Abstract
The central organelle within the secretory pathway is the Golgi apparatus, a collection of flattened membranes organized into stacks. The cisternal maturation model of intra-Golgi transport depicts Golgi cisternae that mature from cis to medial to trans by receiving resident proteins, such as glycosylation enzymes via retrograde vesicle-mediated recycling. The conserved oligomeric Golgi (COG) complex, a multi-subunit tethering complex of the CATCHR (complexes associated with tethering containing helical rods) family, organizes vesicle targeting during intra-Golgi retrograde transport. The COG complex both physically, and functionally, interacts with all classes of molecules maintaining intra-Golgi trafficking, namely SNAREs, SNARE-interacting proteins, Rabs, coiled-coil tethers, vesicular coats and molecular motors. In this report we will review the current state of the COG interactome and analyze possible scenarios for the molecular mechanism of the COG orchestrated vesicle targeting, which plays a central role in maintaining glycosylation homeostasis in all eukaryotic cells.
Keywords: COG, Golgi, SNARE, Rab, Tether, membrane trafficking
Introduction
Protein transport within the secretory pathway is a highly orchestrated process, utilizing many molecular players including SNAREs, Rabs, and vesicular tethering proteins (Cottam and Ungar 2012). Transport within this pathway is carried out by small membranes, termed vesicles, and there are four main steps to their transport including budding, tethering, docking, and fusion (Bonifacino and Glick 2004). The central organelle within the secretory pathway is the Golgi apparatus, a collection of flattened membranes organized in a cis-medial-trans fashion. The main function of the bulk of the Golgi is the posttranslational modification of secretory proteins, and the different cisternae are defined by the residence of different modifying enzyme subsets. The Golgi is responsible for the modifying, sorting, and packaging of secretory macromolecules. The cisternal maturation model is currently the most accepted model describing the transport of cargo within the Golgi apparatus. This model depicts Golgi cisternae that mature from cis to medial to trans by receiving resident proteins, such as glycosylation enzymes, via retrograde vesicle-mediated (COPI coated), recycling the Golgi cisternae resident proteins (Bonifacino and Glick 2004; Glick et al. 1997). In this model new cis cisternae are formed by the fusion of ER derived (COPII coated) vesicles followed by the fusion with retrograde COPI vesicles that deliver cis-Golgi localized enzymes.
Vesicle tethering is a two-step process that involves capturing vesicles and organizing the vesicles on acceptor membranes. The current model postulates that the initial sequestering of a vesicle is achieved by long coiled-coil tethers, which with their extended rod like coiled domains make them ideal for long range vesicle capturing (Cottam and Ungar 2012; Ramirez and Lowe 2009; Munro 2011). The next stage of tethering involves a group of tethers known as multi-subunit tethering complexes (MTCs). During MTC mediated tethering, a captured vesicle is now held at its acceptor compartment to organize the vesicle for critical pre-fusion steps such as v-SNARE/t-SNARE alignment. Tethering complexes, in concert with small Rab GTPases, Sec1/Munc18(SM) proteins, and SNAREs, organize the docking of a vesicle to its acceptor compartment, and thereby (in conjunction with the long coiled tethers) determine the targeting specificity of vesicles. Such targeting is a critical determinant of protein sorting, and thus of maintaining the compartmental identity within the secretory pathway
The conserved oligomeric Golgi (COG) complex, an MTC of the CATCHR (complexes associated with tethering containing helical rods) family, functions as a vesicular tether during retrograde intra-Golgi trafficking (Zolov and Lupashin 2005). The COG complex consists of eight subunits (named COG1–8 (Whyte and Munro 2001; Ungar et al. 2002)) which are separated into two subcomplexes named Lobe A (COGs 1–4) and Lobe B (Cogs 5–8) (Fotso et al. 2005; Ungar et al. 2005) with an interaction between COG1 and COG8 bridging the two lobes together. The COG complex tethers vesicles recycling Golgi resident proteins (such as glycosylation enzymes) and therefore is essential for the proper glycosylation of secretory proteins (Pokrovskaya et al. 2011; Shestakova et al. 2006; Kingsley et al. 1986). Defects in COG subunits manifest into a class of human diseases known as congenital disorders of glycosylation (CDG) type II (Zeevaert et al. 2008).
The COG complex is a peripheral membrane protein complex that exists in cells in both a soluble, mostly octameric form (Fotso et al. 2005; Ungar et al. 2002; Whyte and Munro 2001), as well as in a membrane bound form. Membrane-bound COGs are present in different arrangements, including the complete COG1–8 complex, Lobe A and Lobe B sub-complexes, and possibly other minor assemblies (Willett & Lupashin, unpublished). The COG complex both physically and functionally interacts with all classes of molecules maintaining intra-Golgi trafficking, namely SNAREs, SNARE-interacting proteins, Rabs, coiled-coil tethers, vesicular coats and molecular motors (Figure 1 and Tables 1–4). In this report we will review the current state of the COG interactome and analyze possible scenarios for the role of the COG complex in orchestrating intra-Golgi membrane transport. It is important to note that many published interactions with COG subunits have been observed using yeast two hybrid, overexpression, or in vitro strategies and thus represent a set of potential protein-protein interactions. It is likely that not all these interactions are being utilized in a given cell type. The stringency and specificity of COG subunit interactions with partner proteins may also be affected by the assembly of COG subunits into the COG complex and/or COG sub-complexes.
Figure 1.
COG complex interactome defines subunit specific hubs of regulated trafficking related proteins.
A comprehensive map of interactions between COG subunits and different trafficking related proteins. COG subunits can be classified by the groups of trafficking proteins that they interact with including SNAREs (blue), tethers (purple), Rabs (yellow) and coat and motor proteins (maroon). In this organization scheme we define the COG subunits as hubs for the class of protein that they interact with. COG subunits COG2 and COG3 are defined as “tether hubs”, COG2 COG3 and COG4 are defined as “coat and motor hubs”, COG4, COG5, and COG6 are defined as “Rab hubs”, and COG4, COG6, COG7, and COG8 are defined as “SNARE hubs”. Interestingly, COG1 has no other specific interactions therefore it is designated as the critical subunit for maintaining the structural integrity of the entire complex.
Table 1.
