Abstract
Autophagy is finely regulated at multiple levels and plays crucial roles in development and disease. In the fat body of the silkworm, Bombyx mori, autophagy occurs and Atg gene expression peaks during the nonfeeding molting and pupation stages when the steroid hormone (20-hydroxyecdysone; 20E) is high. Injection of 20E into the feeding larvae upregulated Atg genes and reduced TORC1 activity resulting in autophagy induction in the fat body. Conversely, RNAi knockdown of the 20E receptor partner (USP) or targeted overexpression of a dominant negative mutant of the 20E receptor (EcRDN) in the larval fat body reduced autophagy and downregulated the Atg genes, confirming the importance of 20E-induction of Atg gene expression during pupation. Moreover, in vitro treatments of the larval fat body with 20E upregulated the Atg genes. Five Atg genes were potentially 20E primary-responsive, and a 20E response element was identified in the Atg1 (ortholog of human ULK1) promoter region. Furthermore, RNAi knockdown of 4 key genes (namely Br-C, E74, HR3 and βftz-F1) in the 20E-triggered transcriptional cascade reduced autophagy and downregulated Atg genes to different levels. Taken together, we conclude that in addition to blocking TORC1 activity for autophagosome initiation, 20E upregulates Atg genes to induce autophagy in the Bombyx fat body.
Keywords: 20-hydroxyecdysone, fat body, autophagy, Atg genes, Atg1, ATG8, transcriptional regulation, TORC1, Bombyx mori
Introduction
Macroautophagy (hereafter referred to as autophagy) is the prevalent form of autophagy, which mediates a highly regulated self-degradation process that is initiated as an adaptive response in unfavorable conditions, such as nutrient deprivation. During autophagy, unnecessary organelles and dysfunctional proteins are sequestered into autophagosomes and finally delivered to lysosomes for degradation.1 Autophagy is involved in many physiological and developmental processes, such as cell survival, cell death, metabolism and innate immunity.2-4 Defects in autophagy are often associated with diseases such as neurodegradation, cardiomyopathy and tumor progression.5,6
The autophagy process has several distinct steps, including induction, cargo recognition and packaging, vesicle formation and breakdown. The maturation of the autophagosome is governed by a series of Atg genes and at least four important ATG protein complexes. Autophagosome initiation requires the ULK1/ATG1-ATG13 protein kinase complex; autophagosome nucleation is mediated by the BECN1/ATG6-PIK3C3/Vps34 (catalytic subunit of class III PtdIns3K) complex; autophagosome expansion and completion is regulated by two ubiquitin-like conjugation systems: ATG12–ATG5-ATG16L1 and ATG8–II (the cleaved and lipidated form of ATG8) conjugates.1 The regulation of autophagosome initiation by upstream signaling events is conserved in yeast, arthropods and vertebrates.7 In nutrient-rich conditions, a central inhibitor of autophagy is TORC1, which integrates information from multiple upstream signals and inhibits autophagosome initiation by phosphorylating ULK1.8 In nutrient-poor conditions, the energy sensor AMPK not only inhibits TORC1 activity but also associates with and directly phosphorylates ULK1 to induce autophagosome initiation.9,10 Meanwhile, in nutrient-poor conditions, the transcription factor FOXO is able to induce autophagosome formation by upregulating the Atg genes.11
Most of the original discoveries about autophagy were made in yeast. Although the Atg genes are quite well conserved from yeast to insects to humans,7,12 the molecular mechanism by which ATG proteins collaborate to form autophagosomes is largely unknown in higher eukaryotes. For example, autophagy in higher eukaryotes involves much more complex membrane dynamics than in yeast.13 The fruit fly, Drosophila melanogaster, is an excellent insect model for autophagy research, owing in part to the powerful genetic tools that have been developed in this model animal. A very important autophagy-related discovery in Drosophila is Dmel/drpr (Draper), which is an ortholog of C. elegans ced-1, and has been the first identified component that distinguishes autophagy-associated cell death from autophagy-associated cell survival.14 A number of studies have revealed that autophagy plays a predominant role in controlling cell death in the salivary gland15,16 and the midgut17 as well as cell survival in the fat body12 during Drosophila metamorphosis.
By binding its nuclear receptor EcR-USP, the steroid hormone 20-hydroxyecdysone (20E; the molting hormone) plays an important role in inducing autophagy in insects.7,18,19 The ligand-receptor complex, 20E-EcR-USP, triggers a transcriptional cascade, including transcription of the 20E primary-response genes (i.e., transcription factor genes Br-C, E74, E75 and E93) and, subsequently, the 20E secondary-response genes.18,20 The 20E-EcR-USP complex can block TORC1 activity and thereby induce autophagosome initiation in the fat body;21-23 the 20E primary-response gene E93 mediates 20E-induced autophagic cell death in the salivary gland15 and the midgut.24,25 Some Atg genes are upregulated in the salivary gland with a 20E pulse that induces autophagic cell death during pupation,25,26 and mutation of E93 downregulates the Atg genes.25 Therefore, it appears that E93 predominantly mediates 20E signaling to induce autophagy at the transcriptional level in Drosophila.
Some of the earliest work in the autophagy field was performed during Lepidoptera metamorphosis, when the bulk of the larval tissues are removed by autophagy.27-29 The silkworm, Bombyx mori, is now used as a representative model among Lepidoptera for studying physiology, biochemistry, developmental biology and genetics. Bombyx is certainly an excellent insect model for studying autophagy and programmed cell death. For example, in the Bombyx midgut, autophagy and apoptosis gradually occur with autophagy preceding apoptosis,30 and the expression of Atg1 steadily increases during metamorphosis.31
The insect fat body is an organ analogous to vertebrate adipose tissue and liver and functions as a major organ for nutrient storage and energy metabolism. It is considered to be the central organ that integrates and coordinates different hormonal and nutritional signals for regulating insect development and metamorphosis.20,32,33 In the fat body of two lepidopteran species and Drosophila, 20E-induced developmental autophagy progressively occurs during the larval-prepupal transition,21,27,28,34-37 and in the Drosophila fat body, EcR is required for 20E to induce autophagy.21 We have previously determined that in the Bombyx fat body, 20E upregulates apoptosis genes to induce some apoptotic events.38 We here demonstrated that in addition to blocking TORC1 activity to induce autophagosome initiation, 20E upregulates Atg genes involved in all steps of autophagosome formation in the Bombyx fat body during molting and pupation.
Results
Developmental profiles of autophagy
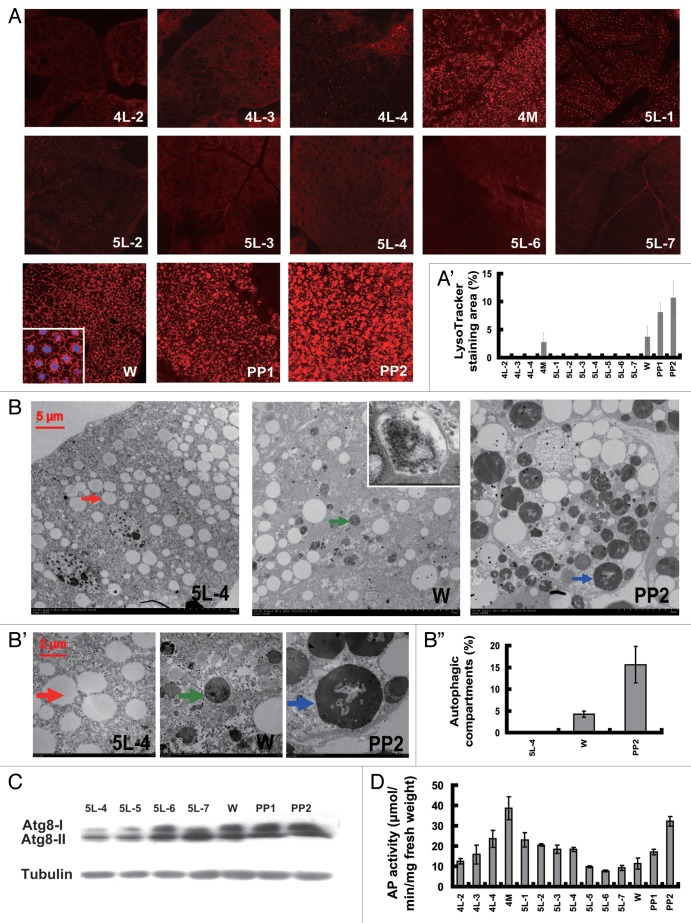
LysoTracker Red has proven to be an effective indicator of autophagy in the Drosophila fat body.21,37 Using LysoTracker Red staining, we first estimated the developmental profile of autophagy in the Bombyx fat body from day 2 of the 4th instar (4L-2) to day 2 of the prepupal stage (PP2). LysoTracker Red staining was undetectable during the feeding stage of the 4th instar (4F) but was detected during the final larval molt (4M). Staining dropped to undetectable levels during the feeding stage of the 5th instar (5F) and was observed again during the wandering stage (W) and steadily increased during pupation, composed of PP1 and PP2 (Fig. 1A and A’).
Figure 1. Developmental profiles of autophagy. (A and A’) LysoTracker Red staining (red, 40×) of the fat body from 4L-2 to PP2. The inset in (W) shows that LysoTracker (red) and DAPI (blue) stain the cytoplasm and the nuclei, respectively. The chart (A’) shows the quantification of LysoTracker Red staining in (A). (B–B”) TEM analysis of the fat body from 5L-4, W and PP2 animals. The arrows indicate a lipid droplet (red, 5L-4) or an autolysosome (green, W; blue, PP2). The inset in (W) shows a double membrane-bound vesicle autophagosome. (B’) (30,000×) shows magnifications of portions of the image in (B) (7,500×). Note the remnants of organelles and the undigested materials in the autolysosomes. The chart (B”) shows the quantification of autophagosomes and autolysosomes in (B). (C) ATG8 protein level in the fat body from 4L-2 to PP2. ATG8-I and ATG8-II are the nonlipidated and lipidated forms of ATG8, respectively. Tubulin was used as a loading control for the western blotting analysis. (D) Measurement of acid phosphatase (AP) activity in the fat body from 4L-2 to PP2. 4L-2, day 2 of the 4th instar, and so on; 4M, the 4th molting; W, the wandering stage; PP1, day 1 of the prepupal stage, and so on.
