Abstract
Autophagy-related 1 (Atg1)/Unc-51-like protein kinases (ULKs) are evolutionarily conserved proteins that play critical physiological roles in controlling autophagy, cell growth and neurodevelopment. RB1-inducible coiled-coil 1 (RB1CC1), also known as PTK2/FAK family-interacting protein of 200 kDa (FIP200) is a recently discovered binding partner of ULK1. Here we isolated the Drosophila RB1CC1/FIP200 homolog (Fip200/CG1347) and showed that it mediates Atg1-induced autophagy as a genetically downstream component in diverse physiological contexts. Fip200 loss-of-function mutants experienced severe mobility loss associated with neuronal autophagy defects and neurodegeneration. The Fip200 mutants were also devoid of both developmental and starvation-induced autophagy in salivary gland and fat body, while having no defects in axonal transport and projection in developing neurons. Interestingly, moderate downregulation of Fip200 accelerated both developmental growth and aging, accompanied by target of rapamycin (Tor) signaling upregulation. These results suggest that Fip200 is a critical downstream component of Atg1 and specifically mediates Atg1’s autophagy-, aging- and growth-regulating functions.
Keywords: autophagy, neurodegeneration, Drosophila, aging, growth
Introduction
A proper balance between cellular anabolism and catabolism is important for maintaining cell physiology and perpetuating life phenomena. Recently, autophagy has been highlighted as one of the critical catabolic mechanisms that degrade toxic protein aggregates, dysfunctional organelles,1 excessive nutrient deposits such as glycogen and lipid droplets2 and invading microorganisms.3 Defective regulation of autophagy can result in diverse pathological phenomena such as neurodegeneration,4 cancer,5 muscular dystrophy,6 cardiac malfunction,7 fat accumulation,8 developmental abnormalities9 and defective responses to starvation and infections.10,11
Autophagy is regulated by diverse signaling pathways to meet the needs of cells or organisms. A family of protein kinases named Atg1 (in yeast and Drosophila), UNC-51 (in worms) or ULK1/2/3/4 (in mammals) is shown to be an evolutionarily conserved component that is essential for autophagy initiation.12 Diverse cell growth-stimulating signals such as nutrients and growth factors suppress autophagy through target of rapamycin complex 1 (TORC1)-mediated inhibitory phosphorylation of ULK1.13 On the other hand, during energetic stress, autophagy is induced through AMP-activated protein kinase (AMPK)-mediated activatory phosphorylation of ULK1.14,15 Activation of ULK1 results in activation of class III PtdIns3K complex16 that subsequently transmits the signal to the core autophagy machinery composed of ATG3, ATG7, ATG10, etc. that conjugates ATG5 and MAP1LC3/LC3 (ortholog of yeast Atg8) into target lipids and proteins, via a mechanism analogous to the ubiquitin conjugating system. The conjugation of LC3 into target lipid membranes induces the formation of autophagosome that is destined to be fused with lysosome and become ultimately degraded.17
Upon activation, ULK1 suppresses cell growth and protein synthesis by inhibiting TORC1 while promoting autophagic degradation.18,19 Reciprocal inhibition between ULK1 and TORC1 is suggested to be an interlocked feedback loop that serves as a switch between growth arrest and proliferation. Drosophila Atg1 mutation indeed abolishes autophagic activities20 and promotes cell growth.21 Interestingly, in addition to problems caused in autophagy and cell growth regulation, Atg1 mutation precipitates various neurodevelopmental defects, such as failures in axonal transport and neuronal wiring,22-24 which could ultimately result in lethality.20,21 Other autophagy mutants, such as Atg8- or Atg7-null mutants, do not display neurodevelopmental defects or lethality,9,11,25 suggesting that Atg1 plays autophagy-independent developmental functions. The role of Atg1 in regulating axonal transport, growth and projection is also well established in mammals.26-29
A proteomic search for ULK1-interacting proteins isolated RB1CC1/FIP200 as an ULK1 binding partner that is critical for the formation of autophagosomes.30 RB1CC1 was originally isolated as a PTK2/FAK-interacting protein31,32 and as a tumor suppressor RB1-inducing protein.33 However, RB1CC1 interacts with many other proteins as well, such as TP53,34 TSC1,35 TRAF2 and MAP3K5/ASK1.36 Most importantly, RB1CC1 forms a complex with ATG1 and ATG13 and is essential for the autophagy regulation.30,37-41 fip200-null mutant mice are lethal at embryonic stage due to heart failure and liver degeneration,36 a phenomenon that is distinct from other autophagy-defective mouse strains that are usually born at a Mendelian ratio like atg5- or atg7-knockout mice.42,43 On the other hand, neuron-specific fip200 mutant mice show motor deficits and progressive neurodegeneration44 similar to neuron-specific atg5- or atg7-knockout mice.4,45 RB1CC1 is also important for fetal hematopoiesis46 in which elimination of mitochondria through ULK1-dependent autophagy is critical.47 However, because there are multiple orthologs of Atg1 in mammalian genome (ULK1/2/3/4) that can compensate for each other, it was far from possible to evaluate the physiological contribution of RB1CC1 to ULK/Atg1 function, for which direct comparison between ULK/Atg1 and RB1CC1 mutant phenotypes in a single model organism was needed.
Here we have identified a single RB1CC1 homolog in the Drosophila melanogaster genome (Drosophila Fip200), which was previously known as CG1347. By biochemically and genetically analyzing Fip200 loss-of-function mutants, we show that Fip200 is an important mediator of Atg1 in controlling autophagy and cell growth, while it is largely dispensable for Atg1- or Fak-dependent neuronal morphogenesis and axonal transport. The Fip200-dependent control of autophagy is important for maintaining protein homeostasis, attenuating age-associated accumulation of ubiquitinated protein and preventing neuronal cell death in adult brain. In addition, although all Fip200-null mutants are developmentally lethal, viable Fip200 hypomorphs show acceleration in both developmental growth and aging, further highlighting the antagonistic physiological relationship between autophagy (Atg1) and cell growth (TORC1)-controlling signaling complexes.
Results
The identification of Drosophila Fip200
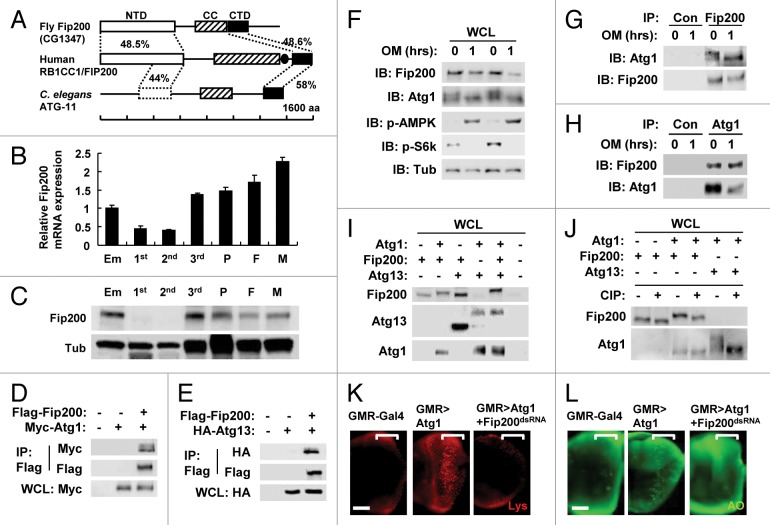
A BLAST search against the Drosophila melanogaster genome identified a protein (annotated as CG1347) as the only homolog of the mammalian RB1CC1 protein, which was named as Drosophila Fip200 (hereafter, Fip200). Strong primary sequence homology (approximately 50% similarity) was detected at both terminal regions of the protein. We subsequently designated the two homologous regions as N-terminal and C-terminal domain (NTD and CTD in Fig. 1A), respectively. However, the central region, which contains coiled-coil and leucine zipper motifs, does not show significant homology between the two proteins. The single RB1CC1/FIP200 homolog in C. elegans, which was previously designated as ATG-11,48 also showed significant homology in both the NTD and CTD (Fig. 1A), suggesting that these domains are evolutionarily conserved. Interestingly, although sequence homology was not apparent in the central region, bioinformatic analyses through COILS algorithm49 demonstrated that both the Drosophila and C. elegans Fip200 homologs contain strong coiled-coil signature motifs around the central region, suggesting that this coiled-coil (CC) domain is also functionally conserved. Therefore, we could infer that most of the primary sequence features in mammalian RB1CC1 are conserved in Fip200. We also analyzed the expression of Fip200 during Drosophila development. Abundant expression of Fip200 mRNA (Fig. 1B) and Fip200 protein (Fig. 1C) was detected in embryos, wandering third-instar larvae, pupae and adults, but the expression decreased significantly in feeding-stage larvae including first and second instar larvae. This pattern shows that Fip200 expression was at its lowest level when the rate of organism growth was at the highest, implicating the inverse correlation between Fip200 activity and developmental growth.
