h. In vitro translated proteins were incubated with the protein-beads at room temperature for 1.5 h, and mixed by rotating. After washing, protein complexes were eluted with 20 mM glutathione (reduced form) twice at room temperature. The protein eluent was analyzed on SDS/PAGE followed by autoradiography. In some assays, in vitro-translated Pelle was incubated with 1 mM ATP in kinase buffer for 1 h, which produced a mixture of unphosphorylated and autophosphorylated Pelle. The kinase reaction was terminated by adding 10 mM EDTA, and proteins were incubated with immobilized GST-fusion proteins as described above.
In Vitro Kinase Assays.
Two hundred nanograms of GST-dTRAF2 proteins, 40 ng of purified HisPelle, and 10 μCi of [γ-32P]ATP were incubated in 30 μl of kinase buffer (23) at 30°C for 30 min. Reactions were terminated by 10% trichloroacetic acid precipitation. After 30 min or more on ice, protein precipitates were recovered by centrifugation, washed with acetone, and dissolved in SDS-sample buffer. Proteins were analyzed by SDS/PAGE followed by autoradiography.
Results
dTRAF2 Activates Dorsal in Cotransfected SL2 Cells.
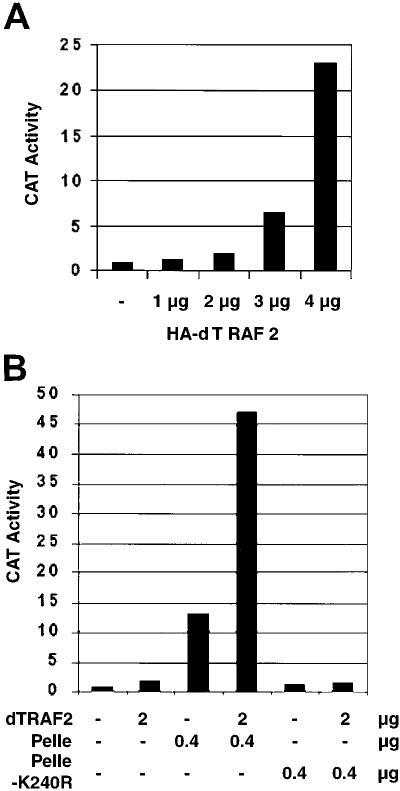
In light of the homology between dTRAF2 and TRAF6, we first tested whether dTRAF2 can activate Dorsal in cotransfected SL2 cells. Such assays were previously used to analyze components of the signaling pathway leading to Dorsal-dependent transcription (e.g., ref. 22). Different amounts of dTRAF2 expression vector were cotransfected with a constant amount of a Dorsal expression vector, a Dorsal-dependent CAT reporter, and copia-lacZ (as an internal control). Dorsal-dependent transcription was quantitated by measuring the ratio of CAT and β-gal activities. Fig. 1A shows that dTRAF2 activated Dorsal in a concentration-dependent manner, ≈20-fold at the highest concentration. Activation was Dorsal dependent, as transfections without Dorsal did not give rise to significant CAT activity (data not shown).
Figure 1.
dTRAF2 activates Dorsal in cotransfected Schneider cells, and functionally interacts with Pelle. (A) Overexpression of dTRAF2 leads to Dorsal activation in SL2 cells. Transfections were performed with the indicated amounts of AcdTRAF2 and additional plasmids as described in Materials and Methods. After 48 h, cells were harvested and extracts were prepared for CAT and β-gal assays. Dorsal activity was quantitated by measuring the ratio of CAT activity to β-GAL activity. (B) Synergy between Pelle and dTRAF2 in Dorsal activation. The indicated amounts of AcdTRAF2 and AcPelle/AcPelleK240R were cotransfected into Schneider cells as described in A. PelleK240R is a kinase inactive Pelle mutant.
dTRAF2 and Pelle Functionally Interact.
Given the observation that IRAK and TRAF6 can be found together in a complex after IL-1 induction (14), we next wished to investigate whether dTRAF2 functions by interacting with Pelle. We first tested whether Pelle and dTRAF2 could interact in a functional assay. Remarkably, in SL2 cells cotransfected with dTRAF2 and Pelle, Dorsal-dependent CAT activity was synergistically enhanced, with the observed activation (>40-fold) being much greater than the sum (or even product) of the activation induced by Pelle (≈12-fold) or dTRAF2 (≈2-fold) alone (Fig. 1B). Coactivation required Pelle kinase activity, as control assays with the kinase-inactive mutant PelleK240R did not produce enhanced CAT activity (Fig. 1B).