The Bombyx fat body was further monitored using transmission electron microscopy (TEM), the classic method for detecting autophagy in most organisms including lepidopteran insects.21,27-30,34-37,39 Neither autophagosomes nor autolysosomes appeared during the feeding larval stages, i.e., 5L-4. Small autolysosomes occurred at W, and double-membrane bound autophagosomes were occasionally observed. Abundant and large autolysosomes formed at PP2, with visible remnants of organelles and undigested materials often detected inside the vesicles (Fig. 1B and B”). The TEM analysis is consistent with the LysoTracker Red results, indicating that autophagy is absent during the feeding larval stages but weakly occurs during larval molts and emerges during pupation.
ATG8 is the most widely monitored autophagy-related protein. The unprocessed form of ATG8, proATG8, is converted into a nonlipidated form, ATG8-I, and finally modified into a cleaved and lipidated form, ATG8-II. ATG8-II is associated with autophagosome expansion and completion.39 An antibody against Bombyx ATG8 was previously shown to detect both ATG8-I and ATG8-II and had a reflection of autophagy in the midgut.30 As detected by western blotting with the Bombyx ATG8 antibody, the ATG8 protein level (including both ATG8-I and ATG8-II) in the Bombyx fat body gradually increased from 5L-4 to PP2 (Fig. 1C). It is necessary to note that a relatively low level of ATG8-II was present at 5L-4, when autophagy was not detected by both LysoTracker Red staining and TEM analysis (Fig. 1A and B’). We assume that the presence of ATG8-II at 5L-4 could be associated with autophagosome membrane dynamics rather than completed autophagosomes.
At the final step of the autophagy process, autophagosomes fuse with lysosomes to form autolysosomes.1 Acid phosphatase (AP) activity is a marker of lysosome activity, and lysosome numbers generally increase when autophagy is induced.39,40 It has been reported that AP activity increases when autophagy occurs in the Bombyx midgut.30 We then measured the developmental profile of AP activity in the Bombyx fat body from 4L-2 to PP2. In general, AP activity was higher during the nonfeeding molting and pupation stages than during the feeding larval stages (Fig. 1D). We suppose that ATG8 protein level and AP activity could reflect autophagy in the Bombyx fat body at certain degrees, but are less evident than TEM analysis. Notably, the dimension of LysoTracker Red spots gradually increases from W to PP1 to PP2, reflecting the gradual increase of autolysosome sizes observed by TEM. Therefore, similar to TEM analysis, LysoTracker Red is also an effective marker for autophagy in the Bombyx fat body.
Considering all the developmental profiles of LysoTracker Red staining, TEM analysis, ATG8 protein level and AP activity (Fig. 1), we conclude that autophagy does not appear during the feeding larval stages but weakly occurs during molting and emerges during pupation, corresponding to the nonfeeding stages when the 20E titer peaks.41 In general, our finding in the Bombyx fat body is consistent with the previous observations that autophagy emerges during insect metamorphosis.14-17,21,27,28,34-37
Atg gene expression profiles
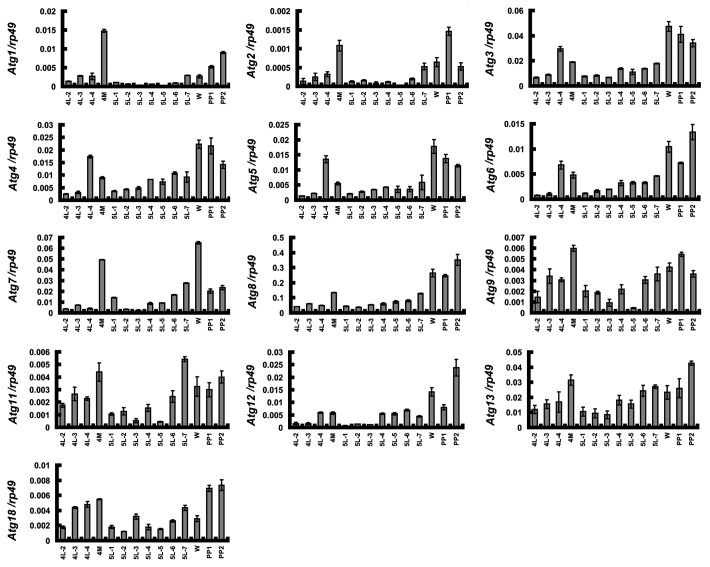
Eleven putative Atg genes, including Atg1, Atg3, Atg4, Atg5, Atg6 (ortholog of yeast VPS30 and human BECN1), Atg7, Atg8, Atg9, Atg12, Atg16 and Atg18, have been identified in the Bombyx genome.30,42 Using the same strategies used in the published reports, we found three additional Atg genes, namely Atg2, Atg11 and Atg13. All three newly predicted Bombyx ATG proteins contain classic ATG domains (Fig. S1). In particular, ATG13 is a component of the ULK1-ATG13 protein kinase complex that plays a key role in regulating autophagosome initiation.1,43
In a previous microarray assay for isolating genes regulated by 20E and juvenile hormone,44,45 we noticed that Atg8 was upregulated in the Bombyx fat body at both 4M and PP in comparison with 5F, suggesting that Atg8 expression might share a similar developmental profile to autophagy and that Atg8 might be upregulated by 20E during molting and pupation. To test this idea, we used quantitative real-time PCR (qPCR) to measure the expression profiles of all 14 Atg genes in the fat body from 4L-2 to PP2. Because the Atg16 mRNA level is beyond the scope of our qPCR detection system, we only measured the remaining 13 Atg genes in the following studies. Surprisingly, the mRNA levels of all 13 Atg genes at 4M and PP are generally higher than at 4F and 5F (Fig. 2) and exhibit similar developmental profiles to 20E titer and autophagy. It is necessary to note that unlike the Bombyx fat body, some, but not all Atg genes are upregulated in the salivary gland with a 20E pulse during pupation in Drosophila.24,46 In addition, the developmental profile of Atg8 mRNA is similar to that of ATG8 protein (Fig. 1C), indicating that ATG8 is regulated at the transcriptional level.
Figure 2. Developmental profiles of Atg gene expression revealed by qPCR. Expression of the 13 Atg genes in the fat body from 4L-2 to PP2. 4L-2, day 2 of the 4th instar, and so on; 4M, the 4th molting; W, the wandering stage; PP1, day 1 of the prepupal stage, and so on.
The similar developmental profiles of 20E titer, autophagy and Atg gene expression in the Bombyx fat body imply that 20E might upregulate the Atg genes during molting and pupation.
Induction of autophagy by injection of 20E
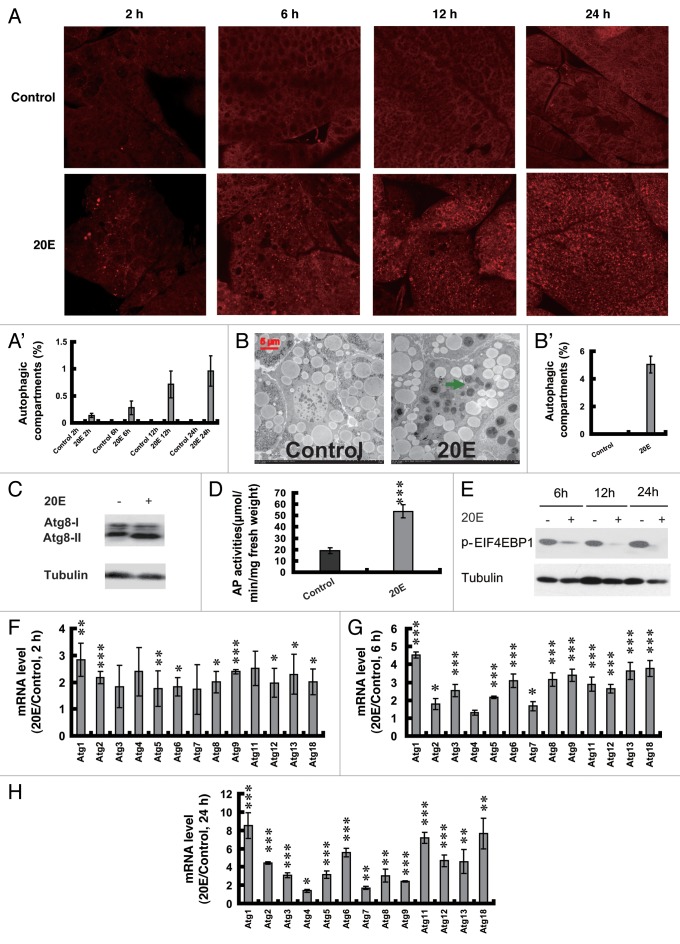
To verify the above hypothesis, we first performed experiments by injecting 20E into 5L-2 larvae, in which the 20E level is low41 and the fat body is sensitive to 20E injection.38,44,45 No LysoTracker Red staining was detected in the control fat body, while slight staining occurred 2 h after 20E injection, and staining progressively became more significant at the later hours (Fig. 3A and A’). TEM analyses revealed that no autophagosomes were detected in the control fat body, but a large number of small autolysosomes were observed 24 h after 20E injection (Fig. 3B and B’). Meanwhile, ATG8 protein level (Fig. 3C) and AP activity (Fig. 3D) in the fat body were increased 24 h after 20E injection.