Figure 1. Conservation of RB1CC1/FIP200 in Drosophila. (A) Schematic representation of the comparison among Drosophila, human and C. elegans RB1CC1 homologs. Amino acid sequence similarity is displayed as a percentage. NTD, N-terminal domain; CC, coiled-coil domain; CTD, C-terminal domain; aa, amino acids. Leucine zipper domain in human RB1CC1 is indicated as a closed circle. NTD is only partially conserved in C. elegans ATG-11. (B and C) Expression of Fip200 at different developmental stages of Drosophila. Em, embryo; 1st, first instar larva; 2nd, second instar larva; 3rd, wandering-stage third instar larva; P, pupa; F, adult female; M, adult male. Quantitative reverse transcriptase-real time polymerase chain reaction was performed, and Fip200 mRNA expression was normalized with ribosomal protein 49 expression (B), n = 3. Quantification data are represented as means ± standard error. Immunoblot analysis was performed to reveal Fip200 and tubulin (Tub) expression (C). (D–J) Physical interaction among Atg1, Fip200 and Atg13. (D, E, I and J) After transfection with the indicated constructs, Drosophila Kc cells were incubated for 48 h. Whole cell lysates (WCL) were prepared and subsequently subjected to immunoprecipitation (IP) with or without (Con) the indicated antibodies. WCL and IP samples were analyzed by immunoblotting with the indicated antibodies. (J) WCL was incubated with or without calf intestine phosphatase (CIP) at 30°C for 3 h. (F–H) Kc cells were treated with 5 μM oligomycin (OM) for 1 h as indicated and were subjected to WCL-IP sample preparation and immunoblotting (IB) with the indicated antibodies. (K and L) Eye imaginal discs from wandering-stage third instar larvae of the indicated genotypes were subjected to LysoTracker Red (Lys, K) and acridine orange (AO, L) staining. Brackets indicate the area where GMR-Gal4 and transgenes were expressed. Scale bar: 50 μm. Approximate molecular weights (observed/predicted): Fip200 (150 to 200/152 kD), Atg1 (120 to 150/92 kD), Atg13 (60 to 70/57 kD), SNF1A/AMPK (70/65 kD), S6k (60/55 kD), Tub (50/52 kD). Endogenous and overexpressed protein bands exhibited similar size.
Fip200 interacts with Atg1 and Atg13
We first tested if Fip200 could physically interact with Drosophila Atg1 and Atg13, in a similar manner as mammalian RB1CC1 interacting with ULK1 and ATG13.30,37-39 In Drosophila Kc cells, we were able to detect physical interaction between overexpressed Fip200 and Atg1 (Fig. 1D) as well as between Fip200 and Atg13 (Fig. 1E) through co-immunoprecipitation (co-IP) experiments. The interaction between Fip200 and Atg1 was also observed at the endogenous level, both under normal conditions and during energetic stress induced by oligomycin, a mitochondrial ATP synthase inhibitor (Fig. 1F–H). As previously reported,50 oligomycin-induced energetic stress provoked Lkb1-dependent activating phosphorylation of SNF1A/AMPK and suppressed TORC1-dependent activatory phosphorylation of S6k and inhibitory phosphorylation of Atg1 (monitored by phospho-specific antibodies or phosphorylation-induced gel shift, Fig. 1F). Interestingly, we found that cotransfection of Atg1 and Fip200 induced a slight shift of the Fip200 band, which was further enhanced by additional cotransfection of Atg13 (Fig. 1I) and diminished by calf intestinal phosphatase treatment (Fig. 1J). This Fip200 shift by Atg1 may reflect Atg1-induced Fip200 phosphorylation,38 but the shift of endogenous Fip200 was not evident (Fig. 1F–H). On the other hand, although Atg13 induced phosphorylation-mediated shift of Atg1 as previously reported,51 Fip200 did not promote this phosphorylation (Fig. 1I and J), suggesting that Fip200 does not regulate Atg1 phosphorylation.
Fip200 is required for Atg1-induced autophagy
Since Atg1 is known to induce autophagy in Drosophila when overexpressed,9,20 we tested the role of Fip200 in Atg1-dependent induction of autophagy. As previously reported, Atg1 overexpression in Drosophila eye imaginal disc through glass multiple reporter (GMR)-Gal4 driver induced prominent accumulation of acidic vesicles in most of the cells posterior to the morphogenetic furrow where GMR-Gal4 was expressed [Fig. 1K, monitored by LysoTracker Red (Lys) staining]. This tendency is well associated with ectopic cell death in the area [Fig. 1L, monitored by acridine orange (AO) staining], as expected from the known association between Atg1 and autophagic cell death.20 However, silencing of Fip200 through Fip200-dsRNA TRiP construct52 almost completely prohibited the induction of both autophagic activity and autophagy-mediated cell death (Fig. 1K and L; Fig. S1). These results suggest that endogenous Fip200 is critical for the autophagy-controlling function of Atg1 in Drosophila.
Generation of Fip200 loss-of-function mutants
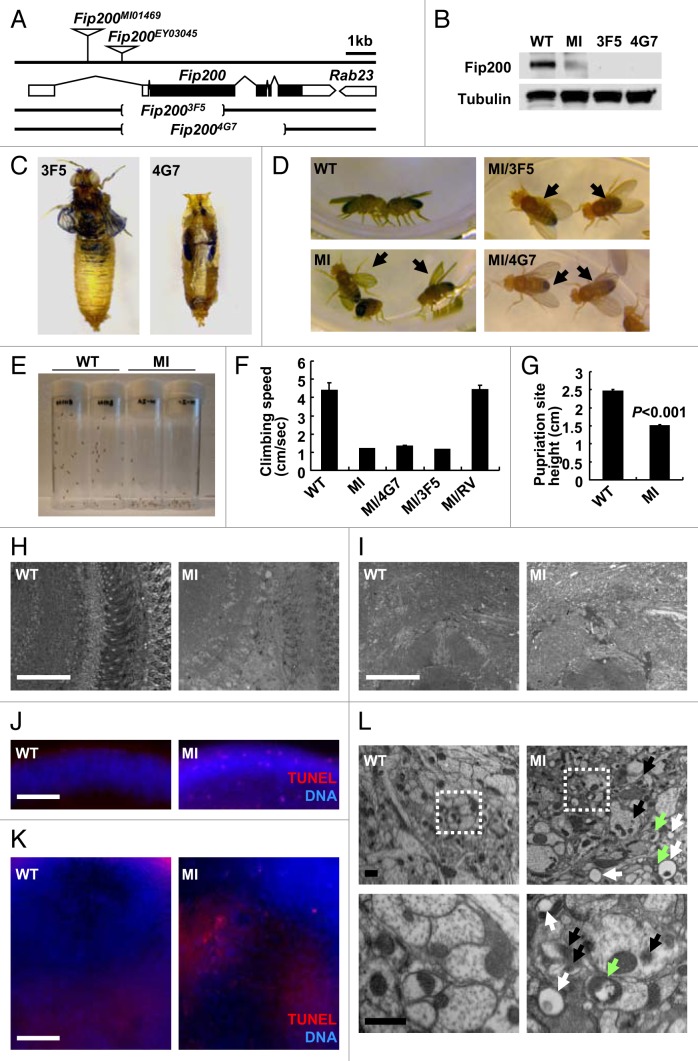
To further study the physiological function of the Fip200 gene, we isolated Fip200 loss-of-function mutants. Flybase search identified two independent transposon insertions in the first intron of the Fip200 gene (Fig. 2A). The Fip200MI01469 (designated as MI) allele was generated by an insertion of a genetically engineered Minos transposon called MIMIC element that contains an enhanced green fluorescent protein (eGFP) open reading frame preceded by a strong splice acceptor sequence and followed by a transcription terminator sequence.53 Therefore, it was expected that the expression of Fip200 mRNA and protein would be suppressed in MI homozygotes. Indeed, Fip200 protein expression was strongly reduced in MI homozygotes, suggesting that MI is a strong hypomorphic allele (Fig. 2B). By removing the MIMIC transgenic cassette in MI allele through phi31C-mediated recombination,53 we obtained a revertant of the MI mutation (designated as RV) as well. We also attempted to generate null mutants of Fip200 gene through imprecise excision of Fip200EY03045 (designated as EY) P-element insertion. Through a large-scale screening, we were able to isolate two deletion mutants, designated as 3F5 and 4G7 alleles, each of which has a deletion in more than 50% of the coding sequence and translation initiation site of the Fip200 gene (Fig. 2A). No Fip200 protein was detected in 3F5 and 4G7 homozygotic flies (Fig. 2B). Mutant alleles of the Fip200 gene used in this study are summarized in Table S1.