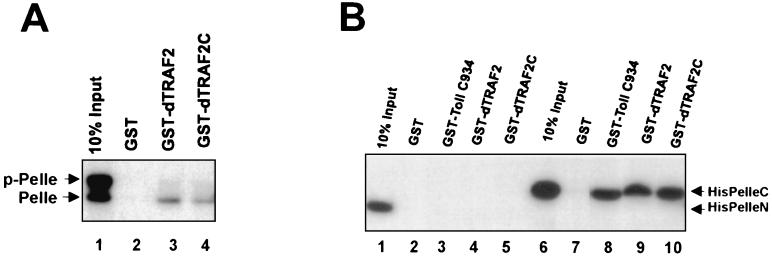
The strong synergistic interaction between Pelle and dTRAF2 in SL2 cells likely reflects a physical interaction between dTRAF2 and Pelle, but to test this directly we used GST “pull-down” assays. Because we have shown that some interactions involving Pelle are dependent on the phosphorylation status of the kinase (23), Pelle produced by in vitro translation was incubated under kinase conditions to allow Pelle autophosphorylation (see Materials and Methods). The resultant Pelle was a mixture of both phosphorylated and unphosphorylated Pelle (based on comparison with assays with PelleK240R; data not shown). This preparation was incubated with glutathione agarose-immobilized GST or GST-dTRAF2. Proteins were eluted with glutathione and analyzed on SDS/PAGE (23). A weak interaction between dTRAF2 and Pelle, predominantly, but not exclusively, with the unphosphorylated form, was observed (Fig. 2A). Domain mapping showed that the Pelle catalytic domain (Fig. 2B, lanes 9 and 10), and not the N-terminal regulatory domain (Fig. 2B, lanes 4 and 5), was responsible for the interaction with dTRAF2. These data suggest that dTRAF2 and Pelle functionally and physically interact in the pathway leading to Dorsal activation.
Figure 2.
dTRAF2 physically interacts with Pelle. (A) Pelle interacts with dTRAF2 in vitro. In vitro-translated HisPelle was first incubated with ATP in kinase buffer, then incubated with 2 μg of purified GST or the indicated GST-dTRAFs fusion protein immobilized on glutathione beads. Protein complexes were eluted with 20 mM glutathione and analyzed by SDS/PAGE followed by autoradiography. Lane 1, 10% of HisPelle/ATP used in binding assays (input); lanes 2, 3, and 4, GST, GST-dTRAF2, or GST-dTRAF2C(265–463), respectively, incubated with HisPelle/ATP. p-Pelle, phosphorylated Pelle. (B) The Pelle catalytic domain interacts with dTRAF2. In vitro-translated PelleN or PelleC was incubated with 2 μg of immobilized GST-fusion and analyzed in A. Lane 1, 10% of PelleN used in binding assays (input); lanes 2–5, GST, GST-Toll IC934, GST-dTRAF2, or GST-dTRAF2C, respectively, incubated with PelleN; lane 6, 10% of PelleC used in binding assays (input); lanes 7–10, GST, GST-Toll IC934, GST-dTRAF2, or GST-dTRAF2C, respectively, plus PelleC.
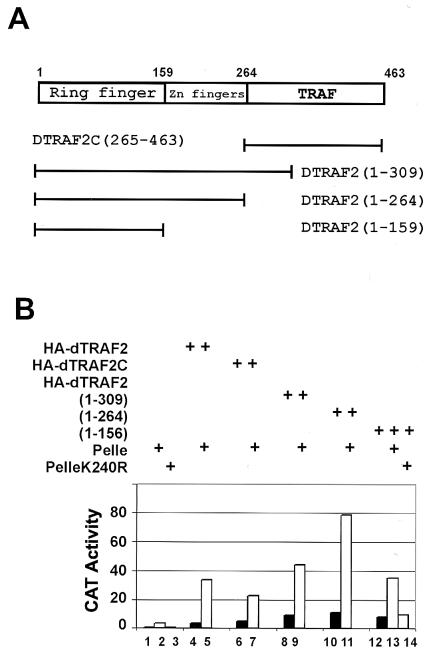
To analyze the functional domains on dTRAF2, we constructed a series of truncated derivatives (Fig. 3A) for both SL2 transfection assays and in vitro binding assays. Fig. 3B shows that, unexpectedly, the TRAF domain (dTRAF2(265–463)) alone was able to activate Dorsal-dependent CAT expression, similar to full-length dTRAF2 (Fig. 3B, lanes 4 and 6). This is in contrast with the behavior of mammalian TRAF6, as expression of the TRAF domain of TRAF6 actually displayed a dominant-negative effect on IL-1 signaling in transfected cells (14). However, without the TRAF domain, the RING-finger plus zinc-finger domains [dTRAF2(1–264)], in fact, displayed enhanced activity in signaling to Dorsal (Fig. 3B, lane 10). As with wild-type dTRAF2, cotransfection of Pelle and dTRAF2(1–264) synergistically activated Dorsal (Fig. 3B, lane 11).
