Abstract
The ca. 275,000 species of flowering plants are the result of a recent adaptive radiation driven largely by the coevolution between plants and their animal pollinators. Identification of genes and mutations responsible for floral trait variation underlying pollinator specificity is critical to understanding how pollinator shifts occur between closely related species. Petunia, Mimulus, and Antirrhinum have provided a high standard of experimental evidence to establish causal links from genes to floral traits to pollinator responses. In all three systems, MYB transcription factors seem to play a prominent role in the diversification of pollinator-associated floral traits.
Introduction
Most flowering plants rely on animal pollination for reproductive success. Flower-pollinator interactions are considered to be a major driver for floral trait diversification and angiosperm speciation [1–3]. A key observation supporting this proposition is that many angiosperm species produce flowers with a particular pollination syndrome, a suite of floral phenotypes that enable specialized associations with the “attraction and utilization of a specific group of animals as pollinators” [2]. These traits include flower size, color, scent, texture, shape, orientation, reward (e.g., nectar, pollen, fragrance), and pistil and stamen arrangements. For example, hummingbird-pollinated species typically have red flowers with a floral tube, copious nectar production, and exserted stamens and pistils, whereas bee-pollinated flowers display various colors (but usually not red), a smaller quantity of nectar, inserted stamens and pistils, and a clear landing platform. A molecular description of the genetic control of such pollinator specificity is crucial to understanding how pollinator shifts occur between closely related species, which are often associated with dramatic floral trait divergence and pollinator-mediated reproductive isolation and speciation.
Since the early 1990s, quantitative trait locus (QTL) analysis of floral traits that affect pollinator preference has been carried out in a few plant lineages such as Mimulus [4], Aquilegia [5], Petunia [6], and Iris [7]. These studies suggest that many pollinator-associated floral traits are controlled by few loci with large effects. However, progress in identifying the specific genes and mutations that are responsible has been quite slow until recently. This is perhaps not surprising because most plant systems for investigating flower-pollinator interactions were not particularly amenable to fine-scale genetic analysis, especially before the advent of massively parallel sequencing technologies.
Another factor that has impeded a deep understanding of the genetic control of flower-pollinator interactions is the admixture of different standards of evidence that have been used in the literature to link genotype to phenotype to pollinator response. Overall, correlative evidence is prevalent in linking genes to floral phenotypes, and pollinator responses to a particular floral trait are often assumed instead of being tested in controlled experiments. Here we first attempt to lay out a common set of experimental evidence that is necessary to establish a causal link from gene to floral trait to pollinator response, and then discuss recent studies that best fit these criteria.
The evidence necessary to link genotype to phenotype to pollinator response
Ideally, a causal link between genotype and phenotype can be established by a combination of fine-scale genetic mapping and functional characterization through transgenic manipulations. We consider that a genotype-phenotype link is formally established if at least one of the two following requirements is fulfilled: (i) fine-scale genetic mapping to the single gene level; (ii) QTL mapping or co-segregation analysis indicates a candidate gene and transgenic manipulations of the candidate gene result in expected phenotypes. Neither of these two lines of evidence has been readily available for most plant systems that are used to study flower-pollinator interactions. However, whole genome sequencing is becoming a routine practice - even for non-model systems - which makes fine-scale genetic mapping feasible, and the development of transformation protocols for a non-model plant is tedious but not always difficult.
Once the genetic basis of a floral trait is determined, pollinator foraging assays (in controlled artificial environments or natural habitats) are required to determine how pollinators respond to alternative phenotypes produced by the different alleles using carefully constructed plant materials. Transgenic lines with manipulation of a single gene would be ideal to test the role of this gene in pollinator preference with absolute confidence. When dealing with the effect of loss-of-function alleles, induced recessive mutants that differ from the wild-type parental line only at the target locus could be equally appropriate. If these materials are not available, a third (suboptimal) alternative is near-isogenic lines (NILs) that differ from the parental line only in a small region of the genome that contains the causal gene, although in this case precautions should be taken to ensure the substituted genomic region does not affect other pollinator-associated floral traits, especially less immediately obvious traits such as scent or texture.