Figure 2. Phenotypes of Drosophila Fip200 mutants. (A) Schematic genomic organization of the Fip200 (CG1347) locus and Fip200 mutants. Brackets indicate the genomic deficiency in Fip200-null alleles. Triangles indicate transposon insertions. Open boxes: untranslated exons; closed boxes: protein-coding exons; size bar: relative length of 1 kb genomic span. (B) Absence of Fip200 protein in Fip200 mutant flies. Protein samples from wandering third-instar larvae of wild type (WT; Fip200+/+), MI (Fip200MI/MI), 3F5 (Fip2003F5/3F5) and 4G7 (Fip2004G7/4G7) were gel separated and immunoblotted with anti-Fip200 and anti-tubulin antibodies. (C) Pupal (pharate adult) lethality of Fip200-null mutants. Most of the 3F5 and 4G7 mutant flies die before or during eclosion. (D) Wing posture defects of Fip200 hypomorphs. WT, MI, MI/3F5 (Fip200MI/3F5) and MI/4G7 (Fip200MI/4G7) adult male flies. Complete phenotypic description of each mutant strain is summarized in Table S1 and S2. (E–G) Mobility defects of Fip200 hypomorphs. (E) A photograph of vials containing 2-week-old WT and MI flies taken 3 sec after negative geotaxis induction. (F) Quantification of climbing speed of the indicated 2-week-old adult male flies (n ≥ 10). Complete statistical comparison between each strain is summarized in Table S3. (G) Quantification of pupariation site height of the indicated fly strains (n > 480). (H and I) Semi-thin section of 2-week-old adult fly brain. Optic lobe (H) and central brain (I) regions were magnified to reveal moderate vacuolization in MI mutant brain. (J and K) TUNEL (red) and DAPI (blue, DNA) staining of 2-week-old adult fly brain. Optic lobe (J) and central brain (K) regions of MI mutant brain shows moderate apoptosis induction. (L) Electron micrographs reveal the presence of ectopic vacuoles (white arrows), inclusion bodies (black arrows) and dysfunctional mitochondria (green arrows) in 2-week-old MI brain (right panels) but not in wild-type control brain (left panels). Lower panels are magnified view of boxed regions in upper panels. Scale bars: 50 μm (white), 2 μm (black). Quantification data are represented as means ± standard error. p value was calculated using Student’s t-test. Approximate molecular weights (observed/predicted): Fip200 (150 to 200/152 kD), Tubulin (50/52 kD).
Fip200 loss-of-function mutants exhibit strong motor deficit
All Fip200 mutant flies progressed well into the pupal stage P15 (also known as pharate adult stage).54 However, after this stage, all Fip200-null mutant flies, both 3F5 and 4G7, were dead before or during the eclosion (Fig. 2C). Whereas artificial removal of puparium allowed wild-type flies to walk at this stage, such manipulation only left 3F5 and 4G7 mutant flies with uncoordinated leg movements. Although most of MI flies successfully completed the eclosion when cultured at 25°C, all were dead before or during the eclosion at 29°C, suggesting that disruptive mRNA splicing caused by the MI insertion is temperature-sensitive. Interestingly, the MI adult flies that eclosed at 25°C were still unable to fly and showed a defective wing posture phenotype (Fig. 2D). Using negative geotaxis assay, we found that MI flies were unable to efficiently climb walls (Fig. 2E and F), further demonstrating their mobility defects. Correspondingly, the pupariation site of MI flies was also lower compared with that of WT flies (Fig. 2G), suggesting that the mobility defects can be observed at both larva and adult stages.
Then we determined if these mobility defects of MI, 3F5, and 4G7 flies were due to loss of Fip200 activity or caused by unknown second-site mutations. The wing posture and climbing ability defects were not observed in RV adult flies in which the MIMIC inhibitory cassette of the MI mutant was removed by PhiC31-mediated recombination.53 This reversion of mutant phenotypes demonstrated that the mobility defects of MI mutants were indeed caused by interruption in Fip200 mRNA transcription. In addition, we performed an array of complementation tests to find out if all of the Fip200 mutants were within a single complementation group (also known as gene). As expected, the MI mobility defects were complemented by RV and EY, but not by 3F5, 4G7 and a deficiency allele covering the Fip200 locus (DF, Df(3L)Exel7283). Similarly, 3F5 and 4G7 lethality was not complemented by each other or by DF, but was complemented by RV and EY alleles. A complete complementation test result is provided in Table S2. Importantly, ubiquitous transgenic expression of Fip200 through daughterless (da)-Gal4 driver (Fig. S2A) suppressed the lethality of 3F5 or 4G7 mutants. These genetic evidences collectively show that the observed phenotypes of Fip200 loss-of-function mutants are indeed due to the absence or reduction of the Fip200 activity.
Fip200 loss-of-function mutants exhibit signs of neurodegeneration
Because the mobility defects could be caused by neurodegenerative brain pathologies, we examined the histology of MI adult brain. Semi-thin sections showed that MI brains suffered from higher levels of vacuolization in both optic lobe (Fig. 2H) and central brain (Fig. 2I) regions, compared with age-matched wild-type flies. This brain vacuolization phenotype is commonly observed in Drosophila mutants with severe neurodegeneration.11,55-57 Terminal deoxynucleotidyl transferase dUTP Nick End Labeling (TUNEL) assay revealed the existence of dying cells in MI brain again in both optic lobe (Fig. 2J) and central brain (Fig. 2K; Fig. S2B) regions. Transmission electron microscopic analysis showed that MI brain contains neurons with various degenerative features like abnormally shaped mitochondria, ectopic vacuoles and dense inclusion bodies (Fig. 2L; Fig. S2C). These data suggest that MI mutants exhibit neurodegeneration in broad parts of the brain.