Figure 3.
Deletion of the TRAF domain of dTRAF2 results in an enhanced Dorsal activation. (A) Functional domains of dTRAF2 and deletion mutations. RING finger, zinc fingers, and TRAF domain are indicated. Numbers indicate amino acid residues. (B) Deletion of the dTRAF2 TRAF domain enhances Dorsal activation in cotransfected SL2 cells. Two micrograms of dTRAF2 or mutant dTRAF2 expression vectors either alone or with 0.05 μg Pelle/PelleK240R expression vectors was transfected into SL2 cells as described in the legend of Fig. 1. Lanes 1–3, AcPA, Pelle or PelleK240R; lanes 4, 6, 8, 10, and 12, dTRAF2, dTRAF2C, dTRAF2(1–309), dTRAF2(1–264), or dTRAF2(1–156), respectively; lanes 5, 7, 9, 11, and 13, different forms of dTRAF2 plus Pelle; lane 14: dTRAF2(1–156) plus PelleK240R.
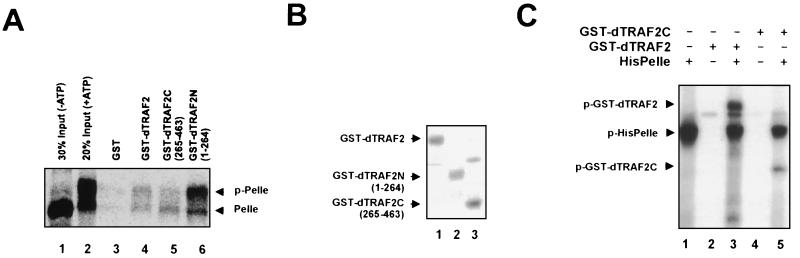
To investigate the possible basis for the above results, we next performed GST pull-down assays using GST derivatives of the different dTRAF2 mutants. Strikingly, dTRAF2(1–264) displayed a much stronger affinity for Pelle in GST-binding assays (Fig. 4A, lane 6) than did wild-type dTRAF2 (lane 4) or dTRAF2(265–463) (lane 5). Interestingly, dTRAF2(1–264) interacted very strongly with phosphorylated, as well as unphosphorylated, Pelle (Fig. 4A, lane 6), but dTRAF2C(265–463) bound preferentially to unphosphorylated Pelle (Fig. 4A, lane 5). Fig. 4B shows that there were equivalent amounts of the purified GST-fusion proteins in the binding assays. These results suggest that dTRAF2 may be a target of phosphorylated (activated) Pelle. Taken together, Pelle and dTRAF2 functionally interact, through the dTRAF2 RING and/or zinc-finger domains, and this interaction is regulated by the TRAF domain (see Discussion).
Figure 4.
The TRAF domain of dTRAF2 regulates the dTRAF2–Pelle interaction. (A) Deletion of TRAF domain stimulates the interaction between dTRAF2 and Pelle. In vitro-translated HisPelle was first incubated with ATP in kinase buffer, then subjected to binding assays with 2 μg of purified, immobilized GST or GST-fusion proteins as in Fig. 2B. Lane 1, 30% of HisPelle (unphosphorylated) used in binding assays; lane 2, 10% of HisPelle/ATP (phosphorylated) used in binding assays (input); lanes 3–6, GST, GST-dTRAF2, GST-dTRAF2C, or GST-dTRAF2N(1–264), respectively, plus HisPelle/ATP. (B) Coomassie blue staining of 2 μg of purified GST-fusion proteins used in binding assays. (C) dTRAF2 is phosphorylated by Pelle in vitro. Two hundred nanograms of GST-dTRAF2, GST-dTRAF2C, and/or 40 ng of HisPelle were incubated in kinase buffer at 30°C for 30 min. Reactions were terminated by trichloroacetic acid precipitation, and precipitates were washed and subjected to SDS/PAGE. Phosphate incorporation was analyzed by autoradiography (see Materials and Methods). Lane 1, HisPelle; lane 2, GST-dTRAF2; lane 3, GST-dTRAF2 plus HisPelle; lane 4, GST-dTRAF2C; lane 5, GST-dTRAF2C plus HisPelle.
dTRAF2 Is Phosphorylated by Pelle.