In recent years considerable efforts have been made to identify genes underlying pollinator-associated floral trait variation in several plant systems, including Petunia, Mimulus, Antirrhinum, Ipomoea, Clarkia, and Phlox. For example, in Ipomoea, down-regulation of the flavonoid 3′-hydroxylase (F3′H) gene due to cis-regulatory change in some species resulted in flower color change from blue/purple to red [8], which is correlated with the transition from bee- to hummingbird-pollination. In Clarkia, up-regulation of the S-LINALOOL SYNTHASE (LIS) gene contributes to the strong scent emission in C. breweri, the only moth-pollinated species in the genus [9]. However, in neither of the two systems has pollinator response to allelic variants of the identified genes been tested, leaving the significance of these individual genes in controlling pollinator preference unresolved. In Phlox, cis-regulatory changes in the flavonoid 3′5′-hydroxylase (F3′5′H) gene and an R2R3-MYB transcription factor gene have been implicated in flower color change leading to pollinator-mediated speciation by reinforcement [10,11]. However, direct evidence from fine-scale mapping or transgenic manipulations to verify the gene identity is still lacking, and the pollinator-mediated selection in this case is due to pollinator constancy rather than the pollinator specificity that is required for a shift between pollinator guilds [11]. It is the other three systems (i.e., Petunia, Mimulus, and Antirrhinum) that have contributed the most rigorous experimental evidence to our current knowledge of the genetic control of flower-pollinator specificity, and these will be discussed in more detail.
Petunia — from flower color to scent
Petunia integrifolia, P. axillaris, and P. exserta are closely related species displaying a typical bee, hawkmoth, and hummingbird pollination syndrome, respectively [12]. Petunia integrifolia has purple, scentless flowers with a short, wide corolla tube and little nectar; P. axillaris has white, fragrant flowers with a long, narrow corolla tube and a large volume of nectar; and P. exserta flowers are bright red, scentless, with exserted stamens and pistils and copious nectar (Figure 1).
Figure 1.

Petunia flowers and their pollinators. (a) Bee-pollinated P. integrifolia. (b) Hawkmoth-pollinated P. axillaris. (c) Hummingbird-pollinated P. exserta. Images are provided by Dr. Cries Kuhlemeier.
A key regulator that controls the flower color difference between P. integrifolia and P. axillaris was identified as ANTHOCYANIN2 (AN2) [13,14••], encoding an R2R3-MYB transcription factor. A typical flowering plant genome harbors >200 MYB genes, ~2/3 of which encode transcription factors with two adjacent MYB domains (i.e., R2R3-MYBs) and ~1/3 with a single MYB repeat [15–17]. AN2 belongs to subgroup 6 of R2R3-MYBs [15,17] that form a protein complex with basic helix-loop-helix (bHLH) and WD repeat proteins to activate anthocyanin biosynthesis in most anthocyanin-pigmented flowers, including P. integrifolia [13,14,18]. The white color of P. axillaris results from loss of AN2 function through multiple independent acquisitions of nonsense or frameshift mutations in the AN2 coding DNA regions [13,14••].
To investigate how alternative AN2 alleles impact pollinator preference, Hoballah et al. [14••] transformed the functional P. integrifolia AN2 allele into the P. axillaris background, converting the white flower to purple, while all other floral traits remained the same as in wild-type P. axillaris. When tested in controlled greenhouse conditions, hawkmoths showed marked preference for the wild-type white flowerover the purple transgenic flowers, whereas bumblebees showed preference in the opposite direction. The AN2 case thus fits our criteria for linking genotype, phenotype, and pollinator response by taking original genetic data from hybrid crosses and transposon tagging, verifying and characterizing the effects of the locus with transgenic plants, and then using those plants to assess pollinator response in a controlled greenhouse environment [13,14••].