Progressive ubiquitin accumulation in Fip200 mutant brain
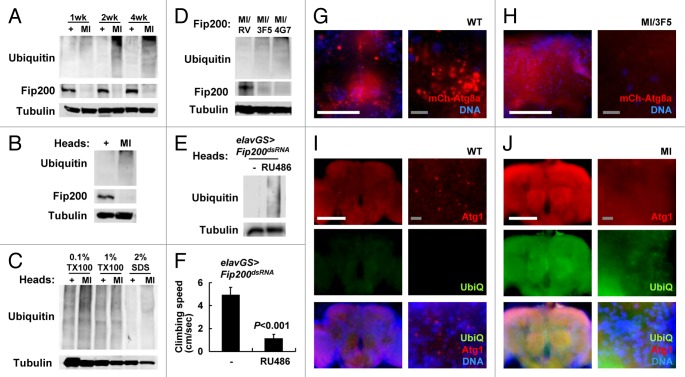
Autophagy has a critical role in regulating cellular protein homeostasis and is especially important for eliminating protein aggregates that are labeled with ubiquitin chains but unable to be degraded by conventional 26S proteasome machinery.1 Previous studies of Atg5 and Atg7 mutant mice4,45 as well as of Atg7 and Atg8 mutant flies11,25 reported an accumulation of ubiquitinated proteins in their brains that are associated with progressive neurodegeneration. We also found that MI mutants age-dependently accumulate ubiquitinated proteins in their brain (Fig. 3A and B). This accumulation was observed in both soluble and insoluble fractions (Fig. 3C). The ubiquitin accumulation by MI mutation was complemented by RV allele but not by 3F5 or 4G7 alleles (Fig. 3D).
Figure 3. Protein homeostasis and autophagy defects in Fip200 mutant brain. (A–D) Accumulation of ubiquitin in Fip200 hypomorphs. (A, B and D) Adult male flies (A and D) or heads (B) of wild-type (+), Fip200MI (MI), Fip200MI/Fip200RV (MI/RV), Fip200MI/Fip2003F5 (MI/3F5) and Fip200MI/Fip2004G7 (MI/4G7) at the indicated ages (A) or at 2 weeks of age (B) were subjected to immunoblot analyses with the indicated antibodies. (C) 50 adult fly heads of indicated 2-week-old adult flies were subjected to serial protein extraction with the indicated concentration of Triton X-100 (TX100) or sodium dodecyl sulfate (SDS) and were analyzed by immunoblotting with the indicated antibodies. (E and F) Temporal silencing of Fip200 in adult neurons is sufficient to induce neurodegenerative phenotypes. 1-week-old elav-GS > Fip200dsRNA flies were cultured for two weeks on standard media (-) or on media supplemented with 200 mM RU486 and were assayed for ubiquitin accumulation (E) and climbing abilities (F). (G and H) Autophagic activities are decreased in MI brain. Fluorescence images of adult fly brain from 2-week-old hs > mCherry-Atg8a/+ (WT) and Fip200MI/hs > mCherry-Atg8a Fip2003F5 (MI/3F5) after eclosion. mCherry-Atg8a (mCh-Atg8a, red) was induced by overnight exposure to low-grade heat shock (29°C), and nuclei in live cells were stained with Hoechst 33258 (DNA, blue). (I an J) Increased ubiquitin (UbiQ, green) and Atg1 (Atg1, red) levels in MI brain. 2-week-old WT and MI fly brains were dissected and subjected to immunostaining with anti-ubiquitin (UbiQ) and anti-Atg1 antibodies. DNA was visualized with DAPI staining. Scale bars: 200 μm (white), 10 μm (gray). Quantification data are represented as means ± standard error. p value was calculated using Student’s t-test. Approximate molecular weights (observed/predicted): Fip200 (150 to 200/152 kD), Tubulin (50/52 kD). As cellular proteins of diverse molecular weights are mono- or polyubiquitinated, ubiquitin immunoblots show a smeared banding pattern toward the top of the gel, as previously reported.11
Temporal neuron-specific Fip200 ablation results in motor deficit with ubiquitin accumulation in brain
We questioned whether ubiquitin accumulation, neurodegeneration and mobility defects were caused by developmental abnormalities or by physiological autophagy failure. To address this, we silenced Fip200 in neurons of adult stage using the elav-GeneSwitch-Gal4 system58 in which Fip200-dsRNA is specifically expressed in neurons only after RU486 feeding. Compared with control flies, Fip200-silenced flies showed a prominent accumulation of ubiquitinated proteins in their brains (Fig. 3E) and exhibited strong motor deficits (Fig. 3F) that were comparable to those of MI mutant flies (Fig. 2F). These results demonstrate that Fip200 function in adult neurons is important for the maintenance of neuronal physiology and preservation of mobility during aging.
Neuronal autophagy is suppressed by Fip200 loss
We attempted to directly measure the autophagic activities by using mCherry-Atg8a (homolog of mammalian LC3) transgene as a marker for autophagosomes.51 While wild-type adults showed a fair number of autophagosomes throughout the brain, adult brains from MI/3F5 mutant flies displayed a dramatically reduced number of autophagosomes (Fig. 3G and H; Fig. S3A), suggesting that the autophagic activity in the brain is generally suppressed by Fip200 loss. Similarly, the immunostaining of Atg1 in wild-type adult brains revealed a punctated staining pattern that is frequently associated with mCherry-Atg8a (Fig. 3I; Fig. S3B),30,59 while this pattern was almost completely absent in MI mutant brains (Fig. 3J; Fig. S3C). Interestingly, the overall Atg1 level was upregulated in MI mutant brains, which may reflect the compensatory upregulation of Atg1 protein to counteract the reduced autophagic activity.51 The level of ubiquitin staining was prominently elevated uniformly throughout the brain (Fig. 3I and J), consistent with the immunoblotting results (Fig. 3B and C). Collectively, these data suggest that neuronal autophagy is suppressed by hypomorphic mutation of Fip200 and that the autophagy defects led neurons to protein homeostasis defects and premature cell death, ultimately resulting in neurodegenerative phenotypes.
Axonal growth, projection and transport are unaffected in Fip200 mutants
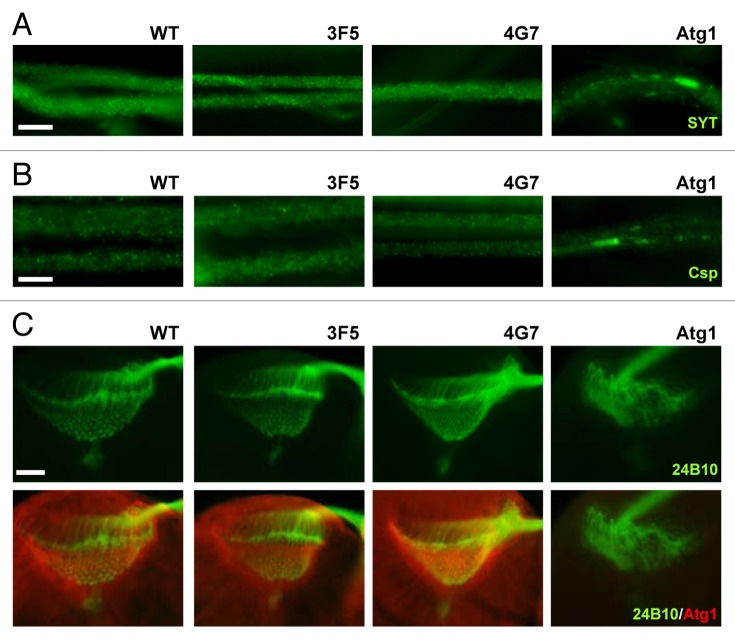
In addition to controlling autophagy, Atg1 has another important physiological role in neurons: Atg1 is required for axonal transport of cargo proteins as well as for axonal growth and projection.22-24 We were curious if Fip200, as a binding partner of Atg1, is also involved in these processes as defective axonal growth and neuronal wiring can contribute to the motor deficits observed in Fip200 mutants. However, the axonal transport in larval motor neurons, monitored by immunostaining of synaptotagmin (SYT, Fig. 4A; Fig. S4A) and cysteine string protein (Csp, Fig. 4B; Fig. S4B) cargo proteins, was completely normal in 3F5 and 4G7 mutants, unlike Atg1-null mutants where cargo proteins showed uneven distribution and formed aggregates.23 Furthermore, the axonal projection of photoreceptor neurons during brain development, monitored by axonal marker 24B10 antibody, was also completely normal in Fip200-null mutant brains (Fig. 4C; Fig. S4C), in contrast to the disorganized projection pattern frequently observed in Atg1 mutants (Fig. 4C; Fig. S4C).22,24 On a side note, Atg1 localization at optic laminal axons were also unaffected by the loss of Fip200 (Fig. 4C). These data demonstrate that although Fip200 is important for Atg1-dependent neuronal autophagy, it is largely dispensable for Atg1’s role in axonal growth, projection and transport.
