Abstract
The identification of inflammatory bowel disease (IBD) susceptibility genes by genome-wide association has linked this pathology to autophagy, a lysosomal degradation pathway that is crucial for cell and tissue homeostasis. Here, we describe autophagy-related 4B, cysteine peptidase/autophagin-1 (ATG4B) as an essential protein in the control of inflammatory response during experimental colitis. In this pathological condition, ATG4B protein levels increase in parallel with the induction of autophagy. Moreover, ATG4B expression is significantly reduced in affected areas of the colon from IBD patients. Consistently, atg4b−/− mice present Paneth cell abnormalities, as well as an increased susceptibility to DSS-induced colitis. atg4b-deficient mice exhibit significant alterations in proinflammatory cytokines and mediators of the immune response to bacterial infections, which are reminiscent of those found in patients with Crohn disease or ulcerative colitis. Additionally, antibiotic treatments and bone marrow transplantation from wild-type mice reduced colitis in atg4b−/− mice. Taken together, these results provided additional evidence for the importance of autophagy in intestinal pathologies and describe ATG4B as a novel protective protein in inflammatory colitis. Finally, we propose that atg4b-null mice are a suitable model for in vivo studies aimed at testing new therapeutic strategies for intestinal diseases associated with autophagy deficiency.
Keywords: ATG4B, autophagin-1, autophagy, colitis, inflammation, intestinal homeostasis, cysteine peptidase, Paneth cell
Introduction
Inflammatory bowel disease (IBD) presents in two major forms, Crohn disease and ulcerative colitis. The precise etiology of IBD is largely elusive but accumulating evidence suggests that genetic, environmental and immunological factors converge to provoke disease initiation and progression.1-3 Although the different forms of IBD have some mechanisms in common, they exhibit distinct pathophysiological features. In ulcerative colitis, inflammation is characterized by a continuous pattern that involves the superficial mucosal and submucosal layers, yet is limited to the colon. In contrast, inflammation in Crohn disease is transmural and discontinuous, and any region of the gut can be affected beyond the ileum, which is most affected.4,5
Genome-wide association studies have led to the identification of several IBD susceptibility genes including three genes implicated in autophagy: the autophagy-related 16-like 1 (ATG16L1) gene, the immunity-related GTPase family M (IRGM) gene and the Unc-51-like kinase 1 (ULK1) gene.6-8 Likewise, a NOD2 mutation associated with Crohn disease impairs autophagy by hampering ATG16L1 recruitment to the plasma membrane at the site of bacterial entry.9 Mutant mice deficient in Atg16l1 possess abnormal Paneth cells and are highly susceptible to dextran sodium sulfate (DSS)-induced colitis.10,11 Collectively, these findings suggest that autophagy is essential for the maintenance of intestinal homeostasis. Autophagy is a degradative process in which portions of the cytoplasm are engulfed by a double-membrane vesicle called the “autophagosome” which then fuses with lysosome to form “autolysosomes.”12,13 Autophagy plays an important role in cell and tissue homeostasis, and has been implicated in multiple processes including cell death, cell differentiation, clearance of intracellular bacteria, antigen presentation and the regulation of innate and adaptive immune responses.14-16 The autophagic process involves a group of evolutionarily conserved proteins, known as Atg proteins, originally described in yeast and required for autophagosome formation.13 Among these autophagic proteins, several studies have focused on yeast Atg4, a cysteine proteinase that proteolytically activates pro-Atg8. This processing event is strictly required for the subsequent conjugation of Atg8 with membrane-bound phosphatidylethanolamine (PE), which is essential for autophagosome completion. Atg4 is also required for deconjugating PE from the C-terminal Gly of Atg8–PE.17 We have previously identified and cloned the four human orthologs of the yeast proteinase Atg4, named ATG4A (autophagy related 4A, cysteine peptidase/autophagin-2), ATG4B (autophagy-related 4B, cysteine peptidase/autophagin-1), ATG4C (autophagy-related 4C, cysteine peptidase/autophagin-3) and ATG4D (autophagy-related 4D, cysteine peptidase/autophagin-4).18 Recently, we have generated mutant mice deficient in ATG4B protease and demonstrated that these animals exhibit a clear reduction of basal- and starvation-induced autophagic flux in all tissues.19 Our previous study revealed that the three other ATG4 proteases (ATG4A, C and D) do not extensively compensate for the lack of ATG4B since very low LC3-I lipidation occurs in most tissues from atg4b-null mice. These findings can be explained on the basis of recent enzymatic studies showing that ATG4B is able to cleave each of the six ATG8 orthologs in mammals (LC3A, LC3B, LC3C, GABARAP, GABARAPL1 and GABARAPL2/GATE-16). In contrast, ATG4A is a potent protease for the GABARAP subfamily of ATG8-orthologs, but not for the LC3 subfamily. Additionally, ATG4C and ATG4D have minimal activities toward the six ATG8-related substrates.20,21 However, this partial autophagy deficiency of atg4b-null mice is compatible with the survival of these animals, a situation which contrasts with other mutant mice deficient in Atg genes such as Atg3, Atg5, BECN1 (yeast VPS30/ATG6) and Atg7, that show a complete autophagy deficiency resulting in embryonic or perinatal lethality.22-24 The availability of atg4b-null mice, which have normal life span and minor pathological alterations, opens the possibility to study the in vivo effect of autophagy impairment on intestinal homeostasis.
In this work, we provided evidence that autophagic activity and ATG4B expression increase during experimental colitis in mice and that the levels of ATG4B are reduced in colon tissues from IBD patients, specifically in inflamed areas. We have also found that deficiency in the ATG4B protease causes important changes in secretory pathways that in turn result in marked alterations in inflammatory responses and in an increased susceptibility to experimental colitis. On this basis, we propose that ATG4B has a crucial role in intestinal homeostasis and plays a protective role in colitis.
Results
Deregulation of autophagy in experimental colitis is linked to changes in ATG4B protein levels
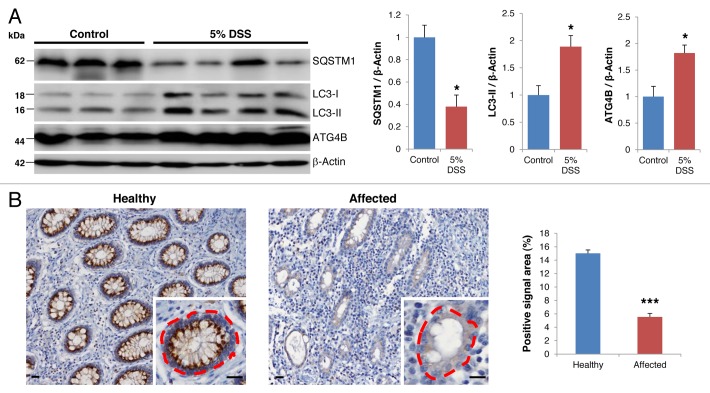
The identification of IBD susceptibility genes related to autophagy has revealed the importance of the autophagic process in the development of this pathology in humans.6,7 Given that autophagy constitutes a protective mechanism by which cells adapt to diverse types of stressors,25 we speculated as to whether autophagy induction in intestinal cells might also serve as a protective mechanism against IBD. To investigate this hypothesis, we treated wild-type (WT) mice with DSS, which induces an experimental colitis that resembles human IBD. As shown in Figure 1A, DSS treatment significantly increased autophagic flux in colon cells, as indicated by the conversion of LC3-I to its cleaved and lipidated form LC3-II, and by a reduction in the cellular levels of the autophagic substrate SQSTM1/p62. Remarkably, DSS treatment also induced a significant increase in the levels of ATG4B protease in colon tissue. ATG4B is known to proteolytically activate LC3 to render possible its lipidation and subsequent incorporation into autophagosome membranes.19 Accordingly, high ATG4B expression levels have been associated with situations of increased autophagic flux.26,27 Prompted by these observations, we analyzed the expression of ATG4B in colon tissues from IBD patients by immunohistochemistry. As shown in Figure 1B, we observed a significant decrease in the levels of ATG4B protease in inflamed areas of the colon when compared with histopathologically normal regions from the same patient. The degree of inflammation inversely correlated with the expression of ATG4B. Based on these results, and taking advantage of the recently generated atg4b-deficient mice,19,28 we decided to investigate the possibility that ATG4B-enabling autophagic activity might protect against colitis.