Given the above interaction between Pelle and dTRAF2, we next wished to determine whether dTRAF2 might be a substrate for Pelle kinase activity. Purified GST-dTRAF2 and GST-dTRAF2C were incubated with or without HisPelle in kinase buffer containing [γ-32P]ATP. Proteins were analyzed by SDS/PAGE and autoradiography (Fig. 5). As observed previously (23), HisPelle was autophosphorylated under these conditions (lanes 1, 3, and 5). More importantly, GST-dTRAF2 was efficiently phosphorylated by HisPelle (lane 3), and GST-dTRAF2C was also phosphorylated, albeit less efficiently (lane 5). As neither GST alone nor GST-dTRAF2(1–264) was phosphorylated detectably (data not shown), we conclude that the phosphorylation of dTRAF2 by HisPelle was specific and likely limited to the C-terminal TRAF domain.
Figure 5.
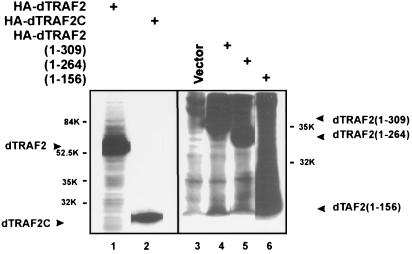
dTRAF2 and RING domain-containing derivatives are modified in cotransfected Schneider cells. Two micrograms of HA-tagged dTRAF2 or mutants was transfected into SL2 cells as described in the legend of Fig. 1. After normalization for variation in transfection efficiency by β-gal assays, whole-cell extracts were subjected to SDS/PAGE and anti-HA Western blotting. Lane 1, dTRAF2; lane 2, dTRAF2(265–463); lane 3, vector; lane 4, dTRAF2(1–309); lane 5, dTRAF2(1–264); lane 6, dTRAF2(1–156). Sizes of molecular weight standards are shown, and the positions of unmodified dTRAF2 and mutants are indicated.
dTRAF2 Is Extensively Modified in Transfected SL2 Cells.
During the course of the above transfection experiments (Fig. 3B), we also measured accumulation of wild-type and mutant dTRAF2 proteins by Western blotting using an antibody directed against the N-terminal influenza virus (HA) epitope present on all of the dTRAF2 derivatives. Unexpectedly, this revealed a ladder-like staining of HA-dTRAF2 (Fig. 5, lane 1). This modification was also detected with all dTRAF2 mutants that retained the RING finger (Fig. 5, lanes 4, 5, and 6), but not with the variant containing only the TRAF domain (dTRAF2C; lane 2). Indeed, these modifications were particularly noticeable with the smaller RING finger-containing mutants. These data suggest that the RING finger domain is both necessary and sufficient for dTRAF2 modification. Given that RING domains frequently function as E3 ubiquitin ligases (27), the ladder-like modification of dTRAF2, which does not reflect phosphorylation (data not shown), likely represents ubiquitination, perhaps signaling subsequent degradation of dTRAF2. However, the functional importance of this modification is not yet clear (see Discussion).
Discussion
We have shown that dTRAF2 is capable of activating Dorsal in transfected Schneider cells. Most importantly, dTRAF2 and Pelle functionally interact to activate Dorsal synergistically, consistent with an observed physical interaction between the two proteins, and with our finding that dTRAF2 is phosphorylated by Pelle in vitro. The N-terminal RING/zinc-finger domains, but not the TRAF domain, interact strongly with both phosphorylated and unphosphorylated Pelle, and activate Dorsal in SL2 cells. That the RING- plus zinc-finger domains interact efficiently with phosphorylated (i.e., activated; B.S. and J.L.M., unpublished data) Pelle supports the idea that dTRAF2 is a functional target of Pelle, consistent with the observed enhanced Dorsal activation. Based on these findings, we describe a model to illustrate the function of dTRAF2 in Toll-induced Dorsal activation.