The genetic basis and functional significance of scent production have also been investigated in Petunia. All wild accessions of P. axillaris produce a substantial amount of methylbenzoate [19••], a volatile that elicits a strong response from hawkmoths in electroantennogram assays [20], whereas the hummingbird-pollinated P. exserta produces no detectable volatile compounds at all. QTL mapping located two major loci underlying the scent production difference between P. axillaris and P. exserta: one on chromosome II and the other on chromosome VII [19••]. The latter locus contains a candidate gene ODORANT1 (ODO1), an R2R3 -MYB gene that belongs to a subgroup with AtMYB42 and AtMYB85 and has been shown to regulate benzenoid volatile production in P. hybrida cv Mitchell [21]. The level of ODO1 transcripts is perfectly correlated with scent production in wild Petunia accessions, and is ~10-fold higher in scented P. axillaris than the background level in scentless P. exserta [19••]. Assaying the relative expression levels of ODO1 alleles in the F1 hybrids indicated that the expression difference between the two species is due to cis-regulatory change in the ODO1 gene. These data strongly implicate ODO1 as the causal gene underlying this QTL, although more definitive evidence will likely come from transgenic rescue experiments to test whether the scented P. axillaris ODO1 allele can restore scent production in the non-scented P. exserta background. The responsible gene underlying the chromosome II QTL has yet to be determined. Another subgroup of R2R3-MYB genes (subgroup 19) has also recently been demonstrated to regulate benzenoid biosynthesis in Petunia [22,23], including EMISSION OF BENZENOIDS I and II (EOBI and EOBII). However, both EOBI and EOBII can be ruled out as candidate genes for the chromosome II QTL, because they are known to positively regulate ODO1, whereas the expression of ODO1 is not dependent on the chromosome II QTL [19••].
By introgressing these two scent loci, Klahre et al. [19••] created a non-scented, white-flowered NIL in the P. axillaris genetic background, and a scented, red-flowered NIL in the P. exserta background. When presented with scented and non-scented white flowers in a dual choice experiment, hawkmoths preferred the scented flowers. Similarly, hawkmoths also preferred scented red flowers to non-scented red flowers. Intriguingly, when confronted with conflicting cues from scented red flowers and non-scented white flowers, the hawkmoths visited both types of flowers equally as a first choice, indicating that color and scent are equally important in driving hawkmoth foraging decisions.
Mimulus — from natural variation to induced mutants
The monkeyflower species Mimulus lewisii and M. cardinalis represent another system for studying the genetics of pollinator-mediated reproduction isolation between sister species [4,24–26]. The bumblebee-pollinated M. lewisii has pale pink flowers with broad, flat petals and contrasting yellow nectar guides (Figure 2a). The hummingbird-pollinated M. cardinalis has red flowers with reflexed petals, exserted stamens and pistils, and copious nectar (Figure 2b). The pale pink color of M. lewisii results from a low concentration of anthocyanins and absence of carotenoids (except in the nectar guides, Figure 2a). The red color of M. cardinalis is produced by a combination of high concentrations of both pigments. Although the two species are interfertile with hand pollination, they rarely hybridize in wild sympatric populations due to pollinator preference [26]. A field experiment using F2 hybrids showed that anthocyanin and carotenoid content, as well as nectar volume, play important roles in pollinator discrimination between the two species [24].
Figure 2.

Mimulus flowers. (a) Bee-pollinated M. lewisii (wild-type). (b) Hummingbird-pollinated M. cardinalis (wild-type). (c) M. lewisii NIL with the recessive M. cardinalis roi1 allele. (d)M. lewisii NIL with the recessive M. cardinalisyup allele. (e) M. lewisii EMS mutant, guideless. All images were taken in the University of Washington Botany Greenhouse.