COG interacting SNAREs and SNARE-interacting (SM) proteins
| COG subunit | Partner protein | organism | Detection method* | Reference |
|---|---|---|---|---|
| COG2 | Sed5 | yeast | PD-W | (Suvorova et al. 2002) |
| COG3 | Sed5 | yeast | PD-W, PD-G | (Suvorova et al. 2002) |
| STX5 | human | Co-IP | (Sohda et al. 2010) | |
| YKT6 | yeast | PD-W | (Suvorova et al. 2002) | |
| Gos1 | yeast | PD-W | (Suvorova et al. 2002) | |
| Sec22 | yeast | PD-W | (Suvorova et al. 2002) | |
| GS28 (GOSR1) | human | Co-IP | (Zolov and Lupashin 2005; Sohda et al. 2010) | |
| COG4 | Sed5 | yeast | Y2H, Co-IP | (Shestakova et al. 2007) |
| STX5 | human | Y2H, Co-IP | (Shestakova et al. 2007; Willett et al. 2013; Sohda et al. 2010; Laufman et al. 2009; Laufman et al. 2013; Kudlyk et al. 2013) | |
| STX16 | human | Co-IP, PD-W | (Laufman et al. 2013) | |
| Vti1a | human | Co-IP, PD-W | (Laufman et al. 2013) | |
| GS28 (GOSR1) | human | Co-IP, PD-W | (Laufman et al. 2013) | |
| Sly1 | human | Co-IP, PD-W | (Laufman et al. 2009; Laufman et al. 2013) | |
| Vps45 | human | Co-IP, PD-W | (Laufman et al. 2013) | |
| COG6 | STX5 | human | Y2H, Co-IP | (Shestakova et al. 2007; Kudlyk et al. 2013; Laufman et al. 2013) |
| STX6 | human | Y2H, PD-W, Co-IP | (Laufman et al. 2011; Kudlyk et al. 2013; Willett et al. 2013) | |
| GS27 (GOSR2) | human | Y2H, Co-IP | (Kudlyk et al. 2013) | |
| SNAP29 | human | Y2H, Co-IP | (Willett et al. 2013; Kudlyk et al. 2013) | |
| COG7 | STX6 | human | Co-IP | (Laufman et al. 2013) |
| STX16 | human | Co-IP | (Laufman et al. 2013) | |
| GS28 (GOSR1) | human | Co-IP | (Laufman et al. 2013) | |
| GS15 (BET1L) | human | Co-IP | (Laufman et al. 2013) | |
| Vps45 | human | Co-IP | (Laufman et al. 2013) | |
| COG8 | STX5 | human | Y2H, Co-IP | (Willett et al. 2013) |
| STX16 | human | Y2H, Co-IP | (Laufman et al. 2013; Willett et al. 2013) | |
| GS27 (GOSR2) | human | Y2H, Co-IP | (Willett et al. 2013) |
- Y2H – yeast two hybrid
- Co-IP – co-immunoprecipitation
- PD-W – pull-down with subsequent Western Blot
- AC-MS – affinity capture with subsequent mass-speck analysis
- PD-G – pull-down with subsequent Gel staining
- PCA – protein fragment complementation assay
Table 4.
Other trafficking proteins that interact with COG
| COG subunit | Partner protein | organism | Detection method* | Reference |
|---|---|---|---|---|
| COG1 | COPI | yeast | PD-W | (Suvorova et al. 2002) |
| COG2 | COPI | yeast | PD-W | (Suvorova et al. 2002) |
| Sec21 | yeast | Y2H | (Suvorova et al. 2002) | |
| COPB | human | Y2H, Co-IP | (Miller et al. 2013) | |
| KLP4 | worm | Y2H | (Li et al. 2004) | |
| COG3 | COPI | yeast | PD-W | (Suvorova et al. 2002) |
| MYH14 (Myosin heavy chain 14) | human | AC-MS | (Kristensen et al. 2012) | |
| MYL12A (Myosin, light chain) | human | AC-MS | (Kristensen et al. 2012) | |
| ACTR1A (ARP1 actin-related protein) | human | AC-MS | (Kristensen et al. 2012) | |
| AP3B1 (clathrin assembly protein) | human | AC-MS | (Kristensen et al. 2012) | |
| ARP66B | fly | Y2H | (Giot et al. 2003) | |
| COG4 | COPI | yeast | PD-W | (Suvorova et al. 2002) |
| p97/Cdc48 (VCP) | human | AC-MS | (Yu et al. 2013) | |
| GEA2 | yeast | Y2H | (Chen et al. 2011) | |
| Chc-1 | worm | Y2H | (Boxem et al. 2008) | |
| COG5 | BLOC-1 | human | AC-MS | (Gokhale et al. 2012) |
| COPB | human | Y2H | (Miller et al. 2013) | |
| p97/Cdc48 (VCP) | human | AC-MS | (Yu et al. 2013) | |
| COG6 | COPI | yeast | PD-W | (Suvorova et al. 2002) |
| Blos-4 (BLOC1-sub4) | worm | Y2H | (Simonis et al. 2009) | |
| COG7 | BLOC-1 | human | AC-MS, Co-IP | (Gokhale et al. 2012) |
| COG8 | COPB | human | Y2H | (Miller et al. 2013) |
SNAREs
Soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs) are the proteins responsible for driving membrane fusion (Weber et al. 1998). During SNARE mediated fusion a v-SNARE, carried on a donor membrane (or vesicle), will bind to a preformed t-SNARE complex on an acceptor membrane (Parlati et al. 2000). SNARE proteins generally have four domains (Ungar and Hughson 2003). First, the N terminal peptide that is the region implicated in SNARE regulation by SM proteins. This is followed by a larger N-terminal domain (NTD), which in some syntaxin family SNAREs forms a three helix bundle and is then referred to as the Habc domain. The SNARE domain is the business end of the fusion machinery, while a transmembrane domain may or may not be present based on the specific SNARE in question. SNAREs can be classified by the structure of their SNARE domain into Qa, Qb, Qc and R categories, with Q SNAREs typically corresponding to t-SNAREs and R SNAREs corresponding to v-SNAREs (Kloepper et al. 2007). Membrane fusion is mediated via the SNARE domains that will form a four-helix bundle (or SNARE-pin) (Sutton et al. 1998). It is the zippering action during the formation of the helix bundle that will drive the membranes into close enough proximity that the two adjacent membranes will fuse (McNew et al. 2000; Söllner et al. 1993). Membrane fusion results in the formation of a the cis-SNARE complex that is then disassembled by NSF, thus recycling the SNAREs for subsequent rounds of membrane fusion.
Interaction with SNAREs and SNARE complexes is the most common feature of CATCHR complexes. The Dsl1/ZW10 complex interacts with the ER-located STX18 SNARE complex (Arasaki et al. 2006; Hirose et al. 2004), the GARP/Vps51–54 complex interacts with the trans-Golgi STX16 SNARE complex (Siniossoglou and Pelham 2002; Perez-Victoria and Bonifacino 2009), and the Exocyst complex interacts with the plasma membrane SNARE Sec9 (Morgera et al. 2012).
There are several SNARE complexes that operate at the Golgi including the STX5/GS27/Sec22/Bet1 complex, which in mammalian cells mostly operates at the ERGIC compartment (Xu et al. 2000), the STX5/GS27/GS28/GS15 complex, which is found throughout the entire Golgi (Volchuk et al. 2004), and the STX16/STX6/Vti1a/Vamp4 complex, which operates between the endosomal compartments and the trans side of the Golgi (Tang et al. 1998). The COG complex has been shown to extensively interact with both intra-Golgi and trans-Golgi localized SNARE complexes, but not with the ERGIC SNAREs (Suvorova et al. 2002; Laufman et al. 2013, 2011; Laufman et al. 2009; Shestakova et al. 2007; Willett et al. 2013). All published interactions between COG subunits and SNARE proteins are summarized in Table 1.
Potential COG-SNARE interactions have been screened using a multitude of different assays including yeast two hybrid, Co-IP, and pull-down assays. Using a directed yeast two hybrid screen, all eight COG subunits were probed for interaction with 14 Golgi localized SNAREs (Willett et al. 2013). From this, it was concluded that COG4 is capable of interaction with STX5, COG6 is capable of interacting with STX5, STX6, GS27, and SNAP29, and finally that COG8 is capable of interacting with STX5, GS27, and STX16. Also suggesting potential interactions between COG and SNAREs was a study using in vitro pull down of COG4 followed by western blotting to detect interactions with the SNAREs GS28, Vti1a, and STX16 (Laufman et al. 2013), indicating the possibility of COG4 to interact directly with both the STX5 containing SNARE complex and the STX16 containing SNARE complex. Similarly, the interaction between COG6 and STX6 was also detected by this method (Laufman et al. 2011). Testing the COG-SNARE interactions in an in vivo system verified the potential interactions detected in the in vitro systems both in a transient over-expression model (Laufman et al. 2011; Laufman et al. 2009; Willett et al. 2013; Kudlyk et al. 2013), and with endogenous proteins (Laufman et al. 2013). Most recently, a comprehensive co-IP (Laufman et al. 2013) revealed previously undetected interactions between COG7 and the SNAREs STX6, STX16, GS28 and GS15.