Figure 3. Induction of autophagy by injection of 20E into larvae on day 2 of the 5th instar. (A and A’) LysoTracker Red staining (red, 40×) in the fat body 2, 6, 12 and 24 h after 20E injection. The chart (A’) shows the quantification of LysoTracker Red staining in (A). (B and B’) TEM analysis (7,500×) 24 h after 20E injections. The green arrow denotes an autolysosome. The chart (B’) shows the quantification of autophagosomes and autolysosomes in (B). (C) Increase of ATG8 protein level 24 h after 20E injection. (D) Increase of AP activity 24 h after injections. (E) EIF4EBP1 phosphorylation (p-EIF4EBP1; a representation of TORC1 activity) was progressively decreased 6, 12 and 24 h after 20E injection. Tubulin was used as a loading control. (F–H) Upregulation of Atg genes 2 (F), 6 (G) and 24 (H) h after 20E injection. 20E/Control refers to as fold change after 20E injection compared with the controls.
RPS6KB/S6K phosphorylation and EIF4EBP1/4EBP phosphorylation could represent TORC1 activity.8 In our preliminary western blotting experiments, EIF4EBP1 phosphorylation was detectable but RPS6KB phosphorylation was undetectable in the Bombyx larval fat body. EIF4EBP1 phosphorylation was significantly reduced 6 h after 20E injection into 5L-2 larvae, and the reduction became stronger with increasing time (Fig. 3E), indicating that 20E injection reduces TORC1 activity. Moreover, as determined by qPCR, 9 Atg genes were upregulated less than 3 fold 2 h after 20E injection (Fig. 3F), 12 Atg genes (all except Atg4) were upregulated 1.8 to 4.5 fold 6 h after 20E injection (Fig. 3G), while all of the 13 Atg genes were upregulated 1.5 to 8.5 fold 24 h after 20E injection (Fig. 3H). In general, Atg gene expression was gradually increased from 2 to 24 h after 20E injection. Among the 13 Atg genes, the expression of Atg1 is most sensitive to 20E injection, and Atg4 is the least sensitive genes to 20E. The composite data suggest that injection of 20E into the feeding larvae reduces TORC1 activity and upregulates Atg genes resulting in autophagy induction in the fat body.
Reduction of autophagy by RNAi knockdown of Atg1
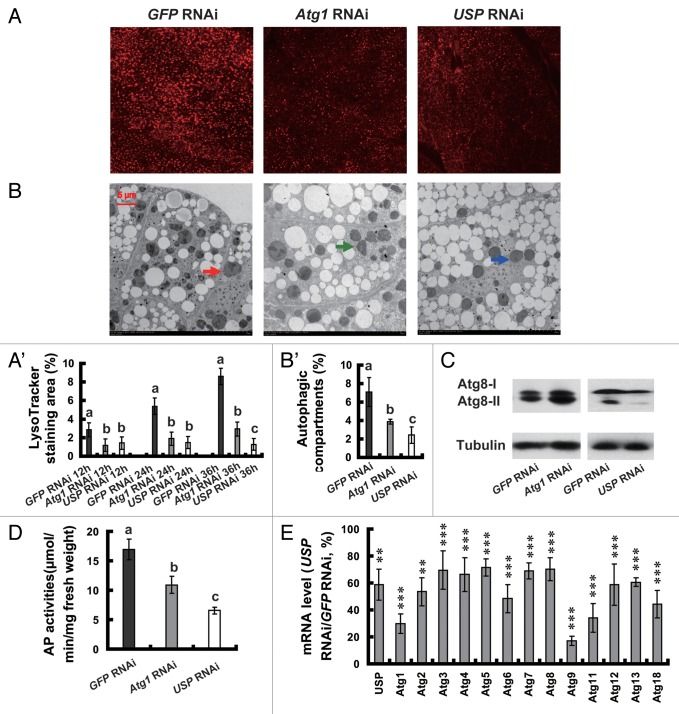
In Drosophila, autophagy is absent in an Atg1 null mutant, Atg1Δ3D,37 and reduced in an Atg8a mutant, Atg8aKG07569,16 and by RNAi knockdown of several key Atg genes.37 We thus investigated whether upregulation of Atg1 during pupation in Bombyx is important for autophagosome formation by using RNAi to knock down Atg1 at the initiation of the wandering stage (IW). The Atg1 mRNA level in the fat body was decreased to approximately 40% of the control level (GFP RNAi) 24 h after Atg1 RNAi. As shown above, LysoTracker Red staining gradually increased in the control fat body during wandering and pupation, 12, 24 and 36 h after GFP RNAi. There was an apparent reduction of LysoTracker Red staining 12 h after Atg1 RNAi, and the reduction was even stronger 24 and 36 h after Atg1 RNAi (Fig. 4A and A’). TEM analyses revealed that a large number of autophagosomes were present in the control fat body 24 h after GFP RNAi, but the number of autophagosomes decreased dramatically by Atg1 RNAi (Fig. 4B and B’). Similar to the previous reports,39 Atg1 RNAi resulted in Atg8 protein accumulation with a higher increase for Atg8-II than for Atg8-I (Fig. 4C). In addition, Atg1 RNAi decreased about 30% of AP activity (Fig. 4D). Similar results were also obtained when several other Atg genes were knocked down by RNAi (data not shown). Conclusively, RNAi knockdown of Atg genes reduced autophagy, demonstrating that the upregulation of Atg genes by 20E is functionally important to induce autophagy in the fat body during pupation in Bombyx.
Figure 4. Reduction of autophagy by RNAi knockdown of Atg1 and USP and downregulation of Atg genes by USP RNAi at the initiation of the wandering stage. (A and A’) LysoTracker Red staining (red, 40×) 12, 24 and 36 h after RNAi treatment. The chart (A’) shows the quantification of LysoTracker Red staining in (A). (B and B’) TEM analysis (7,500×) 24 h after RNAi treatment. The arrows denote autolysosomes. The chart (B’) shows the quantification of autophagosomes and autolysosomes in (B). (C) Increase and decrease of ATG8 protein level 24 h after RNAi knockdown of Atg1 and USP, respectively. (D) Decrease of AP activity 24 h after RNAi treatment. (E) Downregulation of Atg genes 24 h after USP RNAi. Percentage of control is presented.
Reduction of autophagy and downregulation of Atg genes by RNAi knockdown of USP
In a previous study, we documented that RNAi knockdown of EcR-USP at IW resulted in significant prepupal or pupal lethality and a decrease of 20E signaling in the Bombyx fat body, with USP RNAi exhibiting stronger inhibitory effects than EcR RNAi.44,45
Because exogenous 20E was able to induce autophagy and upregulate Atg genes in the 5L-2 larvae, we further investigated whether USP RNAi at IW was able to reduce autophagy and to downregulate Atg genes in the fat body. As determined by LysoTracker Red staining, TEM analysis, ATG8 protein level and AP activity (Fig. 4A–D), autophagy was significantly decreased by USP RNAi. In general, the inhibitory effect on autophagy by USP RNAi was similar to or slightly stronger than that by Atg1 RNAi. Moreover, all 13 Atg genes in the fat body were downregulated more than 30% by USP RNAi 24 h after treatment. Among the 13 Atg genes, the mRNA levels of Atg9 and Atg1 decreased the most, showing reductions of 80% and 70%, respectively (Fig. 4E). In addition, USP RNAi slightly increased EIF4EBP1 phosphorylation 12 h after treatment, but no significant increases of EIF4EBP1 phosphorylation were observed 24 and 36 h after treatment (data not shown).
The systemic USP RNAi experiment strengthened the hypothesis that by binding EcR-USP, 20E upregulates Atg genes to induce autophagy in the Bombyx fat body during pupation.
Reduction of autophagy and downregulation of Atg genes by targeted overexpression of EcRDN in the larval fat body
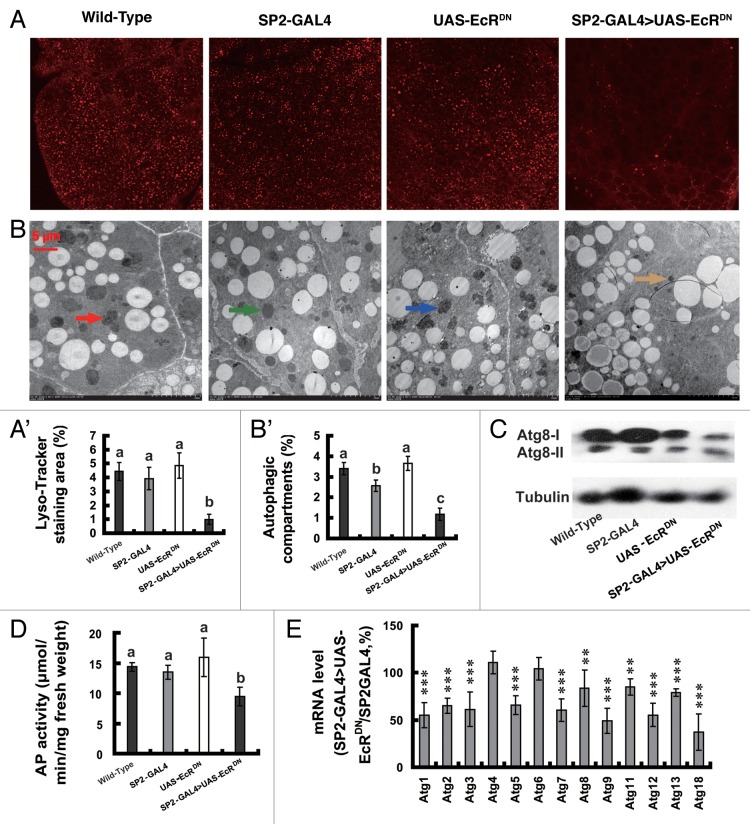
With the binary GAL4/UAS transgenic system established in Drosophila, targeted overexpression of dominant-negative mutants of EcR (EcRDN) is an efficient approach for inhibiting 20E signaling in a tissue-specific manner.47 To avoid the non-tissue, autonomous effects caused by 20E injection and systemic USP RNAi, we intended to inhibit 20E signaling by overexpressing EcRDN specifically in the larval fat body using the Bombyx GAL4/UAS system.48,49 The Bombyx storage protein 2 (SP2) is synthesized and secreted by the larval fat body during the last two feeding larval stages, 4F and 5F.50 Thus, a 2-kb region of the SP2 promoter was isolated to construct a fat body-specific GAL4 line, SP2-GAL4. Meanwhile, in accord with the approach to generate Drosophila UAS-EcR-F645A,47 we mutated the Bombyx EcR-B1 isoform to make a UAS-EcRDN line. The transgenic silkworm SP2-GAL4 > UAS-EcRDN was obtained by crossing SP2-GAL4 with UAS-EcRDN.49
As determined by reverse transcription PCR (RT-PCR), GAL4 was expressed specifically in 5L-4 larval fat body tissues isolated from SP2-GAL4 and SP2-GAL4 > UAS-EcRDN but not in their wild-type and UAS-EcRDN counterparts (Fig. S2A). As determined by qPCR, GAL4 expression in the fat body from the 5L-4 SP2-GAL4 larvae was much more abundant than in other tissues (Fig. S2B), demonstrating the fat body specificity of SP2-GAL4. The EcR/EcRDN transcript in the fat body of SP2-GAL4 > UAS-EcRDN was about 5-fold more abundant than in the other three lines (Fig. S2C), showing the targeted overexpression of EcRDN in the fat body. Moreover, the 20E response genes E75, Br-C, E74, HR3 and βftz-F1, were downregulated by 30 to 70% in SP2-GAL4 > UAS-EcRDN 6 h after IW in comparison with the control SP2-GAL4 (Fig. S2D). These experimental data indicate that targeted overexpression of EcRDN in the larval fat body reduces 20E signaling during pupation.