Fine-scale genetic mapping of the major anthocyanin QTL, ROI1, revealed a single-repeat MYB repressor as the causal gene responsible for the petal anthocyanin content variation between the two species [27••]. The dominant allele in M. lewisii represses anthocyanin biosynthesis, and introgression of the recessive M. cardinalisroi1 allele into the M. lewisii background yields a dark pink flower phenotype in the NIL (Figure 2c). Gene identity was verified by transgenic experiments: knocking down the expression of ROI1 in the wild-type M. lewisiivia RNAi transgenesis recapitulates the dark pink phenotype of the NIL; transformation of the wild-type ROI1 allele into the NIL background restores the light pink phenotype of the wild-type. ROI1 is a small protein (~80 amino acids) that is very similar to the R3 repeat of the anthocyanin-activating R2R3-MYBs (subgroup 6) [15–17,27••], but without a transcriptional activation domain. Like the subgroup 6 R2R3-MYBs, ROI1 possesses the bHLH-interacting motif and presumably competes with the anthocyanin-activating R2R3-MYBs for a limited supply of bHLH proteins, thereby negatively regulating anthocyanin biosynthesis. Subsequent gene expression analysis, in conjunction with site-directed mutagenesis and transgenic experiments, demonstrated cis-regulatory rather than coding DNA change as the causal mutation(s) underlying the allelic difference between M. lewisii and M. cardinalis [27••].
Although field experiments using F2 hybrids showed that increased anthocyanin concentration is correlated with higher hummingbird visitation rate and lower bumblebee visitation rate [24], the more definitive field experiment to test the specific role of ROI1 in pollinator preference still remains to be done using the NIL or transgenic lines.
On the other hand, the ecological significance of YELLOW UPPER (YUP), the locus responsible for the presence or absence of the yellow carotenoids in the petal lobes (Figure 2d), has been demonstrated using NILs in the natural habitat [25]. Substitution of the M. lewisii YUP locus with the recessive M. cardinalisyup allele dramatically increased hummingbird visitation and simultaneously decreased bee visitation. In this case the link between phenotype and pollinator response has been established, but the genotype-phenotype link is still under investigation.
Besides being an excellent model for studying natural variation, Mimulus lewisii is also emerging as a powerful system to study pollinator-associated floral traits using chemically induced mutants [28•]. An ethyl methanesulfonate (EMS) induced mutant, guideless, lacks the yellow color and the trichomes in the ventral petal that serve as nectar guides in the wild-type plant (Figure 2e). The ecological function of this trait in attracting and properly orienting bumblebees has been shown by a pollinator foraging assay under controlled greenhouse conditions. Lack of nectar guides not only decreased visitation rate by ~20%, but led to improper orientation in 55% of the visits as the bumblebees entered the flower upside-down, preventing pollen transfer between flowers via mechanical isolation [28•].
Bulk segregant analysis coupled with deep sequencing has now enabled rapid gene identification in M. lewisii mutants. The GUIDELESS gene encodes a MIXTA-like R2R3-MYB (subgroup 9) [29••]. MIXTA-like genes are known to regulate trichome and epidermal conical cell development in the Asteridae, including Antirrhinum and Petunia [30–32], which is consistent with the aborted trichome elongation in the nectar guides of the guideless mutant. Moreover, the absence of yellow carotenoids in the guideless mutant indicates that MIXTA-like genes might also be involved in regulating carotenoid pigmentation [29••]. Here, an interesting mutant phenotype changes pollinator handling success in controlled greenhouse experiments; the underlying gene has been identified by bulk segregant analyses and verified by transgenic experiments, satisfying the genotype-phenotype-pollinator response criteria.
Antirrhinum — from pigmentation patterns to tactile cues
The bee-pollinated Antirrhinum majus has been an invaluable model system in plant developmental genetics and has contributed to our understanding of the genetic control of floral organ identity, anthocyanin pigmentation, flower symmetry, inflorescence architecture, and petal epidermal cell development [33,34]. The large number of Antirrhinum floral mutants, many transposon-induced, have enabled gene discovery by transposon tagging since the early 1980s [33]. Meanwhile, like the Mimulus mutants, these Antirrhinum mutants provide excellent materials to study pollinator response to various floral traits [34].