Defects in the COG complex were found to be deleterious for the formation and/or stability of both the STX5/GS27/GS28/GS15 and the STX16/STX6/Vti1a/Vamp4 complexes, suggesting that COG-SNARE interactions are essential for the Golgi trafficking (Shestakova et al. 2007; Laufman et al. 2013; Laufman et al. 2009). The COG4 and COG8 interactions with STX5 and STX16 (respectively) were further probed using a mitochondrial knock-sideways assay. In this assay, cells expressing a COG4 or COG8 fusion protein with the mitochondrial targeting sequence ActA (Sengupta et al. 2009) were examined for their ability to recruit interacting proteins from their normal Golgi localization to outer mitochondrial membranes. This demonstrated that specific interacting SNARE proteins are actively recruited to the mitochondria by specific COG subunits, suggesting a role of the COG complex in serving as a specific landmark for SNARE proteins to deliver them to their destination (Willett et al. 2013).
Mapping COG interacting domains with SNARE proteins has also been of interest. The interaction site of the COG4 protein with the SNARE STX5 was narrowed down to the first 222 amino acids of human COG4 and then later further refined to amino acids 1–153 (Shestakova et al. 2007; Laufman et al. 2009). The interaction site of COG6 with the SNAREs GS27 and STX6 has been narrowed down to amino acids 76–150 (Willett et al. 2013; Kudlyk et al. 2013; Laufman et al. 2011). This same part of the protein is essential for COG6's ability to incorporate into the COG complex and for Golgi integrity. The interacting region of COG8 with STX16 has been narrowed down to the C-terminus, since the first 436 amino acids were shown not to be essential for interacting with STX16 (Willett et al. 2013).
It was found that it is the SNARE domains of STX5 and STX16 that are responsible for binding to COG subunits (Willett et al. 2013; Laufman et al. 2013).
SM proteins
Sec1/Munc18 (SM) proteins are a small class of proteins which function to regulate SNARE proteins. SM proteins are “clasp” like shaped proteins that bind to SNAREs in one of three conformations. The first example is the closed conformation, characterized by the Munc18/Syntaxin1 interaction (Misura et al. 2000). In this arrangement, Munc18-1 clamps onto the the SNARE that has its Habc domain folded onto its SNARE domain (closed conformation), thus blocking it from interaction with other SNARE proteins (Fernandez et al. 1998). Munc18-1 will bind to the closed SNARE via its central cavity (Misura et al. 2000), preventing it from interaction with its cognate SNAREs and thus illustrating an inhibitory role of SM regulation of SNARE proteins (Gerber et al. 2008). SM proteins can also bind to the N terminal peptide of SNAREs, and in doing so may act as positive (like Sly1 binding to STX5 (Dulubova et al. 2002)) or negative (like Vps45 binding to STX16 (Dulubova et al. 2002) and Munc18-1 binding to STX1 (Burkhardt et al. 2008)) regulators of SNARE mediated membrane fusion. Finally, SM proteins may bind to the fully assembled trans-SNARE complex (Dulubova et al. 1999; Carr et al. 1999).
The regulation of the direct interaction between COG4 and STX5 by the SM protein Sly1 was affirmed by co-IP and in vitro pull-down experiments (Laufman et al. 2009). In good agreement with this data, the Sly1 mutant dominant allele SLY1-20, is capable of suppressing the phenotyopes of COG subunit mutations in yeast (VanRheenen et al. 1998), further indicating the presence of COG-Sly1 interactions in vivo. COG4 and COG7 were also found to interact directly with another SM protein, the STX16 regulator Vps45 (Laufman et al. 2013).
Rabs
Rab proteins are members of the small Ras like GTPase superfamily and serve as molecular switches, regulating the four steps of vesicular transport at every stage of the secretory pathway. Rab proteins cycle between “active” GTP-bound and “inactive” GDP-bound states, which are regulated by GEFs (guanine nucleotide exchange factors) that exchange the bound GDP for GTP, and GAPs (guanine activating proteins) that accelerate GTP hydrolysis (Hutagalung and Novick 2011). Coupled to the nucleotide exchange is the Rab's localization. When inactive, Rabs are bound by GDIs (GDP dissociation inhibitors) which will mask their hydrophobic isoprenyl moiety, and therefore keep the Rab in the cytosol (Ullrich et al. 1993). GDFs (GDI displacement factors) will dissociate GDI from the Rab, exposing its isoprenyl moiety, as well as making it available for interaction with GEFs, to then activate the Rab and recruit it to a specific membrane compartment where it will function with its effector molecules (Zerial and McBride 2001).
There are several well characterized examples of the function of Rab proteins during membrane trafficking, including sorting, regulating motor proteins, and regulating trafficking steps such as membrane fusion and vesicle tethering. In line with this, there is surmounting evidence of tethering proteins being Rab effectors. One such example is from the yeast system identifying the Exocyst subunits Sec15 and Sec8 as effectors of the Rab Sec4 (Guo et al. 1999; Toikkanen et al. 2003). Exocyst-Rab interactions were also confirmed in the mammalian system with Sec15 functioning as an effector of Rab11 (Zhang et al. 2004).
Of the >60 mammalian Rabs that have been identified to date (Schultz et al. 2000), 12 of them have been shown to interact with different COG subunits including Rab 1 (isoforms 1a and 1b) Rab2a, Rab4a, Rab6 (isoforms 6a and 6b), Rab10, Rab14, Rab30, Rab36, Rab39, and Rab 41 (see table 2). In the yeast system, COG2 (Sec35) and COG3 (Sec34) were shown via GST pull-down from cell lysate to interact with the Rab1 and Rab6 yeast homologs Ypt1 and Ypt6 (respectively) (Suvorova et al. 2002). Subsequent comprehensive yeast two hybrid screens for mammalian COG-Rab interactions highlight multiple potential interactions including COG6 with Rab1a/b, Rab2a, Rab4a, Rab6a/b, Rab10, Rab14, Rab36, and Rab41, COG5 with Rab2a and Rab39, and COG4 with Rab1a, Rab4a, and Rab30 (Miller et al. 2013; Fukuda et al. 2008). Of these interactions, COG4's interactions with Rab1a and Rab30 were detected directly by in vitro pull down, while Rab1a, Rab4a, Rab6a and Rab30 were all shown to interact with the COG complex by an in vitro pull-down approach. In addition, mitochondrial relocalization assays confirmed the interactions between COG5 and Rab2a, as well as between COG6 and Rab1a, Rab2a, Rab4a and Rab6a (Miller et al. 2013). Thus concluding that the COG complex has multiple modes of interactions with Golgi Rab proteins. Importantly, COG interacted in all cases stronger with the GTP-bound Rabs than the GDP versions, indicating that the COG complex and/or COG sub-complexes are true effectors of Rab GTPases.
Table 2.