Figure 4. Axonal growth, projection and transport are unaffected by Fip200 loss. (A and B) Motor neurons from wandering-stage third instar larvae were immunostained with the indicated antibodies. In Fip200-null mutant (3F5 and 4G7) motor neurons, axonal traffic of synaptotagmin (SYT), (A) and cysteine string protein (Csp), (B) cargo molecules were normal and indistinguishable from WT motor neurons. However, the proteins became aggregated in Atg1-null (Atg1Δ3d/Df(3)BSC10) motor neurons. (C) Wandering-stage third instar larval brain was immunostained with the indicated antibodies. In both WT and Fip200-null mutant brains, axons of eye photoreceptor cells, visualized by 24B10 monoclonal antibodies, show stereotyped projection to lamina and medulla region, and Atg1 prominently localizes to axons in lamina of the brain optic lobes. Atg1-null (Atg1Δ3d/Df(3)BSC10) brains show disruption of this structure. Scale bars: 20 μm.
Fip200 mutants are defective in developmental and starvation-induced autophagy
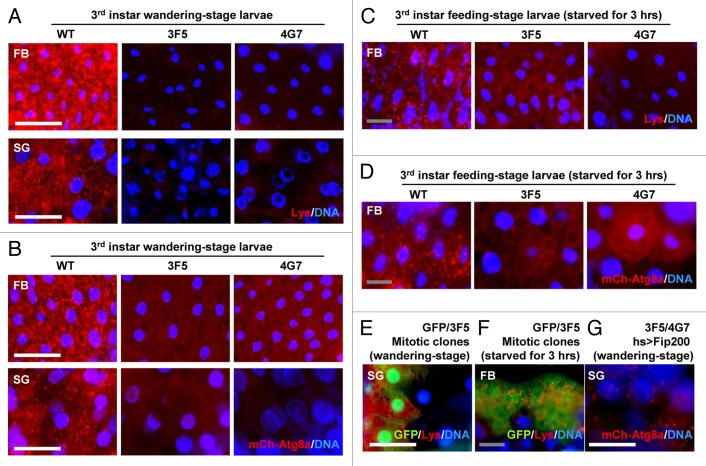
Because Atg1 is essential for developmental and starvation-induced autophagy, we questioned the contribution of Fip200 to these Atg1-dependent processes. As previously reported,9,11 late wandering-stage third instar wild-type larvae exhibited prominent autophagic activities, such as accumulation of acidic vesicles (monitored by LysoTracker Red, Fig. 5A) and autophagosomes (monitored by mCherry-Atg8a, Fig. 5B), in their salivary glands and fat bodies. However, these autophagic activities were almost absent in both of Fip200-null mutant larvae (3F5 and 4G7, Fig. 5A and B; Fig. S5A–D). Fip200-null mutant larvae were also defective in starvation-induced autophagy (Fig. 5C and D; Fig. S5E and S5F). Fip200-null mitotic clones exhibited cell autonomous autophagy defects both during development (Fig. 5E) and upon starvation (Fig. 5F). Transgenic expression of Fip200 in Fip200-null mutant restored developmental autophagy (Fig. 5G; Fig. S5D), showing that the autophagy defects were indeed caused by Fip200 loss. These results collectively indicate that Fip200 is a critical mediator of Atg1-dependent autophagy during development and in response to starvation.
Figure 5. Fip200 is essential for developmental and starvation-induced autophagy. (A) Fat bodies (FB) and salivary glands (SG) of wandering-stage third instar larvae of WT, 3F5 and 4G7 flies were subjected to LysoTracker Red (Lys) and Hoechst 33258 (DNA) staining. (B) FB and SG of wandering-stage third instar larvae of hs > mCherry-Atg8a/+ (WT), hs > mCherry-Atg8a Fip2003F5/Fip2003F5 (3F5) and hs > mCherry-Atg8a Fip2004G7/Fip2004G7 (4G7) flies, heat-shocked at 37°C for 1 h to induce mCherry-Atg8a and recovered at 25°C for 3 h, were stained with Hoechst 33258 (DNA) and observed under fluorescence microscopy. (C) FB from feeding-stage third instar larvae of WT, 3F5 and 4G7, starved for 3 h, were subjected to Lys and DNA staining. (D) FB from feeding-stage third instar larvae of hs > mCherry-Atg8a/+ (WT), hs > mCherry-Atg8a Fip2003F5/Fip2003F5 (3F5) and hs > mCherry-Atg8a Fip2004G7/Fip2004G7 (4G7) flies, heat-shocked at 37°C for 1 h and incubated on 20% sucrose solution at 25°C for 3 h, were subjected to DNA staining. (E) SG from wandering-stage third instar larvae of y w hs-FLP; FRT82B Ubi-eGFP/FRT82B Fip2003F5 (GFP/3F5) flies were subjected to Lys and DNA staining. Absence of GFP marks Fip200-deficient cells, which also have less Lys staining. (F) FB from feeding-stage third instar larvae of y w hs-FLP; FRT82B Ubi-eGFP/FRT82B Fip2003F5 (GFP/3F5) flies, starved for 3 h, were subjected to Lys and DNA staining. (G) SG of wandering-stage third instar larvae of UAS-Fip200/+; hs > mCherry-Atg8a Fip2003F5/Fip2004G7 (3F5/4G7 hs > Fip200) flies, heat-shocked at 37°C for 1 h and recovered at 25°C for 3 h, were subjected to DNA staining. Scale bar: 200 μm (white), 50 μm (gray).
Hypomorphic Fip200 mutation accelerates both aging and development
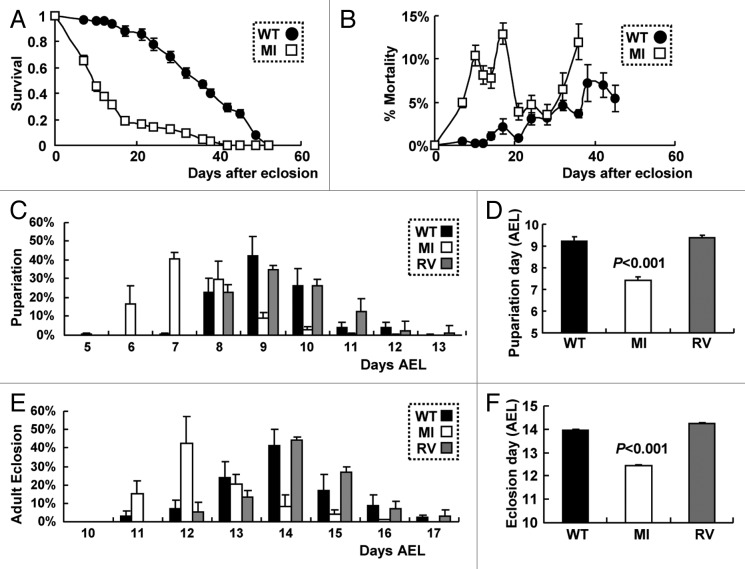
Finally, we tested the effect of Fip200 loss on aging and developmental growth, the two important processes subjected to regulation by autophagy. Associated with neurodegeneration and behavioral defects (Figs. 2–3), Fip200 hypomorphs (MI) showed high mortality in the early ages of their life, making their life span drastically shorter than WT counterparts (Fig. 6A and B). Interestingly, MI mutants were relatively fast in development with the time needed for the laid eggs to pupariate or to become imago reduced by at least 1 to 2 d on average, compared with WT or RV controls (Fig. 6C–F). This is opposite from the previous observation that autophagy defects generally attenuate development.11 However, we noticed that this developmental acceleration was only seen during larval growth, as the time spent between pupariation and adult eclosion was not significantly altered in MI mutants compared with wild-type counterparts. Therefore, considering that the developmental timing of pupariation generally depends on larval growth to critical mass60 and that there is no considerable difference in pupa and adult size between WT and MI flies, the accelerated development of MI mutants seems to be mainly attributable to the enhanced growth rate during larval stage.