The anthocyanin pigment patterning in Antirrhinumis largely determined by three closely related R2R3-MYB genes that belong to subgroup 6, including the tightly linked Rosea1 and Rosea2, and a third gene Venosa [35]. Rosea1 is the major regulator that gives the deep magenta color on both the adaxial and abaxial side of the wild-type flower petal lobes (Figure 3a). Rosea2 generates only weak pigmentation that is mostly restricted to the adaxial side. Venosa controls the “venation” pigmentation pattern that is clearly visible only in the rosea1 mutant background [35,36•]. This pattern probably represents the ancestral state of the genus Antirrhinum, as the early-diverged species mostly produce flowers with purple/pink stripes along the veins in a pale color background [36•]. While the native bumblebee pollinators preferentially visit the wild-type, deep magenta flower morph when compared with white, ivory, or light-pink mutants, they do not show any significant preference between the wild-type and the Venosa/rosea1 mutant, suggesting that the venation pattern in a pale color background plays an important role in bumblebee pollination and can compensate for the loss of overall color intensity [36•].
Figure 3. Antirrhinum majus.
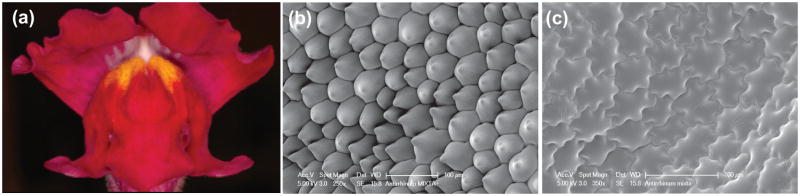
(a) Wild-type flower. (b) Conical cells on the petal epidermis of the wild-type flower. (c) Flat cells on the petal epidermis of the mixta mutant flower. (b) and (c) are provided by Dr. Beverley Glover.
Another Antirrhinum mutant, mixta [37], has shed light on a long-standing puzzle of the adaptive significance of petal epidermal conical cells, which are produced by the majority of insect-pollinated flowers [38]. MIXTA is the founding member of the R2R3-MYB subgroup 9. Lack of MIXTA activity results in flat instead of the pointed wild-type conical cells on the petal surface [37] (Figure 3). Using mutant lines that differ only in this single trait, Whitney et al. [39••] showed that bumblebees could distinguish flowers with or without conical cells by sensing the roughness of the petal epidermis as a tactile cue. When the flowers were presented with the landing surface oriented horizontally, the bumblebees did not display any preference between conical and flat petal surfaces. However, when the landing surface was presented vertically, as in many natural bee-pollinated flowers, the bumblebees showed significant preference for the conical-celled flowers. High-speed videography revealed the reason for this preference: bees were continually slipping and could not find a stable position on the flat-celled surfaces to feed, whereas they were able to use the bumpy conical cells as grips to handle the wild-type flowers [39••].
Over-representation of MYB transcription factors: just a coincidence?
The fully detailed examples discussed above include anthocyanin pigmentation, benzenoid volatile production, and conical cell development, floral traits that serve as visual, olfactory, and tactile cues during flower-pollinator interactions. Curiously, MYB transcription factors seem to play a prominent role in all these cases, even when unbiased search strategies (e.g., QTL mapping between wild taxa, random mutagenesis) were used instead of candidate gene approaches.
The MYB transcription factor family has only a few members in most sequenced green algal genomes [40], and has been expanded to contain a few dozen members in the early-diverged land plant lineages (e.g., Physcomitrella, Selaginella). A second massive expansion led to the hundreds of MYB genes in all sequenced angiosperm genomes. A similar pattern of gene family expansions is also apparent to the MADS-box genes [40], which are master regulators for floral organ identity [41]. From this parallel gene expansion pattern emerges an intriguing possibility — the MADS-box genes provided the basic genetic toolkit to make floral organs (i.e., sepals, petals, stamens, pistils) in the first place, and the MYB genes are the major players elaborating these floral organs, thereby creating the tremendously diverse flowers that interact with different guilds of pollinators.