COG interacting Rab proteins
| COG subunit | Partner protein | organism | Detection method* | Reference |
|---|---|---|---|---|
| COG2 | Ypt1 | yeast | PD-W | (Suvorova et al. 2002) |
| Ypt6 | yeast | PD-W | (Suvorova et al. 2002) | |
| COG3 | Ypt1 | yeast | PD-W | (Suvorova et al. 2002) |
| Ypt6 | yeast | PD-W | (Suvorova et al. 2002) | |
| COG4 | Rab1A | human | Y2H, PD-W | (Miller et al. 2013) |
| Rab4A | human | Y2H, PD-W | (Miller et al. 2013) | |
| Rab30 | human | Y2H, CO-IP, PD-W | (Fukuda et al. 2008; Miller et al. 2013) | |
| COG5 | Rab2A | human | Y2H | (Miller et al. 2013) |
| Rab39 | human | Y2H | (Miller et al. 2013) | |
| COG6 | Rab1A/B | human | Y2H | (Fukuda et al. 2008; Miller et al. 2013) |
| Rab2A | human | Y2H | (Miller et al. 2013) | |
| Rab4A | human | Y2H | (Miller et al. 2013) | |
| Rab6A/B | human | Y2H | (Fukuda et al. 2008) | |
| Rab10 | human | Y2H | (Miller et al. 2013) | |
| Rab14 | human | Y2H | (Miller et al. 2013) | |
| Rab36 | human | Y2H | (Miller et al. 2013) | |
| Rab41 | human | Y2H | (Fukuda et al. 2008) | |
| Ypt1 | yeast | Y2H | (Yu et al. 2008) | |
| Ypt6 | yeast | Y2H | (Yu et al. 2008) |
Tethers
As described above, tethers come in two flavors, long coiled-coil tethers and MTCs. In addition to COG subunits interacting with the aforementioned vesicular trafficking machinery, COG subunits also interact with coiled-coil tethers and other tethering complexes (Table 3). Co-IP of Lobe A subunits revealed an interaction between COG2 and the golgin p115, and COG3 with the golgins p115, GM130, and Giantin (Sohda et al. 2007). Similarly, co-IP analysis also revealed interactions between the golgin Golgin84 and COG subunits COG3 and COG7 (Sohda et al. 2010). Furthermore, in vitro pull-down of recombinant COG7 and Golgin84 affirm that the two proteins are directly interacting.
Table 3.
COG interacting vesicular Tethers
| COG subunit | Partner protein | organism | Detection method* | Reference |
|---|---|---|---|---|
| COG2 | P115 (USO1) | human | Y2H, Co-IP, PD-W | (Sohda et al. 2007) |
| CASP (CUX1) | human | Y2H | (Miller et al. 2013) | |
| GM130 (GOGA_2) | human | Y2H | (Miller et al. 2013) | |
| Golgin84 (GOLGA5) | human | Y2H | (Miller et al. 2013) | |
| TMF (TMF1) | human | Y2H | (Miller et al. 2013) | |
| EXO70 (EXOC_7) | plant | Y2H | (Arabidopsis Interactome Mapping 2011) | |
| COG3 | GM130 (GOGA_2) | rat | Co-IP | (Shestakova et al. 2007; Sohda et al. 2007; Miller et al. 2013) |
| P115 (USO1) | rat | Co-IP | (Shestakova et al. 2007; Sohda et al. 2007) | |
| Giantin (GOGB1) | rat | Co-IP | (Sohda et al. 2007) | |
| Vps52 | yeast | PCA | (Tarassov et al. 2008) | |
| COG4 | Tip20 | yeast | Y2H | (Uetz et al. 2000) |
| Uso-1 | worm | Y2H | (Boxem et al. 2008) | |
| COG5 | GM130 (GOGA_2) | human | Y2H | (Miller et al. 2013) |
| COG6 | TMF (TMF1) | human | Y2H | (Miller et al. 2013) |
| Sec6 | fly | Y2H | (Giot et al. 2003) | |
| COG7 | Golgin84 (GOLGA5) | human | Y2H, Co-IP | (Sohda et al. 2010; Miller et al. 2013) |
| COG8 | CASP (CUX1) | human | Y2H | (Miller et al. 2013) |
In addition to these direct interactions, potential interactions between COG subunits and the golgins TMF, CASP, GM130, and Golgin84 have also been identified. In a yeast two hybrid screen with all eight COG subunits TMF was found to be capable of interacting with COG2 and COG6, CASP was capable of interacting with COG2 and COG8, GM130 was capable of interacting with COG2 and COG5, and Golgin84 (in addition to its verified interaction with COG7) was capable of interacting with COG2 (Miller et al. 2013). These studies revealed COG2 as a major potential link between the COG complex and coiled-coil tethers. The interactions with TMF were also confirmed with co-IPs, and it could be shown that the complex is able to interact with both ends of this golgin (Miller et al. 2013).
COG interactions with other tethers are not restricted to coiled-coil tethers. In yeast, indirect interactions between COG3 and the GARP complex subunit Vps52 (Tarassov et al. 2008) and COG4 and the Dsl1 complex subunit Tip20 (Uetz et al. 2000) were reported using yeast two hybrid assays. Indirect interactions with the Exocyst complex have also been identified by yeast two hybrid screens showing interactions of the subunits COG2 and Exo70 (Arabidopsis Interactome Mapping 2011; Giot et al. 2003), and COG6 and Sec6 (Giot et al. 2003), indicating the possibility of interactions between the COG and the Exocyst complexes.
COG interaction with other trafficking related proteins
Intriguingly, seven out of the eight COG subunits (COG7 being excluded) have shown direct and/or indirect interactions with the COPI vesicle coat proteins. The COPI vesicle protein coat consists of 7 soluble proteins which are recruited to a membrane by the small GTPase Arf1 (Rothman and Wieland 1996). Pull-down of the yeast COG subunits COG1, 2, 3, 4, and 6 followed by western blotting revealed specific interactions with the entire COPI complex (Suvorova et al. 2002). Similar results were obtained in co-IPs with solubilized rat liver Golgi membranes (Zolov and Lupashin 2005). Comprehensive yeast two hybrid analysis of human COG-COPI interactions revealed that human βCOP is the only COPI subunit that can directly interact with three COG subunits COG2, COG5 and COG8, with COG2 being the strongest βCOP partner (Miller et al. 2013). In yeast COG4 may also interact with GEA2, a protein that facilitates guanine nucleotide exchange for the small GTPase regulating formation of COPI coated vesicles, Arf1 (Chen et al. 2011).
In addition to the COPI vesicular coat, the COG complex is likely to interact with the clathrin coat as well, since COG3 was found to be co-precipitated with the clathrin assembly protein AP3B1 (Kristensen et al. 2012).
Finally, COG interactions have also been identified with molecular motor proteins. Affinity capture of COG3 followed by mass spec analysis found potential interactions with the motor proteins myosin heavy chain 14 (MYH14) and myosin light chain 12 (MYL12), as well as with the actin related proteins ACTR1A and ARP66B (Giot et al. 2003; Kristensen et al. 2012). In addition to actin-related motors COG complex may also interact with microtubule motors. A yeast two hybrid screen in worms revealed a direct interaction of COG2 with the kinesin-like protein KLP4 (Li et al. 2004).