Importantly, as determined by LysoTracker Red staining, TEM analysis, ATG8 protein level and AP activity (Fig. 5A–D), autophagy was significantly reduced in the fat body 6 h after IW in SP2-GAL4 > UAS-EcRDN, compared with its wild-type, SP2-GAL4 and UAS-EcRDN counterparts. Moreover, except for Atg4 and Atg6, the other 11 Atg genes in the fat body of SP2-GAL4 > UAS-EcRDN were downregulated by 30 to 70% of the control level (Fig. 5E). The transgenic silkworm experiment confirmed the above hypothesis that by binding EcR-USP, 20E upregulates Atg genes to induce autophagy in the Bombyx fat body during pupation.
Figure 5. Reduction of autophagy and downregulation of Atg genes by targeted overexpression of EcRDN in the larval fat body 6 h after initiation of the wandering stage. (A and A’) LysoTracker Red staining (red, 40×). The chart (A’) shows the quantification of LysoTracker Red staining in (A). (B and B’) TEM analysis (7,500×). The arrows denote autolysosomes. The chart (B’) shows the quantification of autophagosomes and autolysosomes in (B). (C) Decrease of ATG8 protein level 24 h. (D) Decrease of AP activity. (E) Downregulation of Atg genes in SP2-GAL4 > UAS-EcRDN in comparison with SP2-GAL4.
Five Atg genes are potentially 20E primary-responsive
To confirm the in vivo experiments, we investigated whether 20E was able to induce Atg gene expression in vitro. Fat body tissues isolated from 5L-2 larvae were cultured in vitro with or without 20E addition. Two hours after 20E addition, 10 Atg genes were upregulated more than 150%, with the highest upregulation for Atg18 (approximately 3-fold) and Atg1 (more than 2- fold) (Fig. 6A). Six hours after 20E addition, all 13 Atg genes were upregulated more than 2-fold, with the highest upregulation for Atg1 (approximately 18-fold) and Atg18 (approximately 11-fold) (Fig. 6B). The addition of 20E also upregulated Atg genes in a Bombyx cell line, DZNU-Bm-12,51 but the stimulating effect on Atg gene expression in the cell line was relatively weaker than that in the fat body (Fig. S3A and S3B).
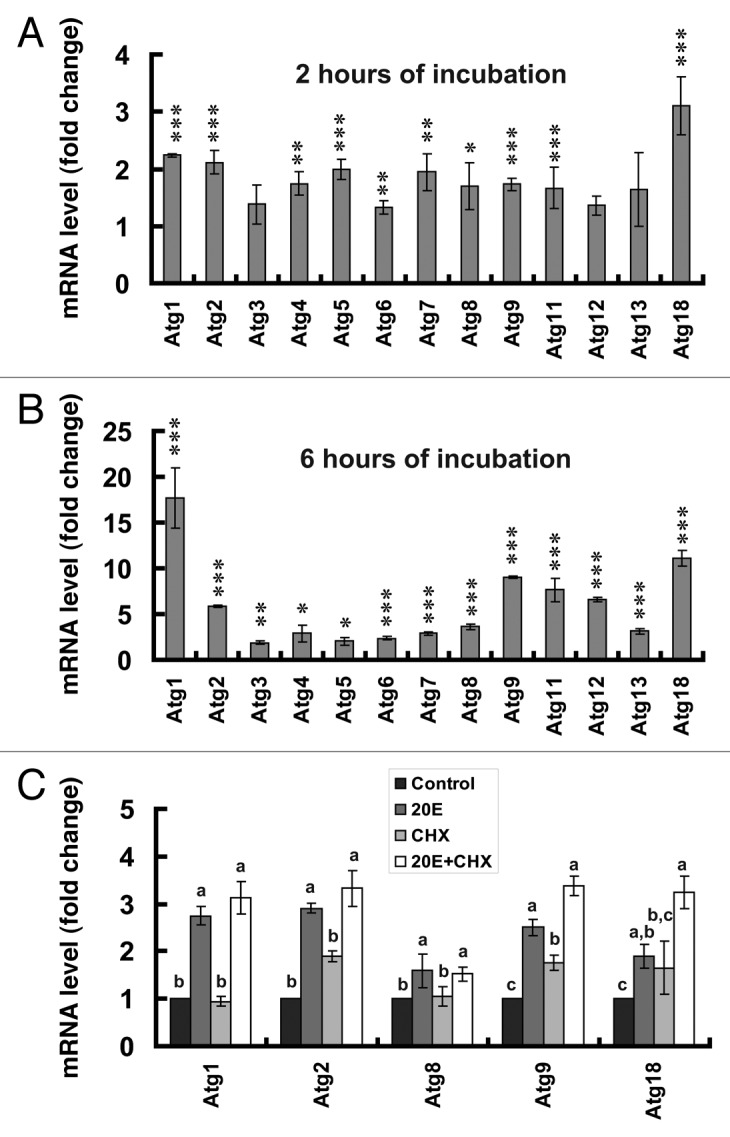
Figure 6. Five Atg genes are potentially 20E primary-responsive. Fat body tissues isolated from larvae on day 2 of the 5th instar (5L-2) were cultured in vitro with or without 20E addition at a final concentration of 5 μM. (A) Upregulation of Atg genes 2 h after incubation with 20E. (B) Upregulation of Atg genes in the fat body 6 h after incubation with 20E. (C) Atg1, Atg2, Atg8, Atg9 and Atg18 are potential 20E primary-response genes. The control fat body tissues were cultured in Grace’s medium (Control), while the experimental fat body tissues were cultured in Grace’s medium containing 5 μM 20E (20E), 10 μg/ml cycloheximide (CHX), or 5 μM 20E and 10 μg/ml CHX (20E + CHX).
We further investigated whether some Atg genes were potentially 20E primary-responsive in the presence and absence of the protein synthesis inhibitor cycloheximide (CHX). Two h after addition of 20E to the cultured fat body tissues isolated from 5L-2 larvae, Atg1 was upregulated more than 2-fold. CHX had no effect on Atg1 expression, while addition of 20E and CHX together resulted in similar accumulation of the Atg1 transcript to addition of 20E alone (Fig. 6C), indicating that Atg1 is a potential 20E primary-response gene. In addition, the 4 other Atg genes (Atg2, Atg8, Atg9 and Atg18) proved to be potentially 20E primary-responsive (Fig. 6C). Similar but less significant results were obtained with cultured DZNU-Bm-12 cells (Fig. S3C).
These in vitro culture experiments suggest that Atg1 and the 4 other Atg genes are potentially 20E primary-response genes.
Identification of an EcRE in the Atg1 promoter region
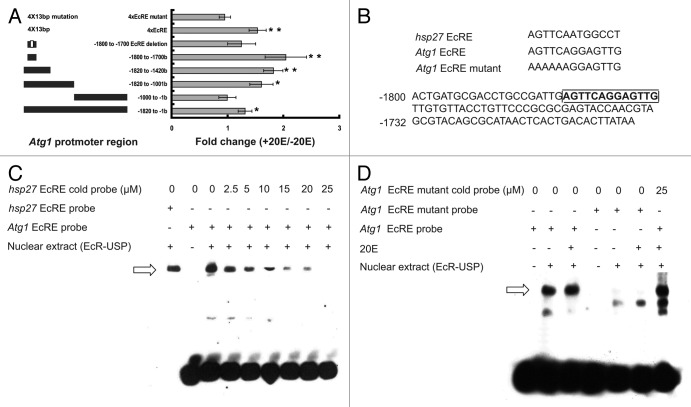
Among the 13 Atg genes, Atg1 expression was most sensitive to 20E treatment in vivo and in vitro, to systemic USP RNAi, and to EcRDN overexpression (Fig. 3–6; Fig. S3). Moreover, Atg1 proved to be a potential 20E primary-response gene (Fig. 6C; Fig. S3C). As the protein kinase ATG1 plays a crucial role in autophagosome initiation, we attempted to identify regions of the Atg1 promoter that are essential for 20E-mediated transcription by using a combined method of dual luciferase assay and electrophoretic mobility shift assay (EMSA).