Conclusion
It is obvious that our current knowledge on the genetic control of floral traits that affect pollinator specificity is overall still quite meager. However, the recent development of ultra-high-throughput-sequencing based methods (e.g., bulk segregant analysis, genome wide association studies, transcriptome profiling) and experimental tools (e.g., chemical mutagenesis, transgenic protocols) in a growing number of plant systems will soon change the slow pace of progress. We expect that our understanding of the genetic basis of many other pollinator-associated floral traits, including flower size [42], carotenoid pigmentation [43], floral tube formation and elaboration, stamen and pistil arrangements, nectar spur development [44–46], and nectar volume and composition [47], will be significantly advanced in the coming years. It will be interesting to see whether the MYB transcription factors also play a prominent role in the diversification of a wider range of floral traits.
Highlights.
Standards of evidence linking genes to floral traits to pollinator responses.
MYB genes underlie flower color and scent change in Petunia pollinator shifts.
MYB genes control both natural and induced floral trait variation in Mimulus.
MYB genes affect Antirrhinum pollination by regulating flower color and texture.
Acknowledgments
We thank Cris Kuhlemeier and Beverley Glover for kindly providing flower images. Work in the Bradshaw laboratory is supported by the US National Science Foundation FIBR grant 0328636 and National Institute of Health grant 5R01GM088805. KJRPB is supported by a National Science Foundation Graduate Research Fellowship, an NSF Doctoral Dissertation Improvement Grant (DEB 1209340,) and an ARCS Fellowship from the Seattle Chapter of the ARCS Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Faegri K, Pijl Lvd. The principles of pollination ecology. Pergamon Press; 1966. [Google Scholar]
- 2.Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. Pollination syndromes and floral specialization. Annu Rev Ecol Evol Syst. 2004;35:375–403. [Google Scholar]
- 3.Kay KM, Sargent RD. The Role of Animal Pollination in Plant Speciation: Integrating Ecology, Geography, and Genetics. Annu Rev Ecol Evol Syst. 2009;40:637–656. [Google Scholar]
- 4.Bradshaw HD, Wilbert SM, Otto KG, Schemske DW. Genetic mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus) Nature. 1995;376:762–765. [Google Scholar]
- 5.Hodges SA, Whittall JB, Fulton M, Yang JY. Genetics of floral traits influencing reproductive isolation between Aquilegia formosa and Aquilegia pubescens. Am Nat. 2002;159:S51–S60. doi: 10.1086/338372. [DOI] [PubMed] [Google Scholar]
- 6.Stuurman J, Hoballah ME, Broger L, Moore J, Basten C, Kuhlemeier C. Dissection of floral pollination syndromes in petunia. Genetics. 2004;168:1585–1599. doi: 10.1534/genetics.104.031138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouck A, Wessler SR, Arnold ML. QTL analysis of floral traits in Louisiana Iris hybrids. Evolution. 2007;61:2308–2319. doi: 10.1111/j.1558-5646.2007.00214.x. [DOI] [PubMed] [Google Scholar]
- 8.Des Marais DL, Rausher MD. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution. 2010;64:2044–2054. doi: 10.1111/j.1558-5646.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- 9.Dudareva N, Cseke L, Blanc VM, Pichersky E. Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopkins R, Rausher MD. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature. 2011;469:411–414. doi: 10.1038/nature09641. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins R, Rausher MD. Pollinator-mediated selection on flower color allele drives reinforcement. Science. 2012;335:1090–1092. doi: 10.1126/science.1215198. [DOI] [PubMed] [Google Scholar]