Interaction hubs of COG
The resulting interaction diagram (Figure 1) offers itself to some generalization on major COG “interaction hubs”. COG1 does not have any verified interactions outside of the COG complex and thus is likely to play a major role in regulating the interaction between different COG sub-complexes (Fotso et al. 2005; Ungar et al. 2005). As proposed by Miller et al. (Miller et al. 2013), COG2 is likely to be a major “tether hub”, connecting the COG complex to coiled-coil tethers. In addition to tethers, COG2 may also serve as a minor hub for vesicular coat components and/or kinesin motor (s). COG3 shows some interactions with tethers serving as a minor “tether hub”. More uniquely COG3 shows interaction with the actin cytoskeleton and myosin motors, indicating that COG3 is a major “motor hub” of the COG complex. In good agreement with this assignment, the acute interference with COG3 function by antibody microinjection or siRNA treatment causes a pronounced Golgi disruption and a massive accumulation of non-tethered vesicles (Zolov and Lupashin 2005). COG4 is the only Lobe A subunit that interacts with multiple SNARE and Rab proteins and we therefore designate COG4 as a “SNARE and Rab hub”. In agreement with this assignment, COG4 is shown to interact with the VCP protein, a member of the AAA ATPase family that binds the COG4-interacting SNARE STX5 (Uchiyama and Kondo 2005) and is important for Golgi re-assembly (Rabouille et al. 1995). COG4 is also a “coat hub” since it shows a unique interaction with the Arf1 Guanidine exchange factor GEA2 and with clathrin heavy chain. COG5 along with COG6 are two “Rab hubs” in the Lobe B COG sub-complex. In addition COG6 along with COG8 show interactions with β-COP and therefore may serve as minor “coat hubs”. In agreement with Miller et al (Miller et al. 2013), COG6 serves as a major Lobe B “SNARE and Rab hub”. COG7 and COG8 both interact with multiple SNAREs and SNARE-associated proteins and therefore serve as additional “SNARE hubs”.
Models for COG complex involvement in membrane trafficking
Three separate, but interconnected, molecular roles for the COG complex have been proposed: assembly of vesicle docking stations (Willett et al. 2013; Shestakova et al. 2007), direct facilitation of vesicle tethering (Suvorova et al. 2002; Zolov and Lupashin 2005; Miller et al. 2013), and promotion of SNARE complex assembly (Shestakova et al. 2007; Laufman et al. 2011, 2013; Kudlyk et al. 2013). Below we will analyze the COG complex interactions with different protein partners in relation to these proposed functions. The importance of COG mediated trafficking comes from the cargo transported by the retrograde vesicles. This is mainly made up of glycosylation enzymes and other glycosylation associated proteins, such as sugar nucleotide transporters. Thus this vesicle transport route is responsible for ensuring the nonuniform distribution of the glycosylation machinery between Golgi cisternae, a prerequisite for correct glycan biosynthesis. COG's central importance in this process is highlighted by the numerous glycosylation phenotypes found in COG defective model organisms (Kingsley et al. 1986; Whyte and Munro 2001; Struwe and Reinhold 2012), and in COG defective patient or human cell lines (Wu et al. 2004; Zeevaert et al. 2008; Pokrovskaya et al. 2011). Interestingly, different COG mutations can cause very different glycosylation anomalies in the same model. Largely, lobe A defects affect early Golgi glycosylation, while lobe B defects modify late Golgi reactions (Miller and Ungar 2012). Detailed mechanistic knowledge of COG function will be required to understand how the different COG mutations cause such different effects on glycosylation homeostasis.
Docking station assembly model
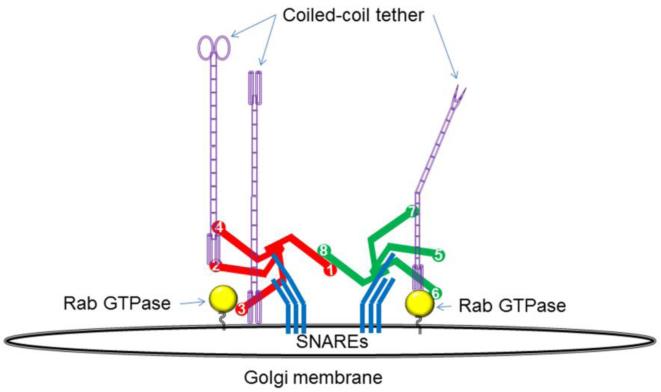
In the docking station assembly model (Figure 2) the entire COG complex, as well as its sub-complexes, may act to orchestrate the assembly of pre-tethering protein complexes on an acceptor membrane. These complexes would include a specific combination of Rabs, coiled-tethers and t-SNAREs, and would transiently assemble at the rims of different Golgi cisternae. These primed assembled complexes would then be ready to accept incoming retrograde transport vesicles. Indeed,immunogold electron microscopy reveals that the COG1 protein is localized both on vesiculat structures and on (or in close proximity) to the tips and rims of the Golgi's cisternae (Vasile et al. 2006). In order to recognize trafficking intermediates with different destination addresses, distinct docking stations must be assembled at different Golgi cisternae. We hypothesize that these assemblies are organized by binary interactions between individual docking components and the multipronged interactions between COG and other docking station components. In this scenario different COG sub-complexes should recognize different protein assemblies. Indeed, specific cis-Golgi interactions (COG2-p115, COG4-Rab1, COG4-STX5) have been observed for Lobe A of the COG complex, while Lobe B components demonstrated interactions with trans-Golgi factors (COG6-Rab6, COG6-STX6, COG8-STX16). In a good agreement with this model COG4 transplanted to the outer mitochondrial membrane was efficient in recruiting other COG subunits, cis-Golgi Rabs and the coiled coil protein p115 to build docking stations that can tether STX5-containing trafficking intermediates, while COG8 was found to recruit a different set of partners including Lobe B components and the trans-Golgi Rab6 to direct the trafficking of STX16-bearing membranes (Willett et al. 2013).
Figure 2.
The COG complex assembles docking stations for retrograde intra-Golgi vesicles
Hypothetical model depicting how the COG complex may serve to assemble docking stations on Golgi membranes for incoming retrograde vesicles. In this model the COG complex (or COG sub-complexes) act to orchestrate the assembly of a pre-tethering protein complex through interactions with Rabs, coiled-coil tethers, and SNAREs. The targeting of a vesicle in this model is defined by the specific COG interactions, meaning the Rab, SNARE, or tether that are assembled into the docking complex.
The major problem with this model is the lack of information about the signal(s) that initiate the formation of the vesicle docking stations in the right place at the right time. One possibility here is that RabGEF's or yet another COG interacting partner could sense environmental queues (abundance of components of the glycosylation machinery, lipid composition, etc) to initiate the formation of docking stations.