Figure 1. Changes in ATG4B expression levels in experimental colitis and IBD. (A) Representative immunoblots (left) and densitometry analysis (right) of endogenous SQSTM1, LC3-I/II and ATG4B in colon tissue extracts from control (n = 3) and DSS-treated mice (n = 4). ACTB was used as loading control. (B) Representative images (left) and quantitative analysis of the fraction of the dashed area with positive signal (n > 50 crypts of each condition; right) of immunohistochemistry analysis against ATG4B in healthy and affected crypts in colon tissue sections from IBD patients. Images are representative of 30 patients and show decreased levels of this protein in affected crypts. Scale bars: 20 μm. The results shown are means ± SEM. Statistical significance was determined by Student’s t-test (*p < 0.05; ***p < 0.005).
Atg4b-deficiency increases mice susceptibility to DSS-induced colitis
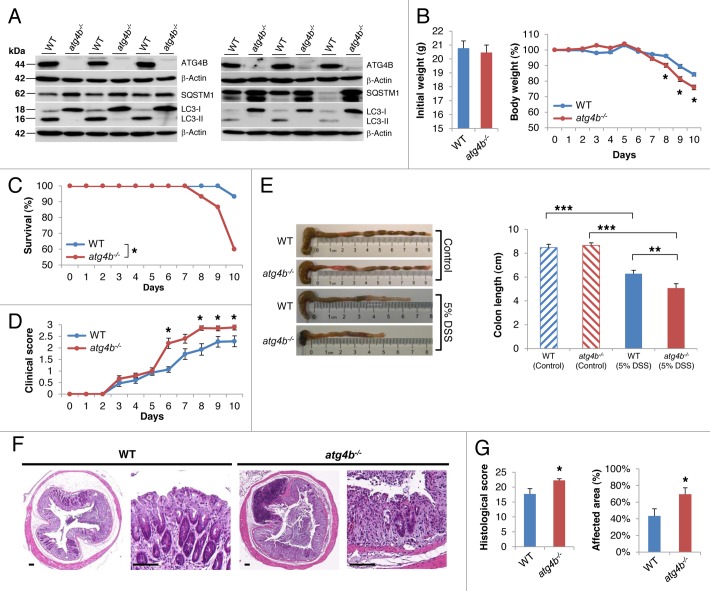
atg4b-null mice are viable and reach adulthood normally, although they show deficient proteolytic processing of LC3 and its paralogues, leading to a systemic decrease in both basal and induced autophagy.19 As shown in Figure 2A, the ileum and colon from atg4b−/− mice showed an impairment in LC3-I to LC3-II conversion together with the accumulation of SQSTM1, indicating reduced autophagic flux.29,30atg4b-deficient mice and control littermates were challenged by oral administration of DSS and the severity of colitis was assessed by evaluating survival, weight loss and clinical illness score. Although the initial weight of both groups was not different, atg4b−/− mice showed a higher weight loss and a significant decrease in survival compared with WT mice (Fig. 2B and C). Consistently, a higher clinical illness score was determined for atg4b−/− mice (Fig. 2D). In fact, mutant mice exhibited more severe colitis symptoms such as abundant bloody diarrhea and rectal bleeding than control animals. Disease severity was also assessed by colon length measurement and macroscopic examination of the colon. While we did not find any differences in colon length from control mice between genotypes, a pronounced colon shortening was observed for DSS-treated atg4b−/− mice as compared with DSS-treated WT mice (Fig. 2E). Additionally, erythema and grossly bloody dark stools were commonly observed in colon from atg4b−/− mice but not in WT mice. Histological analyses after DSS treatment revealed larger areas of ulceration and crypt loss, more massive inflammatory cell infiltration into the submucosa, a more pronounced loss of goblet cells and thickening of the mucosa with abundant edema in the colon from atg4b−/− mice as compared with controls (Fig. 2F). Consequently, both the histological score and the percentage of affected area were significantly higher in atg4b−/− mice relative to WT mice (Fig. 2G). Taken together, these data demonstrate that the absence of ATG4B increases the susceptibility to DSS-induced colitis.
Figure 2. Reduced autophagic flux in the ileum and colon from atg4b−/− mice is linked to their increased susceptibility to DSS-induced colitis. (A) Representative immunoblots of ATG4B, LC3 and SQSTM1 in protein extracts of ileum (left) and colon (right) from age-matched control and mutant mice fed ad libitum. Protein extracts from three mice per genotype are shown. ACTB was used as loading control. (B–E) 5% DSS was administered in drinking water for 7 d and then replaced by regular drinking water as indicated in Materials and Methods. (B) Initial weight and percent body weight after DSS treatment of WT and atg4b−/− mice. (C) Survival of control and mutant mice after DSS administration. (D) Clinical score data (see Materials and Methods) from treated mice. (E) Colon length and representative photographs of colon and cecum from control and DSS-treated mice of each genotype. (F) Representative light microscopy images of H&E stained colon tissue sections from WT and atg4b−/− mice after DSS administration. Scale bars: 100 μm. (G) Histological score data, including the percentage of affected area (see “Methods”). The results shown are mean ± SEM, n = 12 of each group. Statistical significance was determined by Student’s t-test (B, D and G), Log-rank test (C) or one-way analysis of variance (ANOVA; E) (*p < 0.05; **p < 0.01; ***p < 0.005).