First, the 1820-bp Atg1 promoter region or its fragments was cloned into the pGL3 luciferase vector, which was cotransfected with the Bombyx HA-EcR-B1 and FLAG-USP expression constructs into human HEK 293 cells. Addition of 20E increased the luciferase activity by about 130%. The 1820-bp Atg1 promoter region was separated into two portions (the 5′-end portion: −1820 to −1001 bp; the 3′-end portion: −1000 to −1 bp). Addition of 20E increased the luciferase activity by approximately 160% when the 5′-end portion was transfected, but it had no stimulating effects on the 3′-end portion. The 5′-end portion was further separated into two parts: the 5′-end part and the 3′-end part. With the 5′-end part (−1820 to −1421 bp), 20E addition increased the luciferase activity by approximately 180%. Furthermore, 20E addition increased the luciferase activity by about 2.1 fold when a 100-bp fragment (−1800 to −1701 bp) isolated from the 5′-end part was transfected. The 100-bp fragment contains a putative EcRE (−1779 to −1766; AGTTCAGGAGTTG), and little increase of luciferase activity was observed for the mutated 100-bp fragment with the putative EcRE deleted. Intriguingly, when 4 copies of the putative 13-bp Atg1 EcRE were transfected, 20E addition increased the luciferase activity by approximately 150%. In contrast, 20E addition had no effect on 4 copies of the mutated Atg1 EcRE (AAAAAAGGAGTTG) (Fig. 7A and B). When the hsp27 EcRE (AGTTCAATGGCCT) was used as a positive control for the dual luciferase experiments, its luciferase activity was increased approximately 5-fold by 20E addition. The same experiments were performed in the DZNU-Bm-12 cells; 20E addition had similar but stronger stimulatory effects on luciferase activity. For example, when 4 copies of the putative 13-bp Atg1 EcRE was transfected, 20E addition increased the luciferase activity by approximately 4.5-fold (Fig. S4).
Figure 7. Identification of an EcRE in the Atg1 promoter region using human HEK 293 cells. (A) Dual luciferase assay reveals an EcRE in the Atg1 promoter region. The 1820-bp Atg1 promoter region was divided into fragments, cloned into the pGL3 luciferase vector, and cotransfected with the reference pRL vector and the Bombyx HA-EcR-B1 and FLAG-USP expression constructs into human HEK 293 cells. After 36 h of transfection, the HEK 293 cells were treated with 1 μM 20E for 12 h. The relative luciferase activity is defined as the reporter firefly luciferase level/the reference Renilla luciferase level. Luciferase activity fold change is defined as the relative luciferase activity induced by 20E/control. (B) Top: alignment of the hsp27 EcRE, the Atg1 EcRE and the Atg1 EcRE mutant in which AGTTCA was mutated to AAAAAA. Bottom: the nucleotide sequence of the 100-bp fragment (−1800 to −1701 bp in the Atg1 promoter region) containing the Atg1 EcRE (AGTTCAGGAGTTG, boxed). (C and D) The Bombyx HA-EcR-B1 and FLAG-USP expression constructs were transfected into HEK 293 cells, and nuclear extracts were prepared for EMSA. The hsp27 EcRE probe sequence: AGTTCAATGGCCT; the Atg1 EcRE probe sequence: CCTGCCGATTGAGTTCAGGAGTTGTTGTGTTACCTGTTCC; the Atg1 EcRE mutant probe sequence: CCTGCCGATTGAAAAAAGGAGTTGTTGTGTTACCTGTTCC.
To confirm the putative 13-bp Atg1 EcRE, we performed EMSA and competition experiments.52 The GFP or Bombyx HA-EcR-B1 and FLAG-USP expression constructs were transfected into HEK 293 cells, and nuclear extracts were prepared. As expected, the GFP control nuclear extract bound neither the hsp27 EcRE probe (AGTTCAATGGCCT) nor the Atg1 EcRE probe (CCTGCCGATTGAGTTCAGGAGTTGTTGTGTTACCTGTTCC) (data not shown). In contrast, the nuclear extract containing HA-EcR-B1 and FLAG-USP was able to bind the two EcRE probes, and binding of EcR-USP by the Atg1 EcRE probe was gradually abolished in the presence of an increasing amount of the hsp27 EcRE cold probe (Fig. 7C), indicating specific binding of the Atg1 EcRE probe to EcR-USP. Interestingly, 20E had only a weak stimulating effect on the binding. When the first 6 nucleotides of the Atg1 EcRE in the Atg1 EcRE probe were mutated (CCTGCCGATTGAAAAAAGGAGTTGTTGTGTTACCTGTTCC), binding was eliminated. Moreover, binding of the Atg1 EcRE probe to EcR-USP was not attenuated even in the presence of a high amount of Atg1 EcRE mutant probe (Fig. 7D), confirming the specificity of the Atg1 EcRE and of its binding to EcR-USP.
In conclusion, Atg1 is a 20E primary-response gene harboring an EcRE in its promoter region. We then investigated whether the Atg genes are also 20E secondary-response genes.
Reduction of autophagy and downregulation of Atg genes by RNAi knockdown of four key genes in the 20E-triggered transcriptional cascade
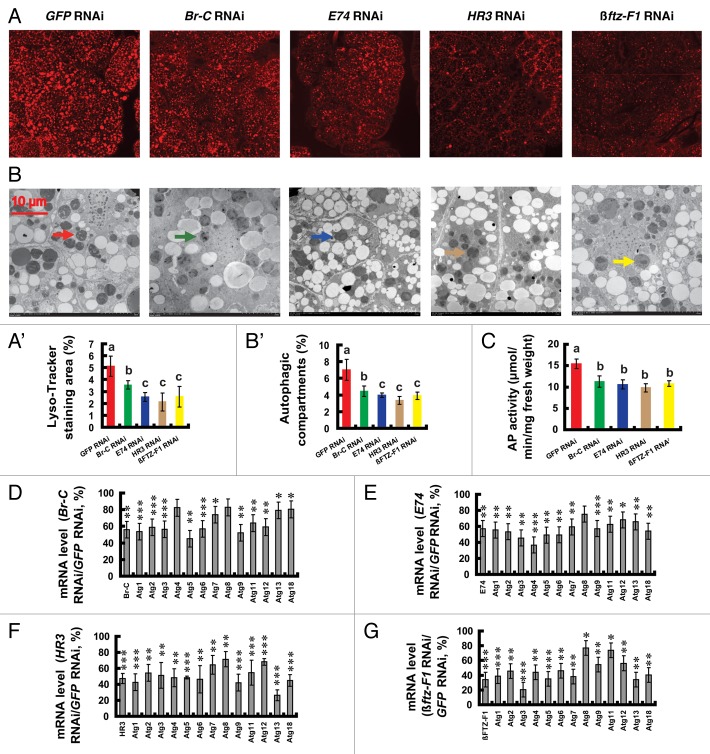
The binding sequences of Br-C, E74, HR3 and βftz-F1 have been identified in Drosophila.20,52 As in Drosophila, Br-C, E74 and HR3 are also key genes in the 20E-triggered transcriptional cascade in Bombyx.53-55 The orphan nuclear receptor βftz-F156 was shown to be essential for 20E signal transduction during Bombyx metamorphosis.57 Using the MATINSPECTOR program (www.genomatix.de), we predicted potential binding sites of Br-C, E74, HR3 and βftz-F1 in the promoter regions of 12 Atg genes; the exception was Atg2, whose promoter region was not found in the silkworm genome database. Many Br-C, E74, HR3 and βftz-F1 binding sites were predicted in the promoter regions of the 12 Atg genes (Fig. S5), suggesting that these 4 proteins might play roles in transducing 20E signaling to induce Atg gene expression.
To test this idea, we investigated whether RNAi knockdown of Br-C, E74, HR3 and βftz-F1 had inhibitory effects on autophagy and Atg gene expression in the fat body during pupation. Similar to USP RNAi and EcRDN overexpression, as determined by LysoTracker Red staining, TEM analysis and AP activity (Fig. 8A–C), autophagy in the Bombyx fat body was decreased to different levels 24 h after the RNAi treatments. Moreover, as determined by qPCR, all 13 Atg genes in the fat body were downregulated to different levels (Fig. 8D–G). In general, the weakest inhibitory effects on autophagy and Atg gene expression were caused by Br-C RNAi, while the inhibitory effects of the other three RNAi treatments were similar to each other. The finding in the Bombyx fat body is quite different from that in the Drosophila salivary gland and midgut, in which E93 predominantly affects autophagic cell death and Atg gene expression.15,24,25,46
Figure 8. Reduction of autophagy and downregulation of Atg genes by RNAi knockdown of key 20E response genes at the initiation of the wandering stage. Fat body tissues were prepared 24 h after RNAi treatment. (A and A’) LysoTracker Red staining (red, 40×). The chart (A’) shows the quantification of LysoTracker Red staining in (A). (B and B’) TEM analysis (7,500×). The arrows denote autolysosomes. The chart (B’) shows the quantification of autophagosomes and autolysosomes in (B). (C) Decrease of AP activity. (D–G) Downregulation of Atg genes by RNAi knockdown of Br-C (D), E74 (E), HR3 (F) and βFTZ-F1 (G).
In summary, via EcR-USP and key genes (Br-C, E74, HR3, βftz-F1 and probably others) in the 20E-triggered transcriptional cascade, 20E upregulates Atg genes to induce autophagy in the Bombyx fat body during pupation.
Discussion
20E upregulates Atg genes in the Bombyx fat body during molting and pupation
It has been shown that in Drosophila, EcR and E93 are critical for 20E signaling to induce autophagy15,22,24,25,46 and E93 affects expression of some, but not all Atg genes.25
Combining a series of in vivo and in vitro experimental results, we conclude that 20E upregulates most, if not all, Atg genes in the Bombyx fat body during molting and pupation. The experimental data for EcRDN overexpression in the transgenic silkworm SP2-GAL4 > UAS-EcRDN (Fig. 5) not only confirm those of 20E injection (Fig. 3) and systemic USP RNAi (Fig. 4), but also demonstrate that 20E signaling could upregulate Atg genes in a tissue-autonomous manner. In general, Atg gene expression was gradually increased from 2 to 24 h after 20E injection, in consistent with the theory that five Atg genes are 20E primary-response genes (Fig. 6; Fig. S3) and all Atg genes are also 20E secondary-response genes (Fig. 8; Fig. S5). Besides EcR-USP, several other transcriptional factors in the 20E-triggered transcriptional cascade, including EcR-USP, Br-C, E74, Hr3 and ftz-F1, together sustain the 20E-induced Atg gene expression to last for a comparatively long time. Therefore, one single transcriptional factor accounts for a portion of 20E signaling and RNAi knockdown of one of these genes (Fig. 8) might result in a relatively weaker inhibitory effect on Atg gene expression than USP RNAi (Fig. 4).