Direct facilitation of vesicle tethering model
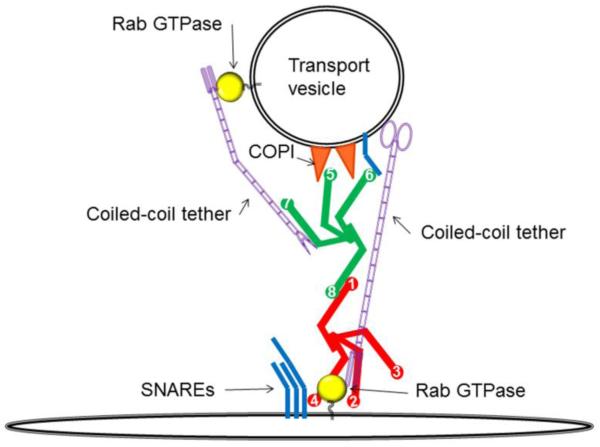
Both yeast and mammalian cells deficient for COG complex subunits accumulate non-tethered transport vesicles that are enriched in recycling Golgi components, indicating that the COG complex, and/or its sub-complexes, are directly facilitating the tethering of transport intermediates (Wuestehube et al. 1996; Zolov and Lupashin 2005; Sohda et al. 2010). Indeed the direct binding of intra-Golgi vesicles to immobilized COG complex was demonstrated (Shestakova et al. 2006). Moreover, several COG subunits are able to bind COPI coat components (Miller et al. 2013; Suvorova et al. 2002; Ram et al. 2002; Zolov and Lupashin 2005), indicating the direct involvement of the COG complex in recognition of COPI-coated intra-Golgi vesicles. In the vesicle tethering model (Figure 3), the COG complex may use its protein-protein recognition skills to establish firm bridges between an acceptor membrane and an incoming retrograde vesicle. In this model, the COG complex may simultaneously establish the connection with the vesicle (via coat, v-SNAREs and Rab proteins) and acceptor membrane (via t-SNAREs and Rabs). The simultaneous binding may stabilize and shorten already existing vesicle-membrane interactions facilitated by coiled-coil tethers like p115, golgin84, or TMF, which form an initial long range link between the Golgi membrane and an incoming vesicle. COG would thus act to reel-in a vesicle and help to read out the targeting information displayed by a specific subset of Rabs and golgins (Miller et al. 2013).
Figure 3.
The COG complex directly facilitates vesicle tethering
Hypothetical model depicting how the COG complex may act to directly facilitate the tethering of a retrograde vesicle with its acceptor membrane. In this model, the interactions between COG subunits and trafficking proteins carried on an incoming vesicle (for example v-SNAREs, Rabs, and/or coat proteins) would establish a connection between the vesicle and an acceptor membrane. Additionally, COG interactions could also serve to shorten or even stabilize the connection between the vesicle and the acceptor membrane achieved by long coiled-coil tethers.
SNARE complexes stabilization model
An important feature of COG-deprived cells is an increased degradation of a sub-set of Golgi SNAREs (Oka et al. 2004; Zolov and Lupashin 2005; Laufman et al. 2011) and a relative decrease in the steady-state levels of Golgi-operating SNARE complexes (Laufman et al. 2013; Laufman et al. 2009; Shestakova et al. 2007). This feature was interpreted as a direct involvement of the COG complex in the formation and/or stabilization of SNARE complexes. Yet caution needs to be exercised with this interpretation, since several Golgi localized proteins (termed GEARs) have been shown to be sensitive to COG mutations (Oka et al. 2004). Thus the instability of the SNAREs could well be a secondary effect.
In the SNARE complex stabilization model (Figure 4) the COG complex or its sub-complexes may enhance the stability of SNAREs by multi-pronged binding to a pre-assembled t-t and/or v-t SNARE complexes. For instance the entire COG complex may bind to the STX5/GS27/GS28 t-t SNARE complex via simultaneous interactions between COG4-STX5 (Shestakova et al. 2007), COG6-GS27 (Kudlyk et al. 2013) and COG7-GS28 (Laufman et al. 2013). Interactions between COG4 and the STX5 partner protein SLY1 (Laufman et al. 2009) may additionally stabilize this complex and protect it from NSF or VCP-mediated disassembly. Interestingly, the VCP/CDC48/p97 ATPase that was specifically implicated in the disassembly of STX5-containing SNARE complexes (Rabouille et al. 1998) can itself bind to COG4 and COG5 (Yu et al. 2013). In addition to stabilization, the COG complex could directly promote the formation of v-t SNARE complexes during vesicle docking. This function has been proposed for other multisubunit tethering complexes including HOPS (Collins et al. 2005) and GARP (Perez-Victoria and Bonifacino 2009). In support to this model COG4 has been shown to facilitate SNARE complex formation in vitro (Laufman et al. 2013).
Figure 4.
The COG complex stabilizes SNARE complexes
Hypothetical model depicting how the COG complex may assist in the stabilization of t-t and or v-t SNARE complexes. In this model multiple COG subunits interacting with the same SNARE complex (COG4-STX5, COG6-GS27, and COG7-GS28 interactions with the STX5/GS27/GS28/GS15 SNARE complex and COG6-STX6, COG8-STX16, and COG4-Vti1a interactions with the STX16/STX6/Vti1a/Vamp4 SNARE complex) may act to stabilize the complex and protect it from NSF mediated disassembly. Furthermore, interactions between COG subunits and SM proteins may aid in the COG mediated stabilization of SNARE complexes.
Finally, the COG complex may act to synchronize and proof-read the assembly of several SNARE complexes via a simultaneous interaction of COG subunits with SNAREs in different sets. For instance STX5 in one SNARE complex can be approached by COG4 while COG6 from the same COG complex would bind to STX5 or GS27 in the nearby SNARE assembly cross-linking both SNARE complexes. It has been postulated that 3–8 SNARE complexes are required for the efficient fusion of one vesicle to an acceptor membrane (Hua and Scheller 2001; Domanska et al. 2009). In this model the COG complex is in a unique position to orchestrate SNARE synchronization during the docking process.
Towards a unified model of COG's molecular function
The above outlined models are not necessarily contradictory to each other, they could in fact represent different stages of COG function. For example, the proposed docking stations could well be formed by sub-assemblies that have been observed on Golgi membranes (for example lobe A (Willett & Lupashin, unpublished)), while other sub-assemblies (for example cytosolic lobe B (Ungar et al. 2002)) could act in bridging the distance between vesicle and target membranes. It is interesting in this respect that excess lobe A may inhibit lobe B mediated tethering at the trans-Golgi (Cottam and Ungar submitted manuscript) implying that a delicate balance of the different COG subassemblies is necessary to facilitate intra-Golgi transport.
Moreover, the SNARE assembly function of COG could easily be incorporated into both the docking station and the membrane bridging models. The complex, or its appropriate subassemblies are in the right spot in the docking station model to promote SNARE assembly once the vesicle is in close enough proximity to the target membrane. In contrast, the bridge formed by COG in the bridging model could well morph into a SNARE assembly helper.
Concluding remarks
It is clear that the COG complex plays a central role in the targeting of vesicles at the Golgi apparatus. This is most likely achieved by a large number of weak interactions and some accompanying conformational changes. The past few years have accumulated a wealth of knowledge about COG's protein interactions, and given us some beginning insight into how these interactions could bring a vesicle close to a specific Golgi cisterna and promote SNARE mediated fusion. Any of the three outlined models – COG acting as a docking station, bridging the membranes, or mediating SNARE assembly – or their combination would fit currently available evidence. The rather large number of interaction partners plausibly reflects several independent targeting steps that COG is involved in, and untangling this mesh will be the most difficult challenge for COG research in the near future. Help could come from the Golgi's main function – glycosylation. Different sets of glycosylation enzymes have to be targeted to the various cisternae. Each of these targeting reactions requires COG, but conceivably different sets of interaction partners. COG dependent CDGs for example exhibit defects in trans-Golgi enzyme sorting, likely due to the inviability of defects earlier in the Golgi. Thus the molecular defects in these diseases can give us some clues about the targeting requirements for the late Golgi region, and similarly, more severe glycosylation defects in model organisms can be used to investigate COG's early Golgi functions. In closing, more detailed structural studies in combination with in vitro reconstitution assays and sophisticated in vivo dissections of the different COG mediated trafficking steps will ultimately be required to fully understand the molecular workings of this intriguing membrane trafficking machine.