- 12.Galliot C, Stuurman J, Kuhlemeier C. The genetic dissection of floral pollination syndromes. Curr Opin Plant Biol. 2006;9:78–82. doi: 10.1016/j.pbi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Arnold M, Kuhlemeier C. Single gene-mediated shift in pollinator attraction in. Petunia Plant Cell. 2007;19:779–790. doi: 10.1105/tpc.106.048694. This study demonstrated that the white color of wild Petunia axilaris accessions resulted from multiple independent acquisitions of loss-of-function mutations in AN2, an R2R3-MYB gene. Pollination foraging assays using introgression lines and transgenic plants showed that AN2 is a major determinant in pollinator preference. As such, this paper set the standard of experimental evidence to link genotype to phenotype to pollinator response in studying the genetic basis of pollinator preference. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 16.Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, et al. The MYB transcription factor superfamily of arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- 17.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 19••.Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PM, Kuhlemeier C. Pollinator choice in Petunia depends on two major genetic loci for floral scent production. Curr Biol. 2011;21:730–739. doi: 10.1016/j.cub.2011.03.059. The authors of this paper identified two QTLs controlling the floral scent production difference between Petunia axillaris and P. exserta. An R2R3-MYB, ORORANT1, was determined as the causal gene underlying one of the QTLs. Near isogenic lines were used to demonstrate the significance of floral scent in hawkmoth pollination. Furthermore, these authors showed that flower color and scent are equally important in hawkmoth foraging decisions when exposing conflicting visual and olfactory cues to these pollinators. [DOI] [PubMed] [Google Scholar]
- 20.Hoballah ME, Stuurman J, Turlings TCJ, Guerin PM, Connetable S, Kuhlemeier C. The composition and timing of flower odour emission by wild Petunia axillaris coincide with the antennal perception and nocturnal activity of the pollinator Manduca sexta. Planta. 2005;222:141–150. doi: 10.1007/s00425-005-1506-8. [DOI] [PubMed] [Google Scholar]
- 21.Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitzer-Rimon B, Marhevka E, Barkai O, Marton I, Edelbaum O, Masci T, Prathapani NK, Shklarman E, Ovadis M, Vainstein A. EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in Petunia. Plant Cell. 2010;22:1961–1976. doi: 10.1105/tpc.109.067280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spitzer-Rimon B, Farhi M, Albo B, Can’ani A, Ben Zvi MM, Masci T, Edelbaum O, Yu Y, Shklarman E, Ovadis M, et al. The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in Petunia. Plant Cell. 2012;24:5089–5105. doi: 10.1105/tpc.112.105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schemske DW, Bradshaw HD. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus) Proc Natl Acad Sci U S A. 1999;96:11910–11915. doi: 10.1073/pnas.96.21.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey J, Bradshaw HD, Schemske DW. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae) Evolution. 2003;57:1520–1534. doi: 10.1111/j.0014-3820.2003.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 27••.Yuan YW, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD. Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics. 2013;194:255–263. doi: 10.1534/genetics.112.146852. Through fine-scale genetic mapping, site-directed mutagenesis, and transgenic experiments, this study identified a single-repeat R3-MYB repressor as the causal gene underlying the major QTL responsible for the petal lobe anthocyanin content variation between M. lewisii and M. cardinalis. This paper also described an efficient stable transformation protocol for M. lewisii. Together with recently developed genomic resources and a large number of EMS-induced floral mutants (see also 28• and 29••), this paper formally established M. lewisii as a powerful genetic model system to study ecologically interesting floral traits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Owen CR, Bradshaw HD. Induced mutations affecting pollinator choice in Mimulus lewisii (Phrymaceae) Arthropod-Plant Interact. 2011;5:235–244. This study used EMS-induced, single-locus floral mutants of M. lewisii to investigate the impact of flower shape and pigmentation pattern on pollinator response. A surprising finding is that the lack of nectar guides not only reduced bumblebee visitation, but also affected the orientation of bumblebee entry, demonstrating that the yellow, hairy nectar guides indeed serve as a “signpost” for bumblebees. [Google Scholar]