For comparison, there are at least three substantial differences on 20E-induction of Atg gene expression between Bombyx and Drosophila. First, nearly all Atg genes are upregulated by 20E signaling in the Bombyx fat body (Fig. 9), while only a few Atg genes are upregulated by 20E in the Drosophila salivary gland.25,26 Second, five 20E primary-response genes are found and an EcRE is identified in the Atg1 promoter in Bombyx, yet it is currently unknown whether there are any 20E primary-response Atg genes in Drosophila. Third, at least four key genes (namely Br-C, E74, HR3 and βftz-F1) in the 20E-triggered transcriptional cascade regulate Atg gene expression in Bombyx, but E93 predominantly transduces 20E signaling to regulate autophagy by affecting a subset of Atg genes in Drosophila.15,24,25,46 The Drosophila E93 paralog has not been cloned in Bombyx yet and whether the Bombyx E93 plays a critical role in regulating Atg gene expression is worthy of further investigation.
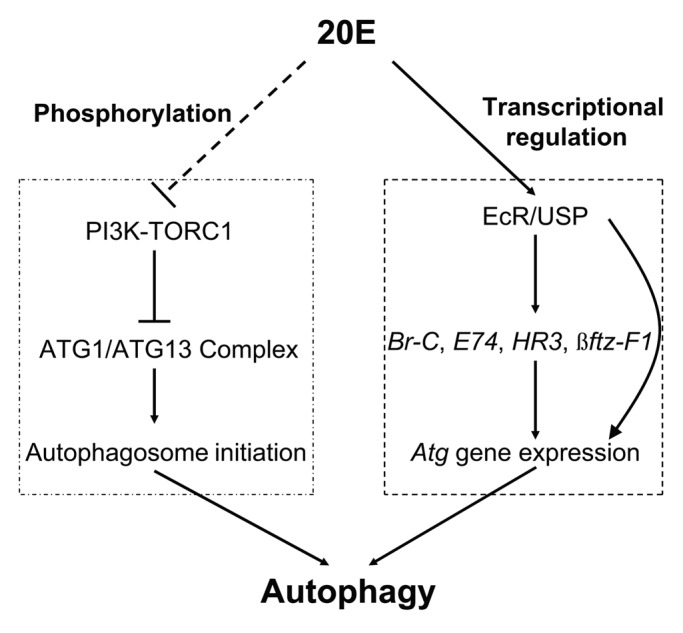
Figure 9. A model for the 20E signal to induce autophagy by two indispensible means. 20E (top: the upstream power source) blocks TORC1 activity to induce autophagosome initiation (left: to break the gate) by phosphorylation and upregulates the Atg genes (right: to provide the flow) by transcriptional regulation resulting in autophagy induction (bottom: the downstream effect).
Taking the data together, we conclude that 20E upregulates the most, if not all, Atg genes in the Bombyx fat body during molting and pupation.
20E reduces TORC1 activity and upregulates Atg genes to induce autophagy in the Bombyx fat body
In Drosophila, 20E induces autophagy by two means. First, 20E blocks TORC1 activity to induce autophagosome initiation.21-23 Second, E93 predominantly transduces 20E signaling to induce autophagy by upregulating a subset of Atg genes.15,24,25,46 However, the relationship between the two means has not been fully addressed in this insect model.
In Drosophila, TORC1 inhibits autophagosome initiation by phosphorylating ATG1 under nutrient-rich conditions; and persistent blockage of the PIK3/PI3K-TORC1 pathway was sufficient to induce autophagy in the fat body.8,16,21,35 Injection of rapamycin into the feeding larvae not only decreased TORC1 activity in the Bombyx fat body, but also moderately induced expression of several Atg genes. However, the resulting induction of autophagy by rapamycin injection is much weaker compared with 20E injection (Fig. S6). This discrepancy should be due to both low expression levels of Atg genes in the Bombyx fat body during the feeding larval stages (Fig. 2) and the insufficient upregulation of some critical Atg genes by rapamycin injection (Fig. S6). We infer that in the absence of sufficient Atg gene expression, acute blocking TORC1 activity has little or no induction of autophagy in the Bombyx fat body during the feeding larval stages.
Based on the results here in Bombyx and the previous reports in Drosophila, we propose a model for the 20E signal in inducing autophagy by two indispensable means (Fig. 9): 20E (the upstream power source) blocks TORC1 activity to induce autophagosome initiation (to break the gate) by phosphorylation and upregulates the Atg genes (to provide the flow) by transcriptional regulation, resulting in autophagy induction (bottom: the downstream effect).
It has been reported that Atg1 overexpression decreases TORC1 activity in Drosophila,16 and RNAi knockdown of Atg1 or/and Atg8 resulted in hyperactivation of TORC1 in the mosquito, Aedes aegypti.58 It will be of interest to investigate whether the 20E-induced Atg gene expression decreases TORC1 activity in Bombyx fat body as well.
Because Bombyx possesses a longer life cycle and larger body size than Drosophila, we are able to inject Bombyx with chemicals and dsRNA and observe the acute effects. However, it is inconvenient to perform similar experiments in Drosophila. This advantage makes Bombyx an ideal Lepidoptera model for autophagy studies compensating Drosophila.
Possible physiological roles of autophagy in insect development and metabolism
In Drosophila, Atg1Δ3D animals die before pupation, suggesting that autophagy is functionally required for Drosophila metamorphosis.15,35 Our preliminary data showed that reduction of autophagy by RNAi knockdown of Atg1 and several other Atg genes in Bombyx caused significant lethality during the prepupal and pupal stages, suggesting that autophagy also plays important roles during Bombyx metamorphosis.
Autophagy promotes cell survival against apoptosis, while extensive autophagy can also cause cell death under certain circumstances.2 During Drosophila metamorphosis, many obsolete larval tissues undergo massive destruction mainly by autophagic cell death.18,19 Both autophagy and caspases, which function in parallel, contribute to cell death in the dying salivary gland, but autophagy plays a more important role than caspases.15,16 Moreover, autophagy governs cell death in the midgut, while caspases do not.17 A different example is the remodeling fat body, where autophagy appears to play a more important role in cell survival than in cell death.14 We have previously determined that in the remodeling Bombyx fat body, 20E upregulates apoptosis genes to induce some apoptotic events,38 and similar apoptotic phenomenon was observed in the remodeling Bombyx midgut30 during Bombyx metamorphosis. How autophagy contributes to cell survival or cell death in different larval tissues of Bombyx remains largely unknown and merits further investigation.
In addition to the essential role of autophagy in regulating cell survival and cell death, recent research has shown that autophagy plays an important role in regulating energy metabolism, particularly in the degradation of lipid droplets and soluble cytosolic proteins in lysosomes.3,59 It is generally accepted that autophagy allows the pupa to fully exploit nutrients remaining in the dying larval tissues, thus supporting the growth and differentiation of the new adult structures. Subsequent cell death then directs the destruction of these doomed larval tissues.18,19 During insect metamorphosis, 20E also induces the endocytotic uptake of the larval serum proteins and the mobilization of lipid droplets in the fat body cells.27,28,34-36,60 Moreover, the 20E-induced autophagy and endocytosis crosstalk with each other and share some regulatory components, including PIK3C3/VPS34.61 The fat body functions as a major organ for nutrient storage and energy metabolism in insects;22,32,33 therefore, the function of autophagy in exploiting remaining nutrients should be particularly important in this insect adipose tissue.
We propose that the insect fat body provides an excellent model system to study autophagy because of its physiological function and hormonal regulation. Taking advantage of the systemic RNAi method to knock down Atg genes in Bombyx and the powerful genetic tools to manipulate Atg genes in Drosophila, we are currently investigating the possible physiological roles of autophagy in both development and metabolism in the two compensate insect models.
Materials and Methods
Silkworms
The silkworms (P50), provided by The Sericultural Research Institute, Chinese Academy of Agricultural Sciences (Zhenjiang, China), were reared with fresh mulberry leaves in the laboratory at 25°C under 14 h light/10 h dark cycles.44,45
LysoTracker Red staining
Newly collected peripheral fat body tissues from the 5th abdominal segment were separated into small pieces by forceps and thoroughly washed with PBS (pH 7.0, 0.1 M). The fragmented fat body tissues were stained with LysoTracker Red DND-99 (Invitrogen, L7528) at a final concentration of 50 nM for 5 min at 37°C and washed with PBS three times. LysoTracker Red staining was monitored under a FV10-ASW confocal microscope (Olympus, Japan). For LysoTracker Red staining and the following experiments, 3 biological replicates were used. In each biological replicate, fat body tissues were always collected from the 5th abdominal segments of 10 animals and combined. The observations were performed under the same conditions.
TEM analysis
Newly collected fat body tissues were fixed in 2.5% glutaraldehyde for 24 h at 4°C, thoroughly washed with PBS, postfixed in 0.5% osmium tetroxide for 2 h, and embedded in Epon resin according to the manufacturer’s recommendations. From the fixed, embedded tissue, 70-nm sections were cut, stained in Reynold’s lead citrate, and viewed on a H7650 transmission electron microscope (Hitachi) to observe autolysosomes and autophagosomes.21,27-30,34-37 An important criterion for autolysosomes is the visible remnants of organelles and the undigested materials.39 The autophagosomes are typically of 1–5 μm in diameter, which gradually increase during pupation.