Unlike the situation observed with mammalian TRAF6 (14), the TRAF domain of dTRAF2 does not show a dominant-negative effect, but instead activates Dorsal to a level comparable to that observed with wild-type dTRAF2. It is possible that an interaction between the exogenously expressed TRAF domain and endogenous dTRAF2 may be responsible for this seemingly anomalous activation of signaling by the isolated TRAF domain. Mammalian TRAF domains have been shown to be involved in both homotypic and heterotypic aggregation (28, 29). In TNF-induced NF-κB activation, the interaction between the TRAF6 N-terminal domain and ζPKC (an atypical member of the protein kinase C family) is important for signaling (30). ζPKC was unable to interact with the N-terminal domain of TRAF6 in its monomeric form, but dimerization of this domain dramatically stimulated interaction. Likewise, the interaction between the dTRAF2 N-terminal region and Pelle could be regulated by TRAF domain dimerization.
We have shown that dTRAF2 is phosphorylated by activated Pelle in vitro. It is not clear how dTRAF2 functions in signaling to Dorsal, and whether phosphorylation is critical for its activity. As mentioned in the introduction, mammalian TRAF6 has been shown to interact with several downstream factors that appear to be required for NF-κB activation (18–20, 31, 32). Because it is unknown whether TRAF6 is phosphorylated by IRAK (or indeed phosphorylated at all), it is unclear how phosphorylation might affect these, or other, interactions. Likewise, additional studies are necessary to determine whether phosphorylated IRAK interacts with TRAF6, as Pelle is capable of binding dTRAF2. If so, TRAF6 could link active IRAK to other downstream targets. In any event, our study has provided evidence that a TRAF can be phosphorylated by an upstream kinase, which extends the possible mechanisms by which these molecules can modulate signaling pathways.
We have shown that dTRAF2 is covalently modified when overexpressed in SL2 cells, and that this modification requires the RING-finger domain. Such domains have been shown to be involved in protein ubiquitination, and indeed it may be that all RING-finger proteins may act as E3 ubiquitin ligases (27, 33). The ladder-like modification of dTRAF2 that we detected in transfected cells is similar to the pattern expected for ubiquitination, and may target dTRAF2 and associated proteins (e.g., Pelle) for degradation. IRAK has been shown to be degraded by the proteasome after phosphorylation (34). We have found that phosphorylated Pelle rapidly disappears from embryos in the 5th hour after egg laying, after completion of Toll signaling and Dorsal activation (B.S. and J.L.M., unpublished data). It is conceivable that activated, phosphorylated Pelle may also be degraded by the ubiquitin–proteasome pathway by binding to dTRAF2, which would thus desensitize Pelle signaling after activation of downstream components. However, it is notable that recent studies have shown that TRAF6 is required for synthesis of an unusual polyubiquitin chain required for IKK activation, by a mechanism not involving degradation (35). Whether this observation is related to the apparent dTRAF2 self-ubiquitination we detected here is not clear.
dTRAF1 has also been shown to interact with Pelle in vitro, and to enhance NF-κB-mediated expression in mammalian cells cotransfected with Pelle (36). Unlike dTRAF2, which interacts with the Pelle catalytic domain and can synergize strongly with Pelle to activate Dorsal, dTRAF1 interacts with Pelle in its N-terminal death domain and gives rise only to weak activation of NF-κB. These results suggest that the two dTRAFs play distinct roles. Supporting this possibility, dTRAF1, but not dTRAF2, was shown to activate the JNK pathway by binding to the Ste20 kinase, Msn (21). In addition, microinjection of mRNA encoding dTRAF1(Δ1–124), which encodes a dominant-negative protein in transfections assays in mammalian cultured cells, into Drosophila embryos failed to affect normal dorsal–ventral patterning (36). These results imply that the different downstream signaling events that activate Dorsal and JNK may bifurcate at the level of the Pelle-dTRAF interaction.
In summary, our data have shown that dTRAF2 functionally and physically interacts with Pelle in Dorsal activation, and is a target of Pelle kinase activity in vitro. Thus our findings suggest that dTRAF2 is an additional component of the Toll signaling pathway in Drosophila. Furthermore, they raise the possibility that TRAF activity can be regulated by phosphorylation. Additional studies using both genetic and biochemical methods are needed to address the developmental function of dTRAF2, to understand further its mechanism of action, and to identify other downstream targets of Pelle in Dorsal activation.
Acknowledgments
We thank Dr. X. Jacq and Z. Chen for helpful comments, and J. Norris for plasmids. This work was supported by National Institutes of Health Grant GM 37971 (to J.L.M.).