Increased inflammation upon DSS-treatment in atg4b−/− mice is linked to alterations in proinflammatory cytokine profiles
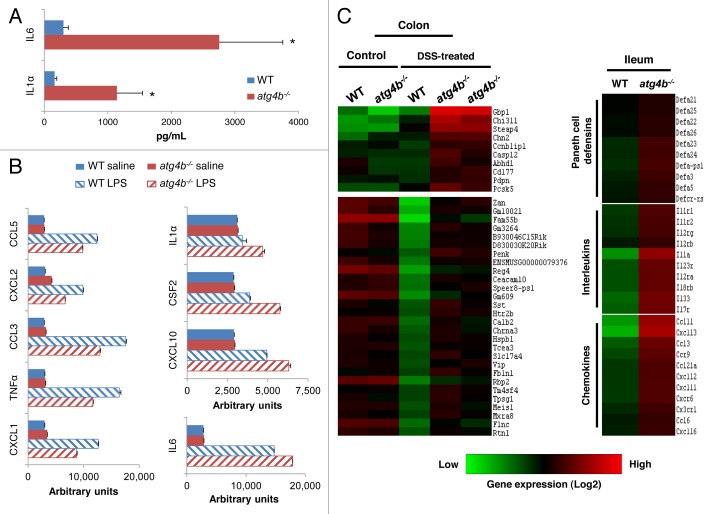
Crohn disease and ulcerative colitis are characterized by an excessive response of the intestinal immune system and a marked overexpression of proinflammatory cytokines.31 Therefore, we comparatively evaluated the pattern of cytokine expression in colon tissue extracts and sera from DSS-treated atg4b−/− and WT mice. IL1A/IL1α and IL6 concentrations were significantly elevated in colons from atg4b−/− mice compared with WT littermates upon DSS treatment (Fig. 3A). However, we did not find differences in serum cytokines between DSS-treated mice from both genotypes (data not shown). The marked changes in IL1A and IL6 levels in colon from atg4b−/− mice are consistent with those observed for both cytokines in cells from patients with Crohn disease and ulcerative colitis.32,33 Additionally, peritoneal macrophages from atg4b−/− mice stimulated with bacterial lipopolysaccharide (LPS) secreted less CXCL1, TNF/TNFα, CCL3, CCL2 and CCL5, while they secreted high levels of CXCL10, CSF2, IL1A and IL6 compared with WT macrophages; however, these differences did not reach statistical significance (Fig. 3B).
Figure 3. Deregulated secretion of proinflammatory cytokines in colon from atg4b−/− mice. (A) Protein level of the proinflammatory cytokines IL1Aand IL6 in colon tissue extracts from DSS-treated WT (n = 8) and atg4b−/− mice (n = 7). (B) Protein level of cytokines secreted by macrophages from WT (n = 5) and atg4b−/− mice (n = 5) after their exposure to LPS in vitro. (C) Microarray analyses of the colon and ileum from atg4b−/− and WT mice. A heat map representing the expression profiles of genes with significant differential expression when comparing WT vs. atg4b−/− colon from control and DSS-treated mice (left) or WT vs. atg4b−/− ileum from control mice (right) is shown, displayed as Log2-transformed expression signals. The results shown are means ± SEM. Statistical significance was determined by Student’s t-test (*p < 0.05).
To further evaluate the mechanism of the severe inflammation phenotype associated with DSS-induced colitis in atg4b−/− mice, we next performed microarray-based comparisons of global transcription in intestinal tissues from atg4b−/− mice vs. WT littermate controls. This analysis revealed the selective upregulation of several genes in the colon from atg4b−/− compared with WT mice in response to DSS (Fig. 3C; Table S1). These include Gbp1, Chi3l1, Steap4, Casp12 and Cd177, which encode proteins of potential interest for the exacerbated colitis developing in atg4b−/− mice. Guanylate binding protein 1, interferon-inducible (GBP1) is a key GTPase in the protective immunity against microbial and viral pathogens34 and is overexpressed in colitis-susceptible mice and in patients with ulcerative colitis.35,36 Chitinase 3-like-1 (CHI3L1) has been proposed as a marker of chronic inflammatory diseases and inflammation-mediated oncogenic processes, and its levels are increased in sera from IBD patients.37 CHI3L1 also plays a role in experimental colitis enhancing intracellular bacterial invasion and adhesion of bacteria to colonic epithelial cells.38 Six-transmembrane epithelial antigen of prostate 4 (STEAP4) is a plasma membrane metalloreductase induced by inflammatory factors such as TNF and IL6.39 CASP12/caspase 12 is implicated in the inflammatory caspase activation pathway and plays a pathogenic role during colitis.40 Finally, CD177 plays a crucial role in neutrophil activation,41 contributes to interactions between neutrophils and endothelial cells and is upregulated during bacterial infections.42 Among the few downregulated genes found in colon from DSS-treated atg4b−/− mice, several interleukins and chemokines were found, perhaps reflecting an antiinflammatory feedback regulation (Table S2). Interestingly, microarray expression analyses of the ileum from untreated mice revealed a deregulation in the expression of genes encoding several interleukins, chemokines and defensins in atg4b−/− mice (Fig. 3C; Table S3). Since we used a small sample size with low statistical power for microarrays, gene expression profiles were integrated with 2D DIGE combined with MALDI-ToF analysis.
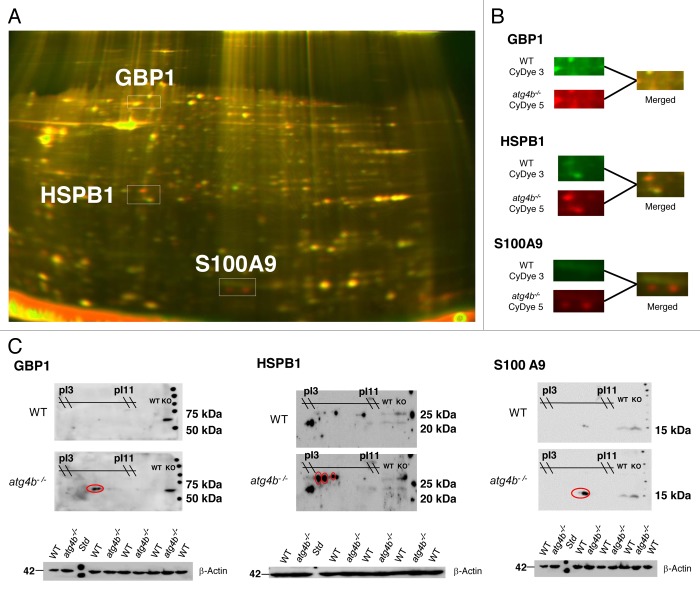
Protein expression patterns between colon tissue from DSS-treated atg4b−/− and WT mice revealed significant differences in three proteins: guanylate binding protein 1, interferon-inducible (GBP1), heat shock protein β 1 (HSPB1) and S100 calcium-binding protein A9 (S100A9) (Fig. 4). The accumulation of GBP1 and HSPB1 in the colon from DSS-treated atg4b−/− mice was consistent with the transcriptional profiling. HSPB1 is a chaperone of the small heat shock protein (sHsp) group with an important role in the control of stress conditions and apoptosis.43 S100A9 is a calcium-binding protein widely associated with inflammatory conditions.44 The upregulation in the three differential proteins found in atg4b−/− compared with WT mice after DSS was confirmed by immunoblot (Fig. 4). In summary, these transcriptomic and proteomic data suggest that a more severe DSS-induced colitis in atg4b−/− mice could be associated with deregulated expression of pro-inflammatory proteins, such as IL1A, IL6, GBP1, HSPB1 and S100A9. However, we cannot rule out that gene expression alterations in intestinal tissue from atg4b-null mice may reflect an increased inflammatory status in colon rather than a causative role in the exacerbated colitis observed in these mutant animals.
Figure 4. Identification of differential proteins in colon from DSS-treated atg4b−/− mice. (A) Differential Gel Electrophoresis (DiGE) was performed with pooled samples from both genotypes: WT CyDye 3 (green, n = 5), KO CyDye 2 (red, n = 4). Differential spots that were analyzed and then validated by 2D western blotting are shown. (B) Closer look on the differential spots. (C) Differential proteins identified by MALDI-ToF were confirmed by 2D western blotting as marked in red ovals. ACTB was used as loading control. pI, isoelectric point; MW, molecular weight.