Measurement of AP activity
After measuring the fresh weight, newly collected fat body tissues were thoroughly washed with PBS, homogenized and extracted for total protein in 0.1 M insect Ringer’s solution. AP activity was measured using p-nitrophenyl phosphate (Biovision, K411-500) as a substrate with a Multiskan Flash Microplate Reader (Thermo Fisher Scientific).30
RT-PCR, qPCR and western blotting
Detailed methods for RT-PCR, qPCR and western blotting were previously described.44,45,49,62 All primers used in this study are listed in Table S1 and TableS2. The primary antibodies, including Bombyx ATG8 (1:3,000),30 phospho-EIF4EBP1 (Thr37/46, Cell Signaling, 9459; 1:2,000) and tubulin (Beyotime, AT819; 1:10,000), were incubated at 4°C overnight.
Injection of 20E and rapamycin and RNAi knockdown in the larvae
Detailed methods for 20E injection and RNAi knockdown in the silkworm were previously described.44,45,62 5L-2 larvae were chosen for injection of 20E (Sigma Aldrich, H5142) (5 μg/larva), rapamycin (Sigma Aldrich, R0395) (10 μg/larva), or 20E (5 μg/larva) and rapamycin (10 μg/larva) and the controls were injected with the same volume of solvent. In some cases, a high dose of rapamycin (50 μg/larva) was used. Fat body tissues were collected for bioassays 2, 6, 12 and 24 h after injection.
Preliminary data showed that the P50 strain of Bombyx (the Chinese strain variation, Dazao) was more sensitive to RNAi treatments than other tested strains,62 so the P50 strain of Bombyx was used throughout this study. The double-stranded RNA (dsRNA) of GFP, Bombyx Atg1, USP, Br-C, E74, HR3 and ßftz-F1 was generated using the T7 RiboMAXTM Express RNAi system (Promega, P1700) according to the manufacturer’s instructions. At 12, 24 and 36 h after RNAi treatments (10 µg USP and 30 µg Atg1, Br-C, E74, HR3, or ßftz-F1 dsRNA per silkworm, or the same amount of GFP dsRNA as a control) at IW, fat body tissues were collected for bioassays. There were no differences in LysoTracker Red staining caused by 10 or 30 µg GFP dsRNA, and only the results of 30 µg GFP dsRNA were presented in the figures.
The binary GAL4/UAS transgenic silkworm system
Using our previously reported strategy,49 we generated the pBac{SP2-GAL4-3XP3-DsRed} (SP2-GAL4) and pBac{UAS-EcRDN-3XP3-EGFP} (UAS-EcRDN) constructs with the transposable element piggyBac as the transgenic vector. A 2-kb region of the SP2 promoter was amplified using forward and reverse primers containing AscI and SacI sites, respectively. The SacI-SacII fragment, which contains the GAL4 gene and the Dmhsp70 terminator, was amplified from phsGAL4. The two PCR fragments above were conjugated, digested with AscI and SacII, and inserted into pBac{3XP3-DsRed} to generate SP2-GAL4. The cDNA of the Bombyx EcR-B1 was amplified using a forward primer containing EcoRI and a reverse primer containing NotI. The Bombyx EcRDN was generated by making a phenylalanine-to-alanine mutation at position 517 of EcR-B1 and deleting from the 525th amino acid residue using the same approach used to generate the Drosophila EcRDN (EcR-B1-F645A).47 The HindIII-EcoRI fragment of the UAS sequence from pUAST was inserted into pEGFP-N1, which contained the EGFP gene and the SV40 terminator, to form an intermediate construct. The EGFP gene in the construct was replaced by the EcoRI-NotI EcRB1DN fragment to generate pUAS-EcRDN. The pUAS-EcRDN fragment was then digested with NheI and AflII and inserted into pBac{3XP3-EGFP} to generate UAS-EcRDN.
For germline transformation, plasmid and helper DNA were injected into preblastoderm eggs, and the embryos were incubated at 25°C with 95 to 100% humidity until hatching. Fluorescence detection of the positive G1 embryos and confirmation of the transgenic lines by inverse PCR were performed as previously described.49 Multiple independent transgenic lines of SP2-GAL4 and UAS-EcRDN were used. In terms of growth and development, SP2-GAL4 and UAS-EcRDN were indistinguishable from the wild-type silkworms. The transgenic silkworm SP2-GAL4 > UAS-EcRDN was obtained by crossing SP2-GAL4 with UAS-EcRDN. SP2-GAL4 > UAS-EcRDN was compared with its wild-type, SP2-GAL4 and UAS-EcRDN counterparts. Fat body tissues were prepared for bioassays 6 h after IW.
Tissue culture and cell culture
Newly collected fat body tissues from 5L-2 larvae were cultured in Grace’s medium (Sigma-Aldrich, 11300-043) at 25°C. DZNU-Bm-12 cells were maintained in TNM-FH medium (Sigma-Aldrich, T3285) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, SH30070.03) at 27°C.51,62 HEK 293 cells were maintained in Dulbecco's Modi□ed Eagle's Medium (Hyclone, SH30022.01B) supplemented with 10% heat-inactivated fetal bovine serum.62
After pre-incubation (1 h for fat body tissue and 1 d for DZNU-Bm-12 cells), the medium was removed and replaced with fresh medium with or without 5 μM 20E. After 2 or 6 h of incubation, mRNA was isolated for qPCR analysis. To determine which Atg genes were the 20E primary response genes, 5 μM 20E or/and 10 μg/ml CHX (Enzo Life Science, ALX-380-269) were used.62
Transient transfection assays in HEK 293 cells were performed using Lipofectamine 2000 (Invitrogen, 11668-019) according to the manufacturer’s instructions.62
Dual luciferase assay
The full-length Bombyx EcR-B1 and USP genes were amplified from the silkworm genome, the HA and FLAG tag sequences were fused to their respective 5′-ends, and the tagged genes were cloned into the pcDNA 3.1(+) vector (Invitrogen, V790-20) to make the HA-EcR-B1 and FLAG-USP expression constructs. The reporter vector pGL3 (Promega, E1751), containing 4 repeated hsp27 (ortholog of human HSPB1/HSP27) EcRE sequences (GACAAGGGTTCAATGCACTTGTC) and the hsp27 mini promoter, was used as a positive control. To identify the Atg1 EcRE, different fragments of the Atg1 promoter (−1820 to −1 bp, −1820 to −1001 bp, −1000 to −1 bp, −1820 to −1421 bp and −1800 to −1701 bp) were cloned. The putative Atg1 EcRE (−1779 to −1766; AGTTCAGGAGTTG) in the −1800 to −1701 bp fragment was mutated to AAAAAAGGAGTTG. The above 6 Atg1 promoter regions were inserted into the pGL3 basic vector containing the hsp27 mini-promoter as the reporter pGL3 vectors. The reference pRL vector (Promega, E2241) carries Renilla luciferase driven by the actin3 promoter.
After cotransfection of the HA-EcR-B1 and FLAG-USP expression constructs, a reporter pGL3 vector and the reference pRL vector into HEK 293 cells or DZNU-Bm12 cells for 36 h, the cells were treated with 20E (1 μM) for 12 h. The relative luciferase activity was calculated by normalizing the reporter firefly luciferase level to the reference Renilla luciferase level. Luciferase activity fold change refers to the relative luciferase activity induced by 20E divided by the control luciferase activity. Dual luciferase assays were performed using the Dual Luciferase Assay System (Promega, E1501) and a Modulus Luminometer (Turner BioSystems).62
EMSA
The HA-EcR-B1 and FLAG-USP constructs were cotransfected into HEK 293 cells, and nuclear extracts were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, 78833). The GFP construct was used as a negative control. The complimentary double-stranded DNA fragments were synthesized based on the minimal hsp27 EcRE probe (AGTTCAATGGCCT), the Atg1 EcRE probe (CCTGCCGATTGAGTTCAGGAGTTGTTGTGTTACCTGTTCC) and the Atg1 EcRE mutant probe (CCTGCCGATTGAAAAAAGGAGTTGTTGTGTTACCTGTTCC). The DNA fragments were biotin-labeled as probes using the Biotin 3′ End DNA Labeling Kit (Pierce, 89818). The EcR-USP protein complexes isolated from the nuclear extracts of the transfected HEK 293 cells were incubated with the biotin-labeled probes. After binding, the nuclear extract containing the biotin-labeled probes and the protein-DNA complexes were separated on a 5% nondenaturing polyacrylamide gel. For the competition experiments, the cold hsp27 EcRE probe or the cold Atg1 EcRE mutant probe was added to the binding reaction. EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Pierce, 89880).62
Statistics
Experimental data were analyzed with the Student’s t-test and ANOVA. t-test: *p < 0.05; **p < 0.01; ***p < 0.001. ANOVA: the bars labeled with different lowercase letters are significantly different (p < 0.05). Throughout the paper, values are represented as the mean ± standard deviation of 3 independent experiments.
Supplementary Material
Acknowledgments
This study was supported by the 973 program (2012CB114605 to S.L.), the Natural Science Foundation of China (31101670 to LT and 31125025 to S.L.), Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (2011KIP303 to L.T.), the Shanghai Municipal Education Commission (10JC1416700 and 12XD1406900 to S.L.), the State Key Laboratory of Silkworm Genome Biology (20120003 to S.L.). LT received a SA-SIBS award. We appreciate the Nature Publishing Group for polishing English for the manuscript. We also appreciate Mr. Xiaoshu Gao, Mr. Xiaoyan Gao, Ms. Jiqin Li and Mr. Zhiping Zhang (Core Facility of the institute) for technique supports.