Acknowledgements
We are thankful to Irina Pokrovskaya and Tetyana Kudlyk for excellent technical support. V.L. was supported by the National Institute of Health (1R01GM083144); D.U. was supported by a BBSRC grant (BB/F006993/1/) and a Marie Curie grant (201098).
References
- Arabidopsis Interactome Mapping C Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333(6042):601–607. doi: 10.1126/science.1203877. doi:10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K, Taniguchi M, Tani K, Tagaya M. RINT-1 regulates the localization and entry of ZW10 to the syntaxin 18 complex. Mol Biol Cell. 2006;17(6):2780–2788. doi: 10.1091/mbc.E05-10-0973. doi:10.1091/mbc.E05-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116(2):153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C, Tipsword M, Drew K, Galli M, Rhrissorrakrai K, Drechsel D, Koller D, Roth FP, Iakoucheva LM, Dunker AK, Bonneau R, Gunsalus KC, Hill DE, Piano F, Tavernier J, van den Heuvel S, Hyman AA, Vidal M. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134(3):534–545. doi: 10.1016/j.cell.2008.07.009. doi:10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. Embo J. 2008;27(7):923–933. doi: 10.1038/emboj.2008.37. doi:10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. Journal of Cell Biology. 1999;146(2):333–344. doi: 10.1083/jcb.146.2.333. doi:DOI 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Cai H, Park SK, Menon S, Jackson CL, Ferro-Novick S. Trs65p, a subunit of the Ypt1p GEF TRAPPII, interacts with the Arf1p exchange factor Gea2p to facilitate COPI-mediated vesicle traffic. Mol Biol Cell. 2011;22(19):3634–3644. doi: 10.1091/mbc.E11-03-0197. doi:10.1091/mbc.E11-03-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KM, Thorngren NL, Fratti RA, Wickner WT. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. Embo J. 2005;24(10):1775–1786. doi: 10.1038/sj.emboj.7600658. doi:10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam NP, Ungar D. Retrograde vesicle transport in the Golgi. Protoplasma. 2012;249(4):943–955. doi: 10.1007/s00709-011-0361-7. doi:10.1007/s00709-011-0361-7. [DOI] [PubMed] [Google Scholar]
- Domanska MK, Kiessling V, Stein A, Fasshauer D, Tamm LK. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J Biol Chem. 2009;284(46):32158–32166. doi: 10.1074/jbc.M109.047381. doi:10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. Embo J. 1999;18(16):4372–4382. doi: 10.1093/emboj/18.16.4372. doi:10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, Rizo J. How Tlg2p/syntaxin 16 `snares' Vps45. Embo J. 2002;21(14):3620–3631. doi: 10.1093/emboj/cdf381. doi:10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94(6):841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J Biol Chem. 2005;280(30):27613–27623. doi: 10.1074/jbc.M504597200. doi:10.1074/jbc.M504597200. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics. 2008;7(6):1031–1042. doi: 10.1074/mcp.M700569-MCP200. doi:10.1074/mcp.M700569-MCP200. [DOI] [PubMed] [Google Scholar]
- Gerber SH, Rah JC, Min SW, Liu X, de Wit H, Dulubova I, Meyer AC, Rizo J, Arancillo M, Hammer RE, Verhage M, Rosenmund C, Sudhof TC. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321(5895):1507–1510. doi: 10.1126/science.1163174. doi:10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr., White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. doi:10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Glick BS, Elston T, Oster G. A cisternal maturation mechanism can explain the asymmetry of the Golgi stack. FEBS Lett. 1997;414(2):177–181. doi: 10.1016/s0014-5793(97)00984-8. doi:Doi 10.1016/S0014-5793(97)00984-8. [DOI] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, Lupashin VV, Smith Y, Faundez V. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci. 2012;32(11):3697–3711. doi: 10.1523/JNEUROSCI.5640-11.2012. doi:10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. Embo J. 1999;18(4):1071–1080. doi: 10.1093/emboj/18.4.1071. doi:DOI 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H, Arasaki K, Dohmae N, Takio K, Hatsuzawa K, Nagahama M, Tani K, Yamamoto A, Tohyama M, Tagaya M. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. Embo J. 2004;23(6):1267–1278. doi: 10.1038/sj.emboj.7600135. doi:10.1038/sj.emboj.7600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Scheller RH. Three SNARE complexes cooperate to mediate membrane fusion. Proc Natl Acad Sci U S A. 2001;98(14):8065–8070. doi: 10.1073/pnas.131214798. doi:10.1073/pnas.131214798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91(1):119–149. doi: 10.1152/physrev.00059.2009. doi:10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM, Kozarsky KF, Segal M, Krieger M. Three types of low density lipoprotein receptor-deficient mutant have pleiotropic defects in the synthesis of N-linked, O-linked, and lipid- linked carbohydrate chains. J Cell Biol. 1986;102(5):1576–1585. doi: 10.1083/jcb.102.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18(9):3463–3471. doi: 10.1091/mbc.E07-03-0193. doi:10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AR, Gsponer J, Foster LJ. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods. 2012;9(9):907–909. doi: 10.1038/nmeth.2131. doi:10.1038/nmeth.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlyk T, Willett R, Pokrovskaya ID, Lupashin V. COG6 interacts with a subset of the Golgi SNAREs and is important for the Golgi complex integrity. Traffic. 2013;14(2):194–204. doi: 10.1111/tra.12020. doi:10.1111/tra.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol. 2011;194(3):459–472. doi: 10.1083/jcb.201102045. doi:10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S. The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. J Cell Sci. 2013;126(Pt 6):1506–1516. doi: 10.1242/jcs.122101. doi:10.1242/jcs.122101. [DOI] [PubMed] [Google Scholar]
- Laufman O, Kedan A, Hong W, Lev S. Direct interaction between the COG complex and the SM protein, Sly1, is required for Golgi SNARE pairing. Embo J. 2009;28(14):2006–2017. doi: 10.1038/emboj.2009.168. doi:10.1038/emboj.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. doi:10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Sollner TH, Rothman JE. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407(6801):153–159. doi: 10.1038/35025000. doi:10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- Miller VJ, Sharma P, Kudlyk TA, Frost L, Rofe AP, Watson IJ, Duden R, Lowe M, Lupashin VV, Ungar D. Molecular insights into vesicle tethering at the Golgi by the conserved oligomeric Golgi (COG) complex and the golgin TATA element modulatory factor (TMF) J Biol Chem. 2013;288(6):4229–4240. doi: 10.1074/jbc.M112.426767. doi:10.1074/jbc.M112.426767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VJ, Ungar D. Re'COG'nition at the Golgi. Traffic. 2012;13(7):891–897. doi: 10.1111/j.1600-0854.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex [see comments] Nature. 2000;404(6776):355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Morgera F, Sallah MR, Dubuke ML, Gandhi P, Brewer DN, Carr CM, Munson M. Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Mol Biol Cell. 2012;23(2):337–346. doi: 10.1091/mbc.E11-08-0670. doi:10.1091/mbc.E11-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3(6) doi: 10.1101/cshperspect.a005256. doi:10.1101/cshperspect.a005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Ungar D, Hughson FM, Krieger M. The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol Biol Cell. 2004;15(5):2423–2435. doi: 10.1091/mbc.E03-09-0699. doi:10.1091/mbc.E03-09-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, McNew JA, Fukuda R, Miller R, Sollner TH, Rothman JE. Topological restriction of SNARE-dependent membrane fusion [see comments] Nature. 