- 29••.Yuan YW, Sagawa JM, Di Stilio VS, Bradshaw HD. A MIXTA-like R2R3 MYB controls nectar guide formation in Mimulus lewisii. Genetics. 2013 doi: 10.1534/genetics.113.151225. Using bulk segregant analysis coupled with deep sequencing, a MIXTA-like R2R3-MYB was identified as the causal gene underlying the EMS-induced mutant, guideless (see also 28•). This paper demonstrates that we can now use M. lewisii mutants to study the developmental genetics of many poorly understood but ecologically important floral traits (e.g., carotenoid pigmentation, corolla tube formation and elaboration, stamen and pistil arrangement) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glover BJ, Perez-Rodriguez M, Martin C. Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development. 1998;125:3497–3508. doi: 10.1242/dev.125.17.3497. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development. 2005;132:359–370. doi: 10.1242/dev.01584. [DOI] [PubMed] [Google Scholar]
- 32.Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin HL, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134:1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz-Sommer Z, Davies B, Hudson A. An everlasting pioneer: the story of Antirrhinum research. Nat Rev Genet. 2003;4:657–666. doi: 10.1038/nrg1127. [DOI] [PubMed] [Google Scholar]
- 34.Glover BJ. Understanding Flowers and Flowering: An Integrated Approach. Oxford University Press; 2007. [Google Scholar]
- 35.Schwinn K, Venail J, Shang YJ, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Shang YJ, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, Martin CR, Davies KM. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011;189:602–615. doi: 10.1111/j.1469-8137.2010.03498.x. The authors of this paper investigated the role of an R2R3-MYB, Venosa, in generating vein-associated pigment stripes in snapdragon flowers. Pollinator foraging assay showed that the venation pattern in a pale color background attracts bumblebee pollinators as effectively as the fully magenta flowers. Phylogenetic analysis suggested that the venation pigmentation pattern probably represents the ancestral state of the snapdragon genus. [DOI] [PubMed] [Google Scholar]
- 37.Noda K, Glover BJ, Linstead P, Martin C. Flower color intensity depends on specialized cell-shape controlled by a MYB-related transcription factor. Nature. 1994;369:661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- 38.Kay QON, Daoud HS, Stirton CH. Pigment distribution, light reflection and cell structure in petals. Bot J Linn Soc. 1981;83:57–83. [Google Scholar]
- 39••.Whitney HM, Chittka L, Bruce TJA, Glover BJ. Conical epidermal cells allow bees to grip flowers and increase foraging efficiency. Curr Biol. 2009;19:948–953. doi: 10.1016/j.cub.2009.04.051. The authors of this paper conducted simple but elegant experiments to first demonstrate that bumblebees could indeed distinguish the fine textures of flat vs. conical cells, using both biological materials (i.e., snapdragon flowers) and epoxy casts of petals with flat or bumpy surfaces; they then revealed that the “adaptive significance” of petal epidermal conical cells is to provide good grip for bees handling the otherwise slippery petal surfaces. [DOI] [PubMed] [Google Scholar]
- 40.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 41.Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot. 2007;100:603–619. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krizek BA. Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol. 2009;12:17–22. doi: 10.1016/j.pbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Águila Ruiz-Sola M, Rodríguez-Concepción M. Carotenoid Biosynthesis in Arabidopsis: A Colorful Pathway. The Arabidopsis Book. 2012;10:e0158. doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittall JB, Hodges SA. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 2007;447:706–709. doi: 10.1038/nature05857. [DOI] [PubMed] [Google Scholar]
- 45.Kramer EM. Aquilegia: a new model for plant development, ecology, and evolution. Annual Review of Plant Biology. 2009;60:261–277. doi: 10.1146/annurev.arplant.043008.092051. [DOI] [PubMed] [Google Scholar]
- 46.Puzey JR, Gerbode SJ, Hodges SA, Kramer EM, Mahadevan L. Evolution of spur-length diversity in Aquilegia petals is achieved solely through cell-shape anisotropy. Proc R Soc B-Biol Sci. 2012;279:1640–1645. doi: 10.1098/rspb.2011.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruhlmann JM, Kram BW, Carter CJ. CELL WALL INVERTASE 4 is required for nectar production in Arabidopsis. J Exp Bot. 2010;61:395–404. doi: 10.1093/jxb/erp309. [DOI] [PMC free article] [PubMed] [Google Scholar]