Increased colitis in atg4b−/− mice depends on both hematopoietic and intestinal tract homeostasis
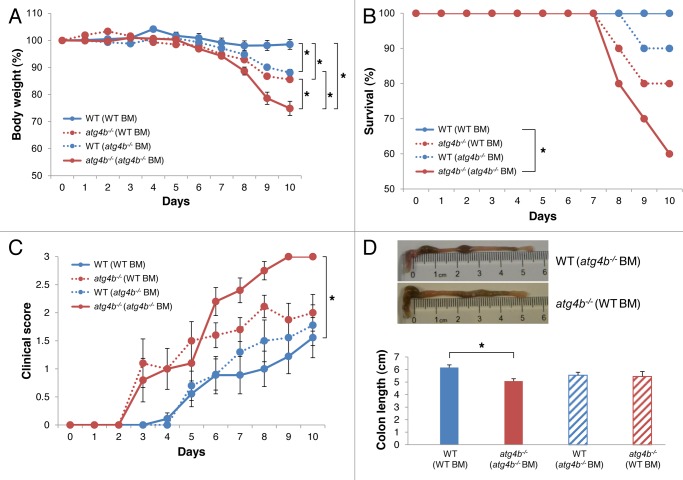
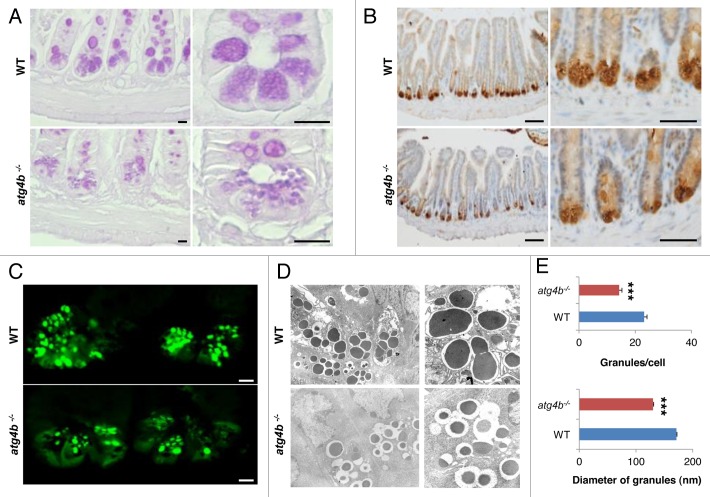
To determine the precise contribution of hematopoietic and nonhematopoietic cells to the exacerbated experimental colitis of atg4b−/− mice, we generated bone marrow chimeric mice and challenged them with DSS at 8 weeks post-transplantation. Atg4b-deficient mice transplanted with WT bone marrow developed significantly less weight loss and milder clinical alterations compared with atg4b−/− mice transplanted with atg4b−/− bone marrow cells (Fig. 5A, C and D). Moreover, we observed an improvement in the survival rate of atg4b−/− mice transplanted with bone marrow from WT animals (Fig. 5B). However, atg4b−/− (WT BM) chimeric mice were still more susceptible to colitis compared with WT mice transplanted with WT bone marrow (Fig. 5C). Interestingly, WT (atg4b−/− BM) chimeric mice were also more susceptible to develop colitis compared with WT (WT BM) chimeric mice. These findings suggest that the atg4b−/− phenotype is partially associated with a deficiency in hematopoietic cells, as WT bone marrow transplantation is not sufficient to fully protect atg4b−/− mice from DSS-induced colitis. Thus, nonhematopoietic cells such as intestinal epithelial or stromal cells could be involved in colitis development in atg4b−/− mice. Therefore, we explored the possibility that Atg4b deletion could lead to defects in intestinal structure. Comparative morphological analyses of intestinal tissue between atg4b−/− and WT mice disclosed substantial abnormalities in the ileum from atg4b−/− mice, including changes in both number and size of secretory granules present in Paneth cells. These cells exhibited a scarce granule number and small granules in atg4b−/− mice, whereas most Paneth cells from WT controls were filled with granules of larger size (Fig. 6A). Additionally, atg4b−/− mice exhibited decreased lysozyme immunostaining compared with WT controls (Fig. 6B and C). Transmission electron microscopy revealed that Paneth cell granules from atg4b−/− mice were usually smaller and composed of an electron-dense core and an electron-lucent peripheral halo that was enlarged compared with WT mice (Fig. 6D and E). Ultrastructural analyses confirmed a reduction in the number of granules in atg4b−/− mice compared with control mice (Fig. 6E). Thus, Atg4b deficiency is associated with a series of Paneth cell alterations that are similar to those observed in Atg16l1HM, Atg5flox/flox-villin Cre and Atg7flox/flox-villin Cre mice.45
Figure 5. Attenuated response to DSS-induced colitis in atg4b−/− (WT) bone marrow chimeric mice. Four groups of bone marrow chimeric mice were used: WT (WT), atg4b−/− (atg4b−/−), WT (atg4b−/−) and atg4b−/− (WT). Chimeric mice were treated with 2.5% DSS in drinking water for 7 d followed by regular drinking water as indicated in Materials and Methods. (A) Percent body weight and (B) survival after DSS administration. (C) Clinical score data (see Materials and Methods). (D) Colon length and representative photographs of colon and cecum from WT (atg4b−/−) (top) and atg4b−/− (WT) (bottom) chimeric mice after DSS-induced colitis. The results shown are means ± SEM, n = 10 of each chimeric mice. Statistical significance was determined on last day of the experiment by one-way analysis of variance (ANOVA; A, C and D), and survival differences between groups were analyzed by Log-rank test (B) (*p < 0.05).
Figure 6.Atg4b deficiency leads to Paneth cell abnormalities. (A) Ileum sections from WT and atg4b−/− mice were stained with PAS to analyze Paneth cell granules by light microscopy. Scale bars: 25 μm. (B) Representative images of anti-lysozyme immunohistochemistry and (C) immunofluorescence in WT and atg4b−/− ileum sections. Scale bars: 100 μm (B) and 50 μm (C). (D) Electron microphotographs of WT and atg4b−/− Paneth cells. Original magnifications: 2500x (left) and 5000× (right). Images are representative of three WT and three atg4b−/− mice. (E) Quantitative analysis of the secretory granules in Paneth cells, revealing a reduction in the number (top) and size (bottom) of these granules. n = 15 of each mouse genotype. The results shown are means ± SEM. Statistical significance was determined by Student’s t-test (***p < 0.005).