Figure 6. Hypomorphic Fip200MI mutation accelerates both aging and development. (A and B) Survivorship (A) and daily mortality (B) of WT and MI male flies (n ≥ 380). (C and D) Pupariation rates of WT, RV and MI flies were recorded at the indicated days after egg laying (C), AEL. Average pupariation day AEL was calculated and presented as a bar graph (D), n ≥ 500. (E and F) Adult eclosion rates of WT, RV and MI flies were recorded at the indicated days AEL (E). Average eclosion day AEL was calculated and presented as a bar graph (F, n ≥ 500). Quantification data are represented as means ± standard error. p values were calculated between WT and MI groups using Student’s t-test.
Fip200 loss provokes TORC1-S6k signaling activation
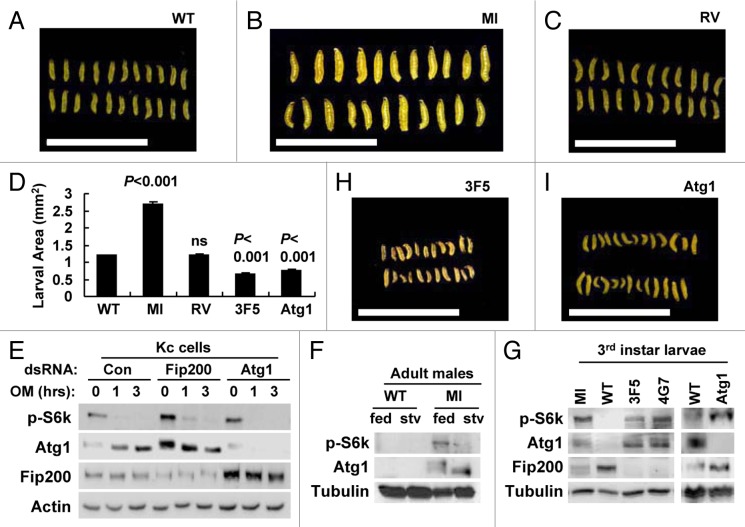
To directly examine the larval growth, we measured the size of larvae at 4 d after egg laying. At that time point, MI mutant larvae were substantially larger than WT or RV controls (Fig. 7A–D), supporting that Fip200 downregulation by MI mutation accelerates larval growth. This is similar to previous findings that reduction of Atg1/ULKs activity promotes cell growth through boosting TORC1-S6k activities.18-21 Therefore, we examined if Fip200 loss could also affect the signaling activity of TORC1-S6k as Atg1 does. In Drosophila Kc cells, silencing of Fip200 or Atg1 increased S6k phosphorylation at Thr398 by TORC1 (Fig. 7E). Similar upregulation of S6k phosphorylation compared with WT counterparts was also observed in MI adults (Fig. 7F) as well as in MI, 3F5, 4G7, Atg1 larvae (Fig. 7G), suggesting that the Atg1-dependent regulation of TORC1-S6k-dependent growth is mediated by Fip200. However, null mutations of Fip200 and Atg1 substantially attenuated larval growth rather than accelerating it (Fig. 7D, H and I), implying that the complete abrogation of Atg1-Fip200 pathway can actually be detrimental to development. Therefore, although Atg1-Fip200 pathway inhibits TORC1-S6k-dependent cell growth, a certain level of its autophagy-controlling activities is essential for proper developmental propagation.
Figure 7. Fip200 loss induces TORC1-S6k activation. (A–C, H and I) Feeding-stage larvae of the indicated strains at 4 d AEL were imaged through dissection microscope. (D) Average larval area from the images was calculated and presented as a bar graph (n ≥ 22). Quantification data are represented as means ± standard error. p values were calculated between WT and indicated groups using Student’s t-test. (E) Drosophila Kc cells treated with the indicated dsRNA for 3 d were treated with 5 μM oligomycin (OM) for the indicated hours and subjected to immunoblotting with the indicated antibodies. (F) 1-week-old adult male flies of the indicated genotypes were incubated on standard media (fed) or 20% sucrose media (stv) for 24 h and subjected to immunoblotting with the indicated antibodies. (G) Wandering-stage third instar larvae of the indicated strains were subjected to immunoblotting with the indicated antibodies. Scale bar: 1 cm. Approximate molecular weights (observed/predicted): Fip200 (150 to 200/152 kD), Atg1 (120 to 150/92 kD), S6k (60/55 kD), Actin (48/42 kD), Tub (50/52 kD).
Interestingly, the level of Fip200 was increased by Atg1 loss (Fig. 7E and G), as the level of Atg1 was increased by Fip200 loss (Fig. 3I and J; Fig. 7E–G), possibly through a compensatory mechanism.51 However, increased Atg1 in Fip200-null mutants does not mean it can induce autophagy as there is no functional Fip200 that is needed for the autophagosome formation. Nonetheless, TORC1-dependent Atg1 phosphorylation51 was still well responsive to cellular energy (Fig. 7E) or nutrition (Fig. 7F) status even in the absence of Fip200, supporting that Fip200 does not play a critical role in the regulation of Atg1 phosphorylation (Fig. 1I and J).
Discussion
In mammalian cells, formation of the ULK1-ATG13-RB1CC1 complex is critical for the initiation of autophagy upon TORC1 inhibition by nutrient deprivation.30,37-39 Although ULK1 and ATG13 homologs in Drosophila and C. elegans have been identified and functionally characterized to play autophagy-related roles, the existence of a functional RB1CC1 homolog in the invertebrate genome had not been determined yet.61,62 Here we isolated Fip200 as a single RB1CC1 homolog in Drosophila. As observed in mammals, Fip200 interacts with Atg1 and Atg13, and this Atg1-Atg13-Fip200 complex constitutively exists regardless of nutrition-dependent Atg1 phosphorylation status. Interestingly, while Atg13 is essential for phosphorylation-dependent regulation of Atg1,51 the loss of Fip200 does not affect the phosphorylation status, as Atg1 phosphorylation is still fully responsive to nutrients and cellular energy status in the absence of Fip200. Furthermore, whereas coexpression of Atg1 and Atg13 in Kc cells induces autophosphorylation-mediated shift of Atg1, Fip200 does not induce this phosphorylation indicating that, unlike Atg13, Fip200 is not an Atg1 activator per se. In consistence with this idea, co-overexpression of Atg1 and Fip200 does not exhibit any genetic interaction in adult eyes while Atg1 and Atg13 synergistically induce eye degeneration (Fig. S6) that results from excessive autophagic cell death.51 Nonetheless, Fip200 is essential for the autophagic output of Atg1-Atg13 because the silencing of endogenous Fip200 strongly suppressed Atg1-Atg13 overexpression phenotype, restoring both eye size and pigmentation (Fig. S6). These results show that Fip200 is strictly epistatic to Atg1 and that the role of Fip200 is distinct from that of Atg13, a regulatory subunit of Atg1 stoichiometrically required for Atg1’s maximum catalytic activity and phosphorylation-dependent regulation. Therefore, our study suggests that Fip200 is the most downstream molecule in the Atg1-Atg13-Fip200 complex and is essentially mediating Atg1 action on the autophagosome formation.