Abbreviations
- TNF
tumor necrosis factor
- dTRAF2
Drosophila TNF receptor-associated factor 2
- TRL
Toll-like receptor
- IL-1R
IL-1 receptor
- IRAK
IL-1R-associated kinase
- ECSIT
evolutionarily conserved signaling intermediate in Toll pathways
- HA
hemagglutinin
- CAT
chloramphenicol acetyltransferase
- GST
glutathione S-transferase
- NF-κB
nuclear factor κB
- I-κB
inhibitory protein κB
- IKK
I-κB kinase
- β-gal
β-galactosidase
References
- 1.Belvin M P, Anderson K V. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 2.Meister M, Lemaitre B, Hoffmann J A. BioEssays. 1997;19:1019–1026. doi: 10.1002/bies.950191112. [DOI] [PubMed] [Google Scholar]
- 3.Imler J L, Hoffmann J A. Curr Opin Microbiol. 2000;3:16–22. doi: 10.1016/s1369-5274(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 4.Aderem A, Ulevitch R J. Nature (London) 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 5.Anderson K V. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 6.Bowie A, O'Neill L A. J Leukoc Biol. 2000;67:508–514. doi: 10.1002/jlb.67.4.508. [DOI] [PubMed] [Google Scholar]
- 7.Grosshans J, Bergmann A, Haffter P, Nusslein-Volhard C. Nature (London) 1994;372:563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- 8.Galindo R L, Edwards D N, Gillespie S K, Wasserman S A. Development. 1995;121:2209–2218. doi: 10.1242/dev.121.7.2209. [DOI] [PubMed] [Google Scholar]
- 9.Grosshans J, Schnorrer F, Nusslein-Volhard C. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 10.Trofimova M, Sprenkle A B, Green M, Sturgill T W, Goebl M G, Harrington M A. J Biol Chem. 1996;271:17609–17612. doi: 10.1074/jbc.271.30.17609. [DOI] [PubMed] [Google Scholar]
- 11.Edwards D N, Towb P, Wasserman S A. Development (Cambridge, UK) 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 12.Karin M, Ben-Neriah Y. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 13.Cao Z, Henzel W J, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 14.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 15.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 16.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 17.Ling L, Cao Z, Goeddel D V. Proc Natl Acad Sci USA. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano H, Shindo M, Sakon S, Nishinaka S, Mihara M, Yagita H, Okumura K. Proc Natl Acad Sci USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. Nature (London) 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 20.Kopp E, Medzhitov R, Carothers J, Xiao C, Douglas I, Janeway C A, Ghosh S. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Su Y C, Becker E, Treisman J, Skolnik E Y. Curr Biol. 1999;9:101–104. doi: 10.1016/s0960-9822(99)80023-2. [DOI] [PubMed] [Google Scholar]
- 22.Norris J L, Manley J L. Genes Dev. 1995;9:358–369. doi: 10.1101/gad.9.3.358. [DOI] [PubMed] [Google Scholar]
- 23.Shen B, Manley J L. Development (Cambridge, UK) 1998;125:4719–4728. doi: 10.1242/dev.125.23.4719. [DOI] [PubMed] [Google Scholar]
- 24.Norris J, Manley J L. Genes Dev. 1996;10:862–872. doi: 10.1101/gad.10.7.862. [DOI] [PubMed] [Google Scholar]
- 25.Han K, Levine M S, Manley J L. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 26.Norris J, Manley J L. Genes Dev. 1992;6:1654–1667. doi: 10.1101/gad.6.9.1654. [DOI] [PubMed] [Google Scholar]
- 27.Freemont P S. Curr Biol. 2000;10:R84–87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 28.Baker S J, Reddy E P. Oncogene. 1998;17:3261–3270. doi: 10.1038/sj.onc.1202568. [DOI] [PubMed] [Google Scholar]
- 29.Arch R H, Gedrich R W, Thompson C B. Genes Dev. 1998;12:2821–2830. doi: 10.1101/gad.12.18.2821. [DOI] [PubMed] [Google Scholar]
- 30.Sanz L, Diaz-Meco M T, Nakano H, Moscat J. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song H Y, Regnier C H, Kirschning C J, Goeddel D V, Rothe M. Proc Natl Acad Sci USA. 1997;94:9792–9796. doi: 10.1073/pnas.94.18.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya-Tsuji J, Matsumoto K. Mol Cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 33.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamin T T, Miller D K. J Biol Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- 35.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen Z J. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 36.Zapata J M, Matsuzawa S, Godzik A, Leo E, Wasserman S A, Reed J C. J Biol Chem. 2000;275:12102–12107. doi: 10.1074/jbc.275.16.12102. [DOI] [PubMed] [Google Scholar]