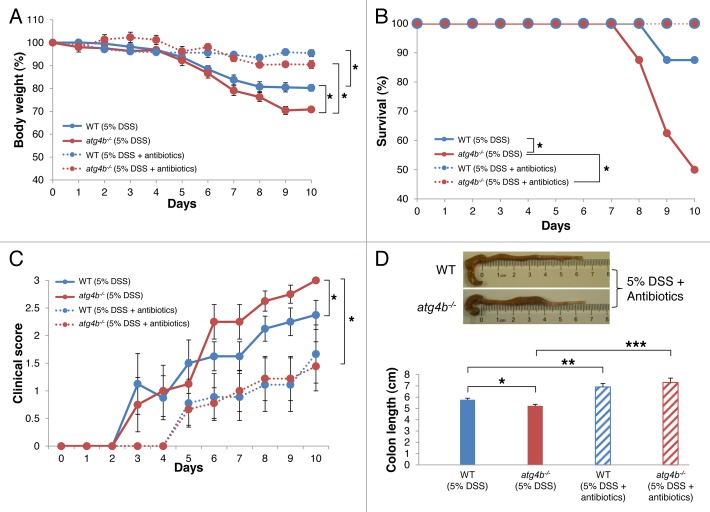
Paneth cells are essential for the maintenance of an adequate population of commensal flora, limiting bacterial penetration of host tissues due to their ability of secreting lysozyme and a variety of antimicrobial peptides.46 Alterations in the commensal microflora, including an increase of bacterial concentration or shifts in bacterial communities, have been described in IBD patients and in experimental models of colitis.47,48 Hypothetically, a deregulation of commensal microflora upon DSS-induced stress, due to defective Paneth cell function, might contribute to the exacerbated intestinal inflammation of atg4b−/− mice. In line with this possibility, antibiotic treatment significantly improved the survival, weight loss and clinical score of atg4b−/− mice (Fig. 7), abolishing the difference in severity of DSS-induced colitis between atg4b−/− and WT mice.
Figure 7. Antibiotic treatment abrogated the susceptibility of atg4b−/− mice to DSS-induced colitis. WT and atg4b−/− mice were treated with antibiotics in drinking water one week before and during the administration of 5% DSS as indicated in Materials and Methods. (A) Percent body weight of WT and atg4b−/− mice and (B) survival after DSS alone or in combination with antibiotic cocktail. (C) Clinical score data (see Materials and Methods). (D) Colon length and representative photographs of colon and cecum from WT (top) and atg4b−/− (bottom) mice after antibiotic treatment during DSS-induced colitis. The results shown are means ± SEM, n = 12 of each mouse genotype. Statistical significance was determined on last day of the experiment by one-way analysis of variance (ANOVA; A, C and D), and survival differences between groups were analyzed by Log-rank test (B) (*p < 0.05; **p < 0.01; ***p < 0.05).
Altogether, these results provide additional evidence for the importance of autophagy in intestinal pathologies and describe ATG4B as a novel protective protein against inflammatory colitis. Finally, we propose that mice deficient in this protease are a suitable model for in vivo studies aimed at testing new therapeutic strategies for intestinal diseases associated with autophagy deficiency.
Discussion
The first hint in favor of a role of autophagy in IBDs has come from the identification of the autophagy-related genes ATG16L1 and IRGM as Crohn disease susceptibility genes.6,7,49 Several mouse models with diminished or null expression of autophagy proteins have illustrated how autophagy defects may lead to intestinal abnormalities and contribute to the development of intestinal inflammation.10,11 The present study has uncovered autophagin-1 (ATG4B) as a novel protein that regulates intestinal homeostasis and plays a crucial role in the control of the inflammatory response occurring during experimental colitis. In this pathological condition, ATG4B protein levels increase paralleling the induction of autophagy in DSS-treated mice. Interestingly, damaged areas of the colon from IBD patients presented a significant decrease in ATG4B expression, as compared with adjacent healthy tissue. Upon DSS treatment, atg4b-deficient mice, which exhibit a general defect in autophagy, developed a more pronounced, often lethal colitis, which was associated with all macroscopic and histopathological hallmarks of IBD, as well as a marked increase of the secretion of two major pro-inflammatory cytokines, IL1A and IL6. These observations agree with previous data showing an upregulation of the expression of IL1A and IL6 in Crohn disease and ulcerative colitis.50,51 The increased susceptibility of atg4b-deficient mice to develop colitis and altered IL1A and IL6 cytokine production seems to be quite unique when compared with other autophagy-deficient models. In fact, Atg16l1 hypomorphic mice only show a clear colitis phenotype when infected with a certain virus strain.52 Moreover, intestinal epithelial deletion of Atg7 in mouse causes alterations in Paneth cells but it does not increase colitis susceptibility in these animals, indicating that this autophagy protein is dispensable for gut immune homeostasis.53 Altogether, these results point to a protective role for ATG4B in the regulation of the immune response in experimental murine colitis.
To explore the role of ATG4B in the production of inflammatory cytokines, we stimulated peritoneal macrophages with LPS. atg4b-deficient macrophages secreted high levels of CXCL10, CSF2, IL1A and IL6 compared with WT macrophages after LPS treatment, although differences did not reach statistical significance.54 Previously, a high IL1B/IL1β production in Atg16l1- and Atg7-deficient macrophages after endotoxin stimulation has been reported. However, we did not find differences in IL1B protein levels in our mutant mice. Remarkably, Il1b mRNA synthesis was not different in Atg16l1-deficient macrophages compared with WT macrophages, indicating that IL1B production could be regulated by autophagy at the post-transcriptional level. Similarly, we did not find IL1B differences at gene expression level in ileum or colon tissue from control or DSS-treated WT and mutant mice. Therefore, it seems that Atg4b deficiency is not playing an essential role in IL1B production when compared with Atg16l1. Other putative difference between Atg16l1- and Atg4b-deficient mice is that Atg16l1-deficiency causes a total impairment in autophagy, whereas Atg4b deletion leads to severe autophagy impairment but not to its total disruption.11
To identify the molecular mechanisms underlying the increased colitis susceptibility of atg4b−/− mice, we performed a series of microarray- and proteomic-based studies, leading to the identification of several potentially relevant biomarkers. Gene expression profile in the inflamed intestine of atg4b−/− mice was characterized by the upregulation of genes indicative of an active inflammatory response including Gbp1, Chi3l1, Steap4, Casp12 and Cd177. Most of these genes have been found overexpressed in models of experimental colitis or in patients with Crohn disease and ulcerative colitis.35,36,38 Similarly, the proteins encoded by these genes have been proposed to participate in the antibacterial immune response or to serve as biomarkers of chronic inflammation.37,40,42,55 Parallel proteomic studies allowed us to identify three differentially regulated proteins. GBP1 and HSPB1 were upregulated in the colon from DSS-treated atg4b−/− mice, consistently with the gene expression data. Interestingly, the expression of GBP1 has been found upregulated in colonic epithelia of individuals with IBD, whereas HSPB1 is overexpressed in colon cancer cells.36 Additionally, S100A9 was also overexpressed in the atg4b−/− inflamed colon. S100A9 plays an important role in innate immune system and its presence at high levels is a hallmark of numerous diseases associated with inflammation, including IBD.44 Collectively, these data suggest that Atg4b deficiency results in a severe colitis phenotype associated with altered regulation of key inflammatory players.
Interestingly, atg4b−/− mice transplanted with bone marrow from WT animals exhibited an improved survival rate after DSS-treatment. However, these chimeric mice were still more susceptible to colitis than control animals indicating that nonhematopoietic cells, such as intestinal epithelial or stromal cells, are also involved in the increased susceptibility of atg4b−/− mice to experimental colitis. In fact, atg4b−/− mice display abnormalities in a specialized intestinal epithelial cell type, the Paneth cells, indicating that ATG4B is required for the normal homeostasis of intestinal tissue. Paneth cells are located at small intestinal crypts and regulate innate immune functions through their ability to produce lysozyme and a broad range of antimicrobial peptides, which are stored in granules for secretion into the crypt lumen.56,57 We found that, similar to other autophagy-deficient models,10 atg4b−/− Paneth cells possess fewer and smaller secretory granules than WT control cells. It is likely that the observed defects in Paneth cells could lead to increased bacterial penetration of intestinal barrier, especially after challenge with DSS. This could explain the incomplete recovery of chimeric atg4b−/− mice transplanted with WT bone marrow. Taken together, these observations indicate that the increased colitis susceptibility of atg4b−/− mice is linked to a defective innate immune response together with an exacerbated inflammatory response to bacterial agents. Additional studies to explore how autophagy regulates granule production and exocytosis pathway in Paneth cells will be important to understand the mechanisms of IBD pathogenesis. Finally, the fact that atg4b−/− mice share many histopathological features and molecular alterations with patients from ulcerative colitis and Crohn disease, strongly supports their utility as an in vivo model to further explore the role of autophagy in intestinal inflammatory diseases, enabling the possibility of developing novel pharmacological strategies for IBD treatment.