2000;407(6801):194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol. 2009;29(19):5251–5263. doi: 10.1128/MCB.00495-09. doi:10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya ID, Willett R, Smith RD, Morelle W, Kudlyk T, Lupashin VV. Conserved oligomeric Golgi complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology. 2011;21(12):1554–1569. doi: 10.1093/glycob/cwr028. doi:10.1093/glycob/cwr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92(5):603–610. doi: 10.1016/s0092-8674(00)81128-9. doi:Doi 10.1016/S0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82(6):905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Ram RJ, Li BJ, Kaiser CA. Identification of Sec36p, Sec37p, and Sec38p: Components of yeast complex that contains Sec34p and Sec35p. Molecular Biology of the Cell. 2002;13(5):1484–1500. doi: 10.1091/mbc.01-10-0495. doi:DOI 10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez IB, Lowe M. Golgins and GRASPs: holding the Golgi together. Semin Cell Dev Biol. 2009;20(7):770–779. doi: 10.1016/j.semcdb.2009.03.011. doi:10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272(5259):227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Schultz J, Doerks T, Ponting CP, Copley RR, Bork P. More than 1,000 putative new human signalling proteins revealed by EST data mining. Nat Genet. 2000;25(2):201–204. doi: 10.1038/76069. doi:10.1038/76069. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J Cell Biol. 2009;186(1):41–55. doi: 10.1083/jcb.200902110. doi:10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J Cell Biol. 2007;179(6):1179–1192. doi: 10.1083/jcb.200705145. doi:10.1083/jcb.200705145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A, Zolov S, Lupashin V. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 2006;7(2):191–204. doi: 10.1111/j.1600-0854.2005.00376.x. doi:10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, Ayivi-Guedehoussou N, Dann E, Bertin N, Szeto D, Dricot A, Yildirim MA, Lin C, de Smet AS, Kao HL, Simon C, Smolyar A, Ahn JS, Tewari M, Boxem M, Milstein S, Yu H, Dreze M, Vandenhaute J, Gunsalus KC, Cusick ME, Hill DE, Tavernier J, Roth FP, Vidal M. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6(1):47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277(50):48318–48324. doi: 10.1074/jbc.M209428200. doi:10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Nakamura N, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Interaction of Golgin-84 with the COG complex mediates the intra-Golgi retrograde transport. Traffic. 2010;11(12):1552–1566. doi: 10.1111/j.1600-0854.2010.01123.x. doi:10.1111/j.1600-0854.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8(3):270–284. doi: 10.1111/j.1600-0854.2006.00530.x. doi:10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway invitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75(3):409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Struwe WB, Reinhold VN. The conserved oligomeric Golgi complex is required for fucosylation of N-glycans in Caenorhabditis elegans. Glycobiology. 2012;22(6):863–875. doi: 10.1093/glycob/cws053. doi:10.1093/glycob/cws053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395(6700):347–353. doi: 10.1038/26412. doi:10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157(4):631–643. doi: 10.1083/jcb.200111081. doi:10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Low DY, Lee SS, Tan AE, Hong W. Molecular cloning and localization of human syntaxin 16, a member of the syntaxin family of SNARE proteins. Biochem Biophys Res Commun. 1998;242(3):673–679. doi: 10.1006/bbrc.1997.8029. doi:10.1006/bbrc.1997.8029. [DOI] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320(5882):1465–1470. doi: 10.1126/science.1153878. doi:10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Toikkanen JH, Miller KJ, Soderlund H, Jantti J, Keranen S. The beta subunit of the Sec61p endoplasmic reticulum translocon interacts with the exocyst complex in Saccharomyces cerevisiae. J Biol Chem. 2003;278(23):20946–20953. doi: 10.1074/jbc.M213111200. doi:10.1074/jbc.M213111200. [DOI] [PubMed] [Google Scholar]
- Uchiyama K, Kondo H. p97/p47-Mediated biogenesis of Golgi and ER. Journal of biochemistry. 2005;137(2):115–119. doi: 10.1093/jb/mvi028. doi:10.1093/jb/mvi028. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. doi: 10.1038/35001009. doi:10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268(24):18143–18150. [PubMed] [Google Scholar]
- Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. doi:10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157(3):405–415. doi: 10.1083/jcb.200202016. doi:10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit architecture of the conserved oligomeric Golgi complex. J Biol Chem. 2005;280(38):32729–32735. doi: 10.1074/jbc.M504590200. doi:10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol. 1998;141(5):1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile E, Oka T, Ericsson M, Nakamura N, Krieger M. IntraGolgi distribution of the Conserved Oligomeric Golgi (COG) complex. Exp Cell Res. 2006;312(16):3132–3141. doi: 10.1016/j.yexcr.2006.06.005. doi:10.1016/j.yexcr.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Volchuk A, Ravazzola M, Perrelet A, Eng WS, Di Liberto M, Varlamov O, Fukasawa M, Engel T, Sollner TH, Rothman JE, Orci L. Countercurrent distribution of two distinct SNARE complexes mediating transport within the Golgi stack. Mol Biol Cell. 2004;15(4):1506–1518. doi: 10.1091/mbc.E03-08-0625. doi:10.1091/mbc.E03-08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92(6):759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Whyte JRC, Munro S. The SeC34/35 golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Developmental Cell. 2001;1(4):527–537. doi: 10.1016/s1534-5807(01)00063-6. doi:Doi 10.1016/S1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Willett R, Kudlyk T, Pokrovskaya I, Schonherr R, Ungar D, Duden R, Lupashin V. COG complexes form spatial landmarks for distinct SNARE complexes. Nat Commun. 2013;4:1553. doi: 10.1038/ncomms2535. doi:10.1038/ncomms2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Steet RA, Bohorov O, Bakker J, Newell J, Krieger M, Spaapen L, Kornfeld S, Freeze HH. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat Med. 2004;10(5):518–523. doi: 10.1038/nm1041. doi:10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- Wuestehube LJ, Duden R, Eun A, Hamamoto S, Korn P, Ram R, Schekman R. New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics. 1996;142(2):393–406. doi: 10.1093/genetics/142.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Joglekar AP, Williams AL, Hay JC. Subunit structure of a mammalian ER/Golgi SNARE complex [In Process Citation] J Biol Chem. 2000;275(50):39631–39639. doi: 10.1074/jbc.M007684200. [DOI] [PubMed] [Google Scholar]
- Yu CC, Yang JC, Chang YC, Chuang JG, Lin CW, Wu MS, Chow LP. VCP phosphorylation-dependent interaction partners prevent apoptosis in Helicobacter pylori-infected gastric epithelial cells. PLoS One. 2013;8(1):e55724. doi: 10.1371/journal.pone.0055724. doi:10.1371/journal.pone.0055724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, deSmet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. doi:10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaert R, Foulquier F, Jaeken J, Matthijs G. Deficiencies in subunits of the Conserved Oligomeric Golgi (COG) complex define a novel group of Congenital Disorders of Glycosylation. Mol Genet Metab. 2008;93(1):15–21. doi: 10.1016/j.ymgme.2007.08.118. doi:10.1016/j.ymgme.2007.08.118. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Ellis S, Sriratana A, Mitchell CA, Rowe T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J Biol Chem. 2004;279(41):43027–43034. doi: 10.1074/jbc.M402264200. doi:10.1074/jbc.M402264200. [DOI] [PubMed] [Google Scholar]
- Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168(5):747–759. doi: 10.1083/jcb.200412003. doi:10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]