Although Fip200 has been revealed to be a critical molecule that mediates autophagy-controlling output of Atg1, it is still possible that both Fip200 and Atg1 play additional physiological roles independent of each other. For instance, Fip200 may interact with many important signaling molecules other than Atg1 and Atg13, such as Fak, Rbf and p53, like its mammalian homolog.32-34 On the other hand, Atg1 also has autophagy-independent functions, especially in neurons, where it controls axonal transport, growth and projection.12 Correspondingly, Atg1 mutation in Drosophila produces strong pleiotropic phenotypes including diminished autophagic activities, cell growth enhancement and defects in axonal transport and projection.20-24 Drosophila Fip200-null mutants, described in this paper for the first time, also exhibited strong autophagy defects and TORC1-S6k activation, which suggests that Fip200 mediates most of Atg1 function in autophagy and cell growth control. However, unlike Atg1-null mutants, Fip200-null mutants developed well up to the late pupal stage without displaying any apparent defect in neurodevelopment and axonal physiology, suggesting that Fip200 is dispensable for the autophagy-independent physiological output of Atg1. Consistently, even though the localization of Atg1 in autophagic vesicles is diminished by Fip200 loss, Atg1 localization in optic lobe laminal axons is completely normal in Fip200-null brains. Furthermore, Fak-dependent optic stalk morphogenesis63 and Rbf-dependent ommatidia array morphogenesis64 are not affected by Fip200 loss, implying that Fip200 is not critically involved in mediating the function of these proteins. Therefore, this study provides the first genetic evidence that Fip200 specifically mediates the autophagy- and cell growth-regulating roles of Atg1 in an organism.
In this study, we isolated a hypomorphic mutant strain of Fip200 (MI) and characterized its phenotypes. The MI mutation strongly accelerated development and aging, both of which can be associated with the effects of downregulated Atg1 activities on autophagy and cell growth inhibition. Because the developmental growth is accelerated only by the MI mutation, not by any other mutations of Atg1 or Fip200 genes, there is still a possibility that this specific phenotype is independent of the Atg1-Atg13-Fip200 complex functionality. In addition to the developmental growth and longevity phenotypes, MI adult flies exhibited obvious deficit in motor abilities such as flying and climbing, very similar to Drosophila models of neurodegenerative diseases such as Alzheimer or Parkinson diseases.65-69 Moreover, MI adult brain exhibited common neurodegenerative histological features such as brain vacuolization, mitochondrial dysfunction and ectopic neuronal cell death. The brain of MI flies accumulated a large amount of ubiquitinated proteins in both soluble and insoluble forms, similar to previously described autophagy-defective mutant flies, such as Atg7−/−, Atg8a−/− and bchs−/− mutants.11,25,57 The accumulation of ubiquitin can be also induced by temporal neuron-specific silencing of Fip200 in the adult brain, demonstrating that the protein homeostasis defect is not due to developmental anomalies. Neuron-specific Fip200 silencing is sufficient to induce mobility defect at the same magnitude as MI mutant, while muscle-specific Fip200 silencing does not produce such a phenotype (Fig. S7A), indicating that the behavioral phenotype of MI mutant is primarily attributable to the neuronal autophagy defects. Further supporting this, transmission electron microscopy showed that microfiber structure of the MI mutant skeletal muscle is largely unaltered (Fig. S7B). Nevertheless, we cannot rule out the possibility that non-neuronal Fip200 function can be important for mobility of flies as well. Collectively, our current results highlight the importance of Fip200 in regulating autophagy and in protecting organisms from premature aging, age-associated neurodegeneration, abnormal developmental growth and metabolic derangements.
Materials and Methods
Fly strains
Atg1Δ3d, UAS-myc-Atg1, UAS-Atg13-GFP and hs > mCherry-Atg8a (hs-Gal4 UAS-mCherry-Atg8a) were gifts from Dr. Neufeld (Univ. of Minnesota). elav-GeneSwitch-Gal4 and Mef2-Gal4 lines were from Wessells lab (UM). GMR-Gal4, Atg1Df(3)BSC10, UAS-Fip200RNAi, Fip200MI01469 and Fip200EY03045 flies, as well as balancer, FLP/FRT, wild-type and transposase strains were obtained from Bloomington Stock Center (Indiana). UAS-Fip200 was made using LD09358 full-length Fip200 cDNA clone obtained from Drosophila Genome Research Center (Indiana). The Fip200 ORF was subcloned into pUAST-attB70 after fusion to an N-terminal 3xFlag epitope and a Kozak sequence. The cloned construct was microinjected into BL-24481 fly strain that has PhiC31 integrase and attP landing site (Rainbow Transgenic Flies, Inc.). Similarly, Fip200MI01469-RV revertants were generated by microinjection of pBS-KS-attB correction plasmid53 with helper integrase into the Fip200MI01469 strain. Fip200-null alleles were generated using imprecise excision by crossing y w; Fip200EY03045 line with y w; Sb Δ2–3/TM6 transposase line. Resulting y w; Fip200EY03045/Sb Δ2–3 F1 males were crossed with y w; TM3/TM6B virgins [~200 single pair mating (s.p.m.)], and F2 males with P element excision were selected and backcrossed with w1118; TM3 Sb Ser/TM6B Tb virgins (~1,000 s.p.m.). Each male from the final s.p.m. was collected 7 d after crosses and screened by PCR for Fip200 deletions using primers that flank the Fip200EY03045 insertion site with 2 to 4kb intervals in the Fip200 locus, as previously described.71 The final screen identified a series of genomic Fip200 deletion alleles including Fip2003F5 and Fip2004G7 that were used in this study. Mosaic clones were generated by placing y w hs-FLP; FRT82B Ubi-eGFP/FRT82B Fip2003F5 first instar larvae in a 37°C water bath for 1 h.
Antibodies
Rabbit anti-Atg1 and guinea pig anti-Fip200 antibodies were made as follows: full-length Atg1 cDNA and a cDNA fragment encoding a.a. 628 to 1358 of Fip200 were cloned into pGEX and transformed into E. coli BL21. Insoluble GST-fusion proteins were purified from SDS-PAGE gel bands and injected to rabbits and guinea pigs, respectively (Pocono Farms, Inc.). Sera were subjected to affinity purification using PVDF-immobilized proteins. 22C10 (DSHB), 24B10 (DSHB), anti-Csp (DSHB, AB49), anti-synaptotagmin (DSHB, 3H2-2D7), anti-myc (DSHB, 9E10), anti-HA (Roche, 1867431), anti-Flag (Sigma, F3165), anti-ubiquitin (Invitrogen, 13-1600), anti-pT398 Drosophila S6k (Cell Signaling, 9209), anti-pT172 AMPK (Santa Cruz, sc-33524), and anti-tubulin (Sigma, T5168) antibodies were used for immunostaining and immunoblot analyses.
Histology
Immunostaining,71 LysoTracker Red staining,72 acridine orange staining,73 TUNEL staining,74 mCherry-ATG8a observation,72 semi-thin histology75 and transmission electron microscopy71 were done as previously described. Quantification of histological phenotypes was done from fluorescence images and electron micrographs using NIH ImageJ software or manually in a blinded manner.
Culture of cells and flies
Drosophila Kc cells were cultured in Schneider’s Drosophila medium (Invitrogen, 21720024) supplemented with 10% FBS (Denville Inc., FB5002-H). Transient transfection of plasmid DNAs was done using Effectene (Qiagen, QIA301425) according to the manufacturer’s recommendation. Expression constructs were carried by pUAST-attB vector and cotransfected with actin-Gal4 driver plasmid. dsRNA transfection was done as previously described.76 The flies were reared on 15% sucrose-yeast medium (for life-span measurement) or standard cornmeal-agar medium (for all other experiments) with humidity (70%), temperature (25°C or as indicated) and light (12/12 h light/dark cycle) control. For negative geotaxis assay, photos were taken 3 sec after negative geotaxis induction, and the climbing speed was calculated from the picture. RU486 was added to the medium as described.58
Biochemical analyses
Total RNA was isolated, reverse transcribed and analyzed as previously described.71 Protein lysate preparation and immunoblotting were done using standard cell lysis buffer or the indicated buffers as previously described.71 Coimmunoprecipitation of proteins was done as described.75 Predicted protein molecular weights were calculated using ExPASy molecular weight computation tool. Observed protein size was estimated by comparison of protein band to protein size marker (Bio-Rad, 161-0374).