Materials and Methods
Animals
The generation of atg4b−/− mice has been previously described.19 In all experiments, homozygous atg4b−/− mice and their corresponding wild-type (WT) littermates were derived from interbreeding of heterozygous C57Bl6/129Sv mice and their genotypes were determined by Southern blot analysis of tail DNA. Mice were bred under specific pathogen-free conditions. All experiments were performed with 8- to 12-week-old mice and were approved by the Committee on Animal Experimentation of Universidad de Oviedo (Oviedo, Spain).
Colitis induction
To induce experimental colitis, WT and atg4b−/− mice were treated with 5% (wt/vol) dextran sodium sulfate (DSS, 36,000 to 50,000 Mw; MP Biomedical, 0216011001) in drinking water ad libitum for 7 d, followed by normal water for 3 d. Control animals were given water only. Weight was monitored daily and rectal bleeding was measured and scored as previously reported.58 After treatment, blood was collected by submandibular bleeding and mice were sacrificed by CO2 inhalation. The colon was removed and its length was measured. To evaluate the effect of antimicrobial agents on experimental colitis, WT and atg4b−/− mice were treated with ampicillin (1 g/L; Sigma A9518-5G), vancomycin (0.5 g/L; Sigma 861987-1G), neomycin sulfate (1 g/L; Sigma N1876-25G) and metronidazole (1 g/L; Sigma M3761-5G) in drinking water for a week. After that, 5% DSS was added to the antibiotic-containing water for 7 d followed by 3 d of antibiotic-containing water. Weight was monitored daily.
Bone marrow transplantation
Bone marrow transplantation experiments were performed as described previously.59 Briefly, to induce bone marrow ablation female WT and atg4b−/− mice were treated with 25 mg/Kg of busulfan for 4 consecutive days, followed by injection of 200 mg/Kg of cyclophosphamide on day 5. Twenty-four hours later, male WT and atg4b−/− donor mice were killed by cervical dislocation and both femurs were removed aseptically. Bone marrow was flushed using PBS and single cell suspension was prepared by gently passing through a 14G needle and 5 x 105 cells were injected in recipient animals through the jugular vein. Eight weeks after transplantation, WT and atg4b−/− mice were treated with 2.5% DSS in drinking water ad libitum for 7 d followed by normal water for 3 d.
Histology and immunohistochemistry
The colon was washed free of stools, divided into proximal and distal portions, fixed in 4% paraformaldehyde and embedded in paraffin. Each sample was serially sectioned 5-μm thick at 100-μm intervals and stained separately with hematoxylin and eosin, periodic acid Schiff (PAS) or Alcian Blue. Colon tissue sections were analyzed for epithelial damage, inflammatory infiltration, crypt loss and ulcerations. The histological score represents the sum of the epithelial damage and inflammatory infiltration scores, and was evaluated as follows, modified from.60 Epithelial damage: 0, normal morphology; 1, loss of goblet cells; 2, loss of goblet cells in large areas; 3, loss of crypts; 4, loss of crypts in large areas; Inflammatory infiltration: 0, no infiltrate; 1, infiltrate around crypt basis; 2, infiltrate reaching to lamina muscularis mucosae; 3, extensive infiltration reaching the lamina muscularis mucosae and thickening of the mucosa with abundant edema; 4, infiltration of the lamina submucosa. To perform immunohistochemistry studies, deparaffined and rehydrated sections were first extensively rinsed in PBS (pH 7.5). Then, endogenous peroxidase activity and nonspecific binding were blocked with peroxidase block buffer (DakoCytomation) and 1% bovine serum albumin, respectively. Sections were incubated overnight at 4°C with a monoclonal antibody anti-lysozyme (Thermo Scientific, PA1-40308) and anti-ATG4B (Sigma, A2981). ATG4B signal was analyzed using the ImageJ software program (version 1.46k). Human tissues were obtained as formalin-fixed, paraffin-embedded tissue sections from the archives of the tumor bank of Hospital Universitario Central de Asturias. They remained anonymous according to guidelines approved by the Hospital’s Research Ethics Board.
Immunoblotting
Colon tissue was immediately frozen in liquid nitrogen after extraction and homogenized in a 20 mM Tris buffer pH 7.4, containing 150 mM NaCl, 1% Triton X-100, 10 mM EDTA and Complete® protease inhibitor cocktail (Roche Applied Science, 05 892 791 001). Then, tissue extracts were centrifuged at 12.000 rpm at 4°C and supernatants were collected. Protein concentration was quantified by bicinchoninic acid technique (BCA protein assay kit, Pierce Biotechnology, 23225). A total of 25 µg of protein sample was loaded on either 8% or 13% SDS-polyacrylamide gels. After electrophoresis, gels were electrotransferred onto polyvinylidene difluoride (PVDF) membranes (Millipore), and then membranes were blocked with 5% nonfat dried milk in PBT (phosphate buffered saline with 0.05% Tween 20) and incubated overnight at 4°C with the following primary antibodies diluted in 3% nonfat dried milk in PBT: anti-ATG4B (Sigma, A2981), anti-SQSTM1/p62 (Abnova, H00008878-M01), anti-LC3 (nanoTools, 0231-100/LC3-5F10), anti-GBP1 (Santa Cruz, sc-10586), anti-HSPB1 (ProteinTech, 18284-1-AP), anti-S100A9 (R&D Systems, MAB2065) and anti-ACTB/β actin (Sigma, A2228). After three washes with PBT, filters were incubated with the corresponding secondary antibody at 1:10.000 dilution in 1.5% milk in PBT, and developed with Immobilon Western Chemiluminescent HRP substrate (Millipore, P36599A).
Multiplex immunoassay
To determine the concentration of mouse CSF2/GMCSF, IFNG/IFNγ, IL1A, IL2, IL4, IL5, IL6, IL10, IL17 and TNF in serum and colon tissue extracts from DSS-treated WT and atg4b−/− mice, an analyte detection system by flow cytometry was used (Th1/Th2 10plex Mouse FlowCytomix, Bender MedSystems, BMS820FF), according to manufacturer's instructions on a Luminex100 LabMap System (Luminex).
Cytokine secretion analysis
Peritoneal macrophages were isolated from WT and atg4b−/− mice 4 d after intraperitoneal injection of 4% thioglycollate broth as described previously.61 Murine macrophages were precultured in DMEM medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 1% penicillin-streptomycin for one day. Adherent macrophages were gently washed with and cultured in DMEM medium without serum 2 h before stimulation with endotoxin (lipopolysaccharide, LPS, 100 ng/mL). Supernatants were collected 16 h after stimulation and levels of several cytokines in the culture medium were determined by Mouse Cytokine Antibody Array Panel A (R&D Systems, ARY006) according to manufacturer's instructions.
RNA isolation and microarray hybridization
Total RNA of ileum and colon from control and DSS-treated WT and atg4b−/− mice was isolated using TRIzol reagent (Invitrogen Life Sciences, 15596-18) following the manufacturer's instructions. Additional cleanup of total RNA was performed using the RNeasy Mini kit (Qiagen). Purity of samples and efficiency of extractions were verified by spectrophotometry (NanoDrop) and using an Agilent 2100 Bioanalyzer.
Double-stranded cDNA synthesis and in vitro transcription to generate cRNA was performed using a commercial kit of Affymetrix (Sense Target Labeling kit). The biotin-labeled cRNA was purified, fragmented and hybridized to a Affymetrix GeneChip® Mouse Gene 1.0 ST Arrays according to the manufacturer's protocol. Washing and scanning were performed with a Fluidics Station 450 and GeneChip Scanner 7G (Affymetrix), and quality control of microarray data was performed using Affymetrix Expression Console™ following standard Affymetrix Exon Array protocols. After scanning, raw data were processed with the RMAExpress program (http://RMAExpress.bmbolstad.com), using default settings. The normalized array data were log2-transformed and averaged. A list of differentially expressed genes was generated for each comparison between genotypes and between control and DSS-treated mice. Array data were clustered using Cluster 3.0 software and the heat maps were created using TreeView 1.1.5 software. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus62 and are accessible through GEO Series accession number GSE 36056 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36056).
2-D DiGE analysis
Colon tissue from WT and atg4b−/− mice was homogenized in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM TRIS-HCl pH 8.5). A total of 50 μg of protein of each sample were covalently labeled with 400 pmol of a specific fluorophore (CyDye, GE Healthcare, 25-8010-65): Cy3 (WT), Cy5 (atg4b−/−) and Cy2 (pooled 1:1 WT and atg4b−/−). Labeled proteins were mixed and set to 450 μL with rehydration buffer (8 M urea, 4% CHAPS, 13 mM DTT, 1% IEF buffer). Samples were applied to IPG strips (24 cm, nonlinear pH gradient 3-11; GE Healthcare, 17-6003-74), and isoelectric focusing (IEF) was performed on an IPGphor Unit (GE Healthcare) for 26 h at a gradient voltage in the dark at 18°C. Once the IEF step was finished, strips were equilibrated for 15 min in SES-DTT buffer (6 M urea, 30% glycerol, 2% SDS, 75 mM TRIS-HCl pH 6.8, 0.5% DTT and bromophenol blue), and for another 15 min in SES-IA (6 M urea, 30% glycerol, 2% SDS, 75 mM TRIS-HCl pH 6.8, 4.5% iodoacetamide and bromophenol blue). Proteins were then resolved in 13% SDS-PAGE gels using a Hoefer S600 instrument (Ettan DALTsix, GE Healthcare). After electrophoresis, cyanine dye-labeled proteins were visualized directly by scanning using a Typhoon™ 9400 imager (GE Healthcare). Cy2 images were scanned using a 488 nm laser and an emission filter of 520 nm band pass (BP) 40. Cy3 images were scanned using a 532 nm laser and a 580 nm BP30 emission filter. Cy5 images were scanned using a 633 nm laser and a 670 nm BP30 emission filter. All gels were scanned at 200 µm resolution, analyzed with Progenesis SameSpots software and stained with SYPRO Ruby (Molecular Probes).
Tryptic digestion and MALDI-ToF analysis
Differential spots were manually excised over a transilluminator. Gel pieces were washed with 25 mM ammonium bicarbonate/acetonitrile (70:30), dried for 15 min at 90°C, and incubated with 12 ng/μL trypsin (Promega) in 25 mM ammonium bicarbonate. The digestion was allowed to proceed for 1 h at 60°C. Peptides were purified with ZipTip C18 (Millipore, ZTC18S960) and eluted with 1 μL of CHCA (α-cyano-4-hydroxycinnamic acid, Sigma, C2020) to be placed onto MALDI-ToF’s plate. Once dried, peptides were analyzed by mass spectrometry on a time-of-flight mass spectrometer equipped with a nitrogen laser source (Voyager-DE STR, Applied Biosystems). Data from 200 laser shots were collected to produce a mass spectrum. Data explorer version 4.0.0.0 software (Applied Biosystems) was used to analyze the spectra.
Statistics
All experimental data are reported as mean ± SEM. Statistical analyses of differences between two groups were performed by the two-tailed Student’s t-test, and survival differences between groups were analyzed by Log-rank test. In experiments with more than two groups, differences were analyzed by multifactorial one-way analysis of variance (ANOVA). The Prism program version 4.0 (Graph-Pad Software Inc.) was used for calculations and p values lower than 0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by grants from Ministerio de Economía y Competitividad-Spain and the European Union (FP7-Microenvimet). The Instituto Universitario de Oncología is supported by Obra Social Cajastur-Asturias and Acción Transversal del Cáncer-RTICC, Spain. S.C. was supported by a fellowship from CONACYT; C.L.-O. is an Investigator of the Botin Foundation.
Glossary
Abbreviations:
- ATG4B
autophagy-related 4B, cysteine peptidase
- IBD
inflammatory bowel disease
- DSS
dextran sodium sulfate
- LPS
lipopolysaccharide
- LC3
microtubule-associated protein 1 light chain 3
- GFP-LC3
green fluorescent protein-LC3
- BM
bone marrow
- WT
wild type
- ATG16L1
autophagy-related 16-like 1
- IRGM
immunity-related GTPase family M
- ULK1
unc-51-like kinase 1
- PE
phosphatidylethanolamine
- NOD2
nucleotide-binding oligomerization domain containing 2
- GABARAP
GABA(A) receptor-associated protein
- GABARAPL1
GABA(A) receptor-associated protein like 1
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- BECN1
Beclin 1, autophagy related
- SQSTM1
sequestosome 1
- IL1A
interleukin 1 alpha
- IL6
interleukin 6
- GBP1
guanylate binding protein 1, interferon-inducible
- HSPB1
heat shock protein beta 1
- S100A9
S100 calcium-binding protein A9
- DIGE
difference gel electrophoresis
- MALDI
matrix-assisted laser desorption/ionization
- ToF
time of flight
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24797
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24797
References
- 1.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 2.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asquith M, Powrie F. An innately dangerous balancing act: intestinal homeostasis, inflammation, and colitis-associated cancer. J Exp Med. 2010;207:1573–7. doi: 10.1084/jem.20101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–70. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- 6.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 7.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Wellcome Trust Case Control Consortium Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K, Rutgeerts P, et al. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm Bowel Dis. 2011;17:1392–7. doi: 10.1002/ibd.21486. [DOI] [PubMed] [Google Scholar]
- 9.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 10.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 12.Mariño G, López-Otín C. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61:1439–54. doi: 10.1007/s00018-004-4012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 16.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 18.Mariño G, Uría JA, Puente XS, Quesada V, Bordallo J, López-Otín C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 19.Mariño G, Fernández AF, Cabrera S, Lundberg YW, Cabanillas R, Rodríguez F, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Chen X, Ye QZ, Vogt A, Yin XM. A high-throughput FRET-based assay for determination of Atg4 activity. Autophagy. 2012;8:401–12. doi: 10.4161/auto.18777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 24.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamart C, Benoit N, Raymackers JM, Kim HJ, Kim CK, Francaux M. Autophagy-related and autophagy-regulatory genes are induced in human muscle after ultraendurance exercise. Eur J Appl Physiol. 2012;112:3173–7. doi: 10.1007/s00421-011-2287-3. [DOI] [PubMed] [Google Scholar]
- 27.Zheng YH, Tian C, Meng Y, Qin YW, Du YH, Du J, et al. Osteopontin stimulates autophagy via integrin/CD44 and p38 MAPK signaling pathways in vascular smooth muscle cells. J Cell Physiol. 2012;227:127–35. doi: 10.1002/jcp.22709. [DOI] [PubMed] [Google Scholar]
- 28.Cabrera S, Mariño G, Fernández AF, López-Otín C. Autophagy, proteases and the sense of balance. Autophagy. 2010;6:961–3. doi: 10.4161/auto.6.7.13065. [DOI] [PubMed] [Google Scholar]
- 29.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 30.Tanida I, Waguri S. Measurement of autophagy in cells and tissues. Methods Mol Biol. 2010;648:193–214. doi: 10.1007/978-1-60761-756-3_13. [DOI] [PubMed] [Google Scholar]
- 31.Scaldaferri F, Correale C, Gasbarrini A, Danese S. Mucosal biomarkers in inflammatory bowel disease: key pathogenic players or disease predictors? World J Gastroenterol. 2010;16:2616–25. doi: 10.3748/wjg.v16.i21.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulze HA, Häsler R, Mah N, Lu T, Nikolaus S, Costello CM, et al. From model cell line to in vivo gene expression: disease-related intestinal gene expression in IBD. Genes Immun. 2008;9:240–8. doi: 10.1038/gene.2008.11. [DOI] [PubMed] [Google Scholar]
- 33.O’Connor PM, Lapointe TK, Beck PL, Buret AG. Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1411–20. doi: 10.1002/ibd.21217. [DOI] [PubMed] [Google Scholar]
- 34.Lubeseder-Martellato C, Guenzi E, Jörg A, Töpolt K, Naschberger E, Kremmer E, et al. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am J Pathol. 2002;161:1749–59. doi: 10.1016/S0002-9440(10)64452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Buhr MF, Mähler M, Geffers R, Hansen W, Westendorf AM, Lauber J, et al. Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses. Physiol Genomics. 2006;25:426–34. doi: 10.1152/physiolgenomics.00022.2005. [DOI] [PubMed] [Google Scholar]
- 36.Schnoor M, Betanzos A, Weber DA, Parkos CA. Guanylate-binding protein-1 is expressed at tight junctions of intestinal epithelial cells in response to interferon-gamma and regulates barrier function through effects on apoptosis. Mucosal Immunol. 2009;2:33–42. doi: 10.1038/mi.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eurich K, Segawa M, Toei-Shimizu S, Mizoguchi E. Potential role of chitinase 3-like-1 in inflammation-associated carcinogenic changes of epithelial cells. World J Gastroenterol. 2009;15:5249–59. doi: 10.3748/wjg.15.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawada M, Chen CC, Arihiro A, Nagatani K, Watanabe T, Mizoguchi E. Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab Invest. 2008;88:883–95. doi: 10.1038/labinvest.2008.47. [DOI] [PubMed] [Google Scholar]
- 39.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–94. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–78. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Jerke U, Rolle S, Dittmar G, Bayat B, Santoso S, Sporbert A, et al. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J Biol Chem. 2011;286:7070–81. doi: 10.1074/jbc.M110.171256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sachs UJ, Andrei-Selmer CL, Maniar A, Weiss T, Paddock C, Orlova VV, et al. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31) J Biol Chem. 2007;282:23603–12. doi: 10.1074/jbc.M701120200. [DOI] [PubMed] [Google Scholar]
- 43.Arrigo AP. The cellular “networking” of mammalian Hsp27 and its functions in the control of protein folding, redox state and apoptosis. Adv Exp Med Biol. 2007;594:14–26. doi: 10.1007/978-0-387-39975-1_2. [DOI] [PubMed] [Google Scholar]
- 44.Foell D, Frosch M, Sorg C, Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clin Chim Acta. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 45.Cadwell K, Stappenbeck TS, Virgin HW. Role of autophagy and autophagy genes in inflammatory bowel disease. Curr Top Microbiol Immunol. 2009;335:141–67. doi: 10.1007/978-3-642-00302-8_7. [DOI] [PubMed] [Google Scholar]
- 46.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–63. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klapproth JM, Sasaki M. Bacterial induction of proinflammatory cytokines in inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:2173–9. doi: 10.1002/ibd.21332. [DOI] [PubMed] [Google Scholar]
- 48.Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–77. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- 49.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andus T, Daig R, Vogl D, Aschenbrenner E, Lock G, Hollerbach S, et al. Imbalance of the interleukin 1 system in colonic mucosa--association with intestinal inflammation and interleukin 1 receptor antagonist [corrected] genotype 2. Gut. 1997;41:651–7. doi: 10.1136/gut.41.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouguen G, Chevaux JB, Peyrin-Biroulet L. Recent advances in cytokines: therapeutic implications for inflammatory bowel diseases. World J Gastroenterol. 2011;17:547–56. doi: 10.3748/wjg.v17.i5.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittkopf N, Günther C, Martini E, Waldner M, Amann KU, Neurath MF, et al. Lack of intestinal epithelial atg7 affects paneth cell granule formation but does not compromise immune homeostasis in the gut. Clin Dev Immunol. 2012;2012:278059. doi: 10.1155/2012/278059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahman FZ, Smith AM, Hayee B, Marks DJ, Bloom SL, Segal AW. Delayed resolution of acute inflammation in ulcerative colitis is associated with elevated cytokine release downstream of TLR4. PLoS One. 2010;5:e9891. doi: 10.1371/journal.pone.0009891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stroncek DF. Neutrophil-specific antigen HNA-2a, NB1 glycoprotein, and CD177. Curr Opin Hematol. 2007;14:688–93. doi: 10.1097/MOH.0b013e3282efed9e. [DOI] [PubMed] [Google Scholar]
- 56.Shi J. Defensins and Paneth cells in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1284–92. doi: 10.1002/ibd.20197. [DOI] [PubMed] [Google Scholar]
- 57.Ouellette AJ. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010;26:547–53. doi: 10.1097/MOG.0b013e32833dccde. [DOI] [PubMed] [Google Scholar]
- 58.Wirtz S, Neurath MF. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev. 2007;59:1073–83. doi: 10.1016/j.addr.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 59.Gutiérrez-Fernández A, Inada M, Balbín M, Fueyo A, Pitiot AS, Astudillo A, et al. Increased inflammation delays wound healing in mice deficient in collagenase-2 (MMP-8) FASEB J. 2007;21:2580–91. doi: 10.1096/fj.06-7860com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–49. [PubMed] [Google Scholar]
- 61.Rendon-Mitchell B, Ochani M, Li J, Han J, Wang H, Yang H, et al. IFN-gamma induces high mobility group box 1 protein release partly through a TNF-dependent mechanism. J Immunol. 2003;170:3890–7. doi: 10.4049/jimmunol.170.7.3890. [DOI] [PubMed] [Google Scholar]
- 62.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.