Glossary
Abbreviations:
- 20E
20-hydroxyecdysone
- EcRDN
dominant-negative mutants of EcR
- 4M
the final larval molt
- 5F
the feeding stage of the 5th instar
- W
the wandering stage
- IW
the initiation of the wandering stage
- PP
the prepupal stage
- TEM
transmission electron microscopy
- qPCR
quantitative real-time PCR
- RT-PCR
reverse transcription PCR
- SP2
storage protein 2
- AP
acid phosphatase
- CHX
cycloheximide
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24731
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24731
References
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 3.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584:1342–9. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–68. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippai M, Csikós G, Maróy P, Lukácsovich T, Juhász G, Sass M. SNF4Agamma, the Drosophila AMPK gamma subunit is required for regulation of developmental and stress-induced autophagy. Autophagy. 2008;4:476–86. doi: 10.4161/auto.5719. [DOI] [PubMed] [Google Scholar]
- 10.Egan DF, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–4. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Vos KE, Gomez-Puerto C, Coffer PJ. Regulation of autophagy by Forkhead box (FOX) O transcription factors. Adv Biol Regul. 2012;52:122–36. doi: 10.1016/j.advenzreg.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Malagoli D, Abdalla FC, Cao Y, Feng Q, Fujisaki K, Gregorc A, et al. Autophagy and its physiological relevance in arthropods: current knowledge and perspectives. Autophagy. 2010;6:575–88. doi: 10.4161/auto.6.5.11962. [DOI] [PubMed] [Google Scholar]
- 13.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–65. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 14.McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–6. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–6. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin VP, Thummel CS. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin Cell Dev Biol. 2005;16:237–43. doi: 10.1016/j.semcdb.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Ryoo HD, Baehrecke EH. Distinct death mechanisms in Drosophila development. Curr Opin Cell Biol. 2010;22:889–95. doi: 10.1016/j.ceb.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2000;60:1–73. doi: 10.1016/S0083-6729(00)60016-X. [DOI] [PubMed] [Google Scholar]
- 21.Rusten TE, Lindmo K, Juhász G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–92. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–70. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- 23.Delanoue R, Slaidina M, Léopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev Cell. 2010;18:1012–21. doi: 10.1016/j.devcel.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Lee CY, Cooksey BAK, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–11. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- 25.Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13:350–7. doi: 10.1016/S0960-9822(03)00085-X. [DOI] [PubMed] [Google Scholar]
- 26.Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, et al. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13:358–63. doi: 10.1016/S0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 27.Sass M, Kovács J. Ecdysterone and an analogue of juvenile hormone on the autophagy in the cells of fat body of Mamestra brassicae. Acta Biol Acad Sci Hung. 1975;26:189–96. [PubMed] [Google Scholar]
- 28.Sass M, Kovács J. The effect of ecdysone on the fat body cells of the penultimate larvae of Mamestra brassicae. Cell Tissue Res. 1977;180:403–9. doi: 10.1007/BF00227604. [DOI] [PubMed] [Google Scholar]
- 29.Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J Morphol. 1977;154:39–57. doi: 10.1002/jmor.1051540104. [DOI] [PubMed] [Google Scholar]
- 30.Franzetti E, Huang ZJ, Shi YX, Xie K, Deng XJ, Li JP, et al. Autophagy precedes apoptosis during the remodeling of silkworm larval midgut. Apoptosis. 2012;17:305–24. doi: 10.1007/s10495-011-0675-0. [DOI] [PubMed] [Google Scholar]
- 31.Casati B, Terova G, Cattaneo AG, Rimoldi S, Franzetti E, de Eguileor M, et al. Molecular cloning, characterization and expression analysis of ATG1 in the silkworm, Bombyx mori. Gene. 2012;511:326–37. doi: 10.1016/j.gene.2012.09.086. [DOI] [PubMed] [Google Scholar]
- 32.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Léopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–49. doi: 10.1016/S0092-8674(03)00713-X. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Liu H, Liu S, Wang S, Jiang RJ, Li S. Hormonal and nutritional regulation of insect fat body development and function. Arch Insect Biochem Physiol. 2009;71:16–30. doi: 10.1002/arch.20290. [DOI] [PubMed] [Google Scholar]
- 34.Sass M, Kovács J. The effect of actinomycin D, cycloheximide and puromycin on 20-hydroxyecdysone induced autophagocytosis in larval fat body cells of Pieris brassicae. J Insect Physiol. 1980;26:568–77. doi: 10.1016/0022-1910(80)90132-8. [DOI] [Google Scholar]
- 35.Butterworth FM, Forrest EC. Ultrastructure of the preparative phase of cell death in the larval fat body of Drosophila melanogaster. Tissue Cell. 1984;16:237–50. doi: 10.1016/0040-8166(84)90047-8. [DOI] [PubMed] [Google Scholar]
- 36.Butterworth FM, Emerson L, Rasch EM. Maturation and degeneration of the fat body in the Drosophila larva and pupa as revealed by morphometric analysis. Tissue Cell. 1988;20:255–68. doi: 10.1016/0040-8166(88)90047-X. [DOI] [PubMed] [Google Scholar]
- 37.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Tian L, Liu S, Liu H, Li S. 20-hydroxyecdysone upregulates apoptotic genes and induces apoptosis in the Bombyx fat body. Arch Insect Biochem Physiol. 2012;79:207–19. doi: 10.1002/arch.20457. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sass M, Komuves L, Csikos G, Kovacs J. Changes in the activities of lysosomal enzymes in the fat body and midgut of two lepidopteran insects (Mamestra brassicae and Pieris brassicae) during metamorphosis. Comp Biochem Physiol. 1989;92A:285–9. doi: 10.1016/0300-9629(89)90565-3. [DOI] [Google Scholar]
- 41.Muramatsu D, Kinjoh T, Shinoda T, Hiruma K. The role of 20-hydroxyecdysone and juvenile hormone in pupal commitment of the epidermis of the silkworm, Bombyx mori. Mech Dev. 2008;125:411–20. doi: 10.1016/j.mod.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Hu ZY, Li WF, Li QR, Deng XJ, Yang WY, et al. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol Biol. 2009;10:50. doi: 10.1186/1471-2199-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–14. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian L, Guo E, Diao Y, Zhou S, Peng Q, Cao Y, et al. Genome-wide regulation of innate immunity by juvenile hormone and 20-hydroxyecdysone in the Bombyx fat body. BMC Genomics. 2010;11:549. doi: 10.1186/1471-2164-11-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian L, Guo E, Wang S, Liu S, Jiang RJ, Cao Y, et al. Developmental regulation of glycolysis by 20-hydroxyecdysone and juvenile hormone in fat body tissues of the silkworm, Bombyx mori. J Mol Cell Biol. 2010;2:255–63. doi: 10.1093/jmcb/mjq020. [DOI] [PubMed] [Google Scholar]
- 46.Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–55. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 47.Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–84. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- 48.Imamura M, Nakai J, Inoue S, Quan GX, Kanda T, Tamura T. Targeted gene expression using the GAL4/UAS system in the silkworm Bombyx mori. Genetics. 2003;165:1329–40. doi: 10.1093/genetics/165.3.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma L, Xu H, Zhu J, Ma S, Liu Y, Jiang RJ, et al. Ras1(CA) overexpression in the posterior silk gland improves silk yield. Cell Res. 2011;21:934–43. doi: 10.1038/cr.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujii T, Sakurai H, Izumi S, Tomino S. Structure of the gene for the arylphorin-type storage protein SP 2 of Bombyx mori. J Biol Chem. 1989;264:11020–5. [PubMed] [Google Scholar]
- 51.Khurad AM, Zhang MJ, Deshmukh CG, Bahekar RS, Tiple AD, Zhang CX. A new continuous cell line from larval ovaries of silkworm, Bombyx mori. In Vitro Cell Dev Biol Anim. 2009;45:414–9. doi: 10.1007/s11626-009-9197-2. [DOI] [PubMed] [Google Scholar]
- 52.Henrich VC. The ecdysteroid receptor, Insect Endocrinology, Academic Press, San Diego, CA, US. 2011:177-206. [Google Scholar]
- 53.Eystathioy T, Swevers L, Iatrou K. The orphan nuclear receptor BmHR3A of Bombyx mori: hormonal control, ovarian expression and functional properties. Mech Dev. 2001;103:107–15. doi: 10.1016/S0925-4773(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 54.Reza AM, Kanamori Y, Shinoda T, Shimura S, Mita K, Nakahara Y, et al. Hormonal control of a metamorphosis-specific transcriptional factor Broad-Complex in silkworm. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:753–61. doi: 10.1016/j.cbpc.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Sekimoto T, Iwami M, Sakurai S. 20-Hydroxyecdysone regulation of two isoforms of the Ets transcription factor E74 gene in programmed cell death in the silkworm anterior silk gland. Insect Mol Biol. 2007;16:581–90. doi: 10.1111/j.1365-2583.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- 56.Sun GC, Hirose S, Ueda H. Intermittent expression of BmFTZ-F1, a member of the nuclear hormone receptor superfamily during development of the silkworm Bombyx mori. Dev Biol. 1994;162:426–37. doi: 10.1006/dbio.1994.1099. [DOI] [PubMed] [Google Scholar]
- 57.Cheng D, Xia Q, Duan J, Wei L, Huang C, Li Z, et al. Nuclear receptors in Bombyx mori: insights into genomic structure and developmental expression. Insect Biochem Mol Biol. 2008;38:1130–7. doi: 10.1016/j.ibmb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Bryant B, Raikhel AS. Programmed autophagy in the fat body of Aedes aegypti is required to maintain egg maturation cycles. PLoS One. 2011;6:e25502. doi: 10.1371/journal.pone.0025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–9. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Liu S, Liu H, Wang J, Zhou S, Jiang RJ, et al. 20-hydroxyecdysone reduces insect food consumption resulting in fat body lipolysis during molting and pupation. J Mol Cell Biol. 2010;2:128–38. doi: 10.1093/jmcb/mjq006. [DOI] [PubMed] [Google Scholar]
- 61.Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–66. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo E, He Q, Liu S, Tian L, Sheng Z, Peng Q, et al. MET is required for the maximal action of 20-hydroxyecdysone during Bombyx metamorphosis. PLoS One. 2012;7:e53256. doi: 10.1371/journal.pone.0053256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.