Online Supplemental Material
Figures S1–S5 show supporting data related to Figures 1–5. Figure S6 shows genetic interaction among Atg1, Atg13 and Fip200 in Drosophila eyes. Figure S7 shows the muscle phenotypes of Fip200-deficient flies. Table S1 shows the list of Fip200 and Atg1 mutant strains and their phenotypes. Table S2 shows complementation test performed between Fip200 alleles. Table S3 shows statistical comparison of the climbing speed between fly strains that appeared in Figure 2F.
Supplementary Material
Acknowledgments
We thank T. Neufeld (UMN), R.J. Wessells, S. Pletcher, B. Richardson, R.A. Miller (UM), DSHB (Iowa), DGRC (Indiana), Cell Signaling Inc., Santa Cruz Biotech. Inc., TRiP (Harvard, GM084947), Bloomington stock centers (Indiana) for cell lines, fly strains, reagents and access to lab equipment. We thank S. Meshinchi for imaging assistance. Work was supported by grants from the Ellison Medical Foundation (AG-NS-0932-12) and by National Institute of Health grants from Research Career Development Core (RCDC) and Pilot and Explorative Studies Core (PESC) of Claude D. Pepper Center (P30-AG024824), Drosophila Aging Core (DAC) of Nathan Shock Center (P30-AG013283), Gut Peptide Center (P30-DK034933), the UM Core Cancer Center (P30-CA46592), and a Research Project Grant (R01-GM052890).
Glossary
Abbreviations:
- TORC1
target of rapamycin complex 1
- S6k
p70 S6 kinase
- Atg
autophagy-related
- UNC
uncoordinated
- ULK
Unc-51-like kinase
- PtdIns3K
phosphoinositide-3-kinase
- UAS
upstream activator sequence
- AMPK
adenosine monophosphate-activated protein kinase
- MIMIC
Minos-mediated integration cassette
- OM
oligomycin
- CIP
calf intestine phosphatase
- dsRNA
double-stranded RNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/24811
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/24811
References
- 1.Yen WL, Klionsky DJ. How to live long and prosper: autophagy, mitochondria, and aging. Physiology (Bethesda) 2008;23:248–62. doi: 10.1152/physiol.00013.2008. [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 5.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 8.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–16. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–6. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alers S, Löffler AS, Wesselborg S, Stork B. The incredible ULKs. Cell Commun Signal. 2012;10:7. doi: 10.1186/1478-811X-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 14.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 18.Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy. 2011;7:1212–21. doi: 10.4161/auto.7.10.16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy. 2011;7:737–47. doi: 10.4161/auto.7.7.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, et al. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–5. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochizuki H, Toda H, Ando M, Kurusu M, Tomoda T, Furukubo-Tokunaga K. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS One. 2011;6:e19632. doi: 10.1371/journal.pone.0019632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toda H, Mochizuki H, Flores R, 3rd, Josowitz R, Krasieva TB, Lamorte VJ, et al. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292–307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahantarig A, Chadwell LV, Terrazas IB, Garcia CT, Nazarian JJ, Lee HK, et al. Molecular characterization of Pegarn: a Drosophila homolog of UNC-51 kinase. Mol Biol Rep. 2009;36:1311–21. doi: 10.1007/s11033-008-9314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonsen A, Cumming RC, Lindmo K, Galaviz V, Cheng S, Rusten TE, et al. Genetic modifiers of the Drosophila blue cheese gene link defects in lysosomal transport with decreased life span and altered ubiquitinated-protein profiles. Genetics. 2007;176:1283–97. doi: 10.1534/genetics.106.065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–46. doi: 10.1016/S0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 27.Tomoda T, Kim JH, Zhan C, Hatten ME. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18:541–58. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajesh S, Bago R, Odintsova E, Muratov G, Baldwin G, Sridhar P, et al. Binding to syntenin-1 protein defines a new mode of ubiquitin-based interactions regulated by phosphorylation. J Biol Chem. 2011;286:39606–14. doi: 10.1074/jbc.M111.262402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci U S A. 2007;104:5842–7. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda H, Abbi S, Zheng C, Guan JL. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol. 2000;149:423–30. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbi S, Ueda H, Zheng C, Cooper LA, Zhao J, Christopher R, et al. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol Biol Cell. 2002;13:3178–91. doi: 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chano T, Ikegawa S, Kontani K, Okabe H, Baldini N, Saeki Y. Identification of RB1CC1, a novel human gene that can induce RB1 in various human cells. Oncogene. 2002;21:1295–8. doi: 10.1038/sj.onc.1205178. [DOI] [PubMed] [Google Scholar]
- 34.Melkoumian ZK, Peng X, Gan B, Wu X, Guan JL. Mechanism of cell cycle regulation by FIP200 in human breast cancer cells. Cancer Res. 2005;65:6676–84. doi: 10.1158/0008-5472.CAN-04-4142. [DOI] [PubMed] [Google Scholar]
- 35.Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. Identification of FIP200 interaction with the TSC1-TSC2 complex and its role in regulation of cell size control. J Cell Biol. 2005;170:379–89. doi: 10.1083/jcb.200411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175:121–33. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganley IG, Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol. 2013;20:144–9. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–12. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 44.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 46.Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–14. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 49.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 50.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 51.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–14. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–7. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–43. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bate M, Hartenstein V. The development of Drosophila Melanogaster. Long island, NY: Cold Spring Harbor Laboratory Press, 1993. [Google Scholar]
- 55.Min KT, Benzer S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science. 1999;284:1985–8. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- 56.Min KT, Benzer S. Spongecake and eggroll: two hereditary diseases in Drosophila resemble patterns of human brain degeneration. Curr Biol. 1997;7:885–8. doi: 10.1016/S0960-9822(06)00378-2. [DOI] [PubMed] [Google Scholar]
- 57.Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, et al. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–64. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirth C, Truman JW, Riddiford LM. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol. 2005;15:1796–807. doi: 10.1016/j.cub.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 61.Zirin J, Perrimon N. Drosophila as a model system to study autophagy. Semin Immunopathol. 2010;32:363–72. doi: 10.1007/s00281-010-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584:1342–9. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami S, Umetsu D, Maeyama Y, Sato M, Yoshida S, Tabata T. Focal adhesion kinase controls morphogenesis of the Drosophila optic stalk. Development. 2007;134:1539–48. doi: 10.1242/dev.001529. [DOI] [PubMed] [Google Scholar]
- 64.Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996;10:1206–18. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- 65.Chakraborty R, Vepuri V, Mhatre SD, Paddock BE, Miller S, Michelson SJ, et al. Characterization of a Drosophila Alzheimer’s disease model: pharmacological rescue of cognitive defects. PLoS One. 2011;6:e20799. doi: 10.1371/journal.pone.0020799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 67.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–8. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 68.Greeve I, Kretzschmar D, Tschäpe JA, Beyn A, Brellinger C, Schweizer M, et al. Age-dependent neurodegeneration and Alzheimer-amyloid plaque formation in transgenic Drosophila. J Neurosci. 2004;24:3899–906. doi: 10.1523/JNEUROSCI.0283-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401. doi: 10.1016/S0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 70.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–7. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–8. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neufeld TP. Genetic manipulation and monitoring of autophagy in Drosophila. Methods Enzymol. 2008;451:653–67. doi: 10.1016/S0076-6879(08)03236-9. [DOI] [PubMed] [Google Scholar]
- 73.Lee JH, Lee E, Park J, Kim E, Kim J, Chung J. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett. 2003;550:5–10. doi: 10.1016/S0014-5793(03)00771-3. [DOI] [PubMed] [Google Scholar]
- 74.Lee JH, Koh H, Kim M, Park J, Lee SY, Lee S, et al. JNK pathway mediates apoptotic cell death induced by tumor suppressor LKB1 in Drosophila. Cell Death Differ. 2006;13:1110–22. doi: 10.1038/sj.cdd.4401790. [DOI] [PubMed] [Google Scholar]
- 75.Kim M, Lee JH, Koh H, Lee SY, Jang C, Chung CJ, et al. Inhibition of ERK-MAP kinase signaling by RSK during Drosophila development. EMBO J. 2006;25:3056–67. doi: 10.1038/sj.emboj.7601180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, et al. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